Abstract
The nematode Caenorhabditis elegans is central to research in molecular, cell, and developmental biology, but nearly all of this research has been conducted on a single strain. Comparatively little is known about the population genomic and evolutionary history of this species. We characterized C. elegans genetic variation by high-throughput selective sequencing of a worldwide collection of 200 wild strains, identifying 41,188 single nucleotide polymorphisms. Unexpectedly, C. elegans genome variation is dominated by a set of commonly shared haplotypes on four of the six chromosomes, each spanning many megabases. Population-genetic modeling shows that this pattern was generated by chromosome-scale selective sweeps that have reduced variation worldwide; at least one of these sweeps likely occurred in the past few hundred years. These sweeps, which we hypothesize to be a result of human activity, have dramatically reshaped the global C. elegans population in the recent past.
Introduction
Caenorhabditis elegans is a globally distributed, free-living nematode that colonizes human-associated habitats, including compost heaps and rotting fruit1. For the past forty years, a single laboratory strain (N2) has proven invaluable to biomedical research as a model for animal development, programmed cell death, and RNA interference2. Studies of a small number of loci suggest that C. elegans has a small effective population size and low diversity compared to closely related species, despite large local population sizes and global gene flow3–14. The factors responsible for this low genetic diversity remain unknown. C. elegans reproduces primarily by hermaphroditic selfing, but the mating system alone is not sufficient to explain the observed reduction in diversity11. Polymorphism rate between the laboratory strain N2 and the wild isolate CB4856 correlates with recombination rate, suggesting that background selection against deleterious mutations also reduces diversity5,15,16. However, CB4856 is genetically isolated from the rest of the C. elegans population17, and analyses based on its divergence alone are subject to significant ascertainment bias and may not fully capture evolutionary processes relevant to the global population. To obtain a more complete description of C. elegans diversity, we sequenced thousands of genome fragments from a globally distributed collection of 200 wild isolates. Our results demonstrate that recent strong sweeps of positive selection have drastically reduced chromosome-wide diversity in this species.
C. elegans genome diversity and strain relationships
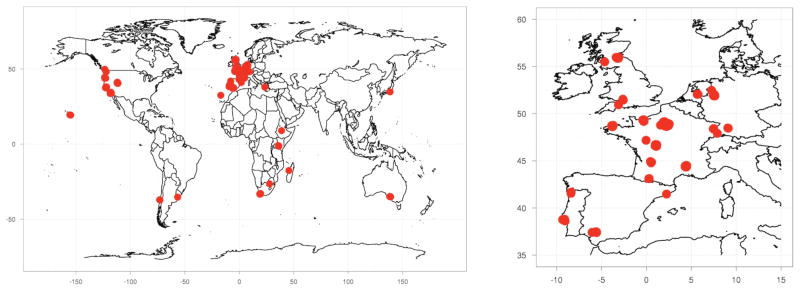
We studied 200 wild strains of C. elegans from 58 collection locations on six continents (Fig. 1, Supplementary Table 1). These strains cover virtually every known collection location, providing the most comprehensive set of C. elegans strains assembled to date. The samples were isolated from a variety of sources, including rotting fruits, compost, mushroom farms, soil, and snails. To characterize genomic variation among these strains, we examined restriction-site associated DNA (RAD)18 covering 8% of the 100 megabase genome. We sequenced 91 bp on both sides of each EcoRI restriction site, yielding a pair of RAD tags every 2.1 kb on average. We achieved a median coverage of 27 reads per tag per strain, allowing SNP identification with a false discovery rate (FDR) less than 0.6% (see Methods). Across all strains, we identified 41,188 SNPs in 8 Mb of sequence (an average of 5.1 SNPs per kb).
Figure 1. Global sampling locations of C. elegans strains.
The isolation locations of wild strains sequenced in this study are shown as red circles on the world map. The right panel is a map of the more densely sampled Western Europe.
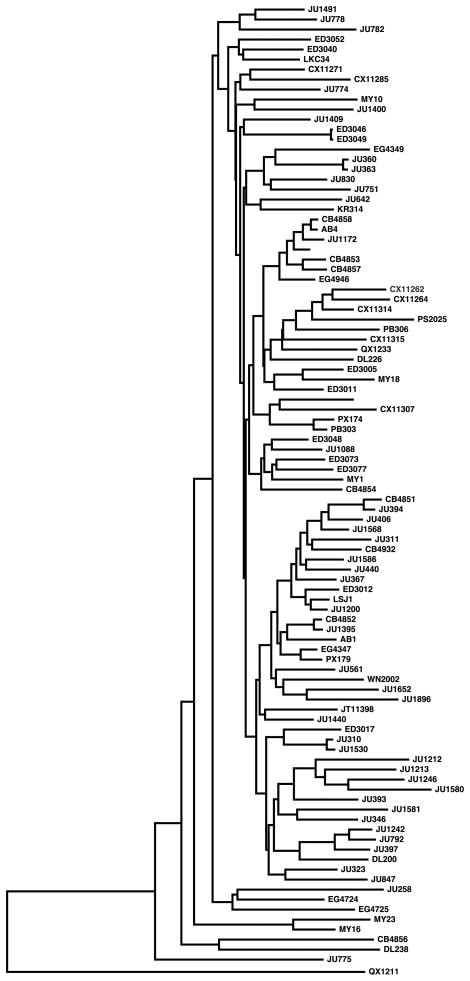
C. elegans reproduces primarily as a selfing hermaphrodite, which can lead to clonal expansions of a single genotype. For this reason, we expected to find identical strains isolated from nearby locations. To find these cases, we examined the number and distribution of discordant genotype calls across all pairwise strain comparisons. Pairs with fewer SNPs than the expected number of false positives given our FDR (250 SNPs) were considered clonal, with the exception of the pair ED3046 and ED3049, for which the SNPs were clustered in a small region on chromosome II. Of the 200 sampled strains, 47 had unique haplotypes. The remaining 153 strains grouped into 50 near-identical sets (Supplementary Table 1). Most of these sets are from a single isolation or from separate samples in close proximity, likely representing strains sampled from a single clonal expansion. However, two sets spanned different continents: AB2 and CX11258 from Australia and the United States, respectively, and JU1171 and MY23 from Chile and Germany, respectively. It is possible that these pairs of strains are the result of recent long-range migrations. However, given previous evidence of strain confusion with wild strains isolated before good record-keeping17,19 and our own results, we conservatively analyzed only one strain from each of these sets. The set of 97 distinct genome-wide haplotypes, referred to as “isotypes” in subsequent analyses, comprises one isolate from each of the 50 near-identical sets and the 47 unique isolates (Supplementary Table 1). Phylogenetic clustering of the isotypes revealed little to no grouping by isolation environment or country of origin (Fig. 2, Supplementary Fig. 1) but identified four highly diverged isotypes: CB4856, DL238, JU775, and QX1211. We identified an average of 3,613 SNPs per isotype compared to the reference strain N2, but these four diverged isotypes had an average of 9,141 SNPs. In particular, 18% of the variants in the full SNP set are found only in QX1211, isolated in San Francisco.
Figure 2. Neighbor-joining tree of 97 C. elegans isotypes.
The tree was constructed using 40,857 polymorphisms in the set of 97 isotypes and pseudo-rooted to QX1211 for visualization reasons. Branch lengths are proportional to the number of polymorphisms that differentiate each pair. Scale bar is the
Linkage disequilibrium and population structure
Among the isotypes, we found several large blocks of strong linkage disequilibrium (LD) (r2 > 0.6) extending several megabases within chromosomes (Supplementary Fig. 2). Substantial LD also exists between chromosomes, with r2 often above 0.2. The population recombination rate (4Nr) on each chromosome, estimated by composite likelihood20, ranged from 90 to 185, suggesting an outcrossing rate between 1/100 and 1/1000 per generation, depending on the estimate of effective population size. To test for population subdivision, we used STRUCTURE21,22 and found statistical support for only one worldwide population (Supplementary Fig. 3). These results suggest that the observed LD is caused mainly by selfing, rather than by separation into distinct subpopulations. Principal Component Analysis (PCA) identified five significant axes that explain 29.7% of the genetic variation (Supplementary Fig. 3). These axes reveal some geographic structure but fail to clearly separate isotypes into distinct subpopulations. There is a weak correlation between geographic distance and genetic relatedness at the local scale (less than 700 km, Supplementary Figure 4), but we found no correlation at larger distances, in agreement with previous analyses3,6,13.
Despite the extensive LD, previous results demonstrated the feasibility of genome-wide association analysis in C. elegans by mapping two qualitative traits, hybrid incompatibility and copulatory plugging, using SNPs between N2 and CB485617. Because the causal variants for these traits are known and exhibit a near-perfect genotype-phenotype correspondence, we genotyped these variants as proxies for the traits, and showed that our set of SNPs can be used to map the variants to the correct genomic regions (Supplementary Fig. 5). We also applied association mapping to two quantitative traits (Supplementary Fig. 5; Methods). Resistance to abamectin, an anthelmintic compound produced by the common soil bacterium Streptomyces avermitilis23, was significantly associated with a 28 kb haplotype on chromosome V, and aversion to the human pathogen Pseudomonas aeruginosa mapped to a 45 kb interval on chromosome IV. Because geographic structure might be observable using association analysis, we mapped the latitude at which a strain was isolated and found a significant locus in the center of chromosome II. This association could reflect subtle population structure, or it might implicate this region in an unknown ecological niche preference, such as temperature.
Using these 97 C. elegans isotypes, association analyses will likely discover alleles only with large phenotypic effects. Additionally, the chromosomal location of the causal variant limits the resolution of mapping. Extensive linkage disequilibrium in the centers of chromosomes results in haplotype blocks over a megabase in size, as shown here for copulatory plugging and latitude. By contrast, causal variants on the more freely recombining chromosome arms can be localized to haplotype blocks smaller than 50 kb, as shown here for hybrid incompatibility, abamectin resistance, and P. aeruginosa avoidance. In this regard, it is worth noting that functional variants in C. elegans are more likely to be located on chromosome arms due to the correlation between rates of recombination and polymorphism15.
Genetic variation and chromosome-wide haplotype sharing
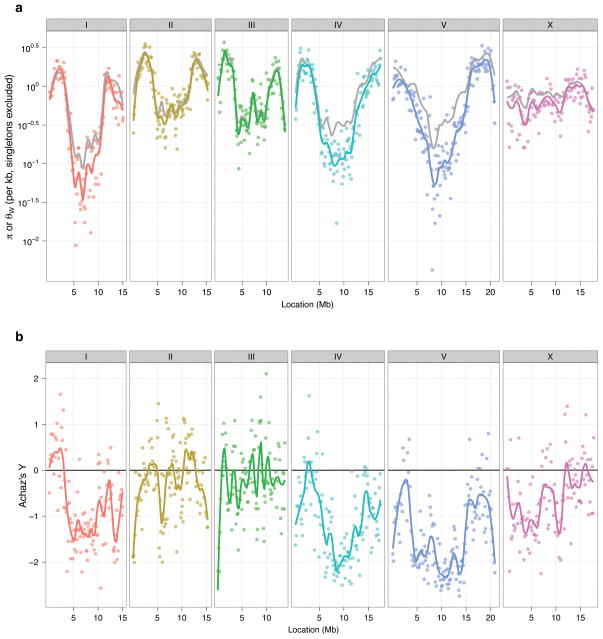
Despite a global distribution, with local populations likely containing millions of individuals6, our results confirm that genetic variation in C. elegans is low. Our genome-wide coverage shows that the level of diversity varies across genomic regions (Fig. 3), as suggested by previous results derived from a small number of loci5,24. The estimated population mutation rate (θW, Methods) varies over two orders of magnitude, from greater than 3.5×10−3 per bp on some chromosome arms to a minimum of 2.5×10−5 per bp in the centers, averaging 8.3×10−4 per bp. The level of polymorphism correlates with the recombination rate on all autosomes17 – diversity is lower in the low-recombination chromosome centers and higher on the more freely recombining arms (Fig. 3, Supplementary Fig. 6). On the X chromosome, this pattern is much weaker, and the level of polymorphism is fairly constant across its entire length (θW ~8.5×10−4), which corresponds to its more uniform recombination rate17. The correlation between rates of polymorphism and recombination is consistent with previous results implicating background selection as a major force shaping patterns of C. elegans diversity5,15. Variation in pairwise diversity (π) follows the same general pattern as θW, but with a larger reduction in π than in θW in the centers of chromosomes I, IV, and V. This difference results in extremely negative values of Achaz’s Y (an analog of Tajima’s D, see Methods) and indicates an excess of low-frequency polymorphism relative to neutral expectation (Fig. 3, Supplementary Fig. 6). The left arm of the X chromosome also shows an excess of rare variants, but unlike on chromosomes I, IV and V, this region does not have a low recombination rate.
Figure 3. Chromosomal patterns of sequence polymorphism.
a, Two estimates of population polymorphism rate, π (colored points and lines) and θW (grey lines), are shown for each chromosome. Each point represents a non-overlapping window of 110 RAD tags (approximately 10 kb of sequence). The lines show a locally weighted polynomial regression.
b, Achaz’s Y, a measure of deviation from the neutral allele frequency spectrum, calculated over the same windows with local polynomial regression. Negative values indicate an excess of rare alleles.
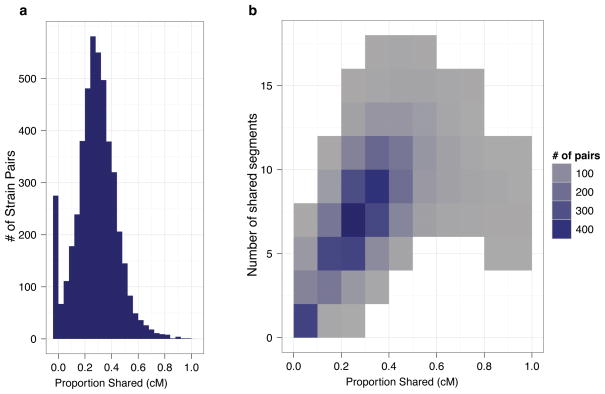
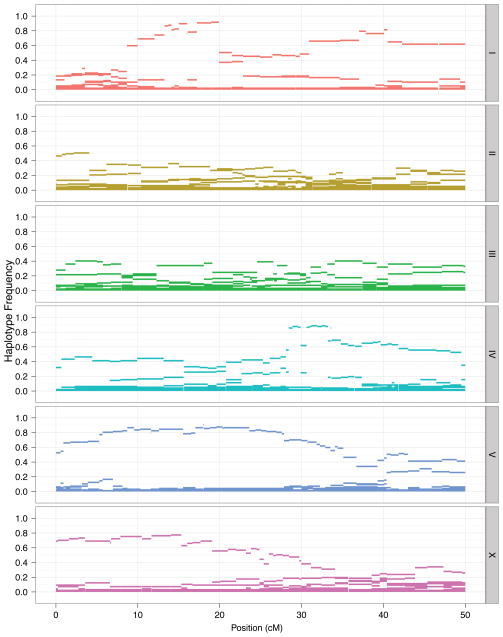
The genome of the wild strain CB4858 appears to contain large haplotypes shared with the reference strain N225, indicating recent common ancestry. To identify whether additional such relationships exist among the 97 isotypes, we used the program GERMLINE26 to search each pair for segments of at least two centimorgans or megabases with no more than two SNP differences, which we defined as “shared” segments (see Methods). Remarkably, we found extensive sharing of large haplotypes among the majority of isotypes, suggesting recent common ancestry (Fig. 4). The average pair shares roughly one third of the genome identical-by-descent when measured on either the genetic (median = 28%) or the physical map (median = 33%). The median block size of the shared segments is roughly a fifth of a chromosome (2.5 Mb). Some blocks span more than a third of a chromosome, indicating that very few generations of outcrossing have occurred since the most recent common ancestor. Most strikingly, the patterns of sharing are unevenly distributed across the genome − 70% to 90% of isotypes share segments that span several megabases on chromosomes I, IV, V, and X (Fig. 5), but such sharing is not observed on chromosomes II and III (Supplementary Fig. 7). In particular, chromosome V shows one common haplotype that spans the majority of its length. These regions of high haplotype homozygosity correspond to the regions with an excess of rare SNPs noted above. Notably, the common haplotypes for chromosomes I, IV, and V are found on all six sampled continents; the chromosome X common haplotype is present on five.
Figure 4. Extensive sharing of large blocks of near-identical haplotypes.
a, Proportion of the genetic map shared (as determined by GERMLINE) for every pairwise comparison of the 97 isotypes is shown as a histogram. Notably, every pairwise comparison with one the most diverged isotypes (CB4856, DL238, and QX1211) shows little to no sharing. By contrast, the average sharing between a pair is one third of the genetic map.
b, Two-dimensional density plot of the number of shared segments and the proportion of the genetic map shared shows that most isotype pairs share about one third of the genome in six to ten segments, indicating that the shared segments are large.
Figure 5. High-frequency extended haplotypes on chromosomes I, IV, V, and X.
The chromosomal region (in Mb) covered by each haplotype block is shown as a bar along the x-axis, with haplotype frequency indicated by height on the y-axis.
Recent strong selective sweeps
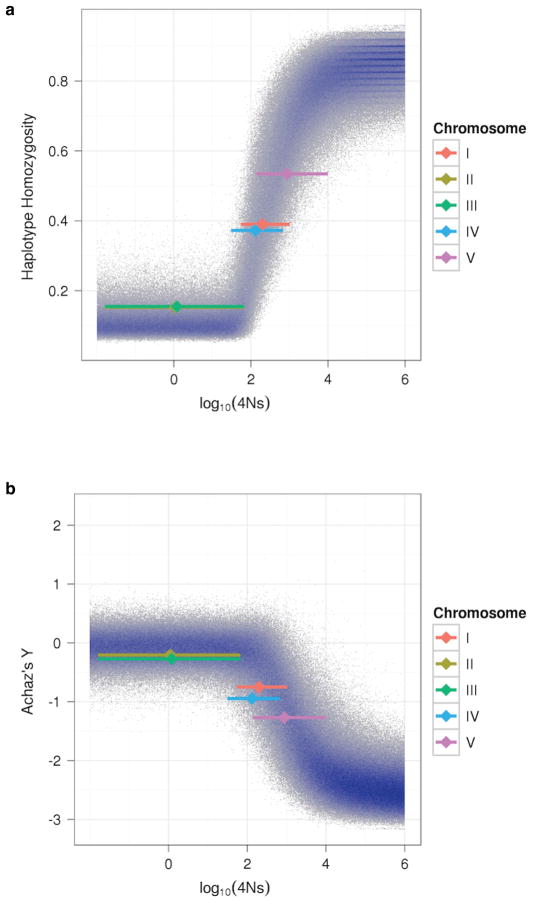
The combination of high haplotype homozygosity extending over large regions and an excess of rare variants is expected after a strong selective sweep, especially when the recombination rate is low27. To estimate the population and selection parameters required to generate the observed patterns, we performed coalescent simulations of entire chromosomes over a range of demographic models, including single and multiple populations with varying migration rates and population sizes. All models incorporated the effects of background selection and recombination on chromosomal diversity patterns. Demographic forces and background selection are expected to affect the patterns of variation on all chromosomes equally, resulting in a single best-fitting model. Contrary to this expectation, the patterns of variation on chromosomes II and III are strikingly different from the patterns on chromosomes I, IV, and V. While the patterns of polymorphism on chromosomes II and III were compatible with models that did not include positive selection, fitting both the excess of rare variants and high haplotype homozygosity observed for chromosomes I, IV, V required incorporating positive selection (Fig. 6; X was not tested, Methods). Our estimates of the population selection parameter 4Ns for these chromosomes ranged from 100 to a maximum of 500. For an effective population size between 10,000 and 25,000, these values of 4Ns correspond to a selective advantage in the range of 0.1% to 1.3% per generation.
Figure 6. Modeled effects of selection.
Results from 106 coalescent simulations of chromosomes with a single positively selected site in the center of the chromosome (Methods) are plotted. Regions with a high density of points are indicated in blue. Achaz’s Y (a) and haplotype homozygosity (b) for the entire chromosome are plotted against the simulated selection coefficient 4Ns. The values observed in our experimental data for each chromosome are indicated by the vertical positions of the colored diamonds. The location of each diamond on the x-axis is the median 4Ns as estimated from the simulated data, with the extent of the bar showing the 90% credible interval.
To estimate the timing of these selective sweeps, we focused on the largest and most highly shared segment found on chromosome V, shared by 84 of the 97 isotypes. Using coalescent simulations with two different models of population growth (Supplementary Fig. 8), we estimated that the haplotype arose between 600 and 1250 generations ago (90% credible interval). In the laboratory, C. elegans can go through 100 generations per year, but the average generation time in nature is likely much longer4. If we assume a conservative estimate of six generations per year28, the common haplotype on chromosome V likely expanded to its current frequency in the past 100 to 200 years. A lower bound is provided by the strain CB4851, which was isolated before 1949 and carries the selected haplotypes on chromosomes I, V, and X, making it likely that those sweeps began no less than 60 years ago. Even if the effective population size and generation time differ by an order of magnitude from our estimated values, the selective sweep still would have occurred in historical times.
Discussion
We report the most comprehensive survey of C. elegans diversity to date. Our results indicate that polymorphism rates are correlated with recombination rates, that linkage disequilibrium extends over long distances and often occurs between loci on different chromosomes, and that there is little detectable subdivision of the global population. Surprisingly, we found extensive sharing of large haplotypes on a subset of chromosomes, accompanied by a paucity of common variation in these regions. The shared haplotypes are distributed throughout the world (Supplementary Figure 9). These observations can only be explained by one or more strong recent global sweeps driven by positive selection, a scenario previously considered unlikely in C. elegans and Caenorhabditis in general5,29. Only QX1211 (recently isolated in California), CB4856, and DL238 (isolated in Hawaii) do not share any large haplotypes with the rest of the isotypes. These observations suggest that Hawaii and the Pacific Rim may be a fruitful ground for discovery of additional highly diverged isolates. Focused searches in these locations, as well as in other poorly sampled parts of the globe, may yield strains that represent the broader C. elegans diversity that existed prior to the selective sweeps that homogenized much of the global population.
Identification of the beneficial alleles that swept through the C. elegans population will be challenging. A selective advantage of 0.1 – 1% per generation is sufficient to drive a rapid selective sweep in nature, but phenotypic differences of that magnitude are difficult to reliably detect in the laboratory. Within each swept region, there are hundreds of genes with potential effects on fitness, and we can also only speculate about the selective forces that drove the sweeps. We know little about C. elegans ecology1, and it is possible that selection occurred for adaptation to a specific, as yet unknown microenvironment.
Positive selection has reduced C. elegans genetic variation on a scale not previously observed in multicellular organisms. The rapid global spread of the selected haplotypes during the past few centuries suggests the possibility that the selected alleles may be related to the association between C. elegans and human activity. Long-range human travel and transportation of agricultural products in this time interval likely contributed to the global spread of the selected haplotypes. Loci that aid human-assisted dispersal and/or confer fitness advantages in human-associated habitats may have driven the observed sweeps. Whether the sweeps resulted in global replacement of endemic populations or de novo colonization of new environments by C. elegans is unclear. The evolution of the parasitic protozoan Toxoplasma gondii provides a striking parallel. Like C. elegans, T. gondii is a small human-associated eukaryote with a selfing life stage. A chromosome-wide selective sweep of a single haplotype that originated around 10,000 years ago spread throughout the world30, suggesting that the dramatic changes in human civilization during this period (such as animal domestication) could have played a role in the rapid evolution of a new lineage. Recent dramatic alterations of global environments by humans, and the creation of human-associated niches, may have made such selective sweeps a common feature of the genomes of many species.
Methods
Strains
Animals were cultured with the bacterial strain OP50 on modified nematode growth medium (NGM)31, containing 1% agar and 0.7% agarose to prevent burrowing of wild C. elegans isolates. Strain information is listed in Supplementary Table 1. These strains represent at least one clone from every known isolation location. For locations with more than one strain, we chose strains isolated from different substrates.
Sequence analysis identified strains for which the true identity is suspect. The CGC versions of strains CB4855 and CB4858 were found to be identical by sequence comparison, even though the strains are reportedly from different isolation locations. Versions of CB4855 and CB4858 from J. Hodgkin are different from each other and from their respective CGC versions but were not used in our analyses. Instead, we treated the two samples as one strain from an unknown location. JU1615 and JU1616 from Melbourne, Australia are likely N2 contaminants as determined by sequence and behavioral assays; they were excluded from our analyses. PX174 and RC301 were found to be identical, despite reported isolations from the United States and Germany, respectively. PX174 was likely mis-frozen from an RC301 stock, and PX174 was excluded from our analyses. JU813 and ED3054 were found to be C. briggsae by sequence32 and mating tests, and were not included in any analyses.
We also sequenced the following strains, but the sequence or mapping qualities were not high enough to include them in downstream analyses: CB4855 (J. Hodgkin version), CX11254, and WN2001.
Restriction-site associated DNA (RAD) marker library construction and sequence determination
We isolated genomic DNA by washing off nearly starved animals from five 10 cm NGM plates to 15 mL conical tubes and settling by gravity for one hour. Genomic DNA was prepared using the DNeasy Blood and Tissue Kit (Qiagen). Seventeen RAD marker libraries were constructed by Floragenex, Inc. Nine additional libraries were constructed using a protocol adapted from previous work18. Illumina Genome Analyzer IIx protocols were used for sequencing at 101 cycles.
SNP determination
Each sequence read was entered into a custom mySQL database. Reads were grouped by strain, checked for the presence of a complete EcoRI cut sequence and mapped to the WS210 version of the N2 genome using bwa33. Loci with sequence from only a single strain or fewer than five reads per strain were excluded, as were locations less than 100 bp from another cut site. Reads that passed these filters were exported to SAMtools34 for SNP identification using the pileup command. Called SNPs not in repetitive regions (defined using RepeatMasker) were imported them into R, where all subsequent analyses were performed.
We determined an optimal SNP calling strategy by comparison of libraries generated from different biological replicates of the same strain. Given a quality threshold, sites that differed between replicate libraries were considered errors, and sites that corresponded in both libraries but differed from the reference were counted as true SNPs. SNPs were called at phred score threshold of 60, requiring at least one of each allele to have a score ≥120. This approach provided the best balance between a low FDR (~0.6%) and power to identify true SNPs, yielding 41,188 SNPs. Any genotype call with a score below 60 was considered missing data, and sites missing in more than 25 strains were removed. We imputed missing genotype calls with NPUTE35. Both the imputed and unimputed datasets were condensed from the 200 strains into the 97 isotypes. This reduction eliminated a small number of segregating polymorphisms, resulting in 40,857 SNPs. This SNP set was used for all analyses, except for association analysis and detection of population structure.
For STRUCTURE and principal component analysis, we constructed a more stringent SNP set. As before, SNPs present in at least one isotype with a quality score of at least 120 then called at a quality score of at least 100 for all other isotypes. SNPs that were missing or low quality in more than six isotypes were removed. The more stringent cutoff resulted in a set of 6,089 high-quality SNPs. The remaining missing calls were imputed using the program NPUTE.
To construct a SNP set for association mapping, we used the 6,089 SNP set but raised the minor allele cutoff to 10 out of 97 isotypes, yielding 4,690 high-quality common SNPs. Missing calls were imputed with NPUTE.
Determination of population structure
For STRUCTURE and Principal Component Analysis (PCA), we pruned the 6,089 SNP set in sliding 25 marker windows at five marker steps, pruning pairs with r2 greater than 0.3 using PLINK36. This reduced the data to 757 SNPs. The results of STRUCTURE and PCA were similar at different levels of pruning or missing data thresholds. We used EIGENSOFT37 for PCA and evaluated significance using Tracy-Widom statistics. Running EIGENSOFT with the “missingmode: YES” option confirmed that the observed patterns are not caused by structure in the missing data.
Association mapping
We used EMMA38 for all association analyses with the default kinship matrix. We ignored significant linkages of single markers, as these results are likely caused by allele frequency skews.
We genotyped the zeel-1 peel-1 and plg-1 loci using genomic PCR for each of the 200 strains (Supplementary Table 2). These presence or absence genotypes might not reflect the phenotypes for these variants.
For abamectin sensitivity, L4 animals were grown for 20 hours on NGM plates freshly seeded with E. coli OP50. Young adults were then transferred onto an unseeded plate and allowed to roam for one minute, then transferred one per well into a 96-well flat-bottom tissue culture treated microtiter plate (Costar) containing 150 μl of M9 buffer with 5 μg/mL abamectin. Animals were monitored at room temperature under a Leica SMZ650 dissecting scope to measure body bends in a 10 second period, either by direct observation or video recordings. A single body bend was defined as bending on either dorsal or ventral side relative to the midline.
For Pseudomonas aeruginosa avoidance, we scored the fraction of animals that crawled off the agar plate during a slow-killing assay. Slow-killing assays were performed as published previously39. Briefly, the standard slow-killing assay40 was performed in the presence of 50 μg/ml 5-fluorodeoxyuridine, using the PA14 strain. A minimum of 80 worms per genotype were assayed in at least two independent trials.
Determination of segment sharing
We ran the program GERMLINE on the imputed 40,857 SNP set to define shared segments as intervals of at least 150 markers and two cM or Mb in length, with no more than two SNPs between isotypes. Shared segments were collapsed into a single haplotype, and we calculated the haplotype frequencies and homozygosity of each interval in the genome.
Calculation of population genetic statistics
To reduce the effects of sequencing errors on standard population genetic statistics (π, Watterson’s θ, Tajima’s D), we excluded all singletons, and calculated Achaz’s Y41 instead of Tajima’s D. Although our error rate is low on a genome-wide scale (less than one false SNP per 10 kb), errors may still account for a large fraction of observed variants in low diversity regions of the genome, substantially biasing Tajima’s D42.
We estimated the population recombination rate (ρ = 4Nr) for each chromosome using composite likelihood20 as implemented in LDhat (version 2.1), using values of ρ between 0 and 250 in increments of five. The outcrossing rate C was estimated as C=ρ/4Nrc where rc = 0.5 is the recombination rate per chromosome per outcross, with effective population size assumed to be between 10,000 and 50,000.
Simulation of population genetic parameters
We performed coalescent simulations of entire chromosomes using the program msms43. To match observed recombination patterns, we adjusted the arms of the simulated chromosomes by dividing the distance between each pair of SNPs by five (increasing the effective recombination rate five-fold) while randomly removing SNPs to maintain SNP density. SNPs in the center of the chromosome were randomly removed with probability 0.9 to match the observed patterns of polymorphism without affecting allele frequencies. The final chromosomes thus contained three regions: two high diversity, high recombination arms covering 20% of the physical chromosome each, and a central region with low recombination and low diversity. The total chromosome length was set at 17 Mb. For all simulations, the population mutation rate (θ) and recombination rate (ρ) were uniformly sampled across a broad range of values [θ = U(4000, 20000), before reductions described; ρ = U(50, 250)]. Simulated chromosomes with a calculated θW (singletons excluded) in the range of the observed data (700 – 1150) were accepted, and 106 such chromosomes were generated for every set of models.
Simulations with selection consisted of a single population with a single selected site in the chromosome center with a final frequency in the population of 90%. We sampled from logarithmic distributions for both selective coefficients [log10(4Ns) = U(−2, 6)] and population sizes [log10(N) = U(2,6)]. Simulations where the calculated values of Achaz’s Y and average haplotype homozygosity differed from the observed value by less than 0.05 were used to construct a distribution of possible values of 4Ns..
Neutral simulations included models of a single population with constant size or a recent period of exponential growth. Models with two ancestral populations were also considered, with individuals sampled from each population in proportion to their relative sizes. Parameters of this model (in addition to θ and ρ) included rates of migration between populations (migration could be asymmetric, and change over time) and the relative population sizes.
Coalescent Simulations to Determine the Age of the Chromosome V Haplotype
We modeled expansion of the largest highly shared segment, Chromosome V at 9.6–11.9 Mb, using coalescent simulations of 84 individuals and no recombination. To model exponential growth, we sampled from uniform distributions of θ and the growth rate parameter, α (Nt = N0e−αt, where Nt is the population size 4Nt generations in the past and N0 is the present size). Values for θ and α were retained from simulated samples with 66–68 segregating sites (observed = 67) and a Tajima’s D between −2.66 and −2.68 (observed = −2.67) (data from the entire population suggest that Tajima’s D is minimally biased in this region). Using the laboratory-derived SNP mutation rate of 9 × 10−9 per bp per generation44 to estimate population size, the median population expansion rate is 0.86% per generation (90% CI: 0.63 – 1.4%, Supplementary Fig. 8). For a 1000-fold population expansion, we then estimate a median of 807 generations (90% credible interval = 636 – 1081).
An alternative simulation approach forced all lineages to coalesce at a given time t (measured in 4N generations). For these “star-like” simulations, we randomly sampled from uniform distributions of θ and t, retaining successful samples as described above. Again using the laboratory-derived mutation rate to estimate population size, we estimate the time to the forced coalescence as 846 generations (90% CI: 630–1158) (Supplementary Fig. 8).
Supplementary Material
Acknowledgments
The authors are grateful to all contributors who provided strains to make this study possible and to the Waksman Genomics Core Facility and the Lewis-Sigler Institute Microarray Facility for Illumina sequencing. The authors also thank P. Andolfatto, M. Rockman, H. Seidel, R. Tanny, and members of the Kruglyak laboratory for helpful discussions and comments on the manuscript. This work was supported by a NIH Ruth L. Kirschstein National Research Service Award (E.C.A.), Merck Fellowship of the Life Science Research Foundation (J.P.G.), National Science Foundation graduate research fellowship (J.S.B.), James S. McDonnell Foundation Centennial Fellowship, Howard Hughes Medical Institute, NIH grant R01-HG004321, NIH R37-MH59520 (L.K.), and NIH grant P50-GM071508 to the Center for Quantitative Biology at the Lewis-Sigler Institute of Princeton University.
Footnotes
Accession number
All reads are archived in the NCBI sequence read archive under the accession number 047839. SNP calls for all strains (including singleton SNPs) have been uploaded to dbSNP and available under the submitter handle “KRUGLYAK”.
URLs
More descriptive methods and additional supplementary information are available on the Lewis-Sigler Institute website (genomics-pubs.princeton.edu/Diversity/). We used RepeatMasker from Smit, A.F.A., Hubley, R., and Green, P. www.repeatmasker.org.
Author Contributions
E.C.A. and J.P.G. conceived and carried out the experiments and data analysis. J.A.S. conceived and carried out data processing, simulations, and analysis. J.R.C. assisted DNA preparations. R.G. performed abamectin sensitivity assays. J.S.B. analyzed and plotted linkage disequilibrium data. M.A.F. provided unpublished wild strains. L.K. supervised all aspects of the study. The manuscript was written by E.C.A., J.P.G., J.A.S., and L.K.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Felix MA, Braendle C. The natural history of Caenorhabditis elegans. Curr Biol. 2010;20:R965–9. doi: 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 2.Riddle DL, Blumenthal T, Meyer BJ, Priess JR. C Elegans II. xvii. Cold Spring Harbor Laboratory Press; Plainview, N.Y: 1997. p. 1222. [PubMed] [Google Scholar]
- 3.Barriere A, Felix MA. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr Biol. 2005;15:1176–84. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 4.Barriere A, Felix MA. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics. 2007;176:999–1011. doi: 10.1534/genetics.106.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutter AD, Payseur BA. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol Biol Evol. 2003;20:665–73. doi: 10.1093/molbev/msg072. [DOI] [PubMed] [Google Scholar]
- 6.Cutter AD. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics. 2006;172:171–84. doi: 10.1534/genetics.105.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutter AD, Baird SE, Charlesworth D. High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics. 2006;174:901–13. doi: 10.1534/genetics.106.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutter AD, Felix MA, Barriere A, Charlesworth D. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics. 2006;173:2021–31. doi: 10.1534/genetics.106.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denver DR, Morris K, Thomas WK. Phylogenetics in Caenorhabditis elegans: an analysis of divergence and outcrossing. Mol Biol Evol. 2003;20:393–400. doi: 10.1093/molbev/msg044. [DOI] [PubMed] [Google Scholar]
- 10.Dolgin ES, Felix MA, Cutter AD. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity. 2008;100:304–15. doi: 10.1038/sj.hdy.6801079. [DOI] [PubMed] [Google Scholar]
- 11.Graustein A, Gaspar JM, Walters JR, Palopoli MF. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics. 2002;161:99–107. doi: 10.1093/genetics/161.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haber M, et al. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol Biol Evol. 2005;22:160–73. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- 13.Sivasundar A, Hey J. Population genetics of Caenorhabditis elegans: the paradox of low polymorphism in a widespread species. Genetics. 2003;163:147–57. doi: 10.1093/genetics/163.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivasundar A, Hey J. Sampling from natural populations with RNAi reveals high outcrossing and population structure in Caenorhabditis elegans. Curr Biol. 2005;15:1598–602. doi: 10.1016/j.cub.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 15.Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330:372–6. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swan KA, et al. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 2002;12:1100–5. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird NA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGrath PT, et al. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–9. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–17. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cully DF, et al. Cloning of an avermectin-sensitive glutamate-gated chloride channel from Caenorhabditis elegans. Nature. 1994;371:707–11. doi: 10.1038/371707a0. [DOI] [PubMed] [Google Scholar]
- 24.Koch R, van Luenen HG, van der Horst M, Thijssen KL, Plasterk RH. Single nucleotide polymorphisms in wild isolates of Caenorhabditis elegans. Genome Res. 2000;10:1690–6. doi: 10.1101/gr.gr-1471r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–8. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 26.Gusev A, et al. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–26. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–7. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 28.Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 2008;25:778–86. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- 29.Phillips PC. One perfect worm. Trends Genet. 2006;22:405–7. doi: 10.1016/j.tig.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Khan A, Taylor S, Ajioka JW, Rosenthal BM, Sibley LD. Selection at a single locus leads to widespread expansion of Toxoplasma gondii lineages that are virulent in mice. PLoS Genet. 2009;5:e1000404. doi: 10.1371/journal.pgen.1000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiontke K, et al. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci U S A. 2004;101:9003–8. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts A, et al. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics. 2007;23:i401–7. doi: 10.1093/bioinformatics/btm220. [DOI] [PubMed] [Google Scholar]
- 36.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patterson N, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–23. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–4. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:715–20. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Achaz G. Frequency spectrum neutrality tests: one for all and all for one. Genetics. 2009;183:249–58. doi: 10.1534/genetics.109.104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Achaz G. Testing for neutrality in samples with sequencing errors. Genetics. 2008;179:1409–24. doi: 10.1534/genetics.107.082198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics. 2010;26:2064–5. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–82. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.