Background: The apoptosome is a major apparatus for intrinsic stimulus-induced apoptosis.
Results: Cdc6, known to assemble prereplicative complexes on replication origins and activate p21CIP1-/p27KIP1-bound Cdk2, obstructs cytochrome c- or some other mechanism-induced apoptosome assembly by forming stable complexes with activated Apaf-1.
Conclusion: Cdc6 is an AAA+ ATPase with three functions, all working for life.
Significance: These results redefine the role of Cdc6 in cell proliferation.
Keywords: Caspase, CDK (Cyclin-dependent Kinase), Cell Cycle, Cell Death, E2F Transcription Factor, Apaf-1, Cdc6, Anoikis, mTORC1
Abstract
Cdc6 is the bifunctional AAA+ ATPase that assembles prereplicative complexes on origins of replication and activates p21CIP1- or p27KIP1-bound Cdk2. During the G1-S transition, the Cdc6 gene essential for chromosomal replication is activated by the E2F transcriptional factor. Paradoxically, Apaf-1 encoding the central component of the apoptosome is also activated at the same time and by E2F. Consequently, genes for antipodal life and death are regulated in the same manner by the same transcriptional factor. Here we report a striking solution to this paradox. Besides performing prereplicative complex assembly and Cdk2 activation, Cdc6 obstructed apoptosome assembly by forming stable complexes very likely with a monomer of cytochrome c-activated Apaf-1 molecules. This function depended on its own ATPase domain but not on the cyclin-binding motif. In proliferating rodent fibroblasts, Cdc6 continued to block apoptosome assembly induced by a non-cytochrome c or some other mechanism, suppressing seemingly unintended apoptosis when promoting cell proliferation. Thus, Cdc6 is an AAA+ ATPase with three functions, all working for life.
Introduction
Cells execute their own death in response to stress-induced intrinsic and death receptor-mediated extrinsic apoptotic stimuli (1, 2). The main apparatus that responds to intrinsic stimuli and initiates the execution of cell death is the apoptosome, which is assembled upon activation by mitochondrial cytochrome c (cytc)2 (3–5). When the outer membrane of mitochondria is permeabilized due to cellular stresses, cytc in the intermembrane space is released into the cytoplasm. Released cytc binds to the repeated WD domain of monomeric Apaf-1, the central component of the apoptosome, and induces its conformational changes into an open shape with ATP/dATP hydrolysis. The resulting open shaped Apaf-1 molecules with cytc attached are allowed to form heptameric complexes, which then bind via their caspase-recruiting domain and activate caspase-9. This ternary complex is the mature apoptosome fully capable of converting procaspase-3 to the active two-chain form. The activated caspase-9 in the apoptosome undergoes a rapid autocatalytic cleavage at Asp315 corresponding to sites in other caspases cleaved for activation. Once the caspase-9 is autocleaved at this site, the apoptosome becomes susceptible to inhibition by XIAP and is facilitated to liberate the cleaved caspase-9, which quickly loses most of its activity (6, 7). Mitochondrial outer membrane permeabilization induced by intrinsic apoptotic stimuli is primarily caused by pore formation resulting from the oligomerization and subsequent outer membrane insertion of mostly Bax and Bak, proapoptotic members of the Bcl-2 family (8, 9). In the absence of apoptotic stimuli, Bax and Bak are sequestered mainly by antiapoptotic Bcl-2 and Bcl-X, which are in turn negatively regulated by the BH3-only proteins.
By contrast, extrinsic apoptotic stimuli originate from the binding of appropriate ligands to the death receptors, such as TNF receptors and Fas (10). Once bound to the ligand, the death receptor molecules aggregate and recruit FADD at their cytoplasmic domain. Receptor-recruited FADD molecules then bind the N-terminal death effector domain of procapase-8 and induce its dimerization to the catalytically active form. The dimerization also promotes autocleavages of the procaspase-8 to the canonical two-chain form and removal of the death effector domain, the latter of which releases the processed caspase-8 into the cytoplasm. Activated caspase-8 then converts procaspase-3 to the active two-chain form for the execution of cell death.
Cell cycle start requires activation of the two G1 cyclin-dependent kinases Cdk4/Cdk6 and Cdk2 (11, 12). Activated Cdk4/Cdk6 phosphorylates Rb at Ser780 to liberate the E2F transcriptional factor from inactivation. Activated E2F induces a subset of genes essential or important for S phase onset, such as Cdc6, cyclin A, and Emi1 (13, 14). Cdc6 protein is the AAA+ ATPase that assembles prereplicative complexes on origin recognition complex-bound replication origins and activates p21CIP1- or p27KIP1-bound Cdk2, the latter only after the bound p27 acquires its C-terminal phosphorylation (15–18). Cyclin A is a noncatalytic partner for Cdk2 implicated to be important for the onset of DNA synthesis, whereas Emi1 directly inhibits the APC/CCDH1 ubiquitin ligase and thereby stabilizes Cdc6 and cyclin A proteins during the G1-S transition (19).
The activity of Cdk4/Cdk6 is strictly regulated by the Tsc1/2-Rheb-mTORC1 pathway that conveys signals for metabolic states and growth factor and nutrient availability to control mRNA translation (20). Recently it was found that in fibroblasts activation of mTORC1 requires a signal for cell anchorage to the extracellular matrix, which is mediated by Rho-associated kinase, which directly inactivates Tsc2 by phosphorylating its Thr1203 (21, 22). When deprived of anchorage to the extracellular matrix, virtually all the cells constituting solid organs of animals arrest in G1 with inactivation of both Cdk4/Cdk6 and Cdk2, facilitated degradation of Cdc6, and induction of apoptosis known as anoikis (23–25). Anchorage deprivation-induced mTORC1 inactivation is responsible for the inactivation of Cdk4/Cdk6 and the degradation of Cdc6, whereas similarly induced FADD-mediated caspase-8 activation is responsible for the initiation of anoikis at least in normal fibroblast and epithelial cells (21, 26).
When cells have committed to proliferation, unintended apoptosis must be ensured not to take place. Otherwise, proliferation would simply be forced to be aborted because cell proliferation and death cannot coexist. However, quite peculiarly, the gene encoding Apaf-1, the key component of the apoptosome, is also activated by E2F, just like the genes required for the onset and progression of S phase (27, 28). Moreover, overexpression of E2F1 induces Apaf-1-dependent apoptosis without an apparent release of cytc (28). Thus, the gene capable of inducing cell suicide even without any known trigger and those required for S phase onset are regulated by the same transcriptional factor.
Recently we found a clue to solving this paradox. To identify the cell cycle-controlling anchorage signal cascade, we have been using rat embryonic fibroblasts (REFs) constitutively activated for mTORC1 by a homozygous Tsc2 gene mutation or active Rheb overexpression with additional overexpression of Cdc6 to monitor its own protein stability (21). In this study, we found that these Cdc6-overexpressing mTORC1-activated cells did not respond to the apoptotic stimuli induced not only by anchorage deprivation but also by proteasome inhibition, although mTORC1-activated cells were reported to be highly susceptible to stress-induced apoptosis (29, 30). Subsequent investigation led us to resolve this discrepancy and discover an entirely new function for Cdc6.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Anti-caspase-3, anti-caspase-9, anti-cleaved caspase-9 (Asp353), anti-cytochrome c, anti-Bad, anti-S6K1, anti-phospho-S6K1(Thr389), and anti-Rheb antibodies were purchased from Cell Signaling Technology; anti-β-actin antibody was from Santa Cruz Biotechnology; anti-Cdc6 antibody was from NeoMarkers; anti-Apaf-1 antibody was from Alexis Biochemicals; anti-phospho-Rb (Ser780) and anti-caspase-9 mouse monoclonal (clone 5B4) antibodies were from MBL International; anti-caspase-8 antibody was from BioVision; anti-XIAP antibody was from R&D Systems; anti-Rb, anti-Bax, anti-Bcl-2, and anti-Bcl-X antibodies were from BD Biosciences; anti-Bak antibody was from Millipore; and anti-p53 antibody was from Oncogene. CNBr-activated Sepharose 4B beads were purchased from GE Healthcare, and bovine cytochrome c was from Sigma.
Cell Culture and Construction
Cells were maintained in DMEM with 10% FCS. REF, Eker, and/or mouse embryonic fibroblast (MEF) cells expressing a human constitutively active mutant Rheb (29) and/or rat Cdc6 from the cytomegalovirus promoter were constructed using the Retroviral Gene Transfer and Expression System (Clontech) with appropriate drug-selectable marker genes. The drugs used for selection were G418, hygromycin, puromycin, and/or blasticidin.
Methylcellulose Culture
Logarithmically proliferating cells were cultured at 35 °C in a semisolid medium composed of 1.17% methylcellulose, DMEM, and 10% FCS.
Cell Viability Assay
Exponentially proliferating cells were cultured in methylcellulose medium (MC) for the specified times. Cells were harvested, washed with DMEM containing 10% FCS, and plated at 500 cells/6-cm dish. After culturing for 2 weeks, the colonies formed were fixed and counted.
Quantitative Real Time RT-PCR
Preparation of RNA, reverse transcription, and quantitative real time PCR of Apaf-1 and cyclin A transcripts were performed as described (21). The primer pairs used for real time PCR were 5′-gccaccgggtcaaatgactcct-3′ (forward) and 5′-tgctgatctcacatcccaaagctt-3′ (reverse) for Apaf-1, 5′-gagaatgtcaaccccgaaaa-3′ (forward) and 5′-gggacgtgctcatcgtttat-3′ (reverse) for cyclin A, and 5′-catcaccatcttccaggagc-3′ (forward) and 5′-attgagagcaatgccagcc-3′ (reverse) for GAPDH.
Construction of Eker REFs Inducible for Cdc6, Cdc6WB, and Cdc6Cy
The pRevTet-Off vector and the pRevTRE response vector (Clontech) inserted with the cDNA for Cdc6, Cdc6WB, or Cdc6Cy (17) were separately transfected into the EcoPack packaging cell line. The resulting Tet-Off virus was used to infect Eker cells. After brief drug selection, stable Tet-Off integrants were infected with pRevTRE-Cdc6 or corresponding mutants and selected for those with integration of both Tet-Off and TRE-Cdc6 or its mutants. Cell clones that exhibited proper induction were isolated and used for experiments. Cells were maintained in DMEM with 10% FCS and 1 μg of doxycycline/ml.
Immunoprecipitation
Cells (107) were lysed in 0.5 ml of ice-cold immunoprecipitation buffer composed of 50 mm HEPES, 150 mm NaCl, 1 mm EDTA, 2.5 mm EGTA, 1% Nonidet P-40, 10% glycerol, 1 mm DTT, 1 mm PMSF, 10 mm β-glycerophosphate, 50 mm NaF, 1 mm sodium orthovanadate, and 1× Protease Inhibitor Mixture (Sigma) and centrifuged to collect supernatants. The supernatants were incubated at 4 °C with anti-Cdc6 or anti-caspase-9 antibody for 2 h and then with Protein G-Sepharose beads (15 μl) (Amersham Biosciences) for 1 h. The bead-bound immunocomplexes were collected, washed five times with the ice-cold glycerol-free immunoprecipitation buffer, and heat-denatured in SDS sample buffer at 90 °C for 2 min.
In Vitro Cytochrome c-triggered Caspase-9 and Caspase-3 Activation Assay
The cytosol fraction was prepared from proliferating Eker cells as described (32). Briefly, cells (2 × 108) were allowed to swell in ice-cold cell extraction buffer for 30 min on ice and then lysed by homogenization with 20 stokes of a B-type pestle. Lysates were centrifuged at 15,000 × g for 15 min at 4 °C. The supernatants were aliquoted and stored at −80 °C until use. The in vitro apoptosome activation assay was performed by incubating 10 μl of the cytosol at 37 °C for 30 min in a 20-μl reaction mixture containing 20 mm HEPES-KOH (pH 7.5), 10 mm KCl, 1 mm EGTA, 6.5 mm MgCl2, 10 μm cytc (Sigma), 1 mm ATP, and either Cdc6, Cdc6WB, or Cdc6Cy prepared with the in vitro transcription and translation system as described (17). The apoptosome activation assay was carried out by adding cytc last.
Cytc-triggered Stable Complex Formation between Bacterially Expressed Apaf-1 and Purified Recombinant Cdc6
The cDNA encoding rat Cdc6 tagged with 3×FLAG and a histidine hexamer at its C terminus was inserted into the pFASTBAC plasmid (Invitrogen) and then converted to a recombinant baculovirus by transfection into SF9 cells (constructed by Q. Kan). The Cdc6 protein expressed in baculovirus-infected SF9 cells was double affinity-purified with nickel-nitrilotriacetic acid beads and anti-FLAG M2 gels. Various amounts of cytc were incubated at 37 °C for 30 min in a 20-μl reaction mixture containing 20 mm HEPES-KOH (pH 7.5), 10 mm KCl, 1 mm EGTA, 6.5 mm MgCl2, 1 mm ATP, Escherichia coli-expressed rat Apaf-1, and the baculovirus-expressed, double affinity-purified Cdc6. The product was immunoprecipitated for Cdc6 and analyzed for coprecipitated Apaf-1 and cytc.
Preparation of Cytochrome c-conjugated Sepharose Beads
The coupling of cytc to CNBr-activated Sepharose beads was performed according to the vendor's instructions (GE Healthcare). Briefly, the Sepharose beads were washed with a cold acidic solution (pH 2–3) and suspended in 1 mm HCl for 30 min to be allow swelling. The swelled Sepharose beads (1 ml) were suspended in 1 ml of 0.1 m NaHCO3 (pH 8.3) (coupling solution) and incubated at 4 °C overnight with 1 ml of coupling solution containing 10 mg of cytc to let coupling take place. After incubation, the beads were washed once with coupling solution and incubated at room temperature for 2–4 h in 0.1 m Tris-HCl (pH 8.0) buffer to block unused activated sites. The beads were washed with 0.1 m Tris-HCl (pH 8) containing 0.5 m NaCl and then with 0.1 m sodium acetate (pH 4) containing 0.5 m NaCl. This set of washing was repeated eight times. The cytc-conjugated Sepharose beads were stored at 4 °C in 2 ml of 20% ethanol until use.
RNAi-mediated Cdc6 Knockdown
Cells were transfected with the Cdc6-specific 23/27-mer RNA duplex (Integrated DNA Technologies) or the universal negative control duplex at 10 nm according to the vendor's instruction. The following rat and mouse Cdc6-specific RNA duplexes were used: 5′-rGrGrUrUrUrArGrArArArGrArUrGrArArArCrGrGrArArUGA-3′ and 3′-rCrUrCrCrArArArUrCrUrUrUrCrUrArCrUrUrUrGrCrCrUrUrArCrU-5′ (rat) and 5′-rArGrUrUrUrArGrArArGrGrArUrGrArArArUrGrGrArArUrGrA-3′ and 3′-CrUrUrCrArArArUrCrUrUrCrCrUrArCrUrUrUrArCrCrUrUrArCrUr-5′ (mouse).
RESULTS
Enforced Expression of Cdc6 Suppresses Anoikis of mTORC1-activated Fibroblasts
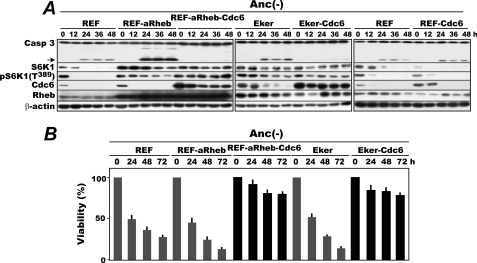
To confirm the initial finding that overexpression of Cdc6 suppresses anoikis of mTORC1-activated rat embryonic fibroblasts irrespective of how mTORC1 is activated, we first compared the anoikis susceptibility of the REFs activated for mTORC1 by a Tsc2 mutation or active Rheb overexpression in the presence or absence of overexpression of Cdc6. Original REFs, REFs overexpressing a constitutively active mutant Rheb (REF-aRheb) (31), original Eker REFs in which the Tsc2 genes are homozygously inactivated by a retrotransposon insertion (Eker) (33, 34), and their Cdc6 overexpressors (REF-aRheb-Cdc6 and Eker-Cdc6 cells, respectively) were cultured in MC, and cells were harvested every 12 h for 48 and 72 h and analyzed for time-dependent activation of caspase-3 and loss of viability, respectively (Fig. 1). In all of the REF, REF-aRheb, and Eker cells, caspase-3 was activated at 24 h of anchorage deprivation with a significantly higher production of its fragmented form in REF-aRheb and Eker cells. In these cells, mTORC1 remained activated despite anchorage loss as shown by the persistent S6 kinase 1 Thr389 phosphorylation. By contrast, in both of the Cdc6 overexpressors, caspase-3 stayed inactive throughout the experiment, confirming the previous results (21). The Cdc6-mediated suppression of caspase-3 activation in anchorage-deprived mTORC1-activated cells is not a local phenotype of rat cells. Virtually identical results were obtained with a set of similarly manipulated MEFs: original MEFs, MEFs activated for mTORC1 by overexpression of active mutant Rheb (MEF-aRheb), and its Cdc6 overexpressor (MEF-aRheb-Cdc6) (supplemental Fig. S1).
FIGURE 1.
Enforced expression of Cdc6 suppresses anoikis of rat embryonic fibroblasts constitutively activated for mTORC1. A, exponentially proliferating REF, REF-aRheb, REF-aRheb-Cdc6, Eker, Eker-Cdc6, and REF-Cdc6 cells were cultured in MC (Anc(-)) with cell sampling every 12 h for 48 h and analyzed for the production of activated caspase-3 and the levels of S6K1 and S6K1 Thr389 phosphorylation, Cdc6, and Rheb by immunoblotting. The arrow indicates the caspase-3-derived fragment generated by its activation. B, the above cells incubated in MC (Anc(-)) for up to 72 h were analyzed for their viability by assaying their colony-forming ability in anchorage-furnished culture plates. The results are shown as averages with S.D. bars in three separate assays. Casp, caspase.
Consistent with the caspase-3 activation, the former three cells (REF, REF-aRheb, and Eker) but not the Cdc6 overexpressors gradually lost viability during MC culture (Fig. 1B). Perhaps reflecting the higher production of activated caspase-3, REF-aRheb and Eker cells lost viability more rapidly than REFs. The anoikis resistance of the overexpressors was not caused by a potential artifact of the retroviral vector system used for Cdc6 overexpression because similarly constructed Cdc6-overexpressing REFs in which Cdc6 protein was completely degraded upon anchorage loss were indistinguishable from original REFs in their susceptibility to anoikis-associated caspase-3 activation (Fig. 1A, rightmost column).
Both Caspase-8 and -9 Are Activated during Anoikis of mTORC1-activated Fibroblasts
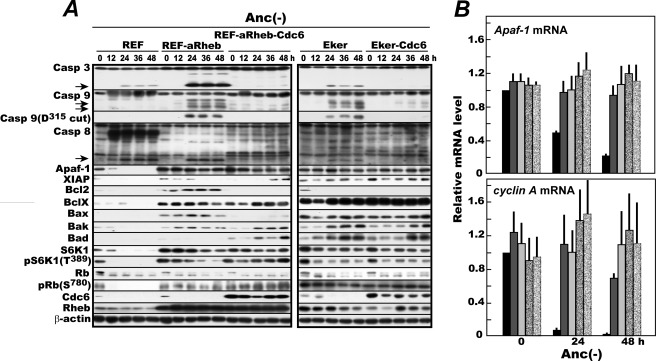
When normal fibroblasts are deprived of anchorage, caspase-8 is activated to initiate anoikis. In those cells, the Apaf-1 gene is turned off because Cdk4/Cdk6 becomes inactive due to a loss of mTORC1 activity (21, 22). By contrast, in the REF-aRheb and Eker cells used above, Apaf-1 is expected to stay turned on without anchorage. Therefore, we next sought to identify which initiator caspase is mainly responsible for executing anoikis of these mTORC1-activated cells. The same set of cells as used for the Fig. 1 analysis were cultured in MC and similarly analyzed for the levels of Apaf-1; activated caspase-3, -8, and -9; anti-apoptotic XIAP; proapoptotic or antiapoptotic Bcl-2 family members; and other proteins reflecting the activities of mTORC1 and Cdk4/Cdk6 (Fig. 2A). Although the procaspase-9 autocleaved at Asp315 and released from the apoptosome is not the active species that processes caspase-3, it serves as a good indicator for caspase-9 activation.
FIGURE 2.
During anoikis of mTORC1-activated fibroblasts, not only caspase-8 and -3 but also caspase-9 are activated, and Cdc6 blocks their activation. A, rapidly proliferating REF, REF-aRheb, REF-aRheb-Cdc6, Eker, and Eker-Cdc6 cells were similarly cultured in MC (Anc(-)) for 48 h and analyzed by immunoblotting for the production of activated caspase-3, caspase-9 (and Asp315-cleaved form), and caspase-8 and for the levels of Apaf-1, proapoptotic and antiapoptotic Bcl-2 family members, S6K1 and Thr389-phosphorylated S6K1 for monitoring mTORC1 activity, and Rb and Ser780-phosphorylated Rb for monitoring Cdk4/Cdk6 activity. Arrows indicate fragments of the corresponding procaspases generated by their activation. B, quantitative real time RT-PCR was performed for the Apaf-1 transcripts with cyclin A mRNA as a reference in the above cells sampled at 0, 24, and 48 h. The data obtained from three independent cell samples are averaged with S.D. bars after normalization to GAPDH mRNA signals and expressed as levels relative to the value of 0-h REFs. Black columns, REFs; dark gray columns, REF-aRheb cells; light gray columns, REF-aRheb-Cdc6 cells; dark dotted columns, Eker cells; light dotted columns, Eker-Cdc6 cells. Casp, caspase.
In anchorage-deprived REFs, activated caspase-8 fragments emerged at 12 h prior to activation of caspase-3. Perhaps due to the cessation of Apaf-1 expression, no detectable amount of autocatalytically cleaved caspase-9 was produced up to 48 h despite its constitutive expression, confirming the generally accepted caspase activation scenario for anoikis. In anchorage-deprived REF-aRheb and Eker cells, however, Apaf-1 expression persisted throughout the experiment because of active Cdk4/Cdk6 resulting from persistently activated mTORC1 in these cells as documented previously and indicated here by the continued Rb Ser780 phosphorylation. Consequently, autocatalytically cleaved procaspase-9 began to emerge at 24 h and remained thereafter with concurrent activation of caspase-3. In addition, in these cells, caspase-8 was activated at 24 h, the same time as the activation of caspase-9 and -3 but slightly behind its activation in REFs. By contrast, neither caspase-8 nor caspase-9 was activated in any of the Cdc6 overexpressors in which activation of caspase-3 was well suppressed.
Meanwhile, antiapoptotic Bcl-X was highly expressed in the mTORC1-activated cells, but its level was not influenced by the enforced expression of Cdc6. On the other hand, Bcl-2 expression was high in REF-aRheb but not in Eker cells. Similarly, the level of XIAP was high in the mTORC1-activated cells but was not influenced by the enforced Cdc6 expression. Consequently, no simple correlation was found between any of these factors and caspase-3 activation. Thus, during anoikis of mTORC1-activated cells in which Apaf-1 continues to be expressed, caspase-9 joined caspase-8 to activate caspase-3, and enforced expression of Cdc6 effectively suppressed activation of both caspase-8 and -9. Consistent with the up-regulated Apaf-1 protein, the Apaf-1 gene was persistently activated in mTORC1-activated cells as shown by real time reverse transcription-coupled PCR quantification of its transcript (Fig. 2B).
Rapamycin Treatment Terminates Both Caspase-9 Activation and Apaf-1 Expression in Anchorage-deprived mTORC1-activated Fibroblasts
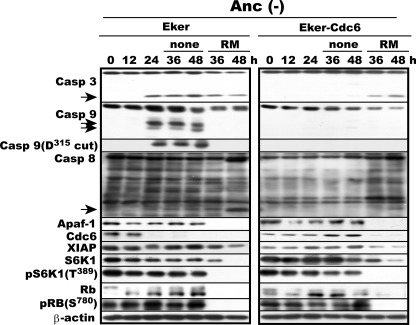
To confirm the responsibility of active mTORC1 for Apaf-1 expression and Cdc6-suppressible caspase-9 activation in anchorage-deprived Eker and Eker-Cdc6 cells, we examined the effects of mTORC1 inactivation on the three caspases in these cells. Proliferating Eker or Eker-Cdc6 cells were cultured in MC for 24 h and then treated with rapamycin, a specific mTORC1 inhibitor, or not for another 24 h, harvested every 12 h from the start of culture, and analyzed for the activation state of caspase-3, -8, and -9 and the levels of Apaf-1 and Cdc6. In Eker cells without treatment, all three caspases were activated at 24 h of anchorage deprivation with continued production of Apaf-1 protein, confirming the Fig. 2 data (Fig. 3). But when the cells were treated with rapamycin, a specific mTORC1 inhibitor, both activated caspase-9 and expressed Apaf-1 vanished, whereas caspase-8 and -3 remained active. On the other hand, when Eker-Cdc6 cells were treated with rapamycin, both caspase-8 and -3 became activated with a rapid loss of Cdc6 and Apaf-1 proteins. However, due to the loss of Apaf-1, caspase-9 remained inactive despite the absence of Cdc6. In this experiment, rapamycin-mediated inactivation of both mTORC1 and Cdk4/Cdk6 was confirmed by the disappearance or marked diminishment of S6K1 and its Thr389 phosphorylation and of Rb and its Ser780 phosphorylation, respectively. Thus, the inhibition of mTORC1 in Eker and Eker-Cdc6 cells terminated Apaf-1 expression and restored the dependence of the initiation of their anoikis only on caspase-8 just like in original REF and REF-Cdc6 cells (Fig. 1).
FIGURE 3.
Inactivation of mTORC1 reverses caspase-8- and -9-dependent anoikis to regular caspase-8-dependent anoikis irrespective of presence or absence of enforced Cdc6 expression. Proliferating Eker and Eker-Cdc6 cells were cultured in MC (Anc(-)) for 24 h and then for another 24 h with or without the addition of rapamycin (RM), a specific inhibitor of mTORC1. Cells were harvested every 12 h and analyzed for the indicated factors by immunoblotting. Arrows indicate fragments of the corresponding procaspases generated by their activation. Casp, caspase.
ATPase Domain but not Cyclin-binding Motif Is Essential for Cdc6 to Suppress Caspase-9 Activation
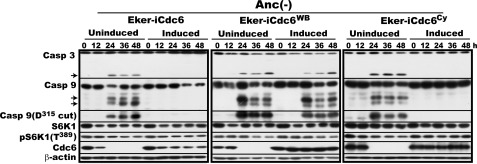
Given the results, we next focused on how Cdc6 quells caspase-9 activation and first sought to understand which domain and motif of Cdc6 are required for this particular ability. Cdc6 contains two domains important for its function: the ATPase domain essential for both prereplicative complex assembly and activation of p21CIP1- or p27KIP1-inactivated Cdk2 and the cyclin-binding (Cy) motif essential only for Cdk2 activation (17, 18, 35). We therefore examined the effects of missense mutations of this domain and this motif on the ability of Cdc6 to suppress caspase-9 activation. Eker cells inducible for the previously prepared and functionally confirmed ATP hydrolysis-deficient Walker B mutant (Eker-iCdc6WB) or Cy motif mutant (Eker-iCdc6Cy) of Cdc6 were constructed with a doxycycline-repressible retroviral system. As a positive control, Eker cells inducible for wild-type Cdc6 (Eker-iCdc6) were also constructed. Without induction, both caspase-9 and -3 were activated in all three cell lines during MC culture as expected (Fig. 4). When wild-type Cdc6 or Cdc6Cy was induced, activation of caspase-9 and -3 was completely suppressed. By contrast, Cdc6WB failed to suppress activation of caspase-9 and -3 as if there was no induction. This result indicates that the ATPase domain but not the Cy motif is required for Cdc6 to block the activation of caspase-9 in sharp contrast to the absolute requirement for both domains in activating p21CIP1- or p27KIP1-bound Cdk2.
FIGURE 4.
ATPase domain but not Cy motif is required for Cdc6 to suppress caspase-9 activation. Rapidly proliferating Eker-iCdc6, Eker-iCdc6WB, and Eker-iCdc6Cy cells were induced for Cdc6 by withdrawal of doxycycline 3 days before analysis or uninduced, incubated in MC Anc(-)) for 24 h with cell sampling at 0 and 24 h, and analyzed for the indicated factors. Arrows indicate fragments of the corresponding procaspases generated by their activation. Casp, caspase.
Cdc6 Forms Stable Complexes with Cytc-bound Apaf-1 and Prevents Apoptosome Assembly in Vivo
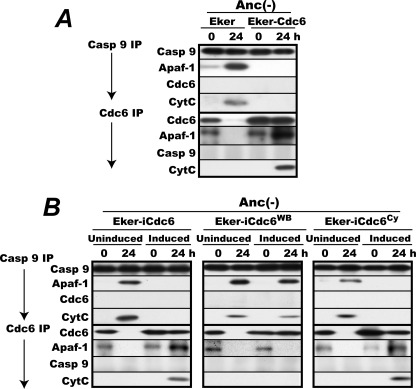
We next sought to identify the molecule(s) with which Cdc6 interacts to block caspase-9 activation. Rapidly proliferating Eker and Eker-Cdc6 cells were incubated in MC for 0 and 24 h, lysed, immunoprecipitated for caspase-9 or Cdc6 with subsequent washes with the ATP-free immunoprecipitation buffer, and analyzed for coprecipitated Apaf-1, cytc, and Cdc6 or caspase-9 (Fig. 5A). Analysis of proteins coprecipitated with Apaf-1 was not possible because no antibody that could effectively immunoprecipitate intact Apaf-1 protein was available. From the lysates of the 0-h incubated Eker and Eker-Cdc6 cells, small amounts of Apaf-1 and cytc were coprecipitated with caspase-9, consistent with no caspase-9 activation in these cells under proliferation. Instead, a small but significant amount of Apaf-1 coprecipitated with Cdc6 from both cell lysates. On the other hand, from the 24-h Eker cell lysate, a large amount of Apaf-1 and cytc coprecipitated with caspase-9, indicating cytc-triggered active apoptosome formation consistent with the caspase-9 activation shown in Fig. 2. In this cell line, Cdc6 protein was markedly diminished or completely disappeared as already shown, and no detectable amounts of Apaf-1 or caspase-9 were precipitated with the anti-Cdc6 antibody. This negative result in turn indicates that the presence of Apaf-1 in the Cdc6 immunoprecipitates from both of the 0-h Eker and Eker-Cdc6 lysates was not an artifact of a possible cross-reaction of the anti-Cdc6 antibody with Apaf-1 itself or some of its interacting proteins.
FIGURE 5.
Cdc6 forms stable complexes with cytc-associated Apaf-1 in ATPase-dependent manner. A, proliferating Eker and Eker-Cdc6 cells were incubated for 0 and 24 h in MC (Anc(-)) lysed, immunoprecipitated (IP) with the anti-caspase-9 or anti-Cdc6 antibody as described under “Experimental Procedures,” and analyzed for immunoprecipitated caspase-9 or Cdc6 and coprecipitated Apaf-1, Cdc6 or caspase-9, and cytc. B, proliferating Eker-iCdc6, Eker-iCdc6WB, and Eker-iCdc6Cy Cells were induced or uninduced for Cdc6 as in Fig. 4, then incubated in MC for 0 and 24 h, and analyzed as in A. Casp, caspase.
By contrast, from the 24-h Eker-Cdc6 lysate in which caspase-9 activation was fully suppressed, neither Apaf-1 nor cytc coprecipitated with caspase-9. Instead, considerable amounts of Apaf-1 and cytc coprecipitated with Cdc6. Thus, the blockade of caspase-9 activation perfectly coincided with the disappearance of coprecipitation of Apaf-1 and cytc with caspase-9 and the reciprocal emergence of coprecipitation of these proteins with Cdc6.
ATP Hydrolysis-deficient Mutant Cdc6 Cannot Form Complexes with Cytc-bound Apaf-1
As shown above, the ATPase domain is essential for Cdc6 to block caspase-9 activation, whereas Cdc6 likely obstructs apoptosome assembly by forming complexes with Apaf-1 or cytc. Given this finding, we next addressed whether the ATPase domain is required for Cdc6 to form the complexes or act at a later step. Eker-iCdc6, Eker-iCdc6WB, and Eker-iCdc6Cy cells induced or uninduced for Cdc6, Cdc6WB, or Cdc6Cy were incubated in MC for 0 and 24 h, lysed, and analyzed as in Fig. 5A (Fig. 5B). Without induction, Apaf-1 and cytc coprecipitated with caspase-9 from each of the 24-h anchorage-deprived cell lysates, reflecting active apoptosome assembly accompanied by caspase-9 activation in these cells. Upon induction, Apaf-1 and cytc coprecipitated with Cdc6 and Cdc6Cy but not with caspase-9 in agreement with the ability of these Cdc6 proteins to block caspase-9 activation. By contrast, Apaf-1 and cytc failed to coprecipitate with Cdc6WB despite its abundant induction. Instead, these proteins coprecipitated with caspase-9, again showing active apoptosome formation, consistent with the inability of this ATPase-deficient mutant to block caspase-9 activation. Thus, the ATP hydrolysis-deficient WB mutant of Cdc6 could not form stable complexes with cytc-activated Apaf-1.
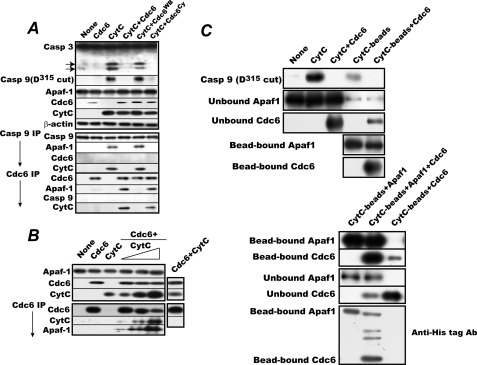
Cdc6 Suppresses Cytc-triggered Caspase-9 Activation by Forming Cdc6·Apaf-1·Cytc Complexes in ATPase-dependent Manner in Vitro
To confirm the above in vivo results, we performed an in vitro reconstitution experiment with a mitochondrion-free preparation of proliferating Eker cell cytosols, bovine cytc, and rat Cdc6 or its mutants produced by a reticulocyte lysate-based expression system (Fig. 6A, upper panel). When cytc was added to the reaction mixture, endogenous caspase-9 and -3 were activated with concurrent apoptosome assembly as indicated by coprecipitation of cytc and Apaf-1 with caspase-9. The addition of Cdc6 without cytc had no effect. The addition of Cdc6 or Cdc6Cy completely suppressed cytc-triggered activation of caspase-9 and -3 with formation of stable complexes between cytc-activated Apaf-1 and Cdc6. By contrast, the same amount of Cdc6WB failed to block their activation and allowed active apoptosome assembly as indicated by the coprecipitation of Apaf-1 and cytc with cappase-9. These results confirm the in vivo findings and demonstrate that Cdc6 forms stable complexes with cytc-activated Apaf-1 to block apoptosome assembly and that this complex formation requires the ATPase of Cdc6.
FIGURE 6.
Cdc6 blocks apoptosome assembly by forming complexes with cytc-bound Apaf-1 in reconstituted system. A, cytc-triggered caspase-9 and caspase-3 activation assay was performed with a mitochondrion-free cytosol preparation in the presence or absence of Cdc6, Cdc6WB, or Cdc6Cy as described under “Experimental Procedures.” The products were directly analyzed for activated caspase-9 and caspase-3 or immunoprecipitated (IP) with the anti-caspase-9 or anti-Cdc6 antibody and analyzed for coprecipitated Apaf-1, Cdc6 or caspase-9, and cytc as in Fig. 5. B, in vitro cytc-triggered stable complex formation between recombinant Apaf-1 and Cdc6. The cytc-triggered complex formation assay was performed with E. coli-expressed affinity-purified Apaf-1, baculovirus-produced double affinity-purified Cdc6, and varying amounts of cytc (1, 2, and 4 μg). The products were analyzed as in A. C, Cdc6 binds to a monomer of cytc-activated Apaf-1. Upper panel, cytc-triggered caspase-9 activation and Cdc6 binding assay was carried out with 40 μg of free or Sepharose bead-conjugated cytc and a mitochondrion-free cytosol preparation in the presence or absence of Cdc6 as in A. After reaction, the production of activated caspase-9 (Asp315-cleaved) and the amounts of unbound and bead-bound Apaf-1 and Cdc6 or those in the reaction mixture were determined by immunoblotting. Lower panel, cytc-triggered Apaf-1 and Cdc6 binding assay was carried out with 40 μg of Sepharose bead-conjugated cytc in the presence or absence of E. coli-produced affinity-purified C-terminally histidine hexamer-tagged Apaf-1 and baculovirus-produced double affinity-purified C-terminally histidine hexamer-tagged Cdc6. The amounts of bead-bound and -unbound Apaf-1 and Cdc6 were determined by regular immunoblotting with specific antibodies. In parallel, the relative amounts of bead-bound Apaf-1 and Cdc6 were quantified by immunoblotting with an anti-histidine hexamer antibody (Ab). Two bands above the Cdc6 are N-terminally viral protein-fused C-terminally histidine hexamer-tagged Cdc6 proteins that were generated by transcription from an upstream promoter(s) in the baculovirus vector and affinity-purified together with Cdc6 protein. Casp, caspase.
Recombinant Cdc6 Forms Stable Complexes with E. coli-expressed Apaf-1 in Presence of Cytc
The next obvious question is whether or not another cellular factor(s) is needed for Cdc6 to form the tertiary complexes. To address this question, we used E. coli-produced rat Apaf-1 and baculovirus-expressed double affinity-purified rat Cdc6 for assessing the ability of Cdc6 to form the complexes. When incremental amounts of cytc were incubated with the Apaf-1 and the Cdc6, incremental amounts of both cytc and Apaf-1 were coprecipitated with Cdc6 (Fig. 6B). The Cdc6 in the ternary complexes appeared to be less efficiently recognized by the antibody because significantly less Cdc6 was immunoprecipitated when a higher amount of cytc was incubated with the proteins. Without the addition of cytc, no Apaf-1 coprecipitated with Cdc6, confirming the requirement for cytc in the binding of Cdc6 to Apaf-1. Reciprocally, in the absence of Apaf-1, no cytc coprecipitated with Cdc6. Thus, no direct interaction between Cdc6 and cytc was detected. These results indicate that no other cellular factor appears to be required for the complex formation between Cdc6 and cytc-activated Apaf-1 and also confirm that Cdc6 cannot form complexes with Apaf-1 molecules unless Apaf-1 is activated by cytc.
Cdc6 Is Likely to Bind Monomer of Cytc-activated Apaf-1
Given the above results, we next investigated how Cdc6 obstructs the assembly of the apoptosome. Two modes are conceivable for the action of Cdc6: it obstructs apoptosome assembly by binding either to a monomer or oligomer of cytc-activated Apaf-1 or alternatively very specifically to the heptameric complex of the activated Apaf-1 that is ready to bind and activate procaspase-9. To distinguish these two, we first prepared Sepharose bead-conjugated cytc, which was used for the activation of Apaf-1, and then examined the binding of Cdc6. If the immobilized cytc can activate Apaf-1 and bind most of the Cdc6 in an amount sufficient to fully block the caspase-9 activation induced by the same amount of free cytc but cannot form the mature apoptosome effectively due to a steric hindrance imposed by the immobilization of the cytc, we can tentatively conclude that Cdc6 obstructs apoptosome assembly very likely by binding to a monomer or oligomer, not specifically to the heptameric complex of cytc-activated Apaf-1. This turned out to be the case. When the bead-bound cytc was incubated with a mitochondrion-free cytosol preparation, it failed to fully activate procaspase-9 as indicated by the much reduced production of the Asp315-cleaved form of caspase-9 in sharp contrast to the strong activation of caspase-9 by the same amount of free cytc (Fig. 6C, upper panel). Nevertheless, when the bead-conjugated cytc was incubated in the presence of the same amount of Cdc6 as that which completely blocked the free cytc-induced caspase-9 activation, almost all the Apaf-1 molecules in the cytosol and most of the Cdc6 were found bound to the beads. This indicates that the form of cytc-activated Apaf-1 that Cdc6 binds is mostly its monomer or oligomer.
To confirm the above results, we estimated the relative amounts of the Apaf-1 and Cdc6 bound to the Sepharose bead-bound cytc. To make such estimation possible, E. coli-produced C-terminally histidine hexamer-tagged Apaf-1 and baculovirus-produced C-terminally histidine hexamer-tagged Cdc6 were used for in vitro binding to the bead-bound cytc in the presence of ATP as in Fig. 6C, upper panel. After incubation, the beads were spun down, washed several times with the reaction buffer, and assayed together with the unbound fraction for the amounts of Apaf-1 and Cdc6 in the bound and unbound fractions by regular immunoblotting. In parallel, the relative amounts of the bead-bound Apaf-1 and Cdc6 were determined by immunoblotting with an anti-histidine hexamer antibody. As shown in Fig. 6C, lower panel, the intensity of the bead-bound Cdc6 immunodetected with the anti-histidine antibody was roughly 2-fold higher than that of the Apaf-1. Two bands above the Cdc6 were actually N-terminally viral protein-fused C-terminally histidine hexamer-tagged Cdc6 proteins that were generated by transcription from an upstream promoter(s) in the baculovirus vector. Considering a low efficiency in transfer of large Apaf-1 molecules (∼140 versus ∼67 kDa for Cdc6) from the SDS gel to a polyvinylidene difluoride sheet for immune detection and the presence of the baculovirus-Cdc6 fusion proteins bound to Apaf-1, we tentatively concluded that Cdc6 protein is likely to associate with cytc-activated Apaf-1 protein roughly at a 1:1 ratio.
Cdc6 Blocks Cytc-less Apoptosome Formation and Caspase-9 Activation in Cells Proliferating in Anchorage-furnished Dishes
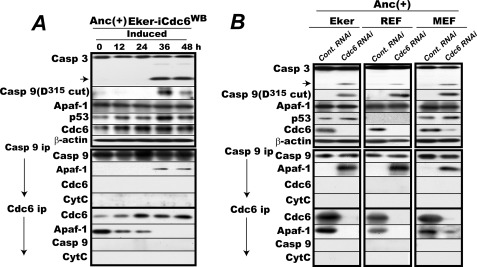
The last question we addressed is that of the nature of the cytc-less Cdc6·Apaf-1 complexes found in proliferating Eker cells: does Cdc6 actually block apoptosome assembly triggered intentionally or unintentionally by some other protein, or are these simply complexes with no functional relevance? We set up two experiments to distinguish these two possibilities: one to examine the putative dominant-negative effect of Cdc6WB expression and the other to examine the effect of endogenous Cdc6 knockdown on the state of both caspase-9 and -3 and cytc-less apoptosome assembly in Eker cells under logarithmic proliferation in culture dishes. If Cdc6 is acting to block apoptosome assembly, both interventions would lead to cytc-less activation of caspase-9 and -3 in proliferating cells. Indeed, this was the case. When Cdc6WB was induced in proliferating Eker cells, both caspase-3 and capase-9 were activated with a cytc-unbound apoptosome assembly reciprocally to the disappearance of cytc-less Apaf-1·Cdc6 complexes (Fig. 7A). Under these experimental conditions, there was only a slight induction of p53, indicating that the cells suffered no excessive stress under the Cdc6WB induction. Cytc-less apoptosome formation with concomitant activation of caspase-3 and -9 was also induced by siRNA-mediated endogenous Cdc6 knockdown in proliferating Eker cells (Fig. 7B, left column).
FIGURE 7.
Enforced expression of Cdc6WB or siRNA-mediated knockdown of Cdc6 induces caspase-9 activation in rodent fibroblasts proliferating in anchorage-furnished culture dishes. A, enforced expression of Cdc6WB induces activation of caspase-9 and -3 with cytc-less apoptosome formation in Eker cells proliferating in anchorage-furnished culture dishes. Proliferating Eker-iCdc6WB cells were transferred into doxycycline-free medium and cultured for 2 days prior to the start of the experiment. The cells were then cultured in the same medium with cell harvests every 12 h for 48 h and analyzed for the indicated factors. In parallel, the lysates were immunoprecipitated (ip) for caspase-9 or Cdc6, and the amounts of coprecipitated Apaf-1, Cdc6 or caspase-9, and cytc were determined. B, siRNA-mediated Cdc6 knockdown induces activation of caspase-9 and -3 with cytc-less apoptosome formation in rodent embryonic fibroblasts cells proliferating in anchorage-furnished culture dishes. Logarithmically proliferating Eker, REF, and MEF cells were transfected with the rat or mouse Cdc6-specific 23/27-mer RNA duplex or a universal negative control (Cont.) and cultured for 48 h. Cells were then harvested and lysed. One-half was analyzed for activation of caspase-3 and -9. The other half was immunoprecipitated for caspase-9 or Cdc6 and analyzed for the amounts of immunoprecipitated caspase-9 or Cdc6 and coprecipitated Apaf-1 and cytc. Casp, caspase. Anc(+), anchorage-furnished.
The blockade of the seemingly inadvertent apoptosome assembly by endogenous Cdc6 and its reversal by Cdc6 knockdown were further confirmed with original REFs and MEFs (Fig. 7B, central and right columns). In REFs and MEFs, the Cdc6 complexed with cytc-less Apaf-1 was present during anchorage-furnished proliferation, and Cdc6 knockdown similarly activated both caspase-9 and -3 with cytc-less apoptosome assembly reciprocally to the disappearance of Cdc6-complexed Apaf-1. Again there was only a marginal elevation in the p53 level (too low to detect in REFs). These results indicate that during regular cell proliferation Cdc6 acts to prevent the active apoptosome assembly either intentionally or unintentionally triggered by some other protein or some other mechanism without apparent apoptotic stimuli that permeabilize the mitochondrial outer membrane.
DISCUSSION
Cdc6 is the AAA+ ATPase that assembles prereplicative complexes on origins of replication and activates p21CIP1- or p27KIP1-inactivated Cdk2. Apaf-1 is another AAA+ ATPase that activates caspase-9 by forming the apoptosome with this initiator caspase and the cytc released from mitochondria upon various intrinsic apoptotic stimuli. Paradoxically, both genes encoding these antipodal life and death AAA+ ATPases are regulated in the same manner by the same transcriptional control system activated by Cdk4/Cdk6 that promotes cell cycle start.
In the present studies, this paradox is solved. The Cdc6 protein expressed at the same time prevents the assembly of the apoptosome by forming complexes with the cytc-activated Apaf-1 molecules. This Cdc6-led complex formation is ATPase-dependent but Cy motif-independent. The ATP binding-proficient but ATP hydrolysis-defective Walker B mutant of Cdc6 could not form such complexes. Once formed, however, these complexes are stable without ATP because similar amounts of cytc-bound Apaf-1 reproducibly coprecipitated with wild-type Cdc6 despite extensive washes of the Cdc6 immunoprecipitants with the ATP-free immunoprecipitation buffer before analysis (see Fig. 5). By contrast, throughout the experiments, no coimmunoprecipitation of Cdc6 with cytc or caspase-9 alone or with the caspase-9 bound to cytc and Apaf-1 was observed. This clearly excludes the possibility that Cdc6 blocks caspase-9 activation by sequestering this caspase or cytc or by physically interfering with the function of properly formed apoptosomes. Thus, the obstruction of mature apoptosome assembly by forming stable complexes with cytc-activated Apaf-1 molecules is the mechanistic basis for the blockade of caspase-9 activation by this AAA+ ATPase. Although three-dimensional structural analysis of the complexes is definitely needed to understand how Cdc6 and Apaf-1 form complexes, an open shape of Apaf-1, such as the one induced by ATP/dATP hydrolysis upon binding of cytc to its WD repeat, always seems to be required for this complex formation because no binding of Cdc6 to Apaf-1 was detected without cytc in the reconstitution systems (Fig. 6, A and B). Furthermore, the experimental data indicate that the cytc-activated Apaf-1 that Cdc6 binds is mostly in the form of monomers or oligomers and not in its heptameric form, which is ready to bind and activate procaspase-9.
Highly intriguingly, in proliferating cells irrespective of the presence or absence of enforced Cdc6 expression, a small but significant amount of cytc-less Apaf-1 was found complexed with Cdc6. This complex could be a representation of either actual obstruction of apoptosome formation by Cdc6 or a mere non-functional binding of Cdc6 to Apaf-1. But subsequent analysis revealed that this complex is indeed the result of obstruction of apoptosome assembly triggered either spontaneously or by some other molecule without apparent apoptotic stimuli during cell proliferation. Both expression of dominant-negative Cdc6WB and RNAi-mediated knockdown of endogenous Cdc6 effectively activated caspase-9 and -3 with cytc-less apoptosome formation. Thus, Cdc6 also appears to protect cells from apoptotic cell death induced by seemingly unintended apoptosome assembly perhaps triggered by some other protein or some other mechanism.
It is well established that anoikis is initiated by FADD-activated caspase 8. But as presented here, the initiator caspase(s) dramatically shifts when the mTORC1 pathway is activated and cytc-activated apoptosomes join to activate the executioner caspase-3. This mechanism involves the well understood cyclin D-dependent kinase cascade. Activated mTORC1 activates Cdk4/Cdk6 despite the absence of anchorage, and this in turn activates E2F by phosphorylating Rb at Ser780 and its cognates, thereby inducing expression of Apaf-1, whereas procaspase-9 is constitutively expressed irrespective of activation of this cascade or not. Upon anchorage deprivation, cytc, presumably released from mitochondria, triggers apoptosome assembly with persistently expressed Apaf-1 and procaspase-9, whereas the endogenous Cdc6 protein produced quickly disappears despite continued transcription because its production is not sufficient to withstand ongoing proteolytic degradation activated by anchorage deprivation. This leads to activation of caspase-9.
During the anoikis of mTORC1-activated cells, both caspase-8 and -9 become activated, and enforced expression of Cdc6 suppresses activation of both caspases. Does Cdc6 have an additional function to independently suppress activation of caspase-8? As shown in the present work, Cdc6 directly blocks activation of caspase-9 by obstructing apoptosome assembly. On the other hand, it is well documented that caspse-8 is also activated by caspase-3 directly or via downstream caspase-6, which forms a positive feedback loop to accelerate apoptosis (36, 37). Consequently, if active mTORC1 suppresses FADD-mediated, but not the feedback-driven, activation of caspase-8, the disappearance of caspase-8 activation in the Cdc6-overexpressing cells might be a mere consequence of suppression of the apoptosome-mediated caspase-3 activation. This possibility may not be so remote because of the following observations. In anchorage-deprived REFs, activation of caspase-8 began at 12 h, whereas in both anchorage-deprived REF-aRheb and Eker cells, its activation occurred at 24 h, the same time as caspase-3 activation (Fig. 2A). In other words, caspase-8 activation in these mTORC1-activated cells was delayed 12 h to coincide with caspase-3 activation. Furthermore, there was no obvious correlation between suppression of caspase-8 activation and Cdc6 expression. In REF-aRheb cells, Cdc6 disappeared at 12 h, but caspase-8 was activated at 24 h concurrently with caspase-3 activation as noted above.
Finally, it is relevant to discuss the biological and potential clinical implications of the integration of antiapoptotic function into the prereplicative complex assembling factor. One biological implication is obviously to protect cells from death induced by an unintended apoptosome assembly once cells have committed to proliferation as discussed above. Another may be to provide a mechanism that aborts proliferation and kills the cells when cells are already set for proliferation by activation of mTORC1 but encounter unfavorable growth conditions that may or may not cause an apparent mitochondrial cytc release but reduces the amount of Cdc6. As shown in Fig. 7B, during cell proliferation, Cdc6 is constantly acting to suppress non-cytc-triggered apoptosome assembly, and when the amount of Cdc6 is reduced below a certain level, this suppression vanishes with consequent activation of caspase-9 and -3. Thus, only a slight down-regulation of Cdc6 expression may be sufficient to activate the executioner caspase in proliferating cells. Consistent with such a mechanism, there are multiple proteolytic systems that degrade Cdc6 when cells are forced to arrest proliferation. During G1 arrest induced by growth factor withdrawal or anchorage loss, Cdc6 is degraded mainly by the APC/CCDH1 ubiquitin ligase but also by another ubiquitin ligase(s) yet to be identified and even likely by a lysosomal cysteine protease(s) (19, 21, 23). In addition, when cell DNA is damaged, Cdc6 is eliminated by the Huwe1 ubiquitin ligase (38). Furthermore, during apoptosis, Cdc6 is fragmented by activated caspase-3 (39). Given our finding, this caspase-3-mediated fragmentation of Cdc6 could be understood as a component of a proapoptotic positive feedback loop in such a mechanism.
To turn to clinical implications, as inferred from the apoptosis-inducing effect of overexpression of the ATP hydrolysis-defective WB mutant of Cdc6, chemicals that specifically inhibit the ATPase of Cdc6 but not that of Apaf-1 would be highly effective in killing any proliferating or proliferation-primed (mTORC1-activated) cells in which Cdc6 is acting to block cytc-less apoptosome assembly, such as proliferating rodent fibroblasts. Based on this inference, we propose a novel anticancer therapeutic strategy utilizing such chemicals.
Supplementary Material
This work was supported by grants-in-aid for scientific research (S) and the Global Center of Excellence from the Ministry of Education, Science and Culture of Japan.

This article contains supplemental Fig. S1.
- cytc
- cytochrome c
- REF
- rat embryonic fibroblast
- Eker
- REF in which the Tsc2 genes are homozygously inactivated by a retrotransposon insertion
- MC
- methylcellulose medium
- MEF
- mouse embryonic fibroblast
- Cy
- cyclin-binding
- WB
- Walker B
- XIAP
- X-linked inhibitor of apoptosis protein
- FADD
- Fas-associated death domain protein
- Cdk
- cyclin-dependent kinase
- APC/C
- anaphase-promoting complex/cyclosome
- S6K1
- S6 kinase 1
- Rb
- retinoblastoma protein.
REFERENCES
- 1. Kerr J. F., Wyllie A. H., Currie A. R. (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vaux D. L., Strasser A. (1996) The molecular biology of apoptosis. Proc. Natl. Acad. Sci. U.S.A. 93, 2239–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zou H., Henzel W. J., Liu X., Lutschg A., Wang X. (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405–413 [DOI] [PubMed] [Google Scholar]
- 4. Schafer Z. T., Kornbluth S. (2006) The apoptosome: physiological, developmental, and pathological modes of regulation. Dev. Cell 10, 549–561 [DOI] [PubMed] [Google Scholar]
- 5. Riedl S. J., Salvesen G. S. (2007) The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8, 405–413 [DOI] [PubMed] [Google Scholar]
- 6. Srinivasula S. M., Ahmad M., Fernandes-Alnemri T., Alnemri E. S. (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1, 949–957 [DOI] [PubMed] [Google Scholar]
- 7. Malladi S., Challa-Malladi M., Fearnhead H. O., Bratton S. B. (2009) The Apaf-1 procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 28, 1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Youle R. J., Strasser A. (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 10. Boatright K. M., Salvesen G. S. (2003) Mechanisms of caspase activation. Curr. Opin. Cell Biol. 15, 725–731 [DOI] [PubMed] [Google Scholar]
- 11. Sherr C. J. (1995) D-type cyclins. Trends Biochem. Sci. 20, 187–190 [DOI] [PubMed] [Google Scholar]
- 12. Pines J. (1994) The cell cycle kinases. Semin. Cancer Biol. 5, 305–313 [PubMed] [Google Scholar]
- 13. Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262 [DOI] [PubMed] [Google Scholar]
- 14. Hsu J. Y., Reimann J. D., Sørensen C. S., Lukas J., Jackson P. K. (2002) E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APCCdh1. Nat. Cell Biol. 4, 358–366 [DOI] [PubMed] [Google Scholar]
- 15. Méndez J., Stillman B. (2000) Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kan Q., Jinno S., Kobayashi K., Yamamoto H., Okayama H. (2008) Cdc6 determines utilization of p21WAF1/CIP1-dependent damage checkpoint in S phase cells. J. Biol. Chem. 283, 17864–17872 [DOI] [PubMed] [Google Scholar]
- 17. Kan Q., Jinno S., Yamamoto H., Kobayashi K., Okayama H. (2008) ATP-dependent activation of p21WAF1/CIP1-associated Cdk2 by Cdc6. Proc. Natl. Acad. Sci. U.S.A. 105, 4757–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uranbileg B., Yamamoto H., Park J. H., Mohanty A. R., Arakawa-Takeuchi S., Jinno S., Okayama H. (2012) Cdc6 protein activates p27KIP1-bound Cdk2 protein only after the bound p27 protein undergoes C-terminal phosphorylation. J. Biol. Chem. 287, 6275–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peters J. M. (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644–656 [DOI] [PubMed] [Google Scholar]
- 20. Sengupta S., Peterson T. R., Sabatini D. M. (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arakawa-Takeuchi S., Kobayashi K., Park J. H., Uranbileg B., Yamamoto H., Jinno S., Okayama H. (2010) Mammalian target of rapamycin complex 1 signaling opposes the effects of anchorage loss, leading to activation of Cdk4 and Cdc6 stabilization. FEBS Lett. 584, 2779–2785 [DOI] [PubMed] [Google Scholar]
- 22. Park J. H., Arakawa-Takeuchi S., Jinno S., Okayama H. (2011) Rho-associated kinase connects a cell cycle-controlling anchorage signal to the mammalian target of rapamycin pathway. J. Biol. Chem. 286, 23132–23141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jinno S., Yageta M., Nagata A., Okayama H. (2002) Cdc6 requires anchorage for its expression. Oncogene 21, 1777–1784 [DOI] [PubMed] [Google Scholar]
- 24. Marconi A., Atzei P., Panza C., Fila C., Tiberio R., Truzzi F., Wachter T., Leverkus M., Pincelli C. (2004) FLICE/caspase-8 activation triggers anoikis induced by β1-integrin blockade in human keratinocytes. J. Cell Sci. 117, 5815–5823 [DOI] [PubMed] [Google Scholar]
- 25. Chiarugi P., Giannoni E. (2008) Anoikis: a necessary death program for anchorage-dependent cells. Biochem. Pharmacol. 76, 1352–1364 [DOI] [PubMed] [Google Scholar]
- 26. Rytömaa M., Martins L. M., Downward J. (1999) Involvement of FADD and caspase-8 signalling in detachment induced apoptosis. Curr. Biol. 9, 1043–1046 [DOI] [PubMed] [Google Scholar]
- 27. Müller H., Bracken A. P., Vernell R., Moroni M. C., Christians F., Grassilli E., Prosperini E., Vigo E., Oliner J. D., Helin K. (2001) E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15, 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Furukawa Y., Nishimura N., Furukawa Y., Satoh M., Endo H., Iwase S., Yamada H., Matsuda M., Kano Y., Nakamura M. (2002) Apaf-1 is a mediator of E2F-1-induced apoptosis. J. Biol. Chem. 277, 39760–39768 [DOI] [PubMed] [Google Scholar]
- 29. Ghosh S., Tergaonkar V., Rothlin C. V., Correa R. G., Bottero V., Bist P., Verma I. M., Hunter T. (2006) Essential role of tuberous sclerosis genes TSC1 and TSC2 in NF-κB activation and cell survival. Cancer Cell 10, 215–226 [DOI] [PubMed] [Google Scholar]
- 30. Karassek S., Berghaus C., Schwarten M., Goemans C. G., Ohse N., Kock G., Jockers K., Neumann S., Gottfried S., Herrmann C., Heumann R., Stoll R. (2010) Ras homolog enriched in brain (Rheb) enhances apoptotic signaling. J. Biol. Chem. 285, 33979–33991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan L., Findlay G. M., Jones R., Procter J., Cao Y., Lamb R. F. (2006) Hyperactivation of mammalian target of rapamycin (mTOR) signaling by a gain-of-function mutant of the Rheb GTPase. J. Biol. Chem. 281, 19793–19797 [DOI] [PubMed] [Google Scholar]
- 32. Slee E. A., Harte M. T., Kluck R. M., Wolf B. B., Casiano C. A., Newmeyer D. D., Wang H. G., Reed J. C., Nicholson D. W., Alnemri E. S., Green D. R., Martin S. J. (1999) Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeung R. S., Xiao G. H., Jin F., Lee W. C., Testa J. R., Knudson A. G. (1994) Predisposition to renal carcinoma in the Eker rat is determined by germ-line mutation of the tuberous sclerosis 2 (TSC2) gene. Proc. Natl. Acad. Sci. U.S.A. 91, 11413–11416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kobayashi T., Hirayama Y., Kobayashi E., Kubo Y., Hino O. (1995) A germline insertion in the tuberous sclerosis (Tsc2) gene gives rise to the Eker rat model of dominantly inherited cancer. Nat. Genet. 9, 70–74 [DOI] [PubMed] [Google Scholar]
- 35. Herbig U., Marlar C. A., Fanning E. (1999) The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol. Biol. Cell 10, 2631–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cowling V., Downward J. (2002) Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death Differ. 9, 1046–1056 [DOI] [PubMed] [Google Scholar]
- 37. Yang S., Thor A. D., Edgerton S., Yang X. (2006) Caspase-3 mediated feedback activation of apical caspases in doxorubicin and TNF-α induced apoptosis. Apoptosis 11, 1987–1997 [DOI] [PubMed] [Google Scholar]
- 38. Hall J. R., Kow E., Nevis K. R., Lu C. K., Luce K. S., Zhong Q., Cook J. G. (2007) Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol. Biol. Cell 18, 3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pelizon C., d'Adda di Fagagna F., Farrace L., Laskey R. A. (2002) Human replication protein Cdc6 is selectively cleaved by caspase 3 during apoptosis. EMBO Rep. 3, 780–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.