Background: The P1B-type ATPase PAA2/HMA8 transports Cu into the thylakoid lumen of the chloroplast for delivery to plastocyanin.
Results: PAA2/HMA8 stability is affected by plastocyanin and by the Cu content within the chloroplast.
Conclusion: The abundance of PAA2/HMA8 is subject to feedback control.
Significance: A newly identified layer of Cu-homeostasis via a novel regulatory mechanism of an organellar Cu-transporter.
Keywords: Chloroplast, Copper, Copper Transport, Plant Physiology, Protein Stability, Copper Homeostasis, P-type ATPase, Plastocyanin, Thylakoid Membrane
Abstract
PAA2/HMA8 (P-type ATPase of Arabidopsis/Heavy-metal-associated 8) is a thylakoid located copper (Cu)-transporter in Arabidopsis thaliana. In tandem with PAA1/HMA6, which is located in the inner chloroplast envelope, it supplies Cu to plastocyanin (PC), an essential cuproenzyme of the photosynthetic machinery. We investigated whether the chloroplast Cu transporters are affected by Cu addition to the growth media. Immunoblots showed that PAA2 protein abundance decreased significantly and specifically when Cu in the media was increased, while PAA1 remained unaffected. The function of SPL7, the transcriptional regulator of Cu homeostasis, was not required for this regulation of PAA2 protein abundance and Cu addition did not affect PAA2 transcript levels, as determined by qRT-PCR. We used the translational inhibitor cycloheximide to analyze turnover and observed that the stability of the PAA2 protein was decreased in plants grown with elevated Cu. Interestingly, PAA2 protein abundance was significantly increased in paa1 mutants, in which the Cu content in the chloroplast is half of that of the wild-type, due to impaired Cu import into the organelle. In contrast in a pc2 insertion mutant, which has strongly reduced plastocyanin expression, the PAA2 protein levels were low regardless of Cu addition to the growth media. Together, these data indicate that plastid Cu levels control PAA2 stability and that plastocyanin, which is the target of PAA2 mediated Cu delivery in thylakoids, is a major determinant of this regulatory mechanism.
Introduction
Copper (Cu) is utilized as a cofactor by the majority of organisms (1). In higher plants cuproproteins can be found in most cell compartments including the apoplast, cytosol, ER, peroxisome, mitochondrion, and chloroplast (for review see Ref. 2). Copper proteins are part of fundamental redox pathways such as respiration (in the form of cytochrome c oxidase), reactive oxygen species metabolism and photosynthesis. The majority of Cu ions in plant leaves are thought to be bound by plastocyanin (PC,3 Ref. 2). PC is an electron carrier for the photosynthetic machinery and located in the thylakoid lumen of the chloroplast. See Fig. 1 for an overview of the major Cu proteins and transporters. The Arabidopsis genome encodes for two PC isoforms (PC1 and PC2; (3)). PC2 accumulation is positively correlated with Cu abundance in the growth media, whereas PC1 is not affected by Cu (4). In a pc2 mutant, Cu abundance in the thylakoids is reduced to less than 20% of that of the wild-type (5). Other abundant cuproproteins are Cu/Zn superoxide dismutases (CSDs), which convert reactive superoxide (O2−) to hydrogen peroxide (H2O2) (6). The major isoforms in Arabidopsis leaves are CSD1 in the cytosol and CSD2 in the stroma (7). Both CSDs receive their cofactor through protein-protein interaction with the copper chaperone for superoxide dismutase (CCS), which is dually targeted to the cytosol and chloroplast stroma (8).
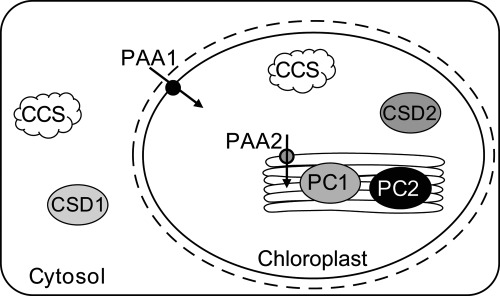
FIGURE 1.
Intracellular localization of cuproproteins. The figure schematically shows the location of major cuproproteins under consideration in this study. CCS isoforms are located in the cytosol and the chloroplast stroma, where they supply Cu to CSD1 and CSD2, respectively. PC1 and PC2 are located in the thylakoid lumen. PAA1 and PAA2 transport Cu over the inner chloroplast envelope and into the thylakoid lumen respectively, as indicated by the black arrows.
PC and CSD2 are translated in the cytosol and subsequently translocated to their respective chloroplastic locations (9). In order for PC to fully mature, Cu must be delivered to the thylakoids separately. This transport is mediated by two P-type ATPases. PAA1/HMA6 (P-type ATPase of Arabidopsis 1/Heavy-metal-associated 6) is a copper transporter located in the inner chloroplast envelope (10), (11) and PAA2/HMA8 is located in the thylakoid membrane, see Fig. 1 (10, 12). The paa1 and paa2 mutant lines exhibit phenotypes that are directly correlated with a lack of Cu in the chloroplast. Both, paa1 and paa2 have decreased photosynthetic activity, which is attributed to a decrease in PC abundance (10, 12). In addition, paa1 mutants lack CSD2 activity and show slow growth in low Cu conditions (10, 12).
In Arabidopsis, moderate Cu deficiency manifests similarly to mutations in paa1 and paa2, resulting in the reduction of photosynthetic efficiency and stunted growth (4, 13). The same chemical properties of Cu that are utilized by cuproproteins can be harmful if Cu is present as a free ion in the cell (2). Toxicity results in chlorosis of the vegetative parts and a reduction of photosynthetic activity, possibly through the impairment of chlorophyll and photosystem II, as reported for aquatic algae (14). Photosynthetic organisms have evolved a central Cu homeostatic machinery that allows a concerted response to low Cu through the Cu-responsive transcription factor SPL7 (SQUAMOSA promoter-binding protein-like 7; (15)). SPL7 activates the transcription of the root plasma membrane transporter COPT1 (16), as well as multiple Cu-miRNAs that down-regulate seemingly dispensable cuproproteins through transcript degradation. CSD1, CSD2, and CCS are targeted by miR398, while several apoplastic laccases and plantacyanin are targeted by miR408 (13, 17, 18). Intracellular Cu transporters such as PAA1 and PAA2 would be ideal control points for cellular Cu homeostasis, but thus far it has not been investigated if these transporters are affected by SPL7 or the Cu status of the plant. We now observed the stabilization of PAA2 protein on low Cu, a process which depends on PC and not SPL7. PAA1 abundance was not altered in response to the Cu status. In two paa1 mutant lines in which Cu transport into the chloroplast is decreased, we observe a significant increase in PAA2 abundance, suggesting that Cu affects PAA2 protein turnover within the chloroplast.
EXPERIMENTAL PROCEDURES
Plant Material, Growth Conditions, and Plant Treatments
The ecotype background for paa1-1 is Ler and Col-0 for paa1-3 (10). The background for paa2-1 is Col-3 gl1 (12). The mutant lines ccs, pc1, and pc2 have been described previously (8, 19) and all have a Col-0 background. A miR408 T-DNA insertion line was obtained from the Arabidopsis Biological Resource Center (ABRC) (Columbus, OH; SALK_023586.22.40.x). A homozygous miR408 knock-out line was isolated through selfing. The presence and localization of the T-DNA was confirmed using gene-specific primers and the T-DNA-specific primer LBb1.3 (supplemental Table S1).
For in vitro plant growth, seeds were surface sterilized by three consecutive 4-min rinses with 70, 90, and 70% ethanol, respectively and air-dried prior to stratification for 3 days at 4 °C. Plants were grown on solidified half-strength MS medium ((20); Caisson Laboratories, North Logan, UT; containing 0.05 μm CuSO4) with 1% sucrose (Sigma-Aldrich), 0.6% agar (Sigma-Aldrich) and additions, as indicated for each experiment. Plants were grown for 18 days at a photon density of 120 μmol m−2 s−1, in a 12-h light/12-h dark cycle at 23 °C, unless specified otherwise. For time courses of PAA2 protein turnover, Col-0 was grown in liquid half-strength MS with 1% sucrose for 10 days in continuous light (120 μmol m−2 s−1) and agitation in the presence of the indicated Cu concentrations. Plants were then treated for the indicated time periods with 100 μm cycloheximide (MP Biomedicals, Solon, OH), added from a 100 mm stock in 100% ethanol (21).
Protoplast and Chloroplast Isolation
Protoplasts were isolated according to (22) with the following modifications. Rosette leaves were cut into 1 mm strips and vacuum infiltrated for 5 min in 10 ml of enzyme solution before overnight digestion in the dark. All subsequent handling was done in the dark and on ice. The next day, protoplasts were diluted with an equal volume of 2 mm MES/KOH (pH 5.7), 154 mm NaCl, 125 mm CaCl2 and 5 mm KCl and filtered through a 215 μm nylon mesh (Component Supply Co., Fort Meade, FL). The flow through was centrifuged in a 50 ml conical tube for 2 min at 200 × g at 4 °C in a pre-cooled Allegra™ 21R centrifuge using the S4180 swing-out rotor (Beckman Coulter Inc., Brea, CA). Pelleted protoplasts were then washed with 10 ml of 11 mm MES/KOH (pH 5.7), 77 mm NaCl, 63 mm CaCl2, 2 mm KCl and 200 mm mannitol and again pelleted as before. Protoplasts were then taken up to a concentration of 1 × 106 intact protoplasts/ml in the same solution. At this step protoplast samples for metal ion determination and immunoblotting were taken. The remaining protoplasts were used for subsequent chloroplast isolation according to (23), with few modifications. Protoplasts were pelleted as before and taken up in 800 μl of chloroplast isolation buffer (CIB; 400 mm mannitol, 5 mm EGTA, 5 mm EDTA, 20 mm HEPES/KOH pH 8, 0.1% BSA, and 10 mm NaHCO3) and subsequently mechanically lysed by forcing them through a 18 μm nylon mesh (Component Supply Co., Fort Meade, FL) held by a syringe filter holder (Pall Life Sciences, Ann Arbor, MI) into a round-bottom 2 ml centrifugation tube. Chloroplasts were then pelleted by a 2 min centrifugation step at 1125 × g. The pellet was resuspended in 400 μl of CIB and loaded onto a 40% Percoll™ cushion (Amersham Biosciences AB, Uppsala, Sweden) in CIB. Intact chloroplasts were recovered after centrifugation for 5 min at 1620 × g in the bottom of the tube and resuspended in CIB containing no BSA. Chlorophyll content of protoplasts and chloroplasts was determined according to (24), and all samples were concentrated to 1 mg chlorophyll/ml in CIB containing no BSA. On average 50–60% of the chloroplasts were recovered from protoplasts.
Cu Content Measurements
Protoplast and chloroplast samples equivalent to 40 μg of chlorophyll were dried for 4 h at 70 °C and subsequently digested with 20 μl of trace metal grade nitric acid overnight in a sand bath at 95 °C. Then, samples were diluted with twice distilled water to a concentration of 10% nitric acid. 60 μl of each sample was further diluted in 540 μl of water and used for metal ion analysis on a Dionex ion chromatography system (Sunnyvale, CA) as described (10).
Plant Sampling, Protein Extraction, and Immunoblot Analysis
Arabidopsis shoots were frozen in liquid nitrogen and stored at −80 °C until use. For protein extraction, plant tissue was ground with mortar and pestle in liquid nitrogen to a fine powder, and its weight was determined while frozen. One part of tissue was homogenized in three parts of sodium dodecyl sulfate (SDS) sample buffer (250 mm Tris/HCl, pH 6.8, 20% glycerol, 4% SDS (w/v), 80 mm dithiothreitol, 0.1% (w/v) bromphenol-blue) in a microcentrifuge tube with the aid of a small pestle. The homogenate was immediately heated to 95 °C for 5 min and the insoluble material removed by centrifugation at 16,000 × g for 15 min at 4 °C. Preliminary experiments showed that flash freezing and preparation of the samples as described above yields the highest extraction of PAA1 and PAA2. The supernatant was fractionated on 10% SDS-PAGE and transferred to nitrocellulose membrane (Trans-Blot® Pure Nitrocellulose, Bio-Rad) by electroblotting. Proteins were immunodetected using the primary antibodies as described for each experiment and a secondary antibody coupled to alkaline phosphatase.
Protein Quantification
For quantification of the immunoblots, signal intensities were analyzed using ImageJ software (NIH, Bethesda, MD) and regression curves were obtained using dilution series on the corresponding membrane. Values are given as averages.
Antibodies
The sequences for the heavy metal binding domains of PAA1 and PAA2 were amplified using the primers listed in supplemental Table S1 and cloned as Ncol/BamHl fragments into the expression vector pCAL-c. The fusion proteins were purified from Escherichia coli strain BL21 (DE3) extracts by chromatography on Calmodulin Sepharose 4B (Amersham Biosciences Pharmacia, Uppsala, Sweden) according to manufacturer's instructions. The calmodulin-binding peptide tag was cleaved using thrombin (Novagen, Madison, WI) and the cleaved products were further purified by ion exchange chromatography on Q-Sepharose (Amersham Biosciences Pharmacia). Purified proteins were used to raise antibodies in rabbits as described (25). Antibodies were subsequently affinity purified with the same peptides they were raised against using AminoLink® Coupling Resin (Pierce) and concentrated by centrifugation (Amicon® Ultra Centrifugal Filters, Millipore, Carrigtohill Co., Cork, Ireland). Antibody for cFBPase, a constitutively expressed protein, was purchased from Agrisera (Vännäs, Sweden). CCS antibody was described previously (18).
RNA Extraction, Quantitative Real-time PCR (qRT-PCR) Analysis, and Mature miRNA Stem-Loop qRT-PCR
Total RNA was extracted from frozen leaf tissue with TRIzol® reagent (Invitrogen, Carlsbad, CA). Manufacturer's instructions for all enzymes and kits were followed if not otherwise specified. 20 μg of RNA were treated with DNase l (Fermentas, Hanover, MD) to remove genomic DNA from the extract. Prior to reverse-transcription, DNase l was removed by phenol-chloroform extraction and subsequent precipitation of mRNA (overnight at −20 °C in 100% EtOH with 3 m NaOAc, pH 5.2). Total RNA concentration was determined and equal amounts per sample were reverse transcribed using the First Strand cDNA Synthesis Kit from Fermentas (Hanover, MD) and random hexamer primers from Promega (Madison, WI). Quantitative PCR and quality control of primers was performed on a Light Cycler® 480 with the Light Cycler SYBR Green l master mix (Roche Applied Science, Indianapolis, IN). Samples without template were used as negative controls. PAA2 and PAA1 transcript abundance was analyzed in biological triplicates and technical duplicates using gene-specific primer pairs (see supplemental Table S1). For normalization of the PAA2 transcript level we used the two control genes yellow-leaf-specific 8 (YLS8) and the gene of a SAND family protein (see supplemental Table S1). Neither transcript is affected by Cu in Arabidopsis (26). The result for YLS8 is presented in Fig. 2B. Comparable results were obtained using SAND for normalization (data not shown). The quantitative PCR results were analyzed using the Light Cycler® 480 software from Roche. Relative transcript levels (2−ΔCt) were calculated as the difference between the threshold cycle (Ct) of the target gene and the Ct of the reference gene for each respective template (27).
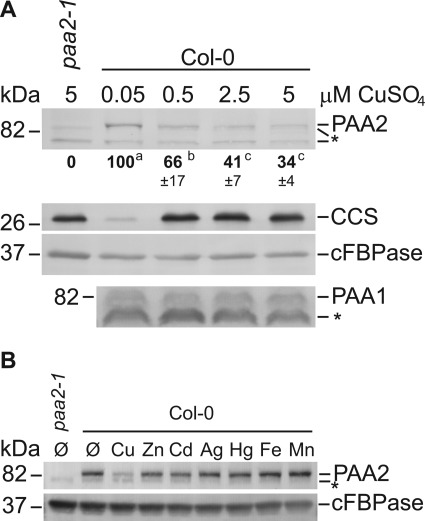
FIGURE 2.
PAA2 protein abundance is modulated by Cu. A, immunoblot analysis of PAA1 and PAA2 in rosette leaves of wild-type (Col-0) and paa2-1 (Col-0) seedlings grown on agar media in the presence of increasing CuSO4 concentrations. From here on out, bold numbers below the PAA2 panels indicate relative protein abundance determined using ImageJ software as described under “Experimental Procedures.” PAA2 abundance in the wild-type grown on 0.05 μm CuSO4 was arbitrarily set at 100%. Numbers in this figure represent an average of four replicates ± S.D. Superscripts indicate statistically significant groups (Student's t test; p < 0.05). In this and following figures, CCS antibody was used as an indicator for the Cu status of the plants and cFBPase as a loading control. * indicates a nonspecific band. B, immunoblot of PAA2 and cFBPase in rosette leaves of wild-type (Col-0) plants cultured for 18 days in the presence of CuSO4 (15 μm), ZnSO4 (150 μm), CdSO4 (20 μm), AgNO3 (30 μm), HgCl2 (10 μm), Fe-EDTA (100 μm), and MnCl2 (300 μm), respectively. paa2-1 seedlings grown on regular ½ MS served as a control. The result is representative of three separate experiments. * indicates an unspecific band.
For the mature miRNA detection, stem-loop pulsed reverse transcription was performed after isolating total RNA as described above. However, ethanol washes were avoided and nucleic acid precipitation steps were performed using 1:1 (v/v) isopropanol and 1:10 (v/v) sodium acetate 3 m pH 5.2 to optimize small RNA molecule retrieval. The stem-loop pulsed reverse transcription and the miRNA qRT-PCR were performed as previously described (28), using the primers listed in supplemental Table S1. Analysis of the data were performed as described above. The relative transcript levels of the mature miRNA were monitored for biological triplicates and technical duplicates, and results were standardized using miR167 expression. Similar results were obtained using miR156 as a reference (data not shown).
Statistical Analysis
JMP software (version 9.0.2; SAS Institute) was used for statistical analysis. Figures and data represent average and S.D. values based on sampling from the indicated amount of biological and technical replicates. The number of total samples is given when appropriate. Student's t test was used to calculate significant differences (p < 0.05), which is reported in the text or figures where appropriate.
RESULTS
PAA2 Protein Abundance Is Affected by Cu
We raised antibodies to the N-terminal domains of PAA1 and PAA2. Immunoblots using total protein extracted from 18-day-old wild-type, paa1-1 and paa2-1 mutant plants indicated that the affinity-purified antibodies recognize their respective antigen (supplemental Fig. S1, A and B). The cytosolic fructose-1,6-bisphosphatase (cFBPase) was probed as a loading control in this and all following blots presented.
To investigate if PAA1 and PAA2 protein levels are affected by Cu, wild-type plants were grown in vitro on agar-solidified half-strength MS media in the presence of CuSO4, ranging from 0.05 μm to 5 μm. The treatments clearly affected the Cu status of the plants as indicated by the abundance of CCS (Fig. 2A). Protein abundance of PAA2 was highest in low Cu and decreased to one-third at 5 μm (Fig. 2A, top panel). In contrast, PAA1 abundance was not affected by Cu.
To test if the abundance of PAA2 could also be affected by other metals, wild-type plants were cultured in vitro in the presence of various metals. To avoid too strong toxicity we used the metals at half the concentration that had been determined to give a 50% root length inhibition in Brassica juncea (29). As a control we quantified the effect of metal ion addition on root length and found that only cadmium reduced root length to more than 50% (supplemental Fig. S2). Importantly, only Cu affected PAA2 protein accumulation (Fig. 2B). We conclude that the effect of Cu on PAA2 abundance is specific.
PAA2 Is Post-transcriptionally Regulated
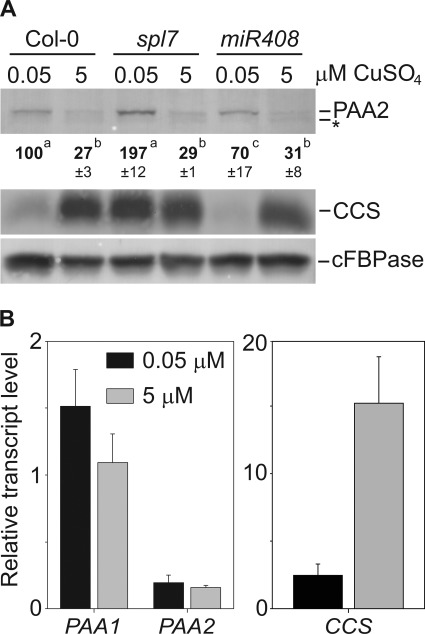
Sequence alignment revealed a potential miR408 targeting site within the PAA2 coding sequence (supplemental Fig. S3A). miR408 is one of the Cu-miRNAs induced by SPL7 in response to low Cu (13). To analyze if miR408 affects the PAA2 protein abundance we characterized a T-DNA knock-out line (supplemental Fig. S3). The T-DNA is inserted within the mature miR408 sequence (supplemental Fig. S3, A and B). The mature miR408 was quantified by stem loop qRT-PCR for plants grown with 0.05 μm or 5 μm CuSO4. In the wild-type, the mature miR408 was only detectable on low Cu. In the knock-out line, mature miR408 was at the detection limit, which confirms that the T-DNA disrupts miR408 (supplemental Fig. S3C). In the miR408 knock-out line, the PAA2 protein abundance still responded to Cu as in the wild-type and therefore miR408 does not affect PAA2 (Fig. 3A, top panel). To examine miR408-independent SPL7 involvement in the regulation of PAA2 abundance, an spl7 mutant (15) was grown in vitro on 0.05 μm and 5 μm CuSO4 and then subjected to immunoblot analysis (Fig. 3A). As expected, CCS was deregulated, but PAA2 protein abundance still decreased on high Cu in this mutant as it did in the wild-type. In addition, we performed transcript analysis of wild-type plants grown in low and high Cu using qRT-PCR (Fig. 3B). Indeed, the transcript level of PAA2 was the same in either Cu condition. The data indicate a post-transcriptional mechanism for the effect of Cu on PAA2.
FIGURE 3.
PAA2 is not regulated on a transcriptional level through SPL7. A, immunoblot analysis of PAA2, CCS, and cFBPase in rosette leaves of wild-type (Col-0), spl7 and miR408 knock-out lines grown on agar media. The immunoblot is representative of three separate experiments. Quantification of band intensities represent an average of three replicates ± S.D. B, mRNA expression levels measured by qRT-PCR for PAA1, PAA2, and CCS in wild-type (Col-0) rosette leaves in the presence of 0.05 μm and 5 μm CuSO4. Shown are the average values of three biological replicates with two technical repeats each. Data are normalized to YLS8 expression. Values are given as averages ± S.D.
Cu Affects PAA2 Protein Turnover
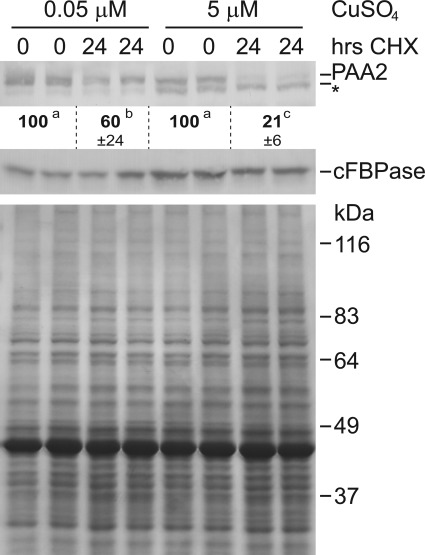
Since Cu does not affect PAA2 mRNA levels it must affect protein accumulation either through protein synthesis or turnover. We analyzed the effect of Cu on PAA2 protein turnover by inhibition of cytosolic protein synthesis with cycloheximide (CHX) (Fig. 4). Wild-type plants were cultured in liquid half-strength MS at 0.05 μm or 5 μm CuSO4 for 10 days and then treated with 100 μm CHX for 24 h. On low Cu, CHX treatment resulted only in about a 40% reduction of PAA2 abundance. In contrast, for plants grown on 5 μm CuSO4 PAA2 abundance was reduced by 80% after 24 h of CHX treatment. Therefore, Cu strongly affects PAA2 protein stability.
FIGURE 4.
Cu affects PAA2 stability. Top panel, immunoblot of PAA2 in wild-type (Col-0) plants grown in liquid half-strength MS in the presence of 0.05 μm CuSO4 and 5 μm CuSO4. 10-day-old plants were treated with 100 μm CHX for the indicated times. Shown are two biological replicates for each time point. To compensate for the reduced abundance of PAA2 in the presence of 5 μm CuSO4, twice the amount of protein extract was loaded in each lane for these samples. Results are representative of four separate biological replicates. Quantification of band intensities represent an average of four replicates ± S.D. * indicates a nonspecific band. Lower panel, Coomassie Brilliant Blue stained 10% SDS-PAGE with the same arrangement of samples shown in the top panel; equal amounts of protein were loaded.
Cu Content in the Chloroplast and Plastocyanin Abundance Determine the Effect on PAA2
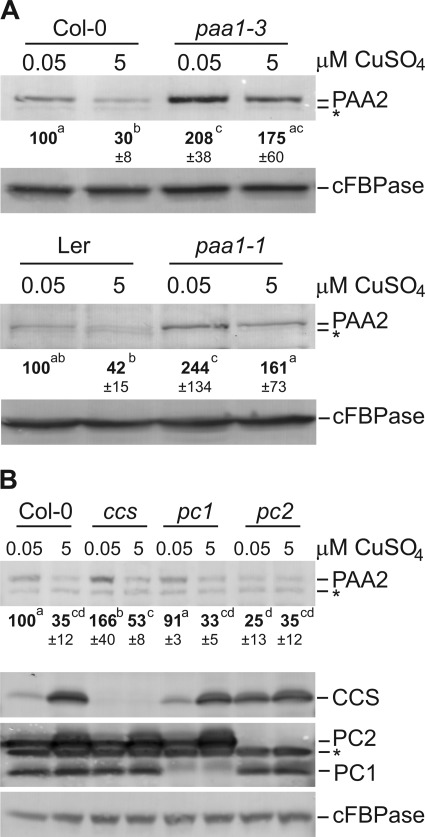
To investigate if the Cu status within the chloroplast is important for the determination of PAA2 abundance, we analyzed paa1-1 and paa1-3 mutants, which are reported to have a lower chloroplastic Cu concentration (10). paa1-1, paa1-3, and their respective isogenic wild-type were grown in vitro at 0.05 μm and 5 μm CuSO4 and probed for PAA2 abundance by immunoblot. Both paa1 alleles showed a significant increase in PAA2 compared with their respective wild-types (Fig. 5A), while a decrease in PAA2 protein is still seen in response to increasing Cu concentration in all lines. The data indicate that Cu levels in the chloroplast affect PAA2 protein accumulation. Known Cu-binding proteins in the chloroplast are the two plastocyanin isoforms, CCS and CSD2. Of the two plastocyanin isoforms, PC1 is expressed at a low but constitutive level whereas PC2 accumulates especially on elevated Cu and is the most abundant plastocyanin isoform in most conditions except under deficiency (4). There is no mutant available for CSD2, but CSD2 activity and accumulation is severely disrupted in a ccs mutant (18). To investigate if the abundance of any of the Cu-binding proteins within the chloroplast could modulate the regulation of PAA2 accumulation in response to Cu, pc1, pc2, and ccs mutants were grown in vitro at 0.05 μm and 5 μm CuSO4 and probed for PAA2 protein. Interestingly, in pc2 mutants the PAA2 protein abundance was reduced by about 75% compared with the wild-type when grown in the presence of low Cu and stayed low at high Cu (Fig. 5B, top panel). In contrast, the PAA2 protein abundance was comparable to the wild-type in pc1 mutants, which still accumulate the PC2 protein when Cu levels are increased. In the ccs mutant, the PAA2 abundance was slightly elevated on low Cu when compared with the wild-type. Importantly, the regulation of PAA2 in response to Cu was maintained in this ccs line. These data indicate that among the chloroplast Cu proteins, PC2 expression is needed to maintain PAA2 protein levels.
FIGURE 5.
PAA2 protein abundance is affected by Cu in the chloroplast and by PC2. A, immunoblot analysis of PAA2 and cFBPase control in paa1-3 (Col-0; top panel), paa1-1 (Ler background; lower panel). To compensate for the reduced abundance of PAA2 in the Ler background, twice the amount of protein extract was loaded in each lane for Ler and paa1-1 in both Cu conditions. B, immunoblot analysis of PAA2, CCS, PC, and the cFBPase control in Col-0 wild-type, ccs, pc1, and pc2 (all Col-0 background) mutant seedlings grown on agar media in the presence of 0.05 μm and 5 μm CuSO4. Results are representative of four separate biological replicates. Quantification of band intensities represent an average of four replicates ± S.D. * indicates a nonspecific band.
We isolated protoplasts and chloroplasts from in vitro grown wild-type, ccs, and paa1-3 plants. Abundance of CCS and PC isoforms were verified as controls (supplemental Fig. S4). The cellular Cu levels were very similar between lines and clearly affected by Cu feeding (supplemental Table S2). No significant change was seen in chloroplast Cu contents in the ccs mutant relative to the wild-type. However, the fraction of cellular Cu that was in the chloroplast in paa1-3 plants was half of the wild-type level, which is in agreement with Shikanai et al. (10).
DISCUSSION
The Arabidopsis P1B-type ATPase PAA2 is a thylakoid located Cu transporter that supplies Cu to PC, an essential protein for photosynthetic activity in higher plants (19). Here we demonstrate that the PAA2 protein is stabilized under Cu limiting growth conditions, leading to its accumulation in the thylakoids (Fig. 2A). We show that PAA2 accumulation is modulated by the Cu levels within the chloroplast (Fig. 5A and supplemental Table S2). Importantly, this regulation is independent of SPL7, the master transcription regulator which controls other responses to Cu deficiency (2, 15). In contrast, it appeared that PC, the sink for Cu in the thylakoid lumen, ultimately controls this regulation (Fig. 5B).
The mRNA expression level of PAA2 is very low (Ref. 12, Fig. 3B). Affinity purification of the antibodies was required for specific detection and allowed us to observe changes in PAA2 protein abundance. The effect of Cu on PAA2 abundance was not only observed for plants on agar media but was also observed for mature plants grown hydroponically (not shown). Only Cu and no other metal affected PAA2 (Fig. 2B). Additional observations underscore the qualitative and quantitative effect of Cu on the transporter abundance. We noted that PAA2 protein accumulated more in the spl7 mutant on low Cu when compared with the wild-type (Fig. 3A). The spl7 mutant phenotype is most evident on low Cu because the SPL7 transcription factor is required to up-regulate Cu transporters under impending deficiency (15). Thus spl7 is specifically deficient for Cu and therefore limited in its capacity to provide Cu for PAA2 and PC in the ½ MS growth conditions (0.05 μm CuSO4) in which wild-type plants show no deficiency symptoms. Addition of 5 μm Cu largely rescues spl7 phenotypes and under this condition, PAA2 protein abundance is the same as in the wild-type (Fig. 3A). In paa1 mutants, Cu also has limited access to PAA2 and PC but compared with spl7 much higher and even toxic Cu levels are required for full rescue of the phenotype (10). Indeed we observed that the abundance of PAA2 was higher in paa1 lines when compared with the wild-type on both regular ½ MS (0.05 μm Cu) and ½ MS with 5 μm CuSO4 present (Fig. 5A). The protein accumulation data in Fig. 5A together with the chloroplast Cu levels reported in supplemental Table S2 support the notion that the regulation of PAA2 is controlled by the Cu content in the chloroplast.
How does PAA2 protein regulation compare with regulation of other metal transporters of the P1B-type class? Post-translational Cu dependent and independent modifications such as phosphorylation and glutathionylation among others have been shown in mammalian cells to modify the activity of the P1B-type Cu transporters ATP7A and ATP7B (30–32). In addition, ATP7A and ATP7B influence Cu homeostasis by being differentially sorted between the trans Golgi network and vesicles (33). Perhaps most reminiscent of PAA2 is the regulation of the Saccharomyces cerevisiae cadmium-exporting P1B-type ATPase Pca1. This transporter is conditionally stabilized in the presence of its substrate, allowing the cell to avoid the cytotoxic effects of cadmium (34, 35).
It is unlikely that PAA2 protein turnover represents an adaptation to toxic Cu excess because the effect is already seen at 0.5 μm CuSO4 (Fig. 2A), which is 40 times lower than the toxicity threshold (2). Instead, the stabilization of PAA2 occurs at very low Cu concentrations similar to those that activate the SPL7 pathway (see CCS abundance in Fig. 2A). Therefore, we propose that the post-translational control of PAA2 contributes to optimal Cu use within the plant. When Cu becomes limiting, SPL7 up-regulates plant Cu acquisition and controls the Cu-microRNA mediated down-regulation of certain cuproproteins. In the chloroplast, CCS and CSD2 are down-regulated so that the Cu pool in this compartment might now be used in “economy mode” for only the most essential functions. In this context, we recently reported that, when Cu is resupplied to Cu-starved Populus trichocarpa, Cu is preferentially allocated to PC and the photosynthetic activity is quickly recovered (36). By contrast CCS and CSD2 recovery in the stroma was delayed (36). The stabilization of PAA2 is likely an additional regulatory mechanism, acting collectively with the SPL7-mediated responses, in order to facilitate the flow of Cu ions into the thylakoid lumen. On the other hand, the destabilization of PAA2 protein on elevated Cu may be part of a feedback mechanism that could allow the plant to quickly adjust to varying Cu statuses that are specific to the chloroplast sub-compartment. On elevated Cu more than enough PC may be active so that it is no longer a priority for cofactor delivery (4). Here, low abundance of PAA2 would retain Cu in the stroma and cytosol for use by cuproproteins in these locations.
The mechanism by which PAA2 is stabilized under Cu limiting conditions represents a future challenge in this research area. Here, three important observations that have bearing on this mechanism are presented. Our data show that the process is not mediated by SPL7 or miRNA408 and therefore involves a new Cu-dependent signaling pathway. The paa1 mutants that have less Cu in the chloroplast, show increased PAA2 accumulation (supplemental Table S2 and Fig. 5A). Finally, our finding that PAA2 is not stabilized under Cu limiting conditions in a pc2 mutant brings genetic evidence for a crucial role of PC in the control of this regulation. PC2 is very abundant in the lumen and thus represents most of the luminal sink for Cu. As suggested above, a feed-back regulatory mechanism may serve to signal that Cu is not preferentially allocated to PC at higher Cu concentrations, resulting in the destabilization of PAA2. This model agrees with the observed roughly 80% reduction of Cu levels in the thylakoid lumen of pc2 mutants compared with the wild-type (5).
In summary, this study shows an exciting new regulation for a P1B-type ATPase transporter and reveals another layer of Cu homeostasis control in Arabidopsis. Low Cu levels in the nucleus and cytosol activate the economy mode via SPL7. This in turn affects cuproproteins, increasing Cu availability for PC in the thylakoid lumen. Our work reveals that intra-organellar Cu pools can also participate in this control. More specifically, plastidial Cu regulates its own transport into the thylakoid lumen by acting on PAA2 stability, a process requiring PC, the ultimate target of Cu delivery.
Supplementary Material
Acknowledgments
We thank Salah E. Abdel-Ghany and Kathryn Gogolin-Reynolds for help with plasmid construction and antibody production and Laura Hantzis for technical assistance with protoplast and chloroplast isolations.
This work was supported by the United States National Science Foundation (Grant Numbers IOS-0847442 and MCB 0950726).

This article contains supplemental Figs. S1–S4 and Tables S1 and S2.
- PC
- plastocyanin
- PAA
- P-type ATPase of Arabidopsis
- CHX
- cycloheximide
- CCS
- copper chaperone for superoxide dismutase
- CSD
- copper zinc superoxide dismutase
- HMA
- heavy metal-associated transporter
- SPL7
- squamosa promoter-binding protein-like 7.
REFERENCES
- 1. Zhang Y., Gladyshev V. N. (2009) Comparative genomics of trace elements: emerging dynamic view of trace element utilization and function. Chem. Rev. 109, 4828–4861 [DOI] [PubMed] [Google Scholar]
- 2. Burkhead J. L., Reynolds K. A., Abdel-Ghany S. E., Cohu C. M., Pilon M. (2009) Copper homeostasis. New Phytol. 182, 799–816 [DOI] [PubMed] [Google Scholar]
- 3. Kieselbach T., Bystedt M., Hynds P., Robinson C., Schröder W. P. (2000) A peroxidase homologue and novel plastocyanin located by proteomics to the Arabidopsis chloroplast thylakoid lumen. FEBS Lett. 480, 271–276 [DOI] [PubMed] [Google Scholar]
- 4. Abdel-Ghany S. E. (2009) Contribution of plastocyanin isoforms to photosynthesis and copper homeostasis in Arabidopsis thaliana grown at different copper regimes. Planta. 229, 767–779 [DOI] [PubMed] [Google Scholar]
- 5. Pesaresi P., Scharfenberg M., Weigel M., Granlund I., Schröder W. P., Finazzi G., Rappaport F., Masiero S., Furini A., Jahns P., Leister D. (2009) Mutants, overexpressors, and interactors of Arabidopsis plastocyanin isoforms: revised roles of plastocyanin in photosynthetic electron flow and thylakoid redox state. Mol. Plant 2, 236–248 [DOI] [PubMed] [Google Scholar]
- 6. McCord J. M., Fridovich I. (1969) Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055 [PubMed] [Google Scholar]
- 7. Kliebenstein D. J., Monde R. A., Last R. L. (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118, 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu C. C., Lee W. C., Guo W. Y., Pan S. M., Chen L. J., Li H. M., Jinn T. L. (2005) A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol. 139, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179, 257–285 [DOI] [PubMed] [Google Scholar]
- 10. Shikanai T., Müller-Moulé P., Munekage Y., Niyogi K. K., Pilon M. (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15, 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Catty P., Boutigny S., Miras R., Joyard J., Rolland N., Seigneurin-Berny D. (2011) Biochemical characterization of AtHMA6/PAA1, a chloroplast envelope Cu(I)-ATPase. J. Biol. Chem. 286, 36188–36197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abdel-Ghany S. E., Müller-Moulé P., Niyogi K. K., Pilon M., Shikanai T. (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17, 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abdel-Ghany S. E., Pilon M. (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Küpper H., Setlik I., Setlikova E., Ferimazova N., Spiller M., Küpper F. C. (2003) Funct. Plant Biol. 30, 1187–1196 [DOI] [PubMed] [Google Scholar]
- 15. Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009) SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21, 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sancenón V., Puig S., Mira H., Thiele D. J., Peñarrubia L. (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 51, 577–587 [DOI] [PubMed] [Google Scholar]
- 17. Yamasaki H., Abdel-Ghany S. E., Cohu C. M., Kobayashi Y., Shikanai T., Pilon M. (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 282, 16369–16378 [DOI] [PubMed] [Google Scholar]
- 18. Cohu C. M., Abdel-Ghany S. E., Gogolin Reynolds K. A., Onofrio A. M., Bodecker J. R., Kimbrel J. A., Niyogi K. K., Pilon M. (2009) Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: regulation and unexpected phenotypes in an Arabidopsis mutant. Mol. Plant 2, 1336–1350 [DOI] [PubMed] [Google Scholar]
- 19. Weigel M., Varotto C., Pesaresi P., Finazzi G., Rappaport F., Salamini F., Leister D. (2003) Plastocyanin is indispensable for photosynthetic electron flow in Arabidopsis thaliana. J. Biol. Chem. 278, 31286–31289 [DOI] [PubMed] [Google Scholar]
- 20. Murashige T., Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum 15, 473–497 [Google Scholar]
- 21. Arnaud N., Murgia I., Boucherez J., Briat J. F., Cellier F., Gaymard F. (2006) An iron-induced nitric oxide burst precedes ubiquitin-dependent protein degradation for Arabidopsis AtFer1 ferritin gene expression. J. Biol. Chem. 281, 23579–23588 [DOI] [PubMed] [Google Scholar]
- 22. Yoo S. D., Cho Y. H., Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 23. Fitzpatrick L. M., Keegstra K. (2001) A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J. 27, 59–65 [DOI] [PubMed] [Google Scholar]
- 24. Bruinsma J. (1961) A comment on the spectrophotometric determination of chlorophyll. Biochim. Biophys. Acta 52, 576–578 [DOI] [PubMed] [Google Scholar]
- 25. Abdel-Ghany S. E., Ye H., Garifullina G. F., Zhang L., Pilon-Smits E. A., Pilon M. (2005) Iron-sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol. 138, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227, 1343–1349 [DOI] [PubMed] [Google Scholar]
- 27. Arrivault S., Senger T., Krämer U. (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46, 861–879 [DOI] [PubMed] [Google Scholar]
- 28. Varkonyi-Gasic E., Wu R., Wood M., Walton E. F., Hellens R. P. (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wangeline A. L., Burkhead J. L., Hale K. L., Lindblom S. D., Terry N., Pilon M., Pilon-Smits E. A. (2004) Overexpression of ATP sulfurylase in Indian mustard: effects on tolerance and accumulation of twelve metals. J. Environ. Qual. 33, 54–60 [DOI] [PubMed] [Google Scholar]
- 30. Singleton W. C., McInnes K. T., Cater M. A., Winnall W. R., McKirdy R., Yu Y., Taylor P. E., Ke B. X., Richardson D. R., Mercer J. F., La Fontaine S. (2010) Role of glutaredoxin1 and glutathione in regulating the activity of the copper-transporting P-type ATPases, ATP7A and ATP7B. J. Biol. Chem. 285, 27111–27121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vanderwerf S. M., Cooper M. J., Stetsenko I. V., Lutsenko S. (2001) Copper specifically regulates intracellular phosphorylation of the Wilson's disease protein, a human copper-transporting ATPase. J. Biol. Chem. 276, 36289–36294 [DOI] [PubMed] [Google Scholar]
- 32. Vanderwerf S. M., Lutsenko S. (2002) The Wilson's disease protein expressed in Sf9 cells is phosphorylated. Biochem. Soc. Trans. 30, 739–741 [DOI] [PubMed] [Google Scholar]
- 33. La Fontaine S., Mercer J. F. (2007) Trafficking of the copper-ATPases, ATP7A and ATP7B: role in copper homeostasis. Arch Biochem. Biophys. 463, 149–167 [DOI] [PubMed] [Google Scholar]
- 34. Adle D. J., Lee J. (2008) Expressional control of a cadmium-transporting P1B-type ATPase by a metal sensing degradation signal. J. Biol. Chem. 283, 31460–31468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adle D. J., Wei W., Smith N., Bies J. J., Lee J. (2009) Cadmium-mediated rescue from ER-associated degradation induces expression of its exporter. Proc. Natl. Acad. Sci. U.S.A. 106, 10189–10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ravet K., Danford F. L., Dihle A., Pittarello M., Pilon M. (2011) Spatiotemporal analysis of copper homeostasis in Populus trichocarpa reveals an integrated molecular remodeling for a preferential allocation of copper to plastocyanin in the chloroplasts of developing leaves. Plant Physiol. 157, 1300–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.