Background: Ca2+-activated nucleotidase 1 (CANT1), an Endoplasmic Reticulum-Golgi resident nucleoside diphosphatase may have a role in protein quality control.
Results: CANT1 has been identified as a novel target of the Ca2+-dependent transcriptional repressor DREAM.
Conclusion: CANT1 down-regulation increased protein degradation. CANT1 overexpression enhances Ca2+ levels in Golgi apparatus, indicating a role in Ca2+ homeostasis.
Significance: Protein degradation represents a novel process modulated by DREAM.
Keywords: Calcium Signaling, ER-associated Degradation, Gene Regulation, Protein Degradation, Protein Folding, CANT1, DREAM, Neuronal Cells
Abstract
DREAM is a Ca2+-dependent transcriptional repressor highly expressed in neuronal cells. A number of genes have already been identified as the target of its regulation. Targeted analysis performed on cerebella from transgenic mice expressing a dominant active DREAM mutant (daDREAM) showed a drastic reduction of the amount of transcript of Ca2+-activated nucleotidase 1 (CANT1), an endoplasmic reticulum (ER)-Golgi resident Ca2+-dependent nucleoside diphosphatase that has been suggested to have a role in glucosylation reactions related to the quality control of proteins in the ER and the Golgi apparatus. CANT1 down-regulation was also found in neuroblastoma SH-SY5Y cells stably overexpressing wild type (wt) DREAM or daDREAM, thus providing a simple cell model to investigate the protein maturation pathway. Pulse-chase experiments demonstrated that the down-regulation of CANT1 is associated with reduced protein secretion and increased degradation rates. Importantly, overexpression of wtDREAM or daDREAM augmented the expression of the EDEM1 gene, which encodes a key component of the ER-associated degradation pathway, suggesting an alternative pathway to enhanced protein degradation. Restoring CANT1 levels in neuroblastoma clones recovered the phenotype, thus confirming a key role of CANT1, and of the regulation of its gene by DREAM, in the control of protein synthesis and degradation.
Introduction
DREAM (downstream regulatory element antagonist modulator)3 is a Ca2+-dependent multifunctional protein that belongs to the KChIP subfamily of neuronal calcium sensors. In the nucleus, DREAM is a Ca2+-dependent transcriptional repressor (1). Outside the nucleus, under the name calsenilin, it regulates the presenilin-mediated Ca2+ release from the lumen of the endoplasmic reticulum (2, 3) and, as KChIP3, regulates the plasma membrane Kv4-type channels (4). Also at the cell membrane, DREAM regulates voltage-dependent calcium channels (5, 6) and modulates downstream signaling of different membrane receptors including NMDA (7, 8) and THSR (9).
All KChIP proteins share a high sequence homology and are highly redundant in terms of their activities in the cell (10, 11). Therefore, gene inactivation of DREAM generally does not produce a strong phenotype. DREAM is highly expressed in a number of tissues, among them the central nervous system, thyroid gland, testis, and thymus and early in vitro studies have described specific target genes for DREAM repression in the brain, the immune system, and in the thyroid gland (1, 10, 12, 13). The transcriptional action of DREAM requires its binding to a downstream regulatory element (DRE) in the promoter of target genes. The binding is regulated by the level of nuclear Ca2+, by the interaction with other nucleoproteins such as the cAMP response element modulator and CREB, and by the PIK3 pathway (14). Mutation of key amino acids within any of the three functional EF hands (a fourth EF hand is not operational) results in a protein insensitive to Ca2+ that will block DRE- and CRE-dependent transcription (1, 15). Introducing a second mutation at the CREB-interacting domain in DREAM results in the double mutant daDREAM that acts as a dominant active repressor blocking specifically DREAM target genes as reported (13).
Cerebellar granule cells from transgenic mice expressing daDREAM show reduced expression level of the plasma membrane Na+/Ca2+ exchanger isoform 3 (NCX3), the major Ca2+ extrusion system in these neurons (16). We have previously shown that in human neuroblastoma clones stably overexpressing wtDREAM or daDREAM, the ER Ca2+ content is drastically reduced, through both transcriptional and non-transcriptional mechanisms. In addition to a modest reduction in NCX3 levels, which was possibly responsible for inhibiting capacitative Ca2+ influx and thus ER store refilling, we observed a marked up-regulation of the InsP3R transcript levels, which could have been responsible for increasing the ER Ca2+ leak. DREAM also acts non-transcriptionally to modulate ER Ca2+ content, and we have shown that it does so by directly interacting with presenilin 2 (PS2) in a Ca2+-independent manner, potentiating the PS2-promoted efflux of Ca2+ from the ER (3).
In the present study we searched for changes in the expression of genes related to protein folding and degradation in cerebella of daDREAM mice to identify other Ca2+-related gene targets of DREAM regulation. We found a strong repression of the transcript of CANT1 (calcium-activated nucleotidase 1), an ER-Golgi resident Ca2+-dependent nucleoside diphosphatase (17–19), that is suggested to have a role in glucosylation reactions related to the quality control of proteins in the ER and the Golgi apparatus (20). The repression was also documented both at the mRNA and at the protein level in wtDREAM and daDREAM stable clones of neuroblastoma cells. qPCR and Western blot also revealed the up-regulation of EDEM1, an α-mannosidase-like protein, that regulates the disposal of misfolded protein from the ER (21, 22). This is in line with previously reported findings that showed an up-regulation of EDEM1 transcript in B cells of daDREAM mice (13).
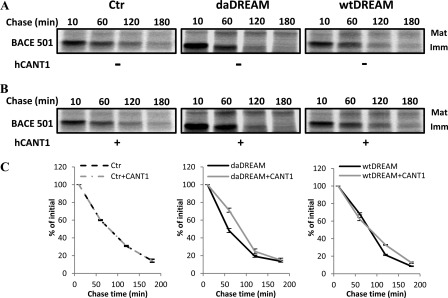
These findings prompted us to study a possible impairment in the protein folding machinery using neuroblastoma clones stably expressing DREAM as cell models. Pulse-chase experiments monitoring the maturation of the folding competent substrates BACE501 and wild type α1-antitrypsin (α1AT-WT) and the folding-defective substrate α1-antitrypsin Null Hong-Kong variant (α1AT-NHK) demonstrated that the reduction of CANT1 levels selectively delayed the secretion rate of the soluble α1AT-WT and, conversely, enhanced ER-associated protein degradation (ERAD) of the α1AT-NHK variant, its folding-defective counterpart. Interestingly, the folding kinetic of the membrane-bound BACE501 was not affected, suggesting the intriguing possibility of a substrate dependent activity of CANT1.
Because CANT1 is a Ca2+-binding protein, it appeared interesting to investigate the possibility that it could directly affect Ca2+ homeostasis. We thus measured intracellular Ca2+ with the recombinant Ca2+-sensitive aequorin targeted to the Golgi apparatus and ER lumen. The measurements have revealed that CANT1 overexpression enhanced the levels of free Ca2+ in the lumen of the Golgi apparatus, suggesting that it may play a role in Ca2+ homeostasis, but it had no effect on ER Ca2+ levels.
In summary, our data, in addition to identifying CANT1 as a novel gene subjected to DREAM regulation, demonstrate that the CANT1 activity is essential to the correct ER protein folding and degradation processes. Interest in these findings is heightened by the recent discovery of CANT1 mutations in Desbuquois dysplasia, a skeletal disorder presenting defects in the endochondral ossification process (23). The molecular mechanism responsible for this phenotype is still unknown, even if it has been proposed to be due to an impairment of proteoglycans synthesis: our evidence of a link between CANT1 deficiency and enhanced protein degradation supports this possibility.
EXPERIMENTAL PROCEDURES
Animals
DREAM transgenic mice (line 33, L33) were previously described (13, 16).
qPCR Analysis
Total RNA from mice cerebella and neuroblastoma cell culture was prepared using TRIzol Reagent (Invitrogen). First-Strand cDNA synthesis was performed by using SuperScript™ II RT (Invitrogen). Briefly, 1 to 5 μg of total RNA were incubated with 1 μl of Oligo(dT) (500 μg/ml), 1 μl of dNTP Mix (10 mm each) and sterile distilled water to a final volume of 12 μl. The mixture was heated at 65 °C for 5 min and quickly chilled on ice before adding 4 μl of 5× First-Strand Buffer, 2 μl of 0.1 m DTT, and 1 μl of RNaseOUT™ (40 units/μl). The mixture was incubated at 42 °C for 2 min before adding 1 μl (200 units) of SuperScript™ II RT to reach a final reaction volume of 20 μl. The reaction was performed at 42 °C for 50 min and then inactivated at 70 °C for 15 min. The cDNA was used as a template for amplification in qPCR.
Quantification of calnexin, calreticulin, Bip, CHOP, EDEM1, and Xbp1 and β-actin in mice cerebella was done using specific primers and Taqman MGB probes (Applied Biosystems, Austin, TX). The results are normalized by parallel amplification of β-actin.
For CANT1 and EDEM1 quantification in neuroblastoma cell clones, qPCR was performed on a Rotor-Gene 3000 platform (Corbet Research, Sydney, Australia). The PCR cycling parameters were: 94 °C for 7 min, 45 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 15 s. An amount of cDNA corresponding to 1–10 ng of total RNA was amplified in 25 μl of a mixture containing 12.5 μl of Platinum SYBR-Green qPCR SuperMix-UGD (Invitrogen) and 2 μl primer mixture (2.5 μm each) for each sample. Primers were designed using Primer3 software. The primers used were: for CANT1, forward, 5′-AACACCGACGACCAGATCATT-3′, and reverse, 5′-CCTTCGTATTTCACGCTTCCG-3′ and for EDEM1, forward, 5′-AGTCAAATGTGGATATGCTACG-3′, and reverse, 5′-ACAGATATGATATGGCCCTCAG-3′.
The relative amount of amplified DNA was calculated as described (24), using β-actin or hypoxantine-guanine phosphoribosyl transferase (HPRT) cDNAs as endogenous control. The HPRT primers used were: forward, 5′-TTGGATACA GGCCAGACTTTGTT-3′ and reverse, 5′-CTG AAGTACTCATTATAGTCAAGGGCATA-3′. The β-actin primers used were: forward 5′-AGAGCTACGAGCTGCCTGAC-3′, reverse 5′-GTAGTTTCGTGGATGCCACAG-3′.
Cell Cultures and Transfection
SH-SY5Y cells were grown in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with 10% fetal bovine serum (FBS), in 75 cm2 flasks; before transfection, the cells were seeded onto 13-mm glass cover slips and allowed to grow to 80% confluence. Transfection with 0.7 μg of plasmid DNA (or 0.5:0.5 μg in co-transfections) was carried out using TransFectin Lipid Reagent (Bio-Rad) according to the manufacturer's instruction. AEQ measurements were performed 36 h later. Cells plated for Western blot were collected 24–36 h after transfection. Stable wtDREAM and daDREAM clones were generated as previously described (3) and maintained in the same conditions as control SH-SY5Y cells.
Establishment of SH-SY5Y shRNA Clonal Population for CANT1
To generate down-regulated CANT1 clones, SH-SY5Y cells were grown in a 6-well tissue culture plate to a 50–70% confluence in antibiotic-free DMEM high glucose medium supplemented with 10% FBS. The day after, cells were either transfected with 4 μg of Control shRNA plasmid (sc-108060, Santa Cruz Biotechnology) or CANT1 shRNA plasmid (sc.94075-SH, Santa Cruz Biotechnology) with Lipofectamine 2000 (Invitrogen) by following the instructions of the manufacturer. 48 h after transfection cells were washed with PBS and then incubated with fresh medium. The day after, the medium has been replaced with fresh medium containing 7 μg/ml puromycin (the lowest concentration that kills 100% of non-transfected cells in 3–5 days after the beginning of puromycin selection) to select stable cells for shRNA plasmid DNA-mediated inhibition of CANT1 expression. Approximately every 2–3 days, the medium was replaced with freshly prepared selective medium. After 2 weeks from the beginning of the selection, shRNA CANT1 plasmid pooled clones were used for subsequent experiments.
Immunocytochemistry
SH-SY5Y cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS, 140 mm NaCl, 2 mm KCl, 1.5 mm KH2PO4, 8 mm Na2HPO4, pH 7.4) for 20 min, washed three times with PBS and then incubated for 10 min in PBS supplemented with 50 mm NH4Cl. Cells were permeabilized for 5 min with 0.1% Triton X-100 in PBS, and washed with 1% gelatin (type IV, from calf skin) in PBS for 1 h. The coverslip was processed for the CANT1 staining with a specific polyclonal antibody (kindly provided by Prof. Terence L. Kirley, University of Cincinnati) at a 1:20 dilution in PBS. Staining was carried out with AlexaFluor488 or AlexaFluor594 secondary antibodies (1:100 dilution in PBS; Molecular Probes, Invitrogen). To reveal calreticulin or GM130 signal the coverslips were also incubated with a rabbit polyclonal anti-calreticulin antibody (Stressgene) or with a mouse monoclonal anti-GM130 antibody (BD Biosciences) and AlexaFluor594 or AlexaFluor488 secondary antibody, respectively. Fluorescence was analyzed with a Zeiss Axiovert microscope equipped with a 12-bit digital cooled camera (Micromax-1300Y, Princeton Instruments Inc., Trenton, NJ) or by Leica SP5 confocal microscope. Cells were excited separately at 488 nm or 594 nm, and the single images were recorded. Images were acquired using Metamorph software (Universal Imaging Corporation, West Chester, PA) or Leica AS software.
Western Blot Analysis
SH-SY5Y cells were washed twice with PBS and harvested from the culture plates in ice-cold Tris/EDTA buffer. Lysis was performed by three cycles of freeze (−70 °C) and thaw (37 °C). Loading of the samples was normalized for the total content of cellular proteins determined by the Bradford assay. 40 μg of each sample were run on a 12% SDS-PAGE Tris/HCl gel and then blotted onto nitrocellulose membrane (GE Healthcare, Little Chalfont, UK). Pure soluble CANT1 (Fig. 2B, lane 1) and overexpressed recombinant CANT1 (Fig. 2B, lane 5) were used as internal controls for antibody specificity and protein migration. Tunicamycin treatment was performed by incubating 1 μg/ml tunicamycin (Sigma) for 15 h in cell culture medium under CO2 atmosphere.
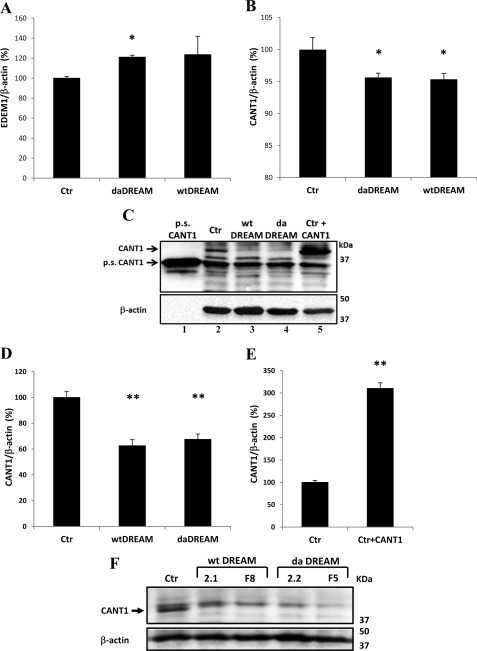
FIGURE 2.
Change in EDEM1 and CANT1 levels in neuroblastoma cells stably overexpressing wtDREAM and daDREAM. A, qPCR for EDEM1 mRNA in control SH-SY5Y cells (Ctr) and in daDREAM and wtDREAM stable clones. B, qPCR for CANT1 mRNA in control SH-SY5Y cells (Ctr) and in daDREAM and wtDREAM stable clones. C, Western blot analysis of CANT1 protein levels. Recombinant purified CANT1 (1 μg, pure soluble CANT1, p.s.CANT1) were loaded in lane 1 as positive control. 40 μg of total protein were loaded in each lane. Lane 2, control cells (Ctr); lanes 3 and 4, wtDREAM and daDREAM neuroblastoma clones; lane 5, protein lysate obtained from control cells transiently transfected with CANT1 expression plasmid. D and E, densitometric analysis of Western blot results. β-Actin levels were used to normalized the amount of total proteins loaded in each lane. D, CANT1 levels in DREAM clones (E) CANT1 overexpression levels in transfected SH-SY5Y cell. F, Western blot showing CANT1 down-regulation in 4 additional neuroblastoma stable clones (2 wtDREAM and 2 daDREAM). The data are representative of three independent experiments. Bars represent means ± S.E., *, p ≤ 0.05; **, p < 0.01.
Western blots were performed using the polyclonal antibody anti-CANT1 (diluted 1: 500 in PBS) and the rabbit polyclonal anti-EDEM1 antibody (Sigma, at 1:1000 dilution in PBS). Detection was carried out by incubation with horseradish peroxidase (HRP) conjugated anti-rabbit or anti-mouse IgG (Santa Cruz Biotechnology) for 1 h and 30 min. The proteins were visualized by the chemiluminescent reagent ImmunStar HRP (Bio-Rad). Densitometric analysis was performed by using the Kodak 1D Image Analysis software (Kodak Scientific Imaging System, New Haven, CT). Means of densitometric measurements of at least four independent experiments, normalized by the endogenous β-actin values, were compared by Student's t test.
Pulse-chase Experiments
Cells were starved for 30 min in Met/Cys free medium, pulsed for 10 min with 100 μCi of 35S-labeled Met/Cys in an 1 ml starvation medium/dish, and chased for the times indicated in the figures with Dulbecco's modified Eagle's medium supplemented with 5 mm cold Met/Cys. Postnuclear supernatants were prepared by solubilization of cells in 800 μl/dish ice-cold 2% CHAPS in Hepes-buffered saline, pH 6.8, containing 20 mm ice-cold N-ethylmaleimide and protease inhibitors (HBS). Cell extracts were prepared by 10 min of centrifugation at 10,000 × g and analyzed by reducing SDS-PAGE. Labeled proteins were immunoprecipitated from cell extracts using an anti-HA antibody (Sigma, 2 μg), which recognizes the HA epitope present in substrates in the case of BACE501 and α1AT-NHK, or using a specific antibody in the case of α1AT-WT substrate (Sigma, 2 μg).
Immunoprecipitations were performed by adding protein A beads (Sigma), 1:10 (w/v) swollen in HBS) and the selected antibody to the cell extracts. The immunoprecipitates were extensively washed three times with HBS, 0.5% CHAPS, and resuspended in sample buffer for SDS-PAGE. Gels were also exposed to BioMax (Eastman Kodak Co.) films and scanned. Relevant bands were quantitated by ImageJ software.
Aequorin (AEQ) Measurements
ER and Golgi apparatus Ca2+ content had to be drastically reduced before the reconstitution of functional low affinity erAEQ and goAEQ (25, 26). To this end the cells were incubated for 1 h at 4 °C in KRB (Krebs Ringer modified buffer: 125 mm NaCl, 5 mm KCl, 1 mm Na3PO4, 1 mm MgSO4, 5.5 mm glucose, 20 mm HEPES, pH 7.4, 37 °C), supplemented with 5 μm coelenterazine n (Molecular Probes, Invitrogen), the SERCA pump inhibitor 2,5-di-tert-butylhydroquinone (tBuBHQ, 10 μm) and 600 μm EGTA. After this incubation, the cells were washed extensively with KRB supplemented with 2% bovine serum albumin (BSA) and 1 mm EGTA, and transferred to the chamber of a purpose-built luminometer. The coverslip with transfected cells was then placed in the thermostated chamber in close proximity to a low-noise photomultiplier. The experiments were terminated by lysing the cells with 100 μm digitonin in a hypotonic Ca2+-rich solution (10 mm CaCl2 in H2O), to discharge the remaining AEQ pool. The light signal was collected and calibrated off-line into Ca2+ concentration values, using a computer algorithm based on the Ca2+ response curve of wt and mutant AEQs as previously described (27, 28).
Statistical Analysis
Data are reported as means ± S.E. Statistical differences were evaluated by two-tailed unpaired Student's t test and p values < 0.05 were considered statistically significant.
RESULTS
Gene-directed Analysis of daDREAM Cerebellum Identifies CANT1 as a Novel Target for DREAM
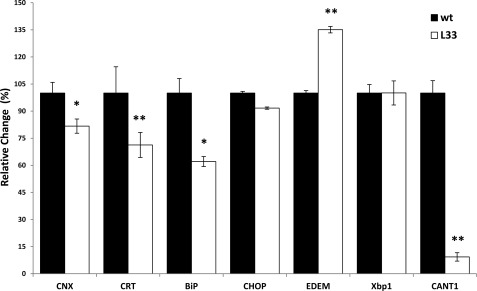
DREAM regulates several genes involved in protein synthesis and protein degradation in transgenic B cells overexpressing the daDREAM mutant (13). To better understand the physiological significance of this regulation we decided to search for changes in the expression of genes related to protein folding and degradation in the cerebellum, an area where DREAM regulates Ca2+ homeostasis and the survival of granule cells in culture by controlling the expression of the NCX3 sodium-calcium exchanger (16). Comparative qPCR analysis of the basal expression of potential target genes in wild type and transgenic cerebellum revealed a significant down-regulation of the transcripts of the protein chaperones calnexin, calreticulin, and BiP (immunoglobulin-binding protein; Grp78) as well as a significant induction of EDEM1 (Fig. 1). Calreticulin and calnexin are key components of the protein folding machinery and BiP and EDEM1 are key components of the ER-associated protein degradation (ERAD) pathway that plays a key role in the recognition and extraction of terminally misfolded polypeptides from the calnexin/calreticulin folding cycle. Enhanced levels of EDEM1 are associated with accelerated degradation of misfolded glycoproteins (21, 29). The expression of CHOP (C/EBP-homologous protein), or Xbp1, two ER stress-inducible factors (30), was instead not significantly modified in transgenic cerebellum (Fig. 1). Another protein involved in protein degradation, CANT1, which hydrolyzes nucleoside 5′-diphosphates in a Ca2+-dependent manner (19), was also considered. The fact that CANT1 is a Ca2+-regulated enzyme made it especially interesting to us. Its transcript levels were reduced by about 90% (Fig. 1). Taken together, these results indicate that DREAM is involved in the regulation of the protein quality control pathway also in neuronal cells.
FIGURE 1.
Changes in gene expression in the cerebella of daDREAM transgenic mice. Quantitative real-time PCR (qPCR) for calnexin (CNX), calreticulin (CRT), BiP, CHOP, EDEM1, Xbp 1, and CANT1 mRNA in wild type (wt) and transgenic daDREAM (L33) mice cerebella. The results are the mean ± S.E. of 8–10 mice and are representative of two experiments. Statistical significance versus corresponding control, *, p < 0.05; **, p < 0.01.
Reduced CANT1 and Increased EDEM1 Levels after DREAM Overexpression in Neuroblastoma Cells
To substantiate the suggestion above and to eventually develop a cell system to test protein folding efficiency, we quantified the EDEM1 and CANT1 levels in neuroblastoma wtDREAM and daDREAM clones. For calnexin, calreticulin, and BiP, we have previously reported that their transcript levels are unchanged (3).
qPCR revealed an increase of about 20% in the level of the EDEM1 transcript in wtDREAM and daDREAM clones with respect to control cells (Fig. 2A). The increase was statistically significant only in the case of the daDREAM clone; nevertheless, Fig. 2A shows the tendency for a similar modest increase of EDEM1 in both clones. The analysis of CANT1 expression was performed both at mRNA and at the protein level. qPCR revealed only minor effects on CANT1 mRNA levels in wtDREAM and daDREAM clones with respect to control cells (Fig. 2B), being the reduction of about 5%. Fig. 2C shows a representative Western blot analysis for CANT1 expression in neuroblastoma DREAM clones. A sample of recombinant purified wt CANT1 (p.s. CANT1, pure soluble CANT1, lane 1; obtained from Prof. T. L. Kirley, University of Cincinnati) was run in parallel with a lysate of SH-SY5Y control cells (lane 2), with lysates obtained from wtDREAM and daDREAM clones (lanes 3 and 4, respectively), and with a lysate of SH-SY5Y cells transiently transfected with the CANT1 expression plasmid (lane 5). Staining with a polyclonal antibody which recognize CANT1 revealed a band at ∼35 kDa corresponding to the recombinant purified CANT1 (lane 1), and another of about 40 kDa corresponding to the endogenous CANT1 in control cells (lane 2), and in DREAM clones (lanes 3 and 4), which matched the overexpressed form (lane 5). A 35-kDa band was also visible in the lanes loaded with cell lysates, but it was probably due to cross reactivity, since it was not increased by CANT1 overexpression (lane 5). The densitometric analysis (Fig. 2D) showed a down-regulation of CANT1 in cells stably expressing either wtDREAM or daDREAM with respect to control cells; the reduction was about 38% in wtDREAM and 33% in daDREAM-expressing clones. The finding is in line with the down-regulation of the CANT1 transcript in the cerebella of daDREAM transgenic mice, and in neuroblastoma clones. The level of CANT1 overexpression in SH-SY5Y-transfected cells was also estimated by densitometric analysis of the whole cell population and the quantification revealed an increase of about 200% (Fig. 2E). However, considering that the average efficiency of transient transfections in SH-SY5Y cells was about 25%, the increase of CANT1 protein in overexpressing cells corrected for the whole cell population would correspond to about 800%, i.e. the total amount of CANT1 would be about 9-fold the endogenous level. To exclude that the reduction of CANT1 protein could be related to clonal peculiarity we analyzed 4 other independent SH-SY5Y clones (2 for wtDREAM and 2 for daDREAM) and we found, as documented by Fig. 2F, that in all the analyzed clones CANT1 protein levels were reduced.
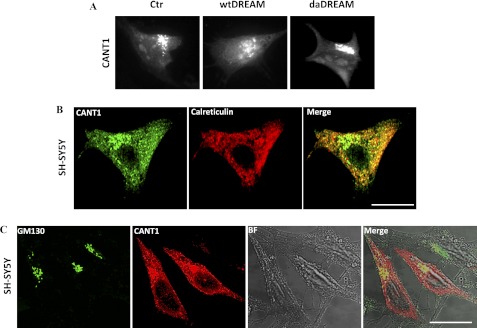
Immunocytochemistry analysis of overexpressed CANT1 in SH-SY5Y cells showed its predominant location in a perinuclear region resembling the Golgi apparatus, with a background reticular pattern, probably corresponding to the ER (Fig. 3A). The intracellular distribution of overexpressed CANT1 was also analyzed in wtDREAM and daDREAM clones and found to show also a predominant localization in the perinuclear region (Fig. 3A). Double immunofluorescence analysis by transfecting CANT1 in SY-SY5Y cells and performing immunostaining against calreticulin as ER marker, or against GM130 (golgin) as Golgi apparatus marker, was performed to demonstrate that CANT1 was expressed both in the ER and in the Golgi apparatus (Fig. 3, B and C, respectively).
FIGURE 3.
Analysis of CANT1 intracellular distribution. A, distribution of overexpressed CANT1 in control (Ctr) and wtDREAM or daDREAM clones was analyzed by epifluorescence microscope and revealed a perinuclear localization. SH-SY5Y cells were transfected with expression plasmid for CANT1 and processed for double immunostaining with CANT1 antibody and anti-calreticulin (B) or anti-GM130 antibody (C) to reveal ER and Golgi apparatus staining. Distribution of CANT1 (green) and calreticulin (red) and of CANT1 (red) and GM130 (green) immunoreactivities was analyzed by confocal microscope. Bright field image was also shown to better visualize analyzed cells. The merge panels on the right show the co-localization. Bars, 10 μm.
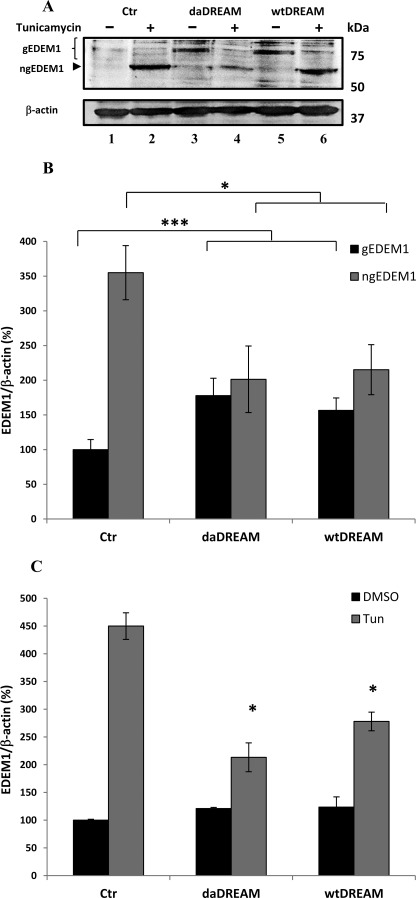
The expression levels of EDEM1 protein were also analyzed in cells stably expressing wtDREAM or daDREAM. Based on EDEM1 glycosylation state, active glycosylated ER-resident forms and an inactive faster migrating de-glycosylated protein have been described (31). Thus, the glycosylation state of EDEM1 was analyzed. Western blot analysis of endogenous EDEM1 was performed using total cell extracts from control cells and from cells stably expressing wtDREAM or daDREAM, either untreated or treated with tunicamycin. Tunicamycin inhibits N-glycosylation, thus increasing the de-glycosylated inactive form of EDEM1. A low basal level of glycosylated EDEM1 was detected in untreated control cells (Fig. 4A, lane 1) that was increased in both wtDREAM and daDREAM clones (Fig. 4A, lanes 3 and 5). Densitometric analysis confirmed that a statistically significant increase of the glycosylated active form of EDEM1 had occurred in both wtDREAM and daDREAM-expressing clones (Fig. 4B), which contained about twice as much EDEM1 protein as the control cells. The tunicamycin treatment induced a decrease in the glycosylated active form of EDEM1, followed by a concomitant increase in the inactive de-glycosylated form (Fig. 4A, lane 2 for control cells and lanes 4 and 6 for daDREAM and wtDREAM clones, respectively). The increase of inactive EDEM1 was less pronounced in the wtDREAM and daDREAM clones than in the control cells as documented by densitometric analysis (Fig. 4B), suggesting that DREAM clones could be subjected to ER stress condition. It must also be mentioned that the tunicamycin treatment induces ER stress, a condition associated with the induction of ER stress markers such as BiP, calreticulin, calnexin, etc., and EDEM1. Fig. 4C shows that the levels of EDEM1 transcript in control cells, in the wtDREAM and the daDREAM clones were increased by the tunicamycin treatment. The enhancement in transcript levels explains the intensity of the band corresponding to non-glycosylated EDEM1 with respect to that of the glycosylated form. The data also show an impairment in the ER stress-activated induction of EDEM1 in wtDREAM and daDREAM clones suggesting that DREAM clones may be more prone to ER stress condition than control cells.
FIGURE 4.
Effects of tunicamycin treatment on the EDEM1 levels in control neuroblastoma cells (Ctr) and in cell clones stably overexpressing wtDREAM or daDREAM. A, Western blot analysis of EDEM1 protein levels. Glycosylated (gEDEM1) and non-glycosylated (ngEDEM1) levels were modified following tunicamycin treatment. B, quantification by densitometric analysis of gEDEM1 and ngEDEM1 protein levels. The data are representative of three independent experiments, and the relative amounts were calculated with respect to the amount of gEDEM1 in control cells which were normalized to 100%. C, qPCR for EDEM1 mRNA in control neuroblastoma cells (Ctr) and in wtDREAM and daDREAM clones in the presence (Tun) or in the absence (DMSO) of tunicamycin treatment. The data are from two independent experiments in duplicate. Bars represent means ± S.E., *, p < 0.05; ***, p < 0.001.
Effects of CANT1 on the Handling of Folding-competent ER Glycoproteins
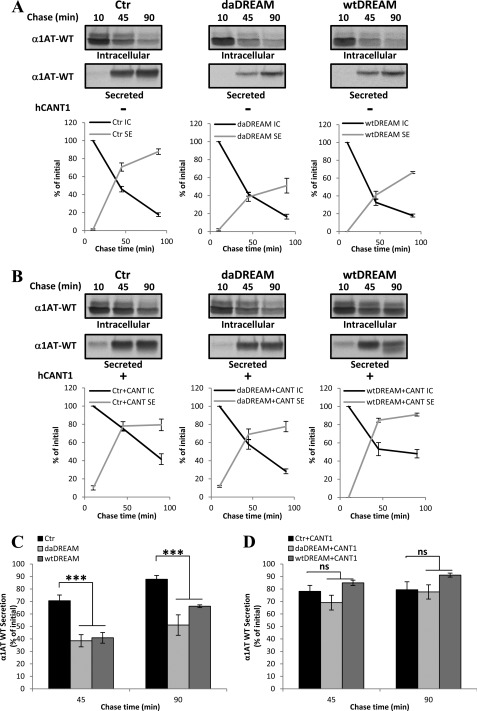
At this point it became interesting to establish whether the CANT1 reduction produced defects in protein folding and maturation, a process also related to the Ca2+ concentration in the ER, which we have previously found to be reduced in DREAM clones (3). Pulse-chase experiments were performed following the maturation of two commonly used folding-competent protein models: BACE (BACE501), a type I transmembrane aspartyl protease, which during its maturation is released from the ER and subjected to complex glycosylation in the Golgi (32) and α1AT-WT, a soluble folding-competent glycoprotein, which is subjected to folding attempts in the calnexin cycle and, once the correct structure is achieved, is exported through the secretory pathway (33). First, SH-SY5Y control cells and cells stably expressing wtDREAM or daDREAM were transiently transfected with a plasmid encoding BACE501 either in the absence (Fig. 5A) or in the presence (Fig. 5B) of ectopically expressed recombinant CANT1. The disappearance of immature un-glycosylated form of native BACE501 occurred with similar kinetics in control cells and in cells stably expressing wtDREAM or daDREAM (Fig. 5A and quantification in Fig. 5C). Interestingly, CANT1 overexpression did not significantly modify immature BACE501 disappearance (Fig. 5B and quantification in Fig. 5C). The normal disappearance of immature BACE501 in wtDREAM and daDREAM stably expressing cells, in which CANT1 was down-regulated, indicated that CANT1 reduction did not affect its correct maturation. Similar pulse-chase experiments were then performed to assess the maturation and secretion efficiency of α1AT-WT (Fig. 6). As a soluble folding-competent substrate, during the chase time, the amount of intracellular α1AT-WT decreased and, simultaneously, the amount of labeled secreted form increased. As shown in Fig. 6A, the secretion efficiency of α1AT-WT was reduced in wtDREAM and daDREAM clones (66 ± 1.12% and 51 ± 8.2%, respectively, at 90 min) as compared with control cells (87.7 ± 3.05%, p < 0.001). This phenotype could either be ascribed to CANT1 down-regulation or to EDEM1 up-regulation since EDEM1 has been shown to play an important role in maintaining protein folding efficiency and secretory capacity (34, 35). Interestingly, restoring CANT1 levels by introducing exogenous CANT1 restored the secretion efficiency in wtDREAM (91 ± 1.6%) as well as in daDREAM (77.6 ± 5.71%) expressing cells to value similar of that obtained in control cells overexpressing CANT1 (79.3 ± 6.39%, at 90 min). CANT1 overexpression in control cells slightly accelerated the secretion efficiency of a soluble folding-competent substrate at early chase times (70 ± 4.55% versus 78 ± 4.8% at 45 min) (Fig. 6B), suggesting that CANT1 could specifically affect this pathway. Fig. 6, C and D show the average % values of α1AT-WT secreted in the different cell batches at the indicated chase time.
FIGURE 5.
CANT1 does not affect the disappearance of the immature form of the folding-competent ER glycoprotein BACE 501. A, pulse-chase analysis showing immunoprecipitated, radiolabeled mature (Mat), and immature (Imm) BACE 501 protein in control neuroblastoma cells (Ctr) and wtDREAM and daDREAM clones. B, same as in A, but after overexpression of CANT1. C, kinetics of experiments in A and B are shown. Data represent mean % ± S.E. from two independent experiments in duplicate.
FIGURE 6.
CANT1 modifies the secretion efficiency of the soluble folding-competent substrate wild type α1-antitrypsin (α1AT-WT). A, radiolabeled α1AT-WT was immunoisolated at the indicated times after the pulse from detergent extract (Intracellular, IC) and from the medium (Secreted, SE) of control cells (Ctr) or wtDREAM and daDREAM expressing clones. B, same as A, but in the presence of CANT1 overexpression. Quantification of secretion kinetics from two independent experiments in duplicate is shown as mean % ± S.E. for each panel. C and D show histograms of the average % values of secreted α1AT-WT at 45 and 90 min chase time. ns, not significant; ***, p < 0.001.
Effects of CANT1 on ERAD of Folding-defective ER Glycoproteins
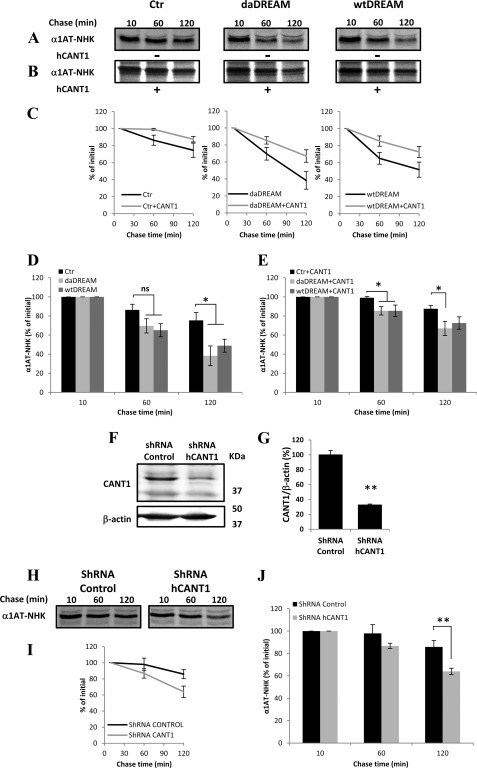
The finding that stable DREAM overexpression, in addition to being responsible for CANT1 down-regulation, also led to the up-regulation of the EDEM1 protein, obviously suggested the possibility of a role of DREAM in the regulation of ERAD. To better clarify this possibility we performed pulse-chase experiments using a certified ERAD folding-defective substrate, the Null-Hong-Kong variant of α-1 antitrypsin (α1AT-NHK) (36). Folding-defective glycoproteins do not normally follow the secretory route, rather, after a series of futile folding attempts, mannose trimming of the N-glycan terminal α1,2-bonded mannose residues occurs as a signal for their degradation (37, 38). As shown in Fig. 7, A–D, the degradation kinetics of the folding defective substrate α1AT-NHK was significantly enhanced in the wtDREAM and daDREAM cell clones, in line with the notion that EDEM1 enhances α1AT-NHK degradation, as previously reported (39). However, to understand whether CANT1 down-regulation also plays a role, we transfected DREAM clones with the CANT1 expression plasmid. Intriguingly, CANT1 transfection reverted the enhanced ERAD phenotype observed in wtDREAM and daDREAM expressing cells (Fig. 7, B and C), and further delayed the degradation kinetics of α1AT-NHK in control cells. Fig. 7, D and E show the average % values of α1AT-NHK degraded in the different cell batches at the indicated chase time.
FIGURE 7.
CANT1 intracellular level affects ER-associated degradation of the soluble folding-defective substrate α1AT-NHK variant. A, radiolabeled α1AT-NHK was immunoisolated after the indicated chase times from detergent extract of control cells (Ctr) or wtDREAM- and daDREAM-expressing clones. B, same as in A but in the presence of overexpressed CANT1. C, quantification of intracellular α1AT-NHK degradation for each cell clone is shown in the absence or in the presence of overexpressed CANT1. D and E show histograms of the average % values at 10, 60, and 120 min chase time. F, Western blot of CANT1 silencing in neuroblastoma cells. G, quantification of CANT1 reduction in selected clonal population. H, radiolabeled α1AT-NHK was immunoisolated after the indicated chase times from detergent extract of shRNA control cells or shRNA CANT1-transfected cells. I, quantification of intracellular α1AT-NHK degradation for clonal population having reduced CANT1 levels is shown. I, shows histograms of the average % values at 10, 60, and 120 min chase time. Data are the mean % ± S.E. of three independent experiments in duplicate. ns, not significant; *, p < 0.05; **, p < 0.01.
The findings show that CANT1 has a role in the glycoprotein retention in the calnexin folding system and that the reduction of its intracellular level could affect protein maturation processes, and direct glycoproteins to faster degradation. This also suggests that folding and ERAD machineries are to some extent competing for newly synthesized polypeptides. To further confirm that the observed effects are due to CANT1 down-regulation we selected a SH-SY5Y clonal population in which CANT1 levels were down-regulated by shRNA plasmid DNA. Fig. 7F shows a representative Western blot showing the reduction of CANT1 band in shRNA CANT1-transfected cells and Fig. 7G refers to the relative densitometric analysis showing a reduction of about 70%. Fig. 7, H and I show that the degradation kinetics of the folding defective substrate α1AT-NHK was significantly enhanced in shRNA CANT1 population in respect to control cells (85.8 ± 5.6% in control shRNA cells and 63.9 ± 2.87% in CANT1 shRNA cells at 120 min, p < 0.01). The average % values at each chase time are reported in Fig. 7J.
Effects of CANT1 on the Intracellular Ca2+ Homeostasis in DREAM Stably Expressing Cells
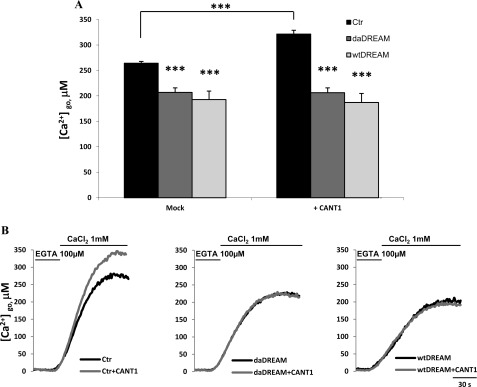
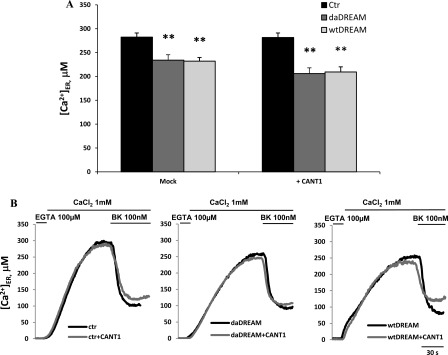
The fact that CANT1 is activated by Ca2+, which is important for the regulation of protein folding and maturation, and that wtDREAM and daDREAM clones have a documented impairment of ER Ca2+ content (3), prompted us to investigate whether CANT1 could be somehow involved in the regulation of Ca2+ homeostasis. Its role in regulating the balance between folding and degradation of glycoproteins could thus be related to the general process of Ca2+ homeostasis. We transfected cells with the Ca2+-sensitive aequorin probes targeted to different intracellular organelles (40). We first evaluated the resting free Ca2+ content in the Golgi apparatus (Fig. 8A) in wtDREAM and daDREAM clones and found that DREAM overexpression reduced it by an extent similar to that we have previously documented for the ER (3). To understand whether CANT1 could have a role in this reduction we transiently co-transfected control cells and DREAM clones with the CANT1 expression plasmid. As shown in Fig. 8A, CANT1 overexpression induced an increase in the free Ca2+ content of the Golgi apparatus in control cells, but not in the wtDREAM and daDREAM stable clones. These findings suggest that CANT1 is involved in the regulation of Golgi Ca2+, but that its reintroduction in the DREAM clones is not sufficient to recover the proper Golgi Ca2+ levels (representative traces are shown in Fig. 8B), thus suggesting that other modifications of ER/Golgi protein levels, i.e. the previously documented InsP3R up-regulation, could be the primary responsible for this phenotype.
FIGURE 8.
Measurements of Ca2+ concentration in the lumen of the Golgi apparatus, [Ca2+]go. Control cells (Ctr) and wtDREAM and daDREAM clones were transfected with goAEQ (26). The effect of CANT1 was monitored by overexpression in the different cell batches. A, bars represent mean resting [Ca2+] values upon stimulation ± S.E., ***, p < 0.001. B, kinetics of Golgi apparatus refilling upon re-addition of 1 mm CaCl2 to Ca2+-depleted cells. The traces are representative of six independent experiments.
Next, we analyzed the effect of CANT1 on ER Ca2+ levels (Fig. 9). As previously reported, the stable overexpression of wtDREAM or daDREAM decreased the resting ER Ca2+ content (3). However, at variance with what was observed for the Golgi Ca2+ level, the overexpression of CANT1 failed to influence the resting free Ca2+ in the lumen of the ER (Fig. 9, A and B), probably reflecting its prevalent intracellular localization to the Golgi compartment (see Fig. 3).
FIGURE 9.
Measurements of Ca2+ concentration in the lumen of ER, [Ca2+]er. Control cells (Ctr), wtDREAM, and daDREAM clones were transfected with erAEQ (25). A, bars represent mean resting [Ca2+] values upon stimulation ± S.E., **, p < 0.01. B, kinetics of ER refilling upon re-addition of 1 mm CaCl2 to Ca2+-depleted cells. Traces are representative of eight independent experiments.
DISCUSSION
DREAM has distinct functions in different cellular compartments, and the overexpression of wt DREAM or daDREAM represses transcription from DRE-containing reporters to a similar extent (1). However, following an increase in the intracellular Ca2+ concentration, the transcription from DRE-reporter becomes derepressed in the presence of DREAM but remains blocked in cells overexpressing daDREAM, which thus acts as a dominant active mutant for the transcriptional repressor function of DREAM (10). Several genes have been found to be regulated through this mechanism that in turn regulates specific physiological processes in brain, in thyroid glands, and in the immune system (1, 10, 12, 13). In this study, we have shown that the expression level of genes involved in protein folding and degradation was altered by DREAM overexpression both in transgenic mice and in model cells. We thus suggest that DREAM modulates the process of protein quality control by regulating the expression of CANT1 and EDEM genes.
Most of the polypeptides emerging in the ER lumen receive N-glycans that become accessible to the glucosidases I and II for the removal of the two outermost glucose residues. The resulting mono-glucosylated trimming intermediate allows the polypeptide chain to enter in cycles of de/re-glucosylation determining substrate dissociation/re-association with calnexin/calreticulin and ERp57 to attain the correct structure (41). Non-native glycoproteins are recognized by the UDP-glucose:glycoprotein glucosyltransferase (GT1) that adds a single glucose residue back to the trimmed oligosaccharide, resulting in a second round of folding (42). The UDP-glucose required for the glucosylation reaction is transported into the ER via a nucleotide sugar/nucleoside monophosphate antiporter (43, 44). The resulting UDP is cleaved to UMP by lumenal nucleoside diphosphatase, and is exchanged via the antiporter system for more nucleotide sugars. A similar topological arrangement exists in the Golgi apparatus (18) and interestingly, the ER protein-folding sensor GT1 is enriched in pre-Golgi intermediates, suggesting that this compartment may participate in protein quality control as well (45).
qPCR analysis of genes related to protein folding and degradation has led us to identify the diphosphate nucleotidase CANT1 as a novel target gene regulated by DREAM. CANT1 has been described as an ER-Golgi resident Ca2+-dependent nucleoside diphosphatase with the catalytic site facing the organelles lumen. (19, 20). It is a member of the ecto-nucleoside trisphosphate diphosphohydrolase (E-NDPases), or apyrase, family cloned from the blood-feeding arthropod Cimex lactularius (bedbug) (46, 47). Sequence related to the Ca2+-dependent apyrases from bloodsucking arthropods were found in other species including mammals, but mammalian proteins differ in cellular localization and also in the catalytic properties (19, 48–50). CANT1 hydrolyzes nucleoside 5′-diphosphates in the order UDP > GDP = IDP ⋙ CDP (but not ADP). Nucleoside 5′-triphosphates are only hydrolyzed to a minor extent and nucleoside 5′-monophosphates are not hydrolyzed. CANT1 prevents the accumulation of diphosphonucleosides that would otherwise cause product inhibition of the glucosyltransferases, and at the same time generates the nucleoside monophosphates needed for the import of new nucleotide sugars (17, 51–53).
CANT1 activity becomes detectable at free Ca2+ concentrations around 100 μm, and becomes maximal in the low millimolar range of Ca2+ (19, 54). The protein is thus fully active in the 200–500 μm Ca2+ concentration range in the ER and the Golgi apparatus (25, 26) and, even if its precise role in ER-folding and quality control pathways is still obscure, it has been proposed to support the calnexin/calreticulin chaperone cycle by preventing the accumulation of UDP and maintaining the flux of UDP-glucose from the cytosol (20).
We have demonstrated that the reduction of CANT1 level is responsible for the impairment of the protein quality control machinery, since secretion efficiency of the soluble folding-competent glycoprotein α1AT-WT is selectively reduced. Folding defective protein model substrate α1AT-NHK undergoes accelerated degradation in neuroblastoma cell clones stably expressing wtDREAM or daDREAM, where the protein levels of CANT1 are reduced, as well as in SH-SY5Y cells, where CANT1 levels were reduced by shRNA CANT1 plasmid transfection. In a previous study we observed that wtDREAM and daDREAM clones also have a reduced ER Ca2+ content (3). We have now better characterized their Ca2+ handling, and we have also found that they have also reduced Golgi Ca2+ levels. We have found that CANT1 overexpression enhanced the intraluminal Ca2+ in the Golgi apparatus in control cells. However, no effect of CANT1 overexpression was observed in wtDREAM and daDREAM clones, possibly suggesting that the observed reduction is secondary to CANT1 down-regulation.
Prompted by the gene profiling studies that identified the up-regulation of two key component of the ERAD pathway, EDEM1 and Derlin, in transgenic B cells (13), we have also identified up-regulation of EDEM1 at the transcript and protein levels in the cerebellum of daDREAM mice as well as in our DREAM cell clones. EDEM1 is an Ire1/Xbp1 dependent ER stress-inducible member of the GH47 family of α1,2-mannosidases that regulates glycoprotein disposal from the ER. It acts by accelerating substrate de-mannosylation and extraction from the calnexin chaperone system, and by chaperoning ERAD candidates to the retro-translocation sites for proteasomal degradation (21, 22, 55). We have found that the tunicamycin treatment, which is responsible for ER stress, significantly enhanced (as expected) the transcript levels of EDEM1 both in control cells and in DREAM clones, even if to a lesser extent, thus suggesting a possible impairment in the induction of ER stress markers in cells where the endogenous CANT1 content was reduced. Accordingly, deletion of APY-1, the C. elegans homolog of CANT1, has been found to sensitize worms to ER stress, and to induce defects in pharynx and muscle organization (50). The findings of up-regulation of EDEM1 in wtDREAM and daDREAM clones, together with that of a reduction of Ca2+ levels in the lumen of the ER and the Golgi apparatus, could at first glance suggest that the CANT1 down-regulation have only a marginal role in determining enhanced protein degradation. However, restoring CANT1 levels in DREAM clones almost completely restored the proper maturation of the substrate model (but not the ER and Golgi apparatus Ca2+ levels), supporting a key role for CANT1 in the pathway of protein maturation.
Overall the data presented here propose a novel role for DREAM in regulating the protein folding machinery by regulating the abundance of two important key proteins, CANT1 and EDEM1. As to EDEM1, previous evidence was already obtained in B cells of daDREAM mice (13), and the results presented here have confirmed the findings in model cells as well.
As for the results on CANT1, their interest is greatly heightened by recent findings showing that nonsense or missense mutations in its gene have been found in a number of cases of severe type 1 and 2 Desbuquois dysplasia, an autosomal recessive chondrodysplasia characterized by severe prenatal and postnatal growth retardation, joint laxity, short extremities, and progressive scoliosis (23, 56, 57). Inclusion bodies containing proteinaceous material within distended ER cisternae have been found in fibroblasts from patients suggesting the possibility of a role for CANT1 in the endochondral ossification process (23, 57–59). One report, actually, postulated that the CANT1 deficiency might interfere with the availability of UDP-sugars needed for proteoglycan synthesis, but no demonstration was provided (23). The work presented here has provided evidence that this could be indeed the case, since pulse-chase analysis of substrate models has established a direct link between CANT1 and protein folding and degradation pathways. It could also be speculated that this defect would be further exacerbated by defects in Ca2+ handling in the Golgi apparatus, since we had found that CANT1 overexpression may control its luminal free Ca2+ concentration in control cells.
In summary, our data have identified a novel target gene for DREAM transcriptional action. Further work will be necessary to fully characterize DREAM action on CANT1 gene promoter. Our results have also demonstrated that CANT1 is a key to the correct glycoprotein maturation process, since the reduction of its intracellular level leads to reduced secretion efficiency and enhanced ERAD of soluble ER-resident glycoproteins. CANT1 thus appears to play a pivotal role in regulating the premature extraction and degradation of glycoproteins from the calnexin cycle, supporting ongoing folding programs. It further suggests that its mutations could impair the correct proteoglycan synthesis in Desbuquois dysplasia patients.
Acknowledgment
We thank Prof. Maurizio Molinari (Bellinzona, Switzerland) for critical reading of the manuscript and providing the plasmids coding for the protein substrates BACE501, α1AT-WT, and α1AT-NHK, and Prof. Terence L. Kirley (Cincinnati, OH) for providing the plasmid coding for CANT1 and the polyclonal antibody.
This work was supported by grants from the Italian Ministry of University and Research [PRIN 2008] and the University of Padova (Progetto di Ateneo 2008) (to M. B.), from Fondazione Cariparo [Progetti di Eccellenza 2008–2009] (to E. C.), from ERA-Net Neuron (grant nEUROsyn 2008) (to E. C. and J. R. N.), from Ministerio Ciencia e Innovacion (SAF2007-62449 (to J. R. N.), SAF2005-04682 and SAF2008-03469, to B. M.), from CIBERNED (to B. M. and J. R. N.), and from the EU 6th Framework Program (NeuroNE) (to E. C. and J. R. N.).
- DREAM
- downstream regulatory element antagonist modulator
- CANT
- Ca2+-activated nucleotidase
- ER
- endoplasmic reticulum
- ERAD
- enhanced ER-associated protein degradation
- DRE
- downstream regulatory element.
REFERENCES
- 1. Carrión A. M., Link W. A., Ledo F., Mellström B., Naranjo J. R. (1999) DREAM is a Ca2+-regulated transcriptional repressor. Nature 398, 80–84 [DOI] [PubMed] [Google Scholar]
- 2. Buxbaum J. D., Choi E. K., Luo Y., Lilliehook C., Crowley A. C., Merriam D. E., Wasco W. (1998) Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat. Med. 4, 1177–1181 [DOI] [PubMed] [Google Scholar]
- 3. Fedrizzi L., Lim D., Carafoli E., Brini M. (2008) Interplay of the Ca2+-binding protein DREAM with presenilin in neuronal Ca2+ signaling. J. Biol. Chem. 283, 27494–27503 [DOI] [PubMed] [Google Scholar]
- 4. An W. F., Bowlby M. R., Betty M., Cao J., Ling H. P., Mendoza G., Hinson J. W., Mattsson K. I., Strassle B. W., Trimmer J. S., Rhodes K. J. (2000) Modulation of A-type potassium channels by a family of calcium sensors. Nature 403, 553–556 [DOI] [PubMed] [Google Scholar]
- 5. Thomsen M. B., Wang C., Ozgen N., Wang H. G., Rosen M. R., Pitt G. S. (2009) Accessory subunit KChIP2 modulates the cardiac L-type calcium current. Circ. Res. 104, 1382–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson D., Mehaffey W. H., Iftinca M., Rehak R., Engbers J. D., Hameed S., Zamponi G. W., Turner R. W. (2010) Regulation of neuronal activity by Cav3-Kv4 channel signaling complexes. Nat. Neurosci. 13, 333–337 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y., Su P., Liang P., Liu T., Liu X., Liu X. Y., Zhang B., Han T., Zhu Y. B., Yin D. M., Li J., Zhou Z., Wang K. W., Wang Y. (2010) The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J. Neurosci. 30, 7575–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu L. J., Mellström B., Wang H., Ren M., Domingo S., Kim S. S., Li X. Y., Chen T., Naranjo J. R., Zhuo M. (2010) DREAM (downstream regulatory element antagonist modulator) contributes to synaptic depression and contextual fear memory. Mol. Brain 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rivas M., Mellström B., Torres B., Cali G., Ferrara A. M., Terracciano D., Zannini M., Morreale de Escobar G., Naranjo J. R. (2009) The DREAM protein is associated with thyroid enlargement and nodular development. Mol. Endocrinol. 23, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Savignac M., Pintado B., Gutierrez-Adan A., Palczewska M., Mellström B., Naranjo J. R. (2005) Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. EMBO J. 24, 3555–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Link W. A., Ledo F., Torres B., Palczewska M., Madsen T. M., Savignac M., Albar J. P., Mellström B., Naranjo J. R. (2004) Day-night changes in downstream regulatory element antagonist modulator/potassium channel interacting protein activity contribute to circadian gene expression in pineal gland. J. Neurosci. 24, 5346–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rivas M., Mellström B., Naranjo J. R., Santisteban P. (2004) Transcriptional repressor DREAM interacts with thyroid transcription factor-1 and regulates thyroglobulin gene expression. J. Biol. Chem. 279, 33114–33122 [DOI] [PubMed] [Google Scholar]
- 13. Savignac M., Mellström B., Bébin A. G., Oliveros J. C., Delpy L., Pinaud E., Naranjo J. R. (2010) Increased B cell proliferation and reduced Ig production in DREAM transgenic mice. J. Immunol. 185, 7527–7536 [DOI] [PubMed] [Google Scholar]
- 14. Sanz C., Mellstrom B., Link W. A., Naranjo J. R., Fernandez-Luna J. L. (2001) Interleukin 3-dependent activation of DREAM is involved in transcriptional silencing of the apoptotic Hrk gene in hematopoietic progenitor cells. EMBO J. 20, 2286–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ledo F., Kremer L., Mellström B., Naranjo J. R. (2002) Ca2+-dependent block of CREB-CBP transcription by repressor DREAM. EMBO J. 21, 4583–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gomez-Villafuertes R., Torres B., Barrio J., Savignac M., Gabellini N., Rizzato F., Pintado B., Gutierrez-Adan A., Mellström B., Carafoli E., Naranjo J. R. (2005) Downstream regulatory element antagonist modulator regulates Ca2+ homeostasis and viability in cerebellar neurons. J. Neurosci. 25, 10822–10830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuhn N. J., White A. (1977) The role of nucleoside diphosphatase in a uridine nucleotide cycle associated with lactose synthesis in rat mammary-gland Golgi apparatus. Biochem. J. 168, 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirschberg C. B., Snider M. D. (1987) Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 56, 63–87 [DOI] [PubMed] [Google Scholar]
- 19. Failer B. U., Braun N., Zimmermann H. (2002) Cloning, expression, and functional characterization of a Ca(2+)-dependent endoplasmic reticulum nucleoside diphosphatase. J. Biol. Chem. 277, 36978–36986 [DOI] [PubMed] [Google Scholar]
- 20. Trombetta E. S., Helenius A. (1999) Glycoprotein reglucosylation and nucleotide sugar utilization in the secretory pathway: identification of a nucleoside diphosphatase in the endoplasmic reticulum. EMBO J. 18, 3282–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. (2003) Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299, 1397–1400 [DOI] [PubMed] [Google Scholar]
- 22. Oda Y., Hosokawa N., Wada I., Nagata K. (2003) EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299, 1394–1397 [DOI] [PubMed] [Google Scholar]
- 23. Huber C., Oulès B., Bertoli M., Chami M., Fradin M., Alanay Y., Al-Gazali L. I., Ausems M. G., Bitoun P., Cavalcanti D. P., Krebs A., Le Merrer M., Mortier G., Shafeghati Y., Superti-Furga A., Robertson S. P., Le Goff C., Muda A. O., Paterlini-Bréchot P., Munnich A., Cormier-Daire V. (2009) Identification of CANT1 mutations in Desbuquois dysplasia. Am. J. Hum. Genet. 85, 706–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montero M., Brini M., Marsault R., Alvarez J., Sitia R., Pozzan T., Rizzuto R. (1995) Monitoring dynamic changes in free Ca2+ concentration in the endoplasmic reticulum of intact cells. EMBO J. 14, 5467–5475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pinton P., Pozzan T., Rizzuto R. (1998) The Golgi apparatus is an inositol 1,4,5-trisphosphate-sensitive Ca2+ store, with functional properties distinct from those of the endoplasmic reticulum. EMBO J. 17, 5298–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brini M., Marsault R., Bastianutto C., Alvarez J., Pozzan T., Rizzuto R. (1995) Transfected aequorin in the measurement of cytosolic Ca2+ concentration ([Ca2+]c). A critical evaluation. J. Biol. Chem. 270, 9896–9903 [DOI] [PubMed] [Google Scholar]
- 28. Barrero M. J., Montero M., Alvarez J. (1997) Dynamics of [Ca2+] in the endoplasmic reticulum and cytoplasm of intact HeLa cells. A comparative study. J. Biol. Chem. 272, 27694–27699 [DOI] [PubMed] [Google Scholar]
- 29. Olivari S., Cali T., Salo K. E., Paganetti P., Ruddock L. W., Molinari M. (2006) EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 349, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 30. Tabas I., Ron D. (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Le Fourn V., Gaplovska-Kysela K., Guhl B., Santimaria R., Zuber C., Roth J. (2009) Basal autophagy is involved in the degradation of the ERAD component EDEM1. Cell Mol. Life Sci. 66, 1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huse J. T., Pijak D. S., Leslie G. J., Lee V. M., Doms R. W. (2000) Maturation and endosomal targeting of beta-site amyloid precursor protein-cleaving enzyme. The Alzheimer's disease beta-secretase. J. Biol. Chem. 275, 33729–33737 [DOI] [PubMed] [Google Scholar]
- 33. Ou W. J., Cameron P. H., Thomas D. Y., Bergeron J. J. (1993) Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature 364, 771–776 [DOI] [PubMed] [Google Scholar]
- 34. Eriksson K. K., Vago R., Calanca V., Galli C., Paganetti P., Molinari M. (2004) EDEM contributes to maintenance of protein folding efficiency and secretory capacity. J. Biol. Chem. 279, 44600–44605 [DOI] [PubMed] [Google Scholar]
- 35. Calì T., Galli C., Olivari S., Molinari M. (2008) Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem. Biophys. Res. Commun. 371, 405–410 [DOI] [PubMed] [Google Scholar]
- 36. Sifers R. N., Brashears-Macatee S., Kidd V. J., Muensch H., Woo S. L. (1988) A frameshift mutation results in a truncated α1-antitrypsin that is retained within the rough endoplasmic reticulum. J. Biol. Chem. 263, 7330–7335 [PubMed] [Google Scholar]
- 37. Hebert D. N., Garman S. C., Molinari M. (2005) The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 15, 364–370 [DOI] [PubMed] [Google Scholar]
- 38. Olivari S., Molinari M. (2007) Glycoprotein folding and the role of EDEM1, EDEM2, and EDEM3 in degradation of folding-defective glycoproteins. FEBS Lett. 581, 3658–3664 [DOI] [PubMed] [Google Scholar]
- 39. Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. (2001) A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brini M. (2008) Calcium-sensitive photoproteins. Methods 46, 160–166 [DOI] [PubMed] [Google Scholar]
- 41. Hammond C., Braakman I., Helenius A. (1994) Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. U.S.A. 91, 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ellgaard L., Molinari M., Helenius A. (1999) Setting the standards: quality control in the secretory pathway. Science 286, 1882–1888 [DOI] [PubMed] [Google Scholar]
- 43. Hirschberg C. B., Robbins P. W., Abeijon C. (1998) Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 67, 49–69 [DOI] [PubMed] [Google Scholar]
- 44. Castro O., Chen L. Y., Parodi A. J., Abeijón C. (1999) Uridine diphosphate-glucose transport into the endoplasmic reticulum of Saccharomyces cerevisiae: in vivo and in vitro evidence. Mol. Biol. Cell 10, 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuber C., Fan J. Y., Guhl B., Parodi A., Fessler J. H., Parker C., Roth J. (2001) Immunolocalization of UDP-glucose:glycoprotein glucosyltransferase indicates involvement of pre-Golgi intermediates in protein quality control. Proc. Natl. Acad. Sci. U.S.A. 98, 10710–10715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valenzuela J. G., Charlab R., Galperin M. Y., Ribeiro J. M. (1998) Purification, cloning, and expression of an apyrase from the bed bug Cimex lectularius. A new type of nucleotide-binding enzyme. J. Biol. Chem. 273, 30583–30590 [DOI] [PubMed] [Google Scholar]
- 47. Valenzuela J. G., Belkaid Y., Rowton E., Ribeiro J. M. (2001) The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J. Exp. Biol. 204, 229–237 [DOI] [PubMed] [Google Scholar]
- 48. Smith T. M., Hicks-Berger C. A., Kim S., Kirley T. L. (2002) Cloning, expression, and characterization of a soluble calcium-activated nucleotidase, a human enzyme belonging to a new family of extracellular nucleotidases. Arch Biochem. Biophys. 406, 105–115 [DOI] [PubMed] [Google Scholar]
- 49. Devader C., Webb R. J., Thomas G. M., Dale L. (2006) Xenopus apyrase (xapy), a secreted nucleotidase that is expressed during early development. Gene 367, 135–141 [DOI] [PubMed] [Google Scholar]
- 50. Uccelletti D., Pascoli A., Farina F., Alberti A., Mancini P., Hirschberg C. B., Palleschi C. (2008) APY-1, a novel Caenorhabditis elegans apyrase involved in unfolded protein response signaling and stress responses. Mol. Biol. Cell 19, 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Novikoff A. B., Goldfischer S. (1961) Nucleosidediphosphatase activity in the Golgi apparatus and its usefulness for cytological studies. Proc. Natl. Acad. Sci. U.S.A. 47, 802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Allen J. M., Slater J. J. (1961) A cytochemical study of Golgi associated thiamine pyrophosphatase in the epididymis of the mouse. J Histochem Cytochem 9, 418–423 [DOI] [PubMed] [Google Scholar]
- 53. Abeijon C., Hirschberg C. B. (1992) Topography of glycosylation reactions in the endoplasmic reticulum. Trends Biochem. Sci. 17, 32–36 [DOI] [PubMed] [Google Scholar]
- 54. Murphy D. M., Ivanenkov V. V., Kirley T. L. (2003) Bacterial expression and characterization of a novel, soluble, calcium-binding, and calcium-activated human nucleotidase. Biochemistry 42, 2412–2421 [DOI] [PubMed] [Google Scholar]
- 55. Kanehara K., Kawaguchi S., Ng D. T. (2007) The EDEM and Yos9p families of lectin-like ERAD factors. Semin. Cell Dev. Biol. 18, 743–750 [DOI] [PubMed] [Google Scholar]
- 56. Laccone F., Schoner K., Krabichler B., Kluge B., Schwerdtfeger R., Schulze B., Zschocke J., Rehder H. (2011) Desbuquois dysplasia type I and fetal hydrops due to novel mutations in the CANT1 gene. Eur. J. Hum. Genet. 19, 1133–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dai J., Kim O. H., Cho T. J., Miyake N., Song H. R., Karasugi T., Sakazume S., Ikema M., Matsui Y., Nagai T., Matsumoto N., Ohashi H., Kamatani N., Nishimura G., Furuichi T., Takahashi A., Ikegawa S. (2011) A founder mutation of CANT1 common in Korean and Japanese Desbuquois dysplasia. J. Hum. Genet. 56, 398–400 [DOI] [PubMed] [Google Scholar]
- 58. Faden M., Al-Zahrani F., Arafah D., Alkuraya F. S. (2010) Mutation of CANT1 causes Desbuquois dysplasia. Am. J. Med. Genet. 152, 1157–1160 [DOI] [PubMed] [Google Scholar]
- 59. Furuichi T., Dai J., Cho T. J., Sakazume S., Ikema M., Matsui Y., Baynam G., Nagai T., Miyake N., Matsumoto N., Ohashi H., Unger S., Superti-Furga A., Kim O. H., Nishimura G., Ikegawa S. (2011) CANT1 mutation is also responsible for Desbuquois dysplasia, type 2 and Kim variant. J. Med. Genet. 48, 32–37 [DOI] [PubMed] [Google Scholar]