Background: Gyrase orthologs differ in supercoiling proficiency for unknown reasons.
Results: Diminished ATPase activity and an inability to couple DNA wrapping with ATP binding differentiate the supercoiling set points of M. tuberculosis and E. coli gyrase.
Conclusion: The shape of a dedicated DNA wrapping domain (the CTD) is not solely responsible for defining gyrase activity.
Significance: Different bacteria fine-tune gyrase by a variety of mechanisms.
Keywords: DNA-binding Protein, DNA Topoisomerase, DNA Topology, Protein-DNA Interaction, Protein Evolution, Chromosome Dynamics
Abstract
DNA topoisomerases are essential enzymes that can overwind, underwind, and disentangle double-helical DNA segments to maintain the topological state of chromosomes. Nearly all bacteria utilize a unique type II topoisomerase, gyrase, which actively adds negative supercoils to chromosomes using an ATP-dependent DNA strand passage mechanism; however, the specific activities of these enzymes can vary markedly from species to species. Escherichia coli gyrase is known to favor supercoiling over decatenation (Zechiedrich, E. L., Khodursky, A. B., and Cozzarelli, N. R. (1997) Genes Dev. 11, 2580–2592), whereas the opposite has been reported for Mycobacterium tuberculosis gyrase (Aubry, A., Fisher, L. M., Jarlier, V., and Cambau, E. (2006) Biochem. Biophys. Res. Commun. 348, 158–165). Here, we set out to understand the molecular basis for these differences using structural and biochemical approaches. Contrary to expectations based on phylogenetic inferences, we find that the dedicated DNA wrapping domains (the C-terminal domains) of both gyrases are highly similar, both architecturally and in their ability to introduce writhe into DNA. However, the M. tuberculosis enzyme lacks a C-terminal control element recently uncovered in E. coli gyrase (see accompanying article (Tretter, E. M., and Berger, J. M. (2012) J. Biol. Chem. 287, 18636–18644)) and turns over ATP at a much slower rate. Together, these findings demonstrate that C-terminal domain shape is not the sole regulatory determinant of gyrase activity and instead indicate that an inability to tightly couple DNA wrapping to ATP turnover is why M. tuberculosis gyrase cannot supercoil DNA to the same extent as its γ-proteobacterial counterpart. Our observations demonstrate that gyrase has been modified in multiple ways throughout evolution to fine-tune its specific catalytic properties.
Introduction
DNA topoisomerases are ubiquitous enzymes that help maintain chromosome superstructure and integrity by counteracting the topological consequences of nucleic acid transactions such as DNA replication and transcription. Topoisomerases fall into two general categories, type I and type II, depending on their respective ability to cleave either one or two DNA strands during catalysis. Although all bacteria sustain a steady-state level of DNA supercoiling by utilizing a combination of both topoisomerase types (1, 2), variations in topoisomerase specialization and composition can greatly affect DNA compaction and gene regulation between species (3–5).
Nearly all bacteria possess a distinct type IIA topoisomerase, termed gyrase, which introduces negative supercoils into DNA using an ATP-dependent DNA duplex strand passage mechanism (4, 6, 7). Gyrase is an A2B2 heterotetramer whose supercoiling activity requires a dedicated DNA binding domain (the CTD)2 that forms the C-terminal portion of the GyrA subunit (8). The GyrA CTD is thought to constrain a positive supercoil by wrapping a DNA duplex around its surface (9–12); this wrap is then converted into two negative supercoils upon strand passage (13). A highly conserved amino acid sequence motif, termed the GyrA box (14), maps to the N-terminal region of the CTD in all gyrases and is required for both wrapping and supercoiling functions (15).
Gyrase is complemented in many bacteria by the presence of a closely related paralog, topo IV, whose ParC subunits bear a divergent variant of the GyrA CTD (4, 16, 17). In contrast to negatively supercoiling DNA, topo IV primarily relaxes positive DNA supercoils and unlinks entangled DNA segments such as catenanes (18–20). Architectural differences between the ParC and GyrA CTDs, such as the deletion of the GyrA box and/or internal subdomains, have been shown to be at least partly responsible for the different activities displayed by gyrase and topo IV on DNA (9, 16, 21). Other considerations, such as CTD position, may also play a role in differentiating between the two enzymes. A number of structural studies in particular have shown that there exists a marked diversity of CTD shapes. Some of these differences, such as whether a CTD is a planar disc or lockwasher-shaped, have been suggested to control the degree to which DNA is wrapped by the GyrA CTD (11). Other features, such as the presence or absence of conserved amino acids at key positions (such as a subdomain interface), have in turn been implicated in controlling CTD shape (9). As yet, there has not been a systematic test of many of these proposals.
The genome of Mycobacterium tuberculosis encodes only a single gyrase ortholog, along with one type IA topoisomerase (22). Because topo IV is the principal agent responsible for unlinking newly replicated chromosomes in Escherichia coli (23), whereas topo IA primarily relaxes negatively supercoiled DNA (24, 25), this combination suggested that M. tuberculosis might employ one of its two topoisomerases in a nonconventional manner. Recent biochemical studies have provided support for this idea, indicating that the decatenation activity of M. tuberculosis gyrase is more robust relative to its supercoiling functions as compared with E. coli gyrase (26). The mechanism underlying this difference likewise has not been elucidated.
In the course of comparing gyrase functions, we noted that the CTD of M. tuberculosis GyrA (MtbGyrA) contains a substitution at the site of a conserved proline that had been proposed to induce a spiral configuration in the domain. Hypothesizing that this change might lead to a new CTD structure that would wrap DNA less effectively, and hence, explain the depressed supercoiling capabilities of M. tuberculosis gyrase, we determined the structure of the M. tuberculosis CTD and characterized its biochemical properties. Surprisingly, we found that the domain adopts a spiral shape nearly indistinguishable from that exhibited by the E. coli GyrA CTD and that the M. tuberculosis CTD was also capable of robustly introducing writhe into DNA. Further investigation revealed that what instead differentiates M. tuberculosis gyrase is that it: 1) bears a naturally truncated version of the unstructured CTD tail that potentiates supercoiling in the E. coli enzyme (see accompanying article (27)) and 2) possesses a much lower DNA-stimulated ATPase activity. Together, these factors appear not to improve decatenation by M. tuberculosis gyrase per se, but rather to alter its supercoiling “set point,” such that the enzyme is incapable of underwinding DNA to the same degree as its E. coli ortholog. Thus, variations in gyrase activity and supercoil addition equilibrium are not solely determined by the CTD and its shape, but can be controlled by a variety of complex factors.
EXPERIMENTAL PROCEDURES
Protein Purification
The coding regions of the M. tuberculosis GyrA CTD (514–838), full-length M. tuberculosis gyrA (1–838), and full-length gyrB (1–714) were amplified from genomic DNA (ATCC) and cloned into a derivative of pET28b behind an N-terminal, tobacco etch virus protease-cleavable hexahistidine tag using an in-house ligation independent cloning vector system (pLIC). The truncated E. coli GyrA CTD (531–853) and full-length E. coli gyrA (1–875) and gyrB (1–804) genes were cloned into pET28b. Proteins were expressed in E. coli BL21-CodonPlus(DE3)-RIL cells (Stratagene) by inducing log-phase cells with 0.25 mm isopropyl-β-d-thiogalactopyranoside either for 4 h at 37 °C or overnight at 18 °C. Full-length M. tuberculosis GyrB was expressed in BL21-CodonPlus(DE3)-pLysS cells by inducing log-phase cells with 1 mm isopropyl-β-d-thiogalactopyranoside for 3 h at 30 °C. Cells were harvested by centrifugation, resuspended in 20 mm Tris-HCl, pH 7.9, 800 mm NaCl, 30 mm imidazole, 10% glycerol, and protease inhibitors (1 μm leupeptin, 1 μm pepstatin A, and 1 mm phenylmethylsulfonyl fluoride), and frozen dropwise in liquid nitrogen for storage at −80 °C.
For purification, cells were sonicated and centrifuged, and the clarified lysate was passed over an Ni2+ affinity column (Amersham Biosciences). His-tagged protein was eluted with 20 mm Tris-HCl, pH 7.9, 100 mm NaCl, 500 mm imidazole, 10% glycerol, and protease inhibitors (1 μm leupeptin, 1 μm pepstatin A, and 1 mm phenylmethylsulfonyl fluoride), concentrated and exchanged into the same buffer containing 30 mm imidazole, and then incubated overnight at 4 °C with 1–1.5 mg of hexahistidine-tagged tobacco etch virus protease (28). Following tobacco etch virus cleavage, the mixture was passed over an Ni2+ affinity column, and the flow-through was collected, concentrated (Millipore Amicon Ultra-10/30), and run over an S-200 or S-300 gel filtration column (Amersham Biosciences) in 50 mm Tris-HCl, pH 7.9, 500 mm KCl, 10% glycerol, and 2 mm 2-mercaptoethanol. Peak fractions were pooled and concentrated by centrifugal filtration (Millipore Amicon Ultra-10/30) and assessed for purity by SDS-PAGE (supplemental Fig. S1). M. tuberculosis GyrB was purified using the above protocol but with the following Ni2+ affinity column buffers: load/wash buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 5 mm imidazole) and elution buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 250 mm imidazole).
Crystallization, Data Collection, and Structure Solution
Purified M. tuberculosis GyrA CTD at 12 mg/ml was dialyzed overnight at 4 °C against 50 mm NaCl and 10 mm Tris-HCl, pH 7.5. Crystals were grown in mosquito drop hanging drop format by mixing 100 nl of protein with 100 nl of well solution containing 0.2 m calcium acetate hydrate and 20% polyethylene glycol 3350. For harvesting, crystals were looped and flash-frozen in liquid nitrogen.
A single native dataset was collected at Beamline 8.3.1 at the Advanced Light Source at Lawrence Berkeley National Laboratory (29). Native data were indexed and reduced as P212121 using HKL2000 (see Table 1) (30). Phasing by molecular replacement using the E. coli GyrA CTD (residues 532–853, Protein Data Bank (PDB) code 1ZI0) revealed one M. tuberculosis GyrA CTD protomer per asymmetric unit (11, 31). Density modification and initial model building were performed by PHENIX (32); subsequent cycles of manual rebuilding and refinement were performed using COOT (33). TLS parameters were analyzed with the TLSMD server (34), and 15 TLS groups were introduced in the later stages of refinement. The final model was refined to 1.65 Å resolution, with a final Rwork/Rfree of 22.2/18.7%. Molprobity analysis (35) showed a total of 99% of residues in the most favored regions of Ramachandran space, with none in disallowed regions. Ribbon structures, electron density, and surface charge images were all prepared in PyMOL (36).
TABLE 1.
Data collection and refinement statistics
| MtbGyrA CTD | |
|---|---|
| Data collection | |
| Resolution (Å) | 50-1.65 |
| Wavelength (Å) | 1.1000 |
| Space group | P212121 |
| Unit cell dimensions (a, b, c) (Å) | 38.87, 82.84, 83.43 |
| Unit cell angles (α, β, γ) (°) | 90, 90, 90 |
| I/σ (last shell) | 27.1 (3.6)a |
| Rmerge (last shell) (%)b | 7.4 (5.6) |
| Completeness (last shell) (%) | 100 (100) |
| Redundancy | 7.1 (7.0) |
| Unique reflectionsc | 33,301 |
| Refinement | |
| Resolution (Å) | 41.72-1.65 |
| No. of reflections | 33,225 |
| Rwork (%) (last shell)d | 18.76 (25.13) |
| Rfree (%) (last shell)e | 22.21 (28.18) |
| Structure and stereochemistry | |
| No. of atoms | 2733 |
| Protein | 2309 |
| Water | 303 |
| Acetate | 1 |
| Calcium | 1 |
| Glycerol | 1 |
| B-factor (Å2) | |
| Protein | 20.72 |
| Water | 29.02 |
| r.m.s.d.f bond lengths (Å) | 0.009 |
| r.m.s.d.f bond angles (°) | 1.182 |
| Ramachandran plot (%)g | |
| Favored region | 99.0 |
| Allowed region | 1.0 |
| Outliers | 0 |
a The values in parentheses indicate the highest resolution shell.
b Rmerge + hkliIi(hkl) I(hkl)/hkliIi(hkl), where Ii(hkl) is the intensity of an observation and I(hkl) is the mean value for its unique reflection. Summations cover all reflections.
c Nobs/Nunique.
d Rwork + Σhkl ‖Fobs| −k|Fcalc‖/Σhkl |Fobs|.
e Rfree was calculated same way as Rwork, but with the reflections excluded from refinement. The Rfree set was chosen using default parameters in PHENIX (32).
f r.m.s.d., root mean square deviation.
g Categories were defined by Molprobity (35).
DNA Binding Assays
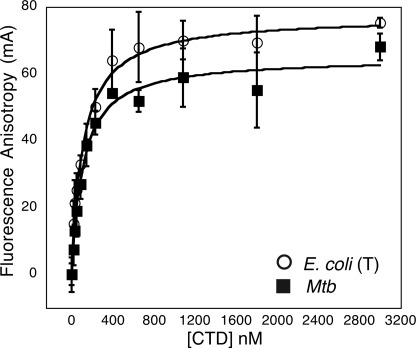
DNA binding was determined by fluorescence anisotropy. The DNA substrate for binding was a random, 37-bp segment with 40% GC content. Annealed oligonucleotides 5′-TAA AGT CTA GAG ACA CGC ATA GTC AAT GAC GGA GTT A-3′ and 5′-|56-FAM|TAA CTC CGT CAT TGA CTA TGC GTG TCT CTA GAC TTT A-3′ (where 56-FAM indicates the position of a carboxyfluorescein dye for visualization) were purchased from Integrated DNA Technologies and resuspended in TE buffer (10 mm Tris, pH 8.0, 1 mm EDTA, pH 8.0). For binding, varying amounts of truncated E. coli GyrA CTD or M. tuberculosis GyrA CTD were incubated with 20 nm of fluorescently labeled duplex oligonucleotide at room temperature in the dark in 20 mm Tris-HCl, pH 7.5, 70 mm KCl, 10% glycerol, and 1 mm MgCl2. Fluorescence anisotropy measurements were performed using a Victor 3V (PerkinElmer Life Sciences) multilabel plate reader. Data points represent the average of three independent measurements on each plate, whereas error bars represent the S.D. between measurements (Figs. 2 and 7). Binding curves were fit to a simplified version of the single site binding equation that holds for our experimental conditions using KaleidaGraph version 4.0 (Synergy software)
 |
where θ represents the fraction of ligand binding sites filled, Kd,app is the apparent dissociation constant, and L is the ligand concentration.
FIGURE 2.
DNA binding by CTD constructs. Binding of fluorescein-tagged, double-stranded oligonucleotides (20 nm), monitored as a function of CTD concentration by fluorescence anisotropy, is shown. The apparent Kd values are as follows: truncated E. coli CTD, 90 ± 21 nm; M. tuberculosis (Mtb) CTD, 80 ± 16 nm. The truncated E. coli CTD is indicated by T.
FIGURE 7.
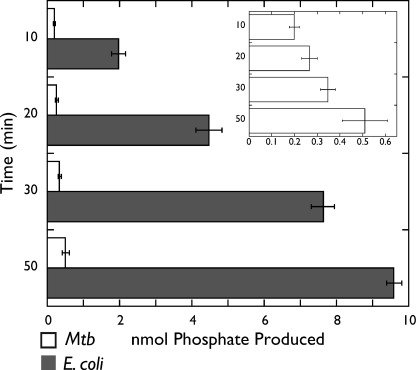
ATPase activity. E. coli and M. tuberculosis (Mtb) gyrase ATPase activities are plotted as a function of nmol of phosphate produced (x axis) and time in minutes (y axis). The graph in the inset provides a magnified version of the M. tuberculosis gyrase ATPase data.
DNA Relaxation, Decatenation, and Supercoiling Assays
pSG483, a derivative of pUC19 containing a unique Nb.BbvCI nicking site (New England Biolabs), was used for supercoiled and relaxed DNA substrates. Negatively supercoiled plasmid was purified from E. coli with a maxiprep kit (Macherey-Nagel). Relaxed plasmid was made using nick ligation by first nicking maxiprepped plasmid DNA with Nb.BbvCI and then ligating with T4 DNA ligase. Kinetoplast DNA (kDNA) from Crithidia fasciculata was purified in-house as described (37). DNA relaxation, decatenation, or supercoiling assays (30 μl) were performed in a buffer containing 15 mm Tris-HCl, pH 7.5, 13% glycerol, 6 mm MgCl2, 0.1 mg/ml BSA, 70 mm KCl, 1 mm dithioerythritol, 2 mm ATP, and 300 ng (6 nm) of DNA substrate. To assess activity, reactions were supplemented with varying amounts of reconstituted holoenzyme, ATP was added to reaction mixtures, and reactions were allowed to proceed for 30 min at 37 °C. Time course reactions contained 4 mm ATP, with an additional 2 mm ATP added at 60 min. Reactions were stopped by the addition of SDS (1% final concentration) and EDTA (10 mm final concentration). Quenched reactions were analyzed by electrophoresis through 1.0% agarose gels with 1× TAE running buffer (40 mm Tris-acetate, 1 mm EDTA) and chloroquine gels containing 3 μg/ml chloroquine (Sigma) in the gel and running buffer. Gels were run at 1.7–5.0 volts/cm for 4–12 h at 25 °C, stained with ethidium bromide (EtBr), and visualized by UV transillumination.
Topological Footprinting Assays
DNA writhe assays (30 μl) were performed in a buffer containing 15 mm Tris-HCl, pH 7.5, 13% glycerol, 6 mm MgCl2, 0.1 mg/ml BSA, 70 mm KCl, and 300 ng (6 nm) of nicked pSG483 as DNA substrate. Varying amounts of CTD were added to reaction mixtures, and the reactions were equilibrated at 37 °C for 20 min, after which 60 units of ligase and 1 mm ATP were added to each reaction mixture. Reactions were then allowed to incubate at 37 °C for an additional 20 min. Reactions were stopped by the addition of SDS (1% final concentration), EDTA (10 mm final concentration), and proteinase K (50 μg/ml final) and incubated at 37 °C for 30 min. Reactions were analyzed by electrophoresis through 1.0% agarose gels with 1× TAE running buffer. Gels were run at 1.7 volts/cm for 19–21 h at 25 °C, stained with ethidium bromide (EtBr), and visualized by UV transillumination.
To capture a gyrase holoenzyme “topology footprint,” we used an ATP-independent assay technique to prevent gyrase from supercoiling the plasmid DNA substrate. In these reactions, gyrase holoenzyme was incubated with relaxed plasmid and eukaryotic topoisomerase IB, which relaxes DNA supercoils in the absence of ATP (38, 39). Holoenzyme writhe assays were performed in the presence and absence of 2 mm AMP-PNP using relaxed pSG483 plasmid and topoisomerase IB purified in-house from wheat germ (40).
ATPase Assays
ATPase assays were performed using a malachite green/molybdate assay for free phosphate. Reactions, run under supercoiling assay conditions, contained 50 nm gyrase, 1 mm ATP, and 50 μm sheared salmon sperm DNA (Sigma). Reactions were initiated by the addition of enzyme and ATP and incubated for varying times at 37 °C. After incubation, 200 μl of malachite green reagent (0.034% malachite green, 10 mm ammonium molybdate, 1 n hydrochloric acid, 3.4% ethanol, and 0.01% Tween 20) was added to each reaction and incubated at room temperature in the dark for 5 min. The malachite green reagent was made fresh daily and sterile-filtered. Absorbance at 620 nm was measured using a PerkinElmer Life Sciences Victor 3V plate reader. All reactions were performed in triplicate.
RESULTS
Structure of MtbGyrA CTD
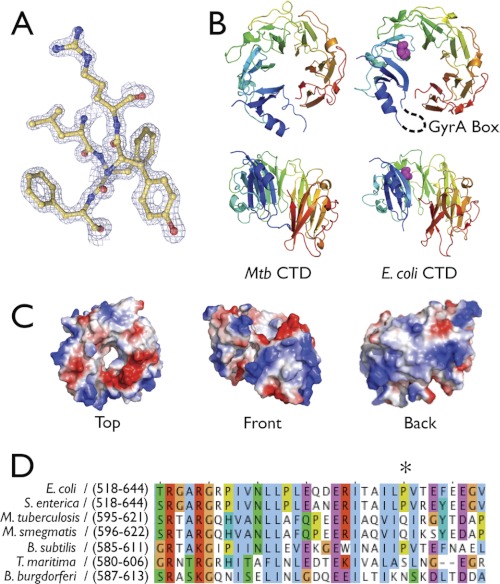
To first assess the physical basis for the activity differences seen between M. tuberculosis and E. coli gyrase, we determined the structure of the MtbGyrA CTD. Crystals of the isolated domain (residues 514–838) were obtained in the space group P212121 and diffracted to 1.65 Å resolution. The structure was solved by molecular replacement using a polyserine model of the E. coli GyrA CTD (residues 532–853, PDB code 1ZI0) (Fig. 1A) (11). The final model, which comprises amino acids 514–538 and 547–819 of the M. tuberculosis CTD, was refined to an Rwork/Rfree of 18.76/22.21% with excellent stereochemistry (Table 1).
FIGURE 1.
M. tuberculosis CTD structure. A, representative electron density from a refined 2Fo − Fc map contoured at 2σ. B, ribbon model of MtbGyrA and E. coli GyrA CTD showing face-on (top panel) and front views (bottom panel). On the E. coli CTD model, the conserved proline is depicted in magenta spheres and the hypothetical location of the GyrA Box is indicated with a dashed black line. Mtb CTD, M. tuberculosis CTD. C, electrostatic surface representation of MtbGyrA CTD. D, sequence alignment of representative CTDs that contain or lack the conserved proline (marked with an asterisk) that follows the GyrA box. M. smegmatis, Mycobacterium smegmatis; B. subtilis, Bacillus subtilis; S. enterica, Salmonella enterica; T. maritima, Thermotoga maritima.
As seen in other GyrA and ParC CTDs, the M. tuberculosis gyrase CTD forms a “β-pinwheel” structure similar to, but topologically distinct from, canonical β-propellers (Fig. 1B). The M. tuberculosis CTD structure turns out to closely mimic that of the E. coli CTD (1.45 Å root mean square deviation) and likewise displays a positively charged band around the perimeter of the disc that is thought to assist with DNA binding and/or bending (Fig. 1C) (12). As with the E. coli CTD, the GyrA box element of the M. tuberculosis domain is also disordered. Interestingly, the M. tuberculosis CTD also adopts a distinctly nonplanar conformation despite lacking a moderately conserved proline that has been suggested to promote the formation of the spiral state adopted by the GyrA CTDs of E. coli and Xanthomonas campestris (9) (Fig. 1D); this finding indicates that the existence of a proline at this position is not responsible for configuring the CTD into a particular shape. Overall, inspection of the M. tuberculosis CTD structure failed to highlight any obvious structural features that might have accounted for the differences in activity reported for M. tuberculosis and E. coli gyrase.
DNA Binding and Wrapping by M. tuberculosis CTD and M. tuberculosis Gyrase
Because biochemical properties are not necessarily evident from structure alone, we next asked whether the M. tuberculosis and E. coli CTDs might associate with DNA differently. We first compared the relative affinities of two CTDs for short DNA substrates using fluorescence polarization. In this assay, a fluorescently labeled, 37-bp duplex oligonucleotide was incubated with varying amounts of purified E. coli or M. tuberculosis gyrase CTD. In the course of conducting these studies, we noted that the E. coli CTD constructs used for biochemical and structural studies by other groups actually employed a variant that lacked some or all of a nonconserved acidic tail located at the C terminus of the domain (11). These constructs bound DNA robustly in our hands, whereas a full-length CTD bearing an intact tail failed to bind DNA at all (see accompanying article (27)). By contrast, the full-length M. tuberculosis CTD bound DNA with an affinity similar to that of the tail-truncated E. coli domain (Kd,app = 80 ± 16 nm and 90 ± 21 nm, respectively) (Fig. 2). An inspection of GyrA amino acid sequences shows that the M. tuberculosis CTD naturally lacks ∼15 residues from its C-terminal tail as compared with E. coli (see Fig. 1B of the accompanying article (27)). Thus, although the innate DNA binding properties of the core CTD are similar between E. coli and M. tuberculosis gyrase, their CTD tails and the regulatory capacities of these elements are distinct.
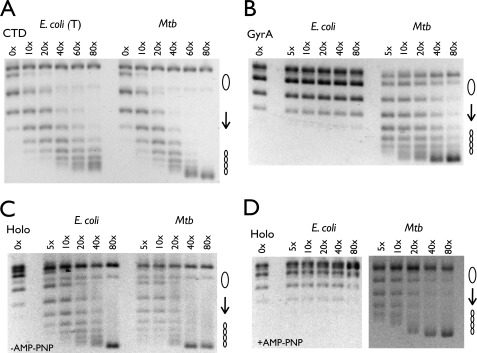
DNA wrapping by the CTD is essential for supporting supercoiling by gyrase (8). Reasoning that binding affinity might not necessarily imply an equal proclivity to introduce writhe into DNA, we set out to compare the wrapping potential of the M. tuberculosis and E. coli CTDs using “topology footprinting” (10, 11). In this assay, each CTD was first individually mixed with nicked plasmid DNA, after which the DNA was ligated, trapping any writhe constrained by the protein. Bound CTD molecules were removed by SDS/proteinase K, and the supercoiling state of the DNA was resolved by native agarose gel electrophoresis. Inspection of the topoisomers resulting from this treatment shows that both the full-length M. tuberculosis and the tail-truncated E. coli CTDs were able to constrain a similar amount of writhe with respect to one another (Fig. 3A). Consistent with its binding defect, the full-length E. coli CTD did not show any ability to wrap DNA (see accompanying article (27)).
FIGURE 3.
Topology footprint assays. A, nicked pSG483 (6 nm) was incubated with varying amounts of CTD and ligase (indicated as a molar excess of CTD over plasmid). The truncated E. coli CTD is indicated by T. Mtb, M. tuberculosis. B, nicked pSG483 (6 nm) was incubated with varying amounts of GyrA dimer and ligase (indicated as a molar excess of CTD over plasmid). C and D, relaxed pSG483 (6 nm) was incubated with varying amounts of reconstituted holoenzyme (Holo) and topo IB in the absence (C) and presence (D) of AMP-PNP (2 mm) (indicated as a molar excess of CTD over plasmid). The positions of relaxed and supercoiled DNA species are labeled with graphic representations on the right.
Because the E. coli gyrase holoenzyme (41), but not the isolated GyrA subunit (10), can constrain writhe, we were curious whether M. tuberculosis gyrase and its GyrA subunit, which naturally lack a portion of the E. coli CTD tail, would behave similarly. When assayed by topology footprinting, M. tuberculosis GyrA was able to wrap DNA to a similar degree as its isolated CTD, whereas E. coli GyrA failed to exhibit any wrapping (Fig. 3B). By contrast, both the E. coli and the M. tuberculosis holoenzymes wrapped DNA nearly equivalently (Fig. 3C). To probe these differences further, we next assessed whether DNA wrapping by M. tuberculosis gyrase could be blocked by the nonhydrolyzable ATP analog AMP-PNP, a behavior seen for E. coli gyrase (42, 43). Strikingly, the ability of M. tuberculosis gyrase to introduce writhe was unaffected by the presence of AMP-PNP, whereas wrapping by E. coli gyrase was all but abolished (Fig. 3D). Taken together, these results demonstrate that the extent of DNA wrapping induced by E. coli and M. tuberculosis gyrase is not appreciably distinct; rather, only E. coli gyrase appears capable of regulating wrapping in an ATP-dependent manner.
Decatenation by M. tuberculosis Gyrase
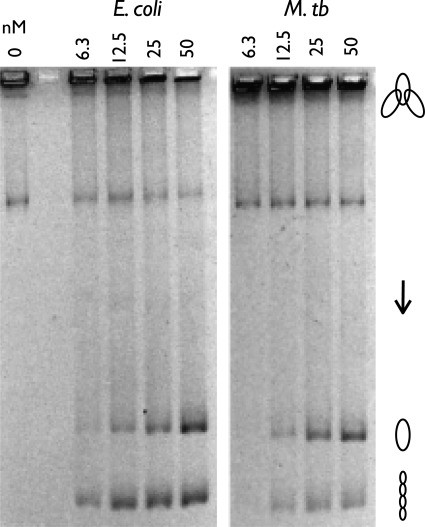
Having established that an ability to switch between wrapped and unwrapped states, rather than an inherent structural or biochemical feature of the GyrA CTD, is a point of divergence for the M. tuberculosis and E. coli orthologs, we turned our attention to other properties of the two enzymes. We first examined DNA decatenation, which has been reported to be more efficient with M. tuberculosis gyrase than with the E. coli protein (26). Decatenation was quantified by incubating each reconstituted holoenzyme with kinetoplast DNA (kDNA) and ATP and then resolving the products using native agarose gel electrophoresis. The large, catenated network of DNA minicircles that comprises kDNA is unable to enter the gel unless individual minicircles are liberated by decatenation by gyrase. Contrary to our expectations, we found that the observed decatenation efficiency of E. coli gyrase was comparable with, or even slightly better than, that seen for the M. tuberculosis enzyme (Fig. 4). Although the basis for this discrepancy was not immediately apparent, it did not arise from the presence of a significant portion of inactive enzyme in our M. tuberculosis gyrase preparations. Indeed, the specific activities of both our gyrase preparations were sufficient to nearly fully supercoil a target plasmid at a >50-fold molar excess of substrate DNA over enzyme under standard published conditions (Fig. 5A). This level of activity, which may arise as a consequence of a more extensive purification procedure than that used in prior studies, is ∼5–10-fold greater than previously observed for the M. tuberculosis enzyme.
FIGURE 4.
E. coli and M. tuberculosis holoenzyme decatenation assay. The positions of the kinetoplast network, relaxed released circles, and supercoiled released circles are labeled with graphic representations on the right. M. tb, M. tuberculosis.
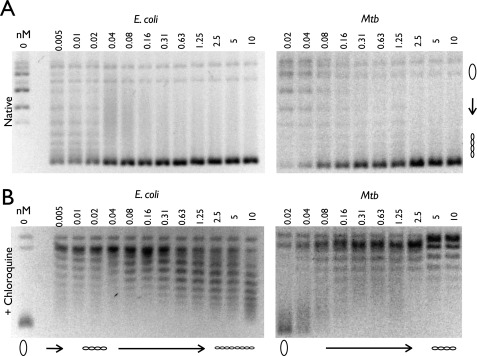
FIGURE 5.
E. coli and M. tuberculosis negative supercoiling holoenzyme assays. A and B, a portion of each sample was run on a 1% agarose gel in the absence (A) and presence (B) of 3 μg/ml chloroquine. Protein concentrations are listed in nm holoenzyme, and DNA topoisomers are labeled with graphic representations across the bottom of the chloroquine gel. Relaxed and supercoiled DNA species are labeled with graphic representations on the right of the native gel. Mtb, M. tuberculosis.
DNA Supercoiling by M. tuberculosis Gyrase
If M. tuberculosis and E. coli gyrase decatenate DNA similarly, then why might E. coli require a topo IV for chromosome segregation, whereas M. tuberculosis does not? To address this question, we re-examined the DNA supercoiling activity of the two proteins in detail. One clue came from inspection of our kDNA decatenation data (Fig. 4), which indicated that the minicircles liberated by E. coli gyrase were supercoiled more readily than those produced by the M. tuberculosis enzyme. Because kDNA is an unusual substrate, we further compared the activities of both gyrases on relaxed circular DNA in the presence of ATP.
Analysis of the resultant products by agarose gel electrophoresis at first appeared to show comparable supercoiling activities between the two orthologs (Fig. 5A). However, we noticed that the band containing the supercoiled DNA species produced by M. tuberculosis gyrase often appeared somewhat “fuzzy,” or less condensed. Because native gels have an innate limit on their ability to resolve topoisomers that are significantly underwound with respect to one another (44), we turned to chloroquine gels to determine whether the degree of DNA supercoiling introduced by M. tuberculosis gyrase might differ from that of E. coli. Chloroquine is a weak intercalating agent that separates supercoiled topoisomers and allows for the resolution of negatively supercoiled DNA bands that would otherwise run together as a single high mobility band on a native gel (44). A portion of each reaction used in our native gel studies was run on an agarose gel in the presence of 3 μg/ml chloroquine, after which the chloroquine was soaked out and replaced with ethidium bromide for visualization. Notably, this analysis revealed that at comparable concentrations, M. tuberculosis gyrase is unable to introduce as many negative supercoils into DNA as E. coli gyrase (Fig. 5B).
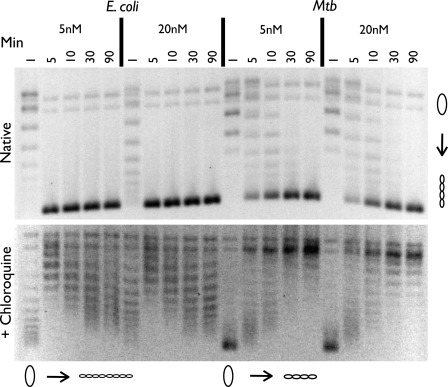
Although this experiment suggested that M. tuberculosis and E. coli gyrase differ in yet another fundamental property, it did not rule out the possibility that strand passage by the M. tuberculosis enzyme might simply be slow, and hence, failed to go to completion during the course of our assay (30 min). To assess this possibility, we assayed supercoiling over a range of times. The reconstituted E. coli and M. tuberculosis gyrase enzymes were added to relaxed plasmid DNA and ATP at two concentrations (5 and 20 nm), both of which completely supercoiled the substrate in our enzyme titration experiments, and incubated at 37 °C for 1, 5, 10, 30, and 90 min. Time points that exceeded 30 min were supplemented with additional ATP to prevent ATP-independent relaxation events from occurring as the nucleotide cofactor was depleted, and the results were again visualized using agarose gel electrophoresis in the absence and presence of 3 μg/ml chloroquine. We found that supercoiling for both enzymes is complete within 30 min and that even at longer times, M. tuberculosis gyrase still does not underwind DNA to the same degree as E. coli (Fig. 6).
FIGURE 6.
Negative supercoiling time course assay using 5 and 20 nm holoenzyme. Time points are listed in minutes. A portion of each sample was run on a 1% agarose gel in the absence (51) and presence (bottom) of 3 μg/ml chloroquine. Protein concentrations are listed in nm holoenzyme, and DNA topoisomers are labeled with graphic representations across the bottom of the chloroquine gel. Relaxed and supercoiled DNA species are labeled with graphic representations on the right of the native gel. Mtb, M. tuberculosis.
ATPase Activity of M. tuberculosis Gyrase
Because M. tuberculosis gyrase is able to robustly bind and wrap DNA, but lacks a CTD control element (the tail) that permits the coordination between nucleotide turnover and strand passage in its E. coli counterpart, we hypothesized that the slower and less extensive supercoiling seen for this mycobacterial enzyme might arise from a similar coupling inefficiency. To test this idea, we examined the ATPase activity of M. tuberculosis and E. coli gyrase using a malachite green colorimetric assay in which ATP hydrolysis is measured by phosphate release (45). We conducted these measurements by incubating reconstituted gyrase in our supercoiling assay conditions with salmon sperm DNA, which is known to dramatically stimulate the ATPase activity of the enzyme (46). The reactions were incubated for different amounts of time using a fixed starting concentration of DNA and ATP. Surprisingly, we found that although both orthologs exhibited ATPase activity, M. tuberculosis gyrase turned over ATP at a significantly lower rate than E. coli gyrase (Fig. 7). Taken together, these data suggest that strand passage is closely linked to ATP hydrolysis in M. tuberculosis gyrase, but that two factors, a lack of coupling between DNA wrapping and ATP binding, as well as an intrinsically slow rate of ATP turnover, contribute to the reduced supercoiling capacity of the holoenzyme.
DISCUSSION
It is known that different bacterial lineages display substantial variation in the number and type of topoisomerases they possess (4). For instance, most bacteria possess two heterotetrameric type IIA topoisomerases, gyrase and topo IV, which divide the labor of DNA supercoiling and chromosome decatenation; however, there exist many species that encode only one paralog or the other. In addition, there is growing evidence of specific functional and architectural differences between type IIA topoisomerase orthologs. A unique domain insertion found in the GyrB subunit of γ-Proteobacteria that impacts DNA binding serves as one example (47), whereas an acidic tail appended to the C terminus of E. coli GyrA that controls DNA wrapping is another (see accompanying article (27)). Despite extensive study, it is often unclear from phylogenetic information alone which modifications impact gyrase function and how.
M. tuberculosis gyrase expands further upon these distinctions. Not only is gyrase the sole type IIA topoisomerase of M. tuberculosis (22), its ability to decatenate DNA is elevated as compared with its supercoiling activity (26) (Fig. 8). Because decatenation is a function generally associated with topo IV, we set out to structurally and biochemically define which elements and characteristics of M. tuberculosis gyrase might differ from its well studied counterpart in E. coli. We were particularly intrigued as to whether the CTD of MtbGyrA might play a role because this domain has been implicated in the controlling the specificity of the gyrase and topo IV strand passage mechanism in numerous contexts (8, 16).
FIGURE 8.
Schematic comparing relative supercoiling set points of a select number of bacterial type IIA topoisomerases. topo IV relaxes DNA. E. coli gyrase can negatively supercoil DNA both rapidly and extensively. M. tuberculosis (Mtb) and S. typhimurium gyrase, as well as an E. coli gyrase lacking the C-terminal tail of the GyrA subunit, can all negatively supercoil DNA, but to a lesser degree than wild-type E. coli gyrase. Some of the mechanistic properties that appear to mediate these differences are listed at bottom.
We began examining these issues by first determining the structure of the MtbGyrA CTD. Phylogenetic and structural analyses had suggested that a buried proline (Pro-636 in E. coli GyrA) imparts a nonplanar, spiral shape to the domain in certain CTDs (9); this lockwasher configuration, when present, has been suggested to enhance DNA wrapping and supercoiling by the gyrase holoenzyme (11). As the M. tuberculosis CTD lacks the proline, we reasoned that the domain might adopt a more flattened conformation that would impair DNA wrapping and thereby the lower the supercoiling propensity of M. tuberculosis gyrase as a whole. Instead, we found that the M. tuberculosis CTD was highly similar structurally to that of E. coli GyrA (Fig. 1) and that the domain could robustly bind and introduce writhe into DNA (Figs. 2 and 3). Thus, the difference in shape between relatively planar CTDs (as with Borrelia burgdorferi GyrA) (12) and more skewed forms is not due to solely to a single amino acid. Likewise, the M. tuberculosis CTD did not contain a particularly distinct shape that might have been responsible for the altered supercoiling functions exhibited by M. tuberculosis gyrase.
The emergence of this result led us to probe the specific activities of M. tuberculosis gyrase further. Comparative biochemical studies with isolated domains and subunits, as well as the reconstituted holoenzyme, eventually highlighted several points of divergence between M. tuberculosis and E. coli gyrase. One was an ability of full-length MtbGyrA and GyrA CTD to wrap DNA (Fig. 3). This activity is exhibited by mutants of E. coli GyrA that lack a nonconserved C-terminal tail (see accompanying article (27)), but not by the full-length E. coli proteins (10). A second difference was that M. tuberculosis gyrase failed to show any coupling between DNA wrapping and nucleotide binding (Fig. 3), an established property of the E. coli enzyme (42). A third variance was the finding that M. tuberculosis gyrase does not possess an enhanced ability per se to decatenate DNA relative to E. coli, but rather exhibits a diminished capacity to underwind DNA to the same extent (Figs. 4–6). Finally, we found that M. tuberculosis gyrase hydrolyzes ATP at a much slower rate than E. coli (Fig. 7).
Together, these distinguishing features have important implications for the mechanism of both M. tuberculosis gyrase and gyrases in general. The ability of gyrase to supercoil DNA derives in part from its DNA wrapping activity (8, 48), which juxtaposes two DNA segments in cis prior to strand transport. This property in turn disfavors the passing of two distal DNA segments through each other, impeding DNA decatenation and unknotting. M. tuberculosis gyrase can wrap DNA efficiently, thereby setting the stage for a single round of supercoiling. However, it does not appear to link the formation or dissolution of this wrapped state to the ATPase cycle (Fig. 3), which in E. coli gyrase occurs through the action of the tail appended to the C terminus of GyrA (see accompanying article (27)). Indeed, removal of the GyrA tail reduces the supercoiling set point of E. coli gyrase significantly, to a level close to that observed here for M. tuberculosis gyrase, yet does not alter the innate decatenation properties of the enzyme (supplemental Fig. S2); these findings suggest that part of the reason M. tuberculosis gyrase does not possess as robust a supercoiling capacity as E. coli is in part because it lacks this evolutionary modification. By failing to stabilize or release a wrapped DNA state at the correct point in time, the M. tuberculosis enzyme may be more prone to “slippage,” particularly as more energy becomes required to supercoil DNA to higher and higher levels. This characteristic would in turn favor strand passage events that do not have as great an energetic consequence, such as those needed to support chromosome disentanglement.
However, an inability to connect DNA wrapping to ATP binding or to supercoil DNA extensively does not imply that M. tuberculosis gyrase is an inefficient enzyme. In E. coli, removal of the GyrA tail dramatically slows the rate of supercoiling, but has little effect on the ATPase activity of gyrase as whole, resulting in a large increase of apparently futile cycles. By contrast, M. tuberculosis gyrase hydrolyzes ATP at a much lower rate, but is also slow to supercoil DNA (Figs. 6 and 7). Thus, ATP turnover rates and strand passage appear to be linked in M. tuberculosis gyrase, although the enzyme lacks the tail found in E. coli GyrA. Why M. tuberculosis gyrase is slow is not clear, but conceivably could be an adaptation to the sluggish life cycle of its host organism (23 h average generation time) (49). This concept is line with recent findings in other bacterial systems; for example, E. coli and Salmonella typhimurium gyrase appear to have evolved different supercoiling set points as a means to accommodate different rates of transcription (5), whereas the gyrase of Aquifex aeolicus appears to have undergone a relatively recent conversion into a topo IV, possibly to accommodate life at hyperthermophilic temperatures (21, 50). The slow rate of turnover exhibited by M. tuberculosis gyrase may also play a role in controlling the likelihood of sampling cis and trans strand passage events; a long dwell time in an inactive state where DNA wrapping and ATPase status are uncoupled would favor unwrapping, and thus loss of the transported segment in cis, disfavoring supercoiling as a nucleic acid substrate becomes progressively more underwound. Further analysis of gyrase orthologs from a range of bacterial species with distinct physiologies and environmental niches will be needed to more fully grasp the myriad of means by which evolution appears to have specialized type IIA topoisomerase function.
Supplementary Material
Acknowledgment
We thank Zev Bryant for critical comments.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-CA077373 from the NCI (to J. M. B.) and PO1-AI068135 from the NIAID (to J. M. B.).
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (code 3UC1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CTD
- C-terminal domain
- kDNA
- kinetoplast DNA
- topo
- topoisomerase
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1. Zechiedrich E. L., Khodursky A. B., Bachellier S., Schneider R., Chen D., Lilley D. M., Cozzarelli N. R. (2000) Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275, 8103–8113 [DOI] [PubMed] [Google Scholar]
- 2. Drlica K. (1990) Bacterial topoisomerases and the control of DNA supercoiling. Trends Genet. 6, 433–437 [DOI] [PubMed] [Google Scholar]
- 3. Champion K., Higgins N. P. (2007) Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. J. Bacteriol. 189, 5839–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forterre P., Gribaldo S., Gadelle D., Serre M. C. (2007) Origin and evolution of DNA topoisomerases. Biochimie 89, 427–446 [DOI] [PubMed] [Google Scholar]
- 5. Booker B. M., Deng S., Higgins N. P. (2010) DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 78, 1348–1364 [DOI] [PubMed] [Google Scholar]
- 6. Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U.S.A. 73, 3872–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schoeffler A. J., Berger J. M. (2008) DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev. Biophys. 41, 41–101 [DOI] [PubMed] [Google Scholar]
- 8. Kampranis S. C., Maxwell A. (1996) Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. U.S.A. 93, 14416–14421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsieh T. J., Yen T. J., Lin T. S., Chang H. T., Huang S. Y., Hsu C. H., Farh L., Chan N. L. (2010) Twisting of the DNA-binding surface by a β-strand-bearing proline modulates DNA gyrase activity. Nucleic Acids Res. 38, 4173–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reece R. J., Maxwell A. (1991) The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 19, 1399–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruthenburg A. J., Graybosch D. M., Huetsch J. C., Verdine G. L. (2005) A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J. Biol. Chem. 280, 26177–26184 [DOI] [PubMed] [Google Scholar]
- 12. Corbett K. D., Shultzaberger R. K., Berger J. M. (2004) The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl. Acad. Sci. U.S.A. 101, 7293–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown P. O., Cozzarelli N. R. (1979) A sign inversion mechanism for enzymatic supercoiling of DNA. Science 206, 1081–1083 [DOI] [PubMed] [Google Scholar]
- 14. Ward D., Newton A. (1997) Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 26, 897–910 [DOI] [PubMed] [Google Scholar]
- 15. Kramlinger V. M., Hiasa H. (2006) The “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J. Biol. Chem. 281, 3738–3742 [DOI] [PubMed] [Google Scholar]
- 16. Corbett K. D., Schoeffler A. J., Thomsen N. D., Berger J. M. (2005) The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 351, 545–561 [DOI] [PubMed] [Google Scholar]
- 17. Hsieh T. J., Farh L., Huang W. M., Chan N. L. (2004) Structure of the topoisomerase IV C-terminal domain: a broken β-propeller implies a role as geometry facilitator in catalysis. J. Biol. Chem. 279, 55587–55593 [DOI] [PubMed] [Google Scholar]
- 18. Crisona N. J., Strick T. R., Bensimon D., Croquette V., Cozzarelli N. R. (2000) Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 14, 2881–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peng H., Marians K. J. (1993) Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 90, 8571–8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404 [DOI] [PubMed] [Google Scholar]
- 21. Tretter E. M., Lerman J. C., Berger J. M. (2010) A naturally chimeric type IIA topoisomerase in Aquifex aeolicus highlights an evolutionary path for the emergence of functional paralogs. Proc. Natl. Acad. Sci. U.S.A. 107, 22055–22059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 23. Zechiedrich E. L., Khodursky A. B., Cozzarelli N. R. (1997) Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 11, 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J. C. (1971) Interaction between DNA and an Escherichia coli protein ω. J. Mol. Biol. 55, 523–533 [DOI] [PubMed] [Google Scholar]
- 25. Liu L. F., Wang J. C. (1979) Interaction between DNA and Escherichia coli DNA topoisomerase I: formation of complexes between the protein and superhelical and nonsuperhelical duplex DNAs. J. Biol. Chem. 254, 11082–11088 [PubMed] [Google Scholar]
- 26. Aubry A., Fisher L. M., Jarlier V., Cambau E. (2006) First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 348, 158–165 [DOI] [PubMed] [Google Scholar]
- 27. Tretter E. M., Berger J. M. (March 28, 2012) Mechanisms for defining supercoiling set point of DNA gyrase orthologs I: a nonconserved acidic C-terminal tail modulates Escherichia coli gyrase activity. J. Biol. Chem. 287, 18636–18644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 29. MacDowell A. A., Celestre R. S., Howells M., McKinney W., Krupnick J., Cambie D., Domning E. E., Duarte R. M., Kelez N., Plate D. W., Cork C. W., Earnest T. N., Dickert J., Meigs G., Ralston C., Holton J. M., Alber T., Berger J. M., Agard D. A., Padmore H. A. (2004) Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J. Synchrotron Radiat. 11, 447–455 [DOI] [PubMed] [Google Scholar]
- 30. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Painter J., Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 35. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 36. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 37. Shapiro T. A., Klein V. A., Englund P. T. (1999) Isolation of kinetoplast DNA. Methods Mol. Biol. 94, 61–67 [DOI] [PubMed] [Google Scholar]
- 38. Shuman S. (1998) Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme. Biochim. Biophys. Acta 1400, 321–337 [DOI] [PubMed] [Google Scholar]
- 39. Champoux J. J., Dulbecco R. (1972) An activity from mammalian cells that untwists superhelical DNA–a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc. Natl. Acad. Sci. U.S.A. 69, 143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dynan W. S., Jendrisak J. J., Hager D. A., Burgess R. R. (1981) Purification and characterization of wheat germ DNA topoisomerase I (nicking-closing enzyme). J. Biol. Chem. 256, 5860–5865 [PubMed] [Google Scholar]
- 41. Rau D. C., Gellert M., Thoma F., Maxwell A. (1987) Structure of the DNA gyrase-DNA complex as revealed by transient electric dichroism. J. Mol. Biol. 193, 555–569 [DOI] [PubMed] [Google Scholar]
- 42. Heddle J. G., Mitelheiser S., Maxwell A., Thomson N. H. (2004) Nucleotide binding to DNA gyrase causes loss of DNA wrap. J. Mol. Biol. 337, 597–610 [DOI] [PubMed] [Google Scholar]
- 43. Kampranis S. C., Bates A. D., Maxwell A. (1999) A model for the mechanism of strand passage by DNA gyrase. Proc. Natl. Acad. Sci. U.S.A. 96, 8414–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clark D. J., Leblanc B. (2009) Analysis of DNA supercoiling induced by DNA-protein interactions. Methods Mol. Biol. 543, 523–535 [DOI] [PubMed] [Google Scholar]
- 45. Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100, 95–97 [DOI] [PubMed] [Google Scholar]
- 46. Maxwell A., Gellert M. (1984) The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 259, 14472–14480 [PubMed] [Google Scholar]
- 47. Schoeffler A. J., May A. P., Berger J. M. (2010) A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Res. 38, 7830–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu L. F., Wang J. C. (1978) Micrococcus luteus DNA gyrase: active components and a model for its supercoiling of DNA. Proc. Natl. Acad. Sci. U.S.A. 75, 2098–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Sullivan D. M., McHugh T. D., Gillespie S. H. (2010) Mapping the fitness of Mycobacterium tuberculosis strains: a complex picture. J. Med. Microbiol. 59, 1533–1535 [DOI] [PubMed] [Google Scholar]
- 50. Forterre P., Bergerat A., Lopez-Garcia P. (1996) The unique DNA topology and DNA topoisomerases of hyperthermophilic archaea. FEMS Microbiol. Rev. 18, 237–248 [DOI] [PubMed] [Google Scholar]
- 51. García-Estrada C., Prada C. F., Fernández-Rubio C., Rojo-Vázquez F., Balaña-Fouce R. (2010) DNA topoisomerases in apicomplexan parasites: promising targets for drug discovery. Proc. Biol. Sci. 277, 1777–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.