Background: Luminescent conjugated polymers (LCPs) are highly specific to amyloid conformations and may represent potential antiprion compounds.
Results: LCPs significantly reduce prion titers, while rendering PrPSc more stable to proteolytic digestion.
Conclusion: LCPs possess pronounced antiprion potential and appear to act by compaction of frangible fibers.
Significance: LCPs may represent novel pharmacophores for the prevention and treatment of prion diseases.
Keywords: Drug Action, Drug Screening, Neurodegenerative Diseases, Pharmacology, Prions, Protein Aggregation, Protein Misfolding, Anti-prion Activity
Abstract
Luminescent conjugated polymers (LCPs) interact with ordered protein aggregates and sensitively detect amyloids of many different proteins, suggesting that they may possess antiprion properties. Here, we show that a variety of anionic, cationic, and zwitterionic LCPs reduced the infectivity of prion-containing brain homogenates and of prion-infected cerebellar organotypic cultured slices and decreased the amount of scrapie isoform of PrPC (PrPSc) oligomers that could be captured in an avidity assay. Paradoxically, treatment enhanced the resistance of PrPSc to proteolysis, triggered the compaction, and enhanced the resistance to proteolysis of recombinant mouse PrP(23–231) fibers. These results suggest that LCPs act as antiprion agents by transitioning PrP aggregates into structures with reduced frangibility. Moreover, ELISA on cerebellar organotypic cultured slices and in vitro conversion assays with mouse PrP(23–231) indicated that poly(thiophene-3-acetic acid) may additionally interfere with the generation of PrPSc by stabilizing the conformation of PrPC or of a transition intermediate. Therefore, LCPs represent a novel class of antiprion agents whose mode of action appears to rely on hyperstabilization, rather than destabilization, of PrPSc deposits.
Introduction
Prion diseases are neurodegenerative infectious disorders characterized by the deposition of β-sheet-rich aggregates. These invariably fatal diseases can be inherited (1), arise spontaneously (2), or develop upon transmission of prion infectivity (3–5). The infectious agent is termed prion and is mainly composed of PrPSc,4 a partially protease-resistant isoform of the physiologically occurring prion protein PrPC (6). PrPSc condenses into macromolecular aggregates that accumulate within the CNS and to a lesser extent in other organs (6, 7). The conversion of PrPC into PrPSc can be triggered by transmitting prions between organisms. Also, it is becoming increasingly evident that many amyloid diseases progress through the trans-cellular propagation of prion-like seeds termed “prionoids” (8, 9).
No efficient treatments against prion diseases are available (10). The central role played by PrPSc in prion diseases has inspired searches for compounds influencing PrPSc stability and/or clearance. Inhibition of PrPSc conversion in prion-infected cultured cells (11) or in animal models (12) was reported for molecules that can cross the blood-brain barrier such as amphotericin B (12), quinacrine, and chlorpromazine. However, clinical trials in humans revealed no significant improvements of the course of chronic disease (13–15).
Several polyanions can interfere with prion replication. Congo red (16) and the fluorescent anthracycline iododoxorubicin (17) prolong incubation time in rodents upon co-inoculation with prions. Congo red interacts with, and stabilizes, PrPSc (18), whereas iododoxorubicin binds to PrPSc deposits in brain sections of Creutzfeldt-Jakob Disease patients (17). It is therefore likely that the interaction of such compounds with infectious prion species is responsible for a decrease of prion infectivity. Therefore, compounds that stabilize prions could represent valuable tools for the treatment or the prophylaxis of prion diseases.
The structural requirements for the antiprion activity of sulfated polyanions in chronically infected cells were investigated in detail (19). These compounds are believed to interfere with the interaction of endogenous glycosaminoglycans with PrPC and/or PrPSc rather than acting directly onto PrPSc aggregates. The most promising compound, pentosan polysulfate (PPS) (20, 21), is currently undergoing clinical trials despite its poor pharmacokinetics and high toxicity (22). Finally, complex polyamines have been shown to disaggregate PrP but suffer poor bioavailability (23). Therefore, it would be desirable to identify new classes of compounds with drug-like properties (including high bioavailability and low toxicity) that directly influence amyloid propagation.
Luminescent conjugated polythiophenes (LCPs) are versatile probes with striking affinity to a variety of amyloids (24–27). Upon binding amyloids, LCPs adopt distinct conformations with characteristic spectral properties (28) specific to distinct amyloid types (27). Newer generation LCPs efficiently cross the blood-brain barrier, enabling visualization of protein aggregates in vivo and, potentially, clinical trials.
Here, we investigate the potential of various LCPs as novel antiprion compounds. All tested LCPs significantly reduced prion infectivity while increasing the protease resistance of PrPSc. We therefore posit that the polythiophene scaffold is a novel generic amyloid-stabilizing pharmacophore that could spawn useful antiprion compounds.
EXPERIMENTAL PROCEDURES
General Methods of Synthesis of PBAT
Organic extracts were dried over anhydrous magnesium sulfate, filtered, and concentrated in vacuo at 40 °C. All chemicals were purchased from Sigma and used as is. NMR spectra were recorded on a Varian 80 instrument (1H 300 MHz, 13C 75.4 MHz, Varian Inc., Santa Clara, CA). Chemical shifts were assigned with the solvent residual peak as a reference according to Gottlieb et al. (29). Thin layer chromatography (TLC) was carried out on precoated 60 F254 plates (Merck) 85 using UV light (l = 254 nm and 366 nm) and charring with ethanol/sulfuric acid/p-anisaldehyde/acetic acid 90:3:2:1 for visualization. Flash column chromatography was performed using Silica Gel 60 (0.040–0.063 mm, Merck). Gradient HPLC-MS was performed on a Gilson system (Middleton, WY). The column used was a Waters X-Bridge C-18 5 μm, 250 × 15 mm, and a Waters X-Bridge C-18 2.5 μm, 150 × 4.6 mm, for semipreparative and analytical runs, respectively, using acetonitrile with 0.05% formic acid and deionized water with 0.05% formic acid as mobile phase. MALDI-TOF-MS was recorded in a linear positive mode with the analyte as matrix. Normal work up means that the organic extracts were dried on MgSO4, filtered, and concentrated in vacuo.
Synthesis of 2-(3-Thienyl)-p-toluenesulfonyl Ethanol (Compound 2)
2-(Thiophene-3-yl)ethanol (0.700 g, 5.43 mmol) was added to a solution of CHCl3 and pyridine (1:1, 12.0 ml). p-Toluensulfonyl chloride (1.24 g, 6.50 mmol) was added. After 16 h, the reaction was quenched by adding water (20 ml) and ether (100 ml). The organic layer was washed with HCl (1 m aq, four times, 25 ml) and water (three times, 25 ml) and subjected to normal workup. The product was purified by flash column chromatography (toluene) to give compound 2 (1.47 g, 97%) as a pale yellow oil. The following were used: TLC (toluene) Rf = 0.09 1H NMR (CDCl3) δ: 2.44 (s, 3H), 2.99 (t, 2H, J = 6.9), 4.21 (t, 2H, J = 6.9), 6.87 (dd, 1H, J = 1.5, 5.1 Hz), 6.97 (m, 1H), 7.23 (dd, 1H, J = 3.0, 5.1 Hz), 7.31 (d, 2H, J = 8.4 Hz), 7.73 (d, 2H, J = 8.4 Hz); 13C NMR (CDCl3) δ: 21.6, 29.8, 69.9, 122.1, 125.8, 127.8, 128.0, 129.8, 133.3, 135.9, 144.9.
Synthesis of (S)-3-[2-(3-Thienyl)-ethoxy]-2-tert-butoxycarbonylaminopropionic Acid (Compound 3)
Compound 2 (1.23 g, 4.36 mmol) was dissolved in dry N,N-dimethylformamide (45 ml), N-butoxycarbonyl-l-Ser-OH (1.79 g, 8.72 mmol), and K2CO3 (1.81 g, 13.1 mmol) was added to the solution. The mixture was heated to 45 °C, and the reaction was quenched after 24 h by pouring the mixture over ice and HCl (2 m aq). The product was extracted with toluene (3 × 40 ml), which was subsequently washed with HCl (2 m aq, 2 × 25 ml) and NaCl (saturated aq, 3 × 50 ml), followed by normal workup. The syrup was purified by flash column chromatography (toluene → toluene/ethyl acetate, 4:1) and gave the colorless syrup product (compound 3) (0.879 g, 64%). For TLC (toluene/ethyl acetate, 4:1) Rf = 0.1, 1H NMR (CDCl3) δ: 1.43 (s, 9H), 2.86 (s, 1H), 2.98 (t, 2H, J = 6.9), 3.84 (dd, 1H, J = 3.5, 11.3 Hz), 3.85 (dd, 1H, J = 3.8, 11.3 Hz), 4.42 (m, 3H), 5.51 (d, 1H, J = 7.8 Hz), 6.95 (dd, 1H, J = 1.5, 4.6 Hz), 7.03 (dd, 1H, J = 1.5, 2.9 Hz), 7.25 (dd, 1H, J = 2.9, 4.6 Hz); 13C NMR (CDCl3) δ: 28.2, 29.4, 55.7, 63.3, 65.2, 80.2, 121.7, 125.7, 128.1, 137.5, 155.7, and 170.7.
Synthesis of (R)-1-Carboxy-N,N,N-trimethyl-2-(2-(thiophen-3-yl)ethoxy)ethanaminium (Compound 4)
For compound 3 (0.692 g, 2.10 mmol), NaHCO3 (0.706 g, 8.41 mmol) and methyl iodide (2.09 ml, 33.6 mmol) were dissolved in dry N,N-dimethylformamide (35 ml). After 3 days, the reaction was diluted with toluene (100 ml) and co-evaporated to give a yellow solid. The precipitate was dissolved in methanol (50 ml) and washed with heptane (2 × 30 ml). The mixture was concentrated, and the product was purified by HPLC (acetonitrile/H2O, 5:95 14 min; flow, 7 ml/min; wavelength, 210 nm; mass, 257.12 (calculated and found)) to provide compound 4 (0.565 g, 70%) as pale yellow crystals. For 1H NMR (methanol-d4) δ: 3.07 (t, 2H, J = 6.6 Hz), 3.31 (s, 9H), 4.16 (dd, 1H, J = 5.4, 13.5 Hz), 4.23 (dd, 1H, J = 3.3, 13.5 Hz), 4.39 (dd, 1H, J = 3.3, 5.4 Hz), 4.48 (dt, 1H, J = 6.6, 10.8 Hz), 4.56 (dt, 1H, J = 6.6, 10.8 Hz), 7.05 (dd, 1H, J = 1.5, 4.8 Hz), 7.20 (dd, 1H, J = 1.5, 2.7 Hz), 7.36 (dd, 1H, J = 2.7, 4.8 Hz); 13C NMR (methanol-d4) δ: 30.2, 54.3, 59.4, 67.6, 76.1, 123.1, 126.9, 129.2, 139.0, and 166.8.
Synthesis of Poly-((R)-1-carboxy-N,N,N-trimethyl-2-(2-(thiophen-3-yl)ethoxy)ethanaminium) (Compound 5)
For compound 4 (0.172 g, 0.666 mmol), FeCl3 (0.486 g, 3.00 mmol) and tetrabutylammonium-trifluoromethanesulfonate (0.521 g, 1.33 mmol) were added to a solution of dry chloroform (13.5 ml). After 2 h the mixture was quenched by adding water (3 ml). The organic layer was extracted by water (2 × 3 ml). The aqueous solution was made acidic by HCl (concentrated aq), and acetone was added to the mixture until the polymer precipitated as red flakes. The mixture was centrifuged, and the yellow water was decanted. The procedure was repeated until the yellow color, from dissolved FeCl3, of the water had disappeared. The polymer was dissolved in water and lyophilized, resulting in a red powder (compound 5) (0.0722 g, 37% calculated with respect to the monomeric unit).
Preparation of LCP Stock Solutions
The synthesis of the other different LCPs has been described elsewhere (24, 26, 30–37). Lyophilized LCPs were resuspended in pure water (B. Braun, Melsungen AG, Germany), and stock solutions at a concentration of 1 mg ml−1 were prepared and stored at 4 °C, protected from light. Serial dilutions of the LCPs were prepared in pure water.
Preparation of CD1 and RML6 Crude Brain Homogenates and Exposure to LCPs and PPS
20% w/v CD1 or RML6 brain homogenates in 0.32 m sucrose in PBS were prepared by three runs in a Precellys24 tissue homogenizer (Bertin) with cooling on ice between each run. Protein concentrations of CD1 or RML6 brain homogenates were determined using the bicinchoninic acid assay (Pierce) and normalized to 1 mg ml−1 total protein with 0.32 m sucrose in PBS.
LCPs and PPS were diluted into aliquots of crude brain homogenates at final concentrations ranging from 10−4 to 5000 μg ml−1 and to final volumes of 45 μl. All samples were incubated for 30 min at 37 °C on a thermoshaker rotating at 700 rpm prior to proteolysis.
Treatment of Slice Cultures with LCPs
Cerebellar slice cultures were prepared and prion-infected according to a protocol described by Falsig et al. (38, 39). Slice culture medium was changed three times per week, and 10 μl of diluted PTAA or PPS (30 μg ml−1, Bene Pharmachem) was added to 1 ml of medium to obtain final concentrations ranging from 0.01 to 60 μg ml−1 PTAA or 0.3 μg ml−1 PPS. Treatment was initiated 3 weeks post-infection or in a time course manner and maintained until the tissue was harvested. Tissue was harvested in PBS and homogenized according to a protocol described by Falsig et al. (38, 39). Protein concentration was determined using the bicinchoninic acid assay (Pierce) and normalized to 1 mg ml−1 total protein with PBS.
Western Blot Analysis
PrPSc was detected by limited proteolysis with PK (Roche Applied Science) and analyzed by Western blotting. Samples of 45 μl of brain homogenate or 20-μl aliquots from slice culture homogenates containing 20 μg of protein were digested with 50 μg ml−1 PK and 25 μg ml−1 PK, respectively, in lysis buffer containing 0.5% w/v sodium deoxycholate, 0.5% v/v Nonidet P-40, and 10% v/v PBS for 60 min at 37 °C and rotating at 700 rpm on a thermoshaker.
PK digestion was terminated by adding 17 μl of 4× lauryl dodecyl sulfate loading buffer (NuPAGE, Invitrogen) and boiling the samples at 95 °C for 5 min. 30 μl of the samples were separated on a 12% BisTris SDS-polyacrylamide gel (NuPAGE, Invitrogen) and blotted onto a nitrocellulose membrane. Membranes were blocked with 5% w/v Topblock (Fluka) in Tris-buffered saline supplemented with Tween (150 mm NaCl, 10 mm Tris-HCl, 0.05% Tween 20 (v/v)) and incubated with POM1 mouse IgG1 antibody to PrPC (anti-PrPC) (200 ng ml−1) as primary antibody. Horseradish peroxidase (HRP)-conjugated rabbit anti-mouse IgG1 (1:10,000, Zymed Laboratories Inc.) was used as a secondary antibody. The blots were developed using SuperSignal West Pico chemiluminescent substrate (Pierce) and detected in a LAS3000 system (FUJI).
Scrapie Cell End Point Assay
SCEPA was performed as described previously (40). Briefly, RML6 samples consisting of 1% brain homogenate containing various concentrations of LCPs were diluted 1–100-fold in cell culture medium to obtain a final dilution of 10−4. These samples were then serially diluted in cell culture medium containing a 10−4 dilution of CD1 brain homogenate and dilutions of RML6 ranging from 10−4 to 10−8 in 96-well plates. Each experimental run included four plates with the first plate containing a 10−4 to 10−8 decadal dilution of untreated RML6 as a control. The other three plates were used to analyze the titer of three dilutions of one LCP or of three distinct LCPs.
For slice culture homogenates, samples normalized to 1 mg ml−1 total protein were diluted 1–100-fold in cell culture medium to obtain a final dilution of 10−4. These samples were then serially diluted in cell culture medium containing a 10−4 dilution of CD1 brain homogenate and dilutions of homogenates from COCS ranging from 10−4 to 10−8 and tested in 96-well plates. Experimental runs included four plates including a homogenate of COCS from 6-week-old prion-infected nontreated tissue as a standard plate. The infection of N2a-PK1 cells and subsequent processing of the SCEPA were performed as described previously (40).
Misfolded Protein Assay
Samples of 10 μg of total protein (brain homogenate prepared as 1% w/v in 0.32 m sucrose in PBS and normalized to 1 mg ml−1) from LCP-treated RML6 or 5 μg of total protein from slice culture homogenates were diluted 100-fold in TBSTT (50 mm Tris-HCl (pH 7.5), 137 mm NaCl, 1% Tween 20, 1% Triton X-100) and incubated for 10 min at 37 °C under permanent agitation (850 rpm). 100 μl was then subjected to precipitation using magnetic beads coupled to the peptoid PSR1 (41) for 1 h at 37 °C under permanent agitation (750 rpm). Beads were washed, and trypsin digestion was performed (12.5 μg ml−1 trypsin in TBST containing 5 mm CaCl2) for 30 min at 37 °C under permanent agitation (750 rpm) and stopped with 2 mm PMSF for 15 min at RT. Beads were washed and denatured with 0.1 n NaOH. After neutralization (0.3 m Na2H2PO4), samples were placed on a magnet, and the supernatant was transferred to POM19 (42)-coated ELISA plates. After incubation (1 h at 37 °C, 300 rpm), plates were washed, and alkaline phosphatase-conjugated POM2 (42) was added. After incubation with substrate (100 μl of Lumiphos Plus substrate (Lumigen, Southfield, MI)), plates were read in a luminometry reader (Luminoskan Ascent; Thermo Fisher Scientific).
Bead-based Capture of PrP41
21 μl of PSR1-conjugated beads (41) were washed five times in 1 ml of PBS (8 mm Na2HPO4, 1.5 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl (pH 7.4)) before incubation with different dilutions of a 10% RML6 brain homogenate ranging from 10−2 to 10−10 overnight at 4 °C upon shaking at 1000 rpm.
Unbound material was removed from the beads by washing five times with 1 ml of PBS. The beads were resuspended in 60 or 120 μl in PBS, and 30 μl of the resuspended beads were intracerebrally inoculated into tga20 mice with groups of at least four mice.
Mice were monitored every 2nd day, and transmissible spongiform encephalopathy was diagnosed according to clinical criteria, including ataxia, wobbling, and hind leg paresis. At the onset of terminal disease, tga20 mice were sacrificed. Mice were maintained under conventional conditions, and all experiments were performed in accordance with the animal welfare guidelines of the Kanton of Zürich.
Histopathology and Immunohistochemical Stains
Two-μm-thick sections were cut onto positively charged silanized glass slides and stained with hematoxylin and eosin or immunostained using antibodies for PrP (SAF84) and for astrocytes (glial fibrillary acidic protein). For PrP staining, sections were deparaffinized and incubated for 6 min in 98% formic acid, then washed in distilled water for 5 min.
Sections were heat-treated and immunohistochemically stained on an automated NEXES immunohistochemistry staining apparatus (Ventana Medical Systems, Switzerland) using an IVIEW DAB Detection kit (Ventana). After incubation with protease 2 (Ventana) for 16 min, sections were incubated with anti-PrP SAF-84 (SPI bio; 1:200) for 32 min. Sections were counterstained with hematoxylin. Glial fibrillary acidic protein immunohistochemistry for astrocytes (rabbit anti-mouse glial fibrillary acidic protein polyclonal antibody 1:1000 for 24 min; DAKO) was similarly performed, however, with antigen retrieval by heating to 100 °C in EDTA buffer (pH 8.0).
For Western blot analysis, 10% brain homogenates were prepared in 0.32 m sucrose using a Precellys24 (Bertin). Extracts of 50–90 μg of protein were digested with 50 μg/ml PK in deoxycholate/Nonidet P-40 0.5% for 45 min at 37 °C. The reaction was stopped by adding 3 μl of complete protease inhibitor mixture and 8 μl of a lauryl dodecyl sulfate-based sample buffer.
MTS Assay
The 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2H-tetrazolium (MTS) assay (Promega) was performed according to the instructions given by the manufacturer.
Conformational Stability Assays
Two aliquots of RML6 brain homogenate were normalized to 2 mg ml−1 protein, and PTAA was added to obtain final concentrations of 0, 10, or 100 μg ml−1 before the samples were incubated for 30 min at 37 °C on a thermoshaker rotating at 700 rpm. Aliquots of 10 μl containing 20 μg of protein each were prepared and two times diluted with either water or different concentrations of GdnHCl. The samples were incubated for 10 min at room temperature prior to analysis by Western blot or MPA. For Western blot analysis, the samples were digested with PK (15 μg/ml), and 20 μl of the samples were separated on a 12% BisTris SDS-polyacrylamide gel (NuPAGE, Invitrogen) followed by blotting onto a nitrocellulose membrane. For MPA analysis, samples without or with 10 μg ml−1 PTAA were diluted 1:100 in TBSTT prior to the capture with peptoid-coated beads, and MPA was performed as described above.
ELISA
Homogenates from COCS were diluted 10-fold in TBSTT to obtain a final concentration of 0.1 mg ml−1 total protein. Digestion with PK was performed in a 96-well plate for 60 min at 37 °C and rotating at 700 rpm on a thermoshaker in a total volume of 50 μl containing 50 μg ml−1 PK. Digestion was stopped by adding 2 mm phenylmethanesulfonyl fluoride (PMSF, Calbiochem) and Complete Mini protease inhibitor mixture (Roche Applied Science). Digested samples were denatured with an equal volume of 3 m GdnSCN for 30 min at 37 °C and diluted with 4 volumes of 0.1 m NaHCO3 (pH 8.9). The samples were then detected by sandwich ELISA. Briefly, ELISA plates were coated with POM1 antibodies (400 ng/well) in coating buffer overnight. After washing with PBST, plates were blocked with blocking buffer (5% Topblock in PBST) for 2 h at room temperature. Next, samples were incubated for 2 h on ELISA plates (Nunc, Maxisorb). Captured samples were detected with biotinylated POM19 antibody (1 μg ml−1, 1 h, diluted in sample buffer) and horseradish peroxidase (HRP)-avidin conjugate (1 μg ml−1, 1 h, diluted in sample buffer) (Pharmingen). Samples were analyzed by ELISA in technical triplicates and washed five times with PBS, 0.05% Tween 20 between each antibody and conjugate incubations. The development was performed either calorimetrically or by chemiluminescence. For the calorimetric detection, 3,3′,5,5′-tetramethylbenzidine substrate (Invitrogen) was added to the wells and incubated for 15 min at room temperature and stopped by adding 0.5 m H2SO4. The absorbance was read at 450 nm by VersaMax microplate reader and validated with SOFTmax PRO software. For chemiluminescence, the chemiluminescent substrate (Thermo Scientific) was added to the wells and incubated for 3 min at room temperature. The relative light units were measured with a luminometer (∼425 nm) and validated with SOFTmax PRO software.
Preparation of Recombinant Mouse mPrP(23–231) and Fibril Formation
Recombinant mouse PrP comprising residues 23–231 was expressed and purified as described elsewhere (43–45). mPrP(23–231) fibers were produced by incubating the protein in 50 μm Tris-HCl, 1 m GdnHCl, 150 mm sodium chloride (pH 7.5), for 48 at 37 °C h and shaking at 600 rpm (46). To study the inhibition of fiber formation by PTAA, mPrP(23–231) fibers were grown in the absence and presence of different concentrations of PTAA ranging from 1 to 50 μg ml−1. The time course of fiber formation was followed by measuring the absorbance at 350 nm on a Tecan Saphire 2 plate reader. After 48 h, the samples were also analyzed by transmission electron microscopy. Preformed fibrils were mixed with PTAA at concentrations of 10, 25, 50, and 100 μg ml−1 PTAA for 30 min at 37 °C and shaking at 600 rpm prior to analysis by transmission electron microscopy.
Transmission Electron Microscopy
Carbon-coated copper grids were placed on 10 μl of the fibril solutions and incubated for 1 min. After removing excess liquid, grids were washed three times with water prior to staining with 2% aqueous uranyl acetate for 1 min. Samples were analyzed on a Philips CM12 electron microscope (transmission electron microscope with an acceleration voltage of 80 keV).
Proteolysis of mPrP(23–231) Fibers in the Presence of PTAA
mPrP(23–231) fibers were incubated in the absence and presence of 10 μg ml−1 PTAA at 37 °C for 60 min on a thermoshaker rotating at 700 rpm. 18 μl of nontreated and PTAA-treated fibers were digested with PK at concentrations of 0.5, 1, 1.5, and 2 μg ml−1 diluted in PBS for 60 min at 37 °C on a thermoshaker rotating at 700 rpm. PK digestion was terminated by adding 7 μl of 4× lauryl dodecyl sulfate loading buffer (NuPAGE, Invitrogen) and boiling the samples at 95 °C for 5 min. Samples were then analyzed by immunoblotting.
His-tagged mPrP(23–231) Purification and Fibril Formation for Fluorescence Assay
His-tagged mPrP(23–231) was purified by affinity chromatography and size exclusion chromatography as described previously (47). Fibrils were generated by unfolding the protein in 6 m GdnHCl followed by partial refolding by dilution to 1 m GdnHCl and 3 m urea and 5 μm mPrP(23–231) at pH 7.3. The protein samples were shaken vigorously at 37 °C for 24 h.
Native and fibrillated mPrP(23–231) was serially diluted 2-fold in several steps in a 96-well plate (Corning 3880), and PTAA was added to a final concentration of 0.2 μm. The samples were excited at 430 nm, and emission spectra were recorded from 470 and 600 using a Tecan Saphire 2 plate reader (Tecan). Fluorescence polarization was determined by excitation at 470 nm and measuring the emission at 520 nm. The anisotropy was calculated as r = (I‖ − I⊥)/(I‖ + 2I⊥), where r is the anisotropy; I‖ is the parallel intensity, and I⊥ is the perpendicular intensity.
Statistical Analysis
For MPA presented in Figs. 2 and 4 and supplemental Fig. S10, technical quadruplicates were compared for each individual sample using a t test. In Fig. 2A, the overall effect of individual LCPs was investigated by comparing a group of four concentrations of each individual polythiophene using a post hoc t test for analysis of variance.
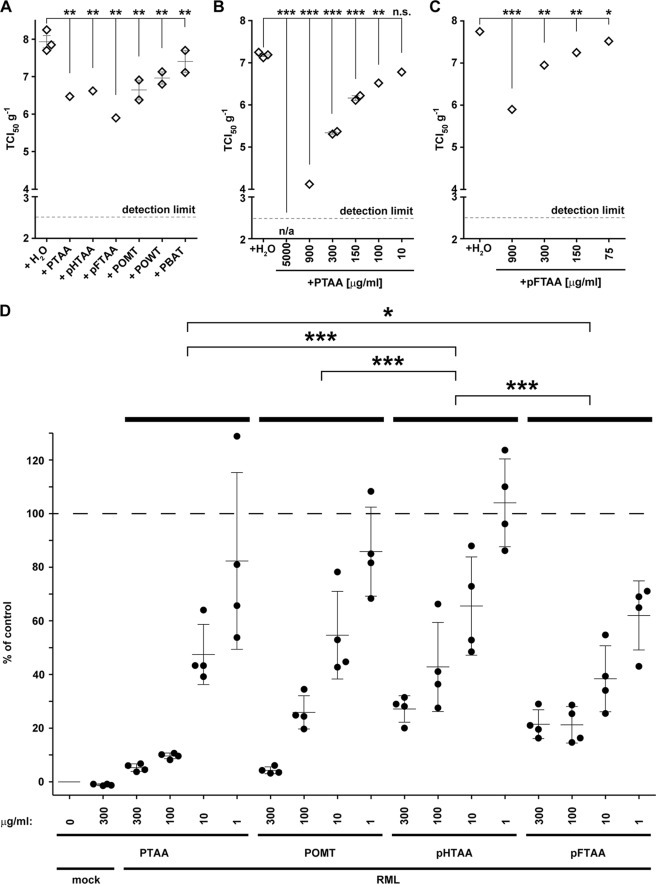
FIGURE 2.
LCPs reduce prion infectivity and the amount of prion aggregates in RML6 brain homogenates. A, SCEPA of RML6 homogenates mixed with the different LCPs at a concentration of 300 μg/ml. B and C, SCEPA of RML6 homogenates mixed with increasing concentrations of PTAA (B) and pFTAA (C). Noninfected (mock) and RML6-infected brain homogenates were incubated with either PTAA or pFTAA, and infectivity was determined on 17,000 cells in the SCEPA. Each data point represents the infectivity of one sample determined by serial 10-fold dilution steps from 10−4 to 10−8 on a 96-well plate. Data are shown as means ± S.E. p values represent statistical difference between polythiophene-treated and nontreated RML homogenates and were calculated with a Mantel-Haenszel χ2 test (supplemental Table S1). Detection limit, theoretical infectivity based on the observation of false positives at concentrations between 10−2 and 10−3 of noninfectious inoculum (mock). ***, p < 0.001; **, p < 0.01; *, p < 0.05. n/a, no positive wells were observed with this sample between concentrations ranging from 10−3 to 10−8. ns, nonsignificant, p > 0.05. D, MPA assay was used to quantify prion aggregates in RML6 brain homogenates treated with increasing concentrations of PTAA, POMT, pHTAA, or pFTAA. Each sample was analyzed in technical quadruplicates (circles) and is represented as the mean ± S.D. All data were corrected for the negative control (mock, CD1 brain homogenate) and represented as relative light units normalized for RML6 prions. Statistical differences were computed to compare LCP-treated RML6 to the control (nontreated RML6) (supplemental Table S1) or groups of four concentrations of each polythiophene (supplemental Table S6).
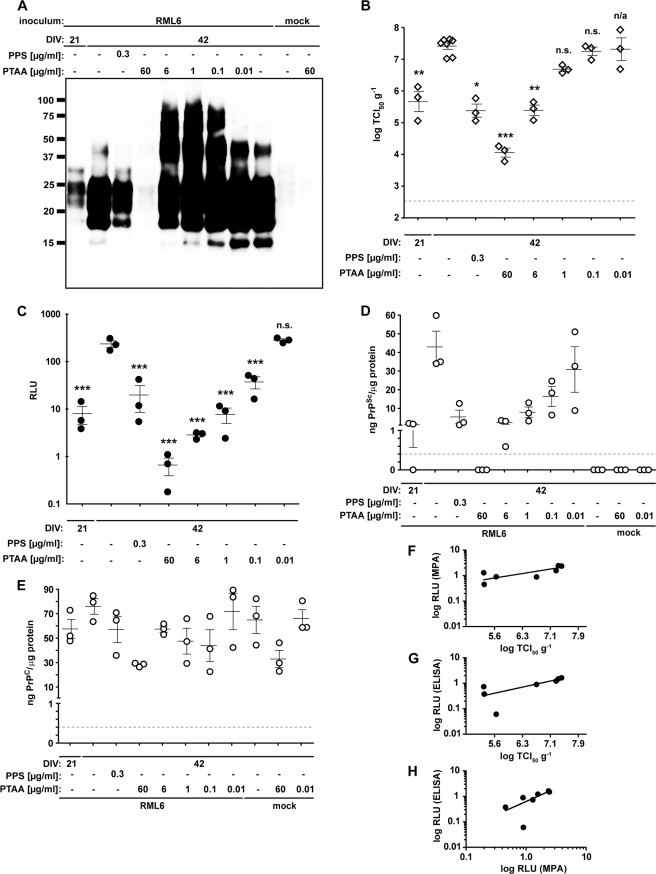
FIGURE 4.
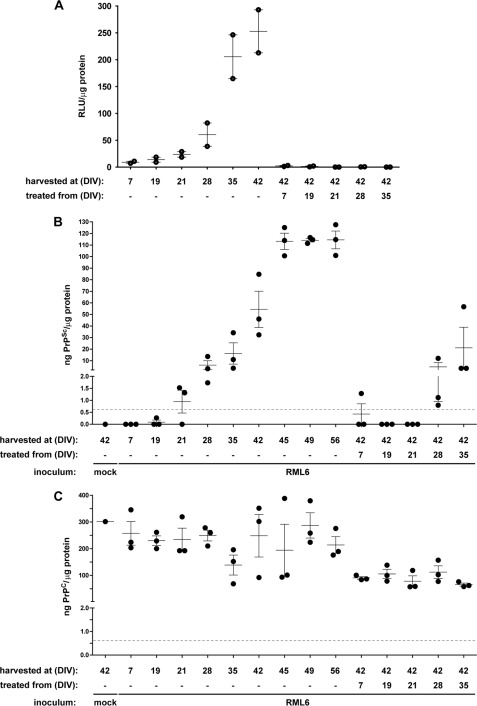
PTAA affects prion replication, accumulation, and neurodegeneration in slice cultures. A, immunoblot from prion-infected slice cultures obtained from tga20 mice treated with different concentrations of PTAA. Cultures were harvested after 21 or 42 DIV, and 20 μg of total protein from all samples was digested with PK. The anti-PrP antibody POM1 was used for detection. Molecular sizes are indicated in kDa. The experiment was performed in biological triplicates, and the immunoblot shown here is representative for the individual experiments. B, SCEPA of noninfected (mock) and RML6-infected slice cultures after exposure to PTAA. Infectivity of noninfected cultures harvested after 21 or 42 DIV are shown for comparison. PPS was used as a positive control. Each diamond represents the TCI50 g−1 of one slice of culture homogenate determined by serial 10-fold dilutions from 10−4 to 10−8 on a 96-well plate. Error bars indicate the mean ± S.E. of biological triplicates. p values represent the statistical differences between PTAA-treated and nontreated homogenates from RML6-infected slice cultures and are shown in supplemental Table S7. ***, p < 0.001; **, p < 0.01; *, p < 0.05. Detection limit (dashed line), theoretical titer based on the observation of false-positives at concentrations between 10−2 and 10−3 of noninfectious inoculum (CD1). n.s., nonsignificant. n/a, these results were not included into the statistical analysis. Because of mathematical complexity, a statistical analysis could not be performed for slice cultures treated with 0.01 μg ml−1 PTAA. C, MPA of the slice culture homogenates shown in A. Each dot represents the average value of technical triplicates for one slice culture homogenate. Error bars represent the mean ± S.E. of biological triplicates. All data were corrected for the negative control (mock) and represented as relative light units (RLU). p values represent the statistical differences between PTAA-treated and nontreated homogenates from RML6 slice cultures and are shown in supplemental Table S8. ***, p < 0.001. D and E, sandwich ELISA of the same homogenates as in A analyzed for PrPSc (D) and PrPC (E). Each dot represents the average value of technical duplicates for one slice culture homogenate. Error bars represent the mean ± S.E. of biological triplicates. All data were normalized to protein concentration of the corresponding sample. p values are shown in supplemental Table S8. The ELISA was developed with a colorimetric substrate. The detection limit of 400 pg ml−1 (dashed line) was determined as the average background levels plus three times S.D. F, correlation diagram to show the relationship between the MPA and SCEPA data presented in B and C. G, correlation diagram to show the relationship between the SCEPA and ELISA data presented in B and D. H, correlation diagram to show the relationship between the MPA and ELISA data presented in C and D.
For the SCEPA presented in Figs. 2 and 4 and supplemental Fig. S7, the proportion of negative wells over dilutions was compared between drugs using a Mantel-Haenszel χ2 test with a Bonferroni-Holm correction. Confidence interval for common odds ratio is given in the respective tables for nonsignificant comparisons.
For Fig. 4B, p values were calculated with a one-sample t test comparing the TCI50 difference to control (homogenates from COCS of nontreated cultures were harvested after 42 dpi). For Fig. 4C, statistical differences were computed to compare each set of biological replicas to the control homogenates from COCS with a one-way analysis of variance.
RESULTS
LCPs Stabilize PrPSc against Proteolysis
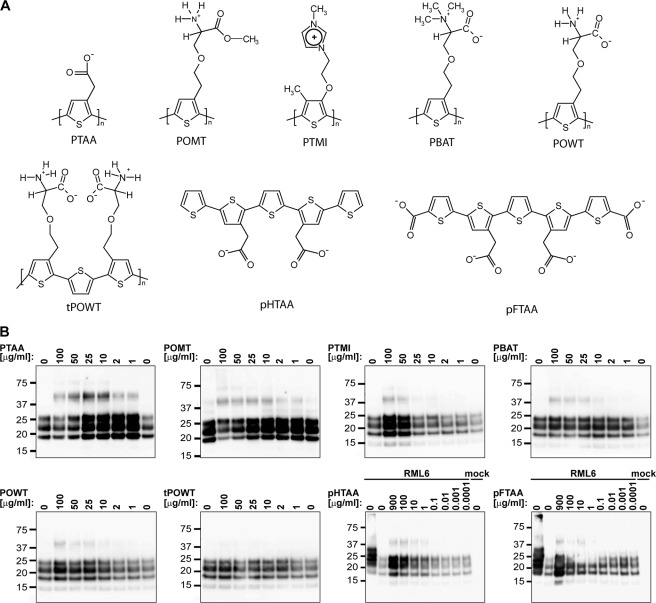
In a first set of experiments, we screened a broad range of LCPs with various side chains and/or polymer lengths for their effects onto the stability of PrPSc to hydrolysis by PK. The compounds tested included polydisperse LCPs such as the anionic PTAA (24, 26, 30, 31), the cationic POMT (26, 30, 31), and polythiophene methyl imidazole (poly(2-(4-methyl-3-thienyloxy)-N-ethane)-N-methylimidazole chloride) (32), the zwitterionic PBAT (supplemental Fig. S1), and POWT (31, 33, 34). We then tested monodisperse LCPs such as the anionic compounds pHTAA and pFTAA (Fig. 1A) (35, 36). All LCPs are charged at physiological pH.
FIGURE 1.
LCPs increase the resistance of PrPSc to PK digestion in RML6 brain homogenates and induce the formation of higher order aggregates. A, molecular structures of LCPs. pHTAA and pFTAA are homodisperse preparations of chemically defined molecules with molecular masses of 573 and 704.7 Da, respectively. All other compounds are polydisperse preparations with average molecular weights ranging from 3000 to 11,000 Da and number of monomers of n = 11–20. B, immunoblots of PK-digested RML6 samples after exposure with increasing doses of the different LCPs. On the 1st lane in the blots of the pHTAA and pFTAA titrations, 2 μg of total protein of undigested RML6 brain homogenate was loaded as a control. For all other lanes, 20 μg of total protein of PK-digested noninfected (mock) or RML6-infected brain homogenate was loaded. The experiment was repeated four times with PTAA and twice with the other LCPs. The immunoblot shown here is representative for the individual experiments. The anti-PrP antibody POM1 was used for detection. Molecular sizes are indicated in kDa.
We mixed a 1% brain homogenate obtained from mice infected with RML6 prions (Rocky Mountain Laboratory strain, passage 6) with each LCP (1–900 μg ml−1). Samples were incubated for 30 min at 37 °C in an orbital shaker (700 rpm). Samples were then digested with 50 μg ml−1 PK and analyzed by immunoblotting. Treatment with each LCP (except tPOWT) was found to increase the PK resistance of PrPSc in a concentration-dependent manner and to induce the formation of SDS-stable aggregates (Fig. 1B). PBAT and POWT were less effective at enhancing the PK resistance of PrPSc but augmented the formation of oligomeric SDS-stable PrPSc aggregates. Longer exposures of the blots revealed that these aggregates were enhanced by all LCPs (Fig. 1B and supplemental Fig. S2). We also used PPS as control. PPS did not show any effect on PK resistance up to a concentration of 10 μg ml−1, whereas the LCPs had a strong effect at the same concentration (supplemental Fig. S3).
To investigate the limit of the LCP-induced enhancement of PK resistance, we performed a digestion with a 40-fold higher concentration of PK on PTAA-treated RML6 and doubled the incubation time of proteolysis. The signal for untreated PrPSc was completely abolished, yet PTAA-treated PrPSc remained detectable even under these harsh conditions (supplemental Fig. S4). Exposure of RML6 to extremely high PTAA and POMT concentrations (>50 μg ml−1) reduced the levels of PrPSc (Figs. 1B and 4A). Hence, PTAA and POMT in extremely high concentrations may partially destabilize the protein aggregates.
We performed several experiments to assess whether the presence of the LCPs compromised the assay used for assessing PK resistance, thereby leading to artifactual results. To investigate whether LCPs interfere with the enzymatic activity of PK, we PK-digested noninfectious CD1 brain homogenates in the absence or presence of PTAA. We did not observe any effect of PTAA onto the efficiency of PrPC proteolysis (supplemental Fig. S5, A and B). Next, we tested whether the addition of PTAA to PK-predigested RML6 homogenate affects the level of PrPSc. The PrPSc signal increased only when proteolysis of RML6 prions was performed after exposure with PTAA, but not when PTAA was added a posteriori (supplemental Fig. S6). We conclude that LCPs indeed increase the resistance of PrPSc to PK by directly modifying the properties of PrPSc.
LCPs Reduce Prion Infectivity in Brain Homogenates
We then asked whether LCPs can reduce prion infectivity titers. We mixed 1% brain homogenate obtained from mice infected with RML6 prions with LCPs (300 μg ml−1) for 30 min at 37 °C. Prion infectivity was then quantified by assessing TCI50 g−1 (number of units transmitting infectivity to tissue cultures with 50% likelihood/g of brain homogenate) with the SCEPA (40). All LCPs significantly reduced infectivity in RML6 homogenates against nontreated RML6 homogenate (log TCI50 g−1 >7.9 ± 0.3; Fig. 2A and supplemental Table S1). Because of their high activity, PTAA and its monodisperse counterpart pFTAA were further investigated in a titration assay. PTAA reduced infectivity in RML6 brain homogenates by 3 logs TCI50 g−1 at 900 μg ml−1, and completely abated infectivity at 5000 μg/ml. The effect of pFTAA on prion infectivity appeared similar to that of PTAA (Fig. 2, B and C).
The validity of the SCEPA is unaffected by LCPs. Next, we asked whether residual LCPs may affect the susceptibility of N2a-PK1 cells to prions, thereby producing artifactually optimistic results. In a first experiment, a 10-fold excess of RML6 homogenate was added to RML6 samples treated, or nontreated, with 500 μg ml−1 PTAA (supplemental Fig. S7). The resulting prion titers were similar to those of nontreated RML6 homogenates, whereas the PTAA-treated control sample exhibited decreased titers (supplemental Tables S2). Hence PTAA did not decrease the susceptibility of N2a-PK1 cells to prion infection or the readout of the SCEPA. In a second experiment, we tested whether preincubation of N2a-PK1 cells with PTAA would affect the susceptibility of cells to prion infection (supplemental Fig. S7). We supplemented the culture medium before infection with 10 μg ml−1 PTAA, which corresponds to the PTAA concentration in the medium used for the RML6 infection treated with 900 μg ml−1 (Fig. 2B). Prion titration by SCEPA yielded comparable TCI50 g−1 values (supplemental Tables S3), confirming that the susceptibility of the cells to prion infection was not affected by the treatment with PTAA. We conclude that the PTAA-treated cells remained fully susceptible to RML6. Hence the observed decrease in infectivity in PTAA-treated sample represented a true PTAA-induced reduction of prion infectivity.
Finally, cell viability was tested to exclude the possibility of acute toxicity of PTAA to N2a-PK1 cells when supplemented into the culture medium (supplemental Fig. S8). PTAA at the highest dose of 45 μg ml−1 used in our study did not affect cell viability. Therefore, LCP can suppress prion infectivity of brain homogenates below the SCEPA detection limit without affecting the sensitivity of the assay.
LCPs Decrease the Concentration of PrP Aggregates
Because PK resistance is often used as a surrogate marker of prion infectivity, the finding that LCPs reduce infectivity while rendering PrPSc more resistant to PK digestion was counterintuitive. We therefore used MPA to further analyze the physical properties of LCP-treated prions. The MPA enables sensitive and quantitative detection of prion aggregates without the use of PK (41, 48, 49). Paramagnetic beads coated with prion-specific binding peptoids selectively capture PrP aggregates, whereas monomeric PrP is not captured. Captured PrP is then eluted and detected by enzyme-linked immunosorbent assay (ELISA) using the PrP-specific antibody POM19 (42).
To evaluate the sensitivity of the MPA, beads were incubated with 10-fold serial dilutions (10−3 to 10−10) of a 10% (w/v) RML6 brain homogenate and intracerebrally inoculated into tga20 mice overexpressing PrPC (supplemental Table S4) (50). Mice developed clinical signs after mean incubation times of 78–106 days (supplemental Table S4). Spongiform encephalopathy was confirmed in all clinically diseased mice by histopathological and immunohistochemical analysis (supplemental Fig. S9). Besides confirming a linear relationship between infectivity and MPA readings, these results show that peptoid-coated beads capture prion infectivity efficiently from RML6 and transmit it to tga20 mice. These data suggest that the MPA readings reflect the number of prion seeds present in a sample.
We then mixed 1% RML6 brain homogenate with PTAA, POMT, pHTAA, or pFTAA. These LCPs were chosen because they showed the strongest effect on infectivity in the SCEPA. All four compounds affected the aggregational state of PrPSc dose-dependently (Fig. 2D). Treatment of RML6 with 10 μg ml−1 PTAA, POMT, and pFTAA significantly reduced the MPA signal (p = 0.004, 0.015, and 0.003, respectively, and supplemental Table S5). PTAA and POMT had the strongest effect at 300 and 100 μg ml−1, respectively, whereas pFTAA was more efficient at 1 and 10 μg ml−1. Already the treatment with 1 μg ml−1 pFTAA caused a significant reduction in the MPA signal (p = 0.028; supplemental Table S5). We then preincubated beads with 100 μg ml−1 PTAA. Pretreatment of the beads had no effect on the capture efficiency of nontreated or PTAA-treated RML6 prions (supplemental Fig. S10 and Table S6), indicating that LCPs do not affect the assay by interacting nonspecifically with peptoid-coated paramagnetic beads.
PTAA Alters the Conformational Stability of PrPSc
We next investigated whether PTAA changes the biochemical properties of RML6 prions by altering the conformational stability of PrPSc. Nontreated and PTAA-treated RML6 brain homogenates were incubated with increasing concentrations of GdnHCl for 60 min, digested with PK to remove soluble PrP, and analyzed by immunoblotting for PrPSc (51). PTAA treatment strongly increased the sensitivity of PrPSc to denaturation with GdnHCl, as reflected by a shift in the concentration of GdnHCl required for loss of PK resistance (3 m for the nontreated sample and 2.4 and 2 m for samples exposed to 10 and 100 μg ml−1 PTAA, respectively; Fig. 3A).
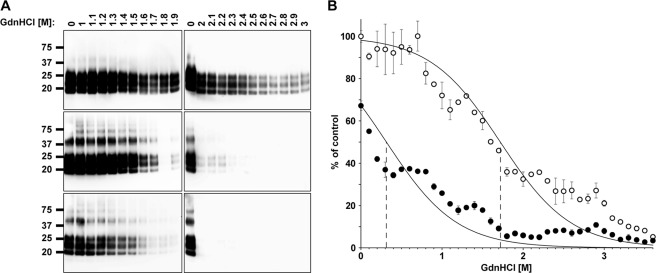
FIGURE 3.
PTAA-treated RML6 prions are more sensitive to denaturation with GdnHCl. A, immunoblots of nontreated (upper panels) and PTAA-treated RML6 brain homogenates at different concentrations of GdnHCl. RML6 brain homogenate was treated with either 10 μg ml−1 (middle panel) or 100 μg ml−1 PTAA (lower panel). Aliquots of 20 μg of protein were denatured with various concentrations of GdnHCl, subsequently digested with PK to remove soluble PrP, and loaded to the gel. The anti-PrP antibody POM1 was used for the detection. Molecular sizes are indicated in kDa. B, MPA of nontreated (open circles) and PTAA-treated RML6 brain homogenate (filled circles) after denaturation with different concentrations of GdnHCl. PTAA was used at a concentration of 10 μg ml−1. Each sample was analyzed in technical duplicates and is represented as the mean ± S.D. All data were corrected for the negative control (CD1 brain homogenate) and represented as relative light units normalized for RML6 prions. Asymptotic curves were fitted to each dataset with the [GdnHCl]½ value at which 50% of the molecules are in a soluble fraction and 50% are in a insoluble fraction indicated by a dotted line at 1.72 m for the nontreated and 0.31 m for the PTAA-treated samples.
We also incubated native and PTAA-treated RML6 with various concentrations of GdnHCl and quantified the amount of aggregated PrP by MPA (Fig. 3B). The inflection point of the transition curve, where 50% of aggregated PrP is captured by the peptoids, was 1.72 m GdnHCl for nontreated RML6 and 0.31 m GdnHCl for PTAA-treated RML6. Hence PTAA-treated RML6 brain homogenate was more sensitive to GdnHCl denaturation than nontreated homogenate, indicating that PTAA physically changes the stability of PrPSc against chaotropic salts.
PTAA Antagonizes Prion Infection in Cerebellar Organotypic Cultured Slices
We then evaluated the antiprion activity of LCPs ex vivo in a complex cellular environment. For that, we used the prion organotypic slice culture assay (38, 39). We again used PTAA as the lead compound to determine the impact of LCPs onto prion replication and accumulation in the prion organotypic slice culture assay. COCS were prepared from 11-day-old PrPC-overexpressing tga20 pups and kept in culture for 42 days (38). Cultures were analyzed for the presence of PrPSc at 21 days post-inoculation (dpi) by assessing PK-resistant materials on immunoblots (Fig. 4A). PTAA was added to the culture medium at 21 dpi at each medium exchange (10 ng to 60 μg ml−1). Slices were harvested at 42 dpi, and the impact of PTAA was analyzed with the tools described above.
We first compared the amounts of PrPSc between nontreated and PTAA-treated slices by PK digestion and immunoblotting (Fig. 4A). As in PTAA-treated RML6 brain homogenates, we observed a substantial dose-dependent increase in PrPSc (Fig. 4A) and high molecular weight PrP aggregates. Treatment with the highest PTAA concentration (60 μg ml−1) again lowered the amount of PrPSc below detectability. PPS, which effectively inhibited PrPSc replication in prion-infected COCS (38), was used as control. In contrast to slices exposed to PTAA, PPS (0.3 μg ml−1) strongly reduced PrPSc when compared with nontreated controls. Untreated or PTAA-treated slices exposed to noninfected brain homogenates from CD1 mice, did not contain any PrPSc.
We next investigated the effect of PTAA on prion replication in COCS (38). During the interval between 21 and 42 dpi, prions were amplified in RML6-infected slices, as evidenced by an increase in infectivity from 5.7 to 7.4 log TCI50 g−1 (1 TCI50 unit being the dose required to infect 50% of PK1 culture wells; Fig. 4B). When slices were exposed to PTAA, a dose-dependent reduction in infectivity was observed. Infectivity was reduced by >3 log TCI50 g−1 with the highest PTAA concentration of 60 μg ml−1, which is >1.5 log TCI50 g−1 below the initial infectivity at day 21 dpi (p = 0.012, p = 0.001 and p < 0.001; respectively, supplemental Table S7). Treatment with PPS at a concentration of 0.3 μg ml−1 was as efficient as treatment with 6 μg ml−1 PTAA and resulted in a reduced infectivity of >2 log TCI50 g−1 when compared with nontreated cultures at 42 dpi. A comparison of MPA readings (Fig. 4C) and SCEPA results (Fig. 4B) shows that the quantification of PrP aggregates in the various samples exactly reflected the data obtained from the SCEPA, again confirming a close relationship between the number of PrP aggregates and prion infectivity (Fig. 4F).
To further validate the potency of PTAA on RML6-infected slice cultures, we developed a sandwich ELISA using the antibodies POM1 for coating and POM19 for detection. This allowed us to simultaneously quantify the levels of total PrP (PrPC + PrPSc) and PrPSc in slice homogenates (Fig. 4D). Homogenates from nontreated slices harvested at days 21 and 42 dpi contained 1.2 ± 1.0 and 42.9 ± 14.6 ng PrPSc/g of total protein. The levels of PrPSc in PTAA-treated slices significantly declined with increasing concentrations of PTAA and strongly correlated with the SCEPA and MPA data (Fig. 4, G and H). A comparison of the ELISA data with the immunoblot indicated that the high molecular weight aggregates may not be detected in the ELISA, probably due to their resistance to denaturation with GdnSCN that prevents the exposure of buried epitopes.
Treatment with PTAA (60 μg ml−1) suppressed PrPSc to values below detectability by ELISA (supplemental Table S8). Homogenates from cultures treated with 0.3 μg ml−1 PPS showed levels of PrPSc similar to the homogenates at day 21 dpi and to COCS treated with PTAA at 6 μg ml−1. No PrPSc was detectable in uninfected controls. To test whether PTAA acts directly on PrPC, we quantified the PrPC levels in the various samples by subtracting the amount of PrPSc from total PrP (Fig. 4E). The amount of PrPC was unaffected in all samples except for slices treated with 60 μg ml−1 PTAA, which showed a significant reduction in PrPC levels.
PTAA Treatment Efficiently Inhibits Prion Replication
To evaluate the efficiency of PTAA as a post-exposure regimen in slice cultures, we first generated a time-dependent standard curve for prion accumulation. Prion-infected slice cultures were harvested at days 0, 7, 19, 21, 28, 35, and 42 dpi, and prion aggregates were analyzed by MPA and by sandwich ELISA after PK digestion. Prion aggregates and PrPSc were hardly detectable until 21 dpi (Fig. 5, A and B), and newly formed PrPSc accumulated gradually thereafter and reached a plateau after 45 days. Prion accumulation was completely suppressed by 10 μg ml−1 PTAA if treatment was initiated before day 21 dpi. If treatment was started at 28 dpi, a conspicuous inhibitory effect of PTAA was observed with reduction of PrP aggregates below the detection limit of the MPA and a strong decline in PrPSc when compared with nontreated prion-infected homogenates.
FIGURE 5.
PTAA inhibits prion accumulation in RML6-infected slice cultures from different time points. A, MPA of homogenates from RML6-infected slices harvested at days 7, 19, 21, 28, 35, and 42 DIV. PTAA was administered at a concentration of 10 μg ml−1 at the time of a fresh medium change from days 7, 19, 21, 28, or 35 DIV. Slice cultures were harvested at 42 DIV and analyzed for PrP aggregates. Each dot represents the average value of technical triplicates for one slice culture homogenate. Error bars represent the mean ± S.E. of biological duplicates. All data were corrected for noninfectious brain homogenates obtained from CD1 mice as negative control (mock) and represented as the ratio between relative light units (RLU) and protein concentration for each sample. p values are shown in supplemental Table S9. B, sandwich ELISA of the same homogenates as in A analyzed for PrPSc. The data are represented as biological triplicates. The signal for the negative control (mock) is shown as a single replica. C, sandwich ELISA of the same homogenates as in A analyzed for PrPC. The ELISA was developed by chemiluminescence. The detection limit of 600 pg ml−1 (dashed line) was determined as the mean background levels plus three times S.D.
A direct comparison of MPA and ELISA shows that the quantification of PrP aggregates with the prion-specific binding peptoids was less sensitive than the sandwich ELISA in this paradigm. Perhaps the proximal coating of PrP aggregates with PTAA interfered with the interaction between peptoids and binding sites on PrP aggregates. Alternatively, PTAA might induce a structural rearrangement of the aggregates into larger associations that reduces the surface available for binding to the peptoid-coated beads.
We also evaluated the levels of PrPC in homogenates (Fig. 5C). PrPC levels were reduced about 2-fold compared with nontreated samples independent from the time point at which treatment was started. This confirms further that PTAA treatment might affect prion accumulation by reducing the amount of PrPC or of a transition intermediate on-path toward PrPSc.
PTAA Inhibits Fibrillogenesis of Recombinant PrP and Affects Pre-formed Recombinant PrP Fibers
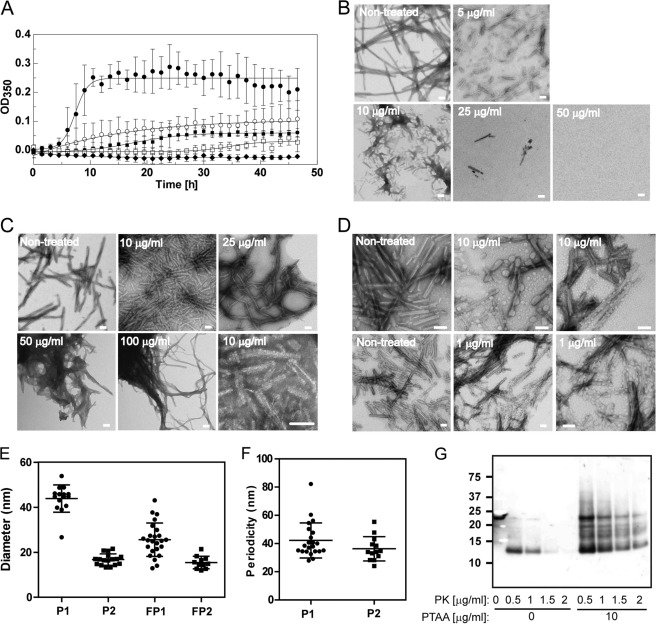
We used an in vitro conversion assay to address the mode of action of the LCPs on prion replication and accumulation in a well defined simple environment. We first investigated whether PTAA inhibits amyloid fiber formation of the recombinant mouse prion protein mPrP(23–231). Monomeric mPrP(23–231) was grown under slightly denaturating conditions (46) in the presence of PTAA, and the time course of fiber formation was assessed by absorbance measurements at 350 nm for 48 h. mPrP(23–231) formed amyloid-like fibers by nucleation-dependent polymerization with a lag phase followed by an exponential growth phase and a plateau (Fig. 6A). The addition of PTAA significantly reduced the growth rate of fibers in a concentration-dependent manner (Fig. 6A); incubation with 50 μg ml−1 resulted in a complete inhibition of fiber formation.
FIGURE 6.
Effect of PTAA on mPrP(23–231) fiber formation and preformed fibers. A, time course of mPrP(23–231) fiber formation grown in the absence (filled circles) and presence of PTAA at different concentrations of 5 μg ml−1 (open circles), 10 μg ml−1 (filled squares), 25 μg ml−1 (open squares), and 50 μg ml−1 (filled diamonds) monitored by measuring the A350 over a period of 48 h. Error bars represent the S.D. of technical triplicate measurements. B, TEMs taken after 48 h of control fibers grown in the absence of PTAA and in the presence of different concentrations of PTAA. C, TEM of preformed mPrP(23–231) fibers incubated with different concentrations of PTAA. The electron micrograph in the lower right corner displays fibers incubated with 1 μg ml−1 PTAA at higher magnification. D, TEM of fibers recorded from another independent experiment grown in the presence of 10 μg ml−1 PTAA (upper panel) and of preformed mPrP(23–231) fibrils mixed with 1 μg ml−1 PTAA (lower panel). E, PTAA formed two different populations of small aggregates at concentrations between 1 and 10 μg ml−1 that were associated at the fibers with mean diameters of 43.9 ± 6.1 nm for population 1 (P1) and 16.8 ± 2.5 nm for population 2 (P2). Free aggregates had mean diameters of 25.6 ± 7.4 nm for population 1 (FP1) and 15.4 ± 2.7 nm for population 2 (FP2). Data are represented as the mean ± S.D. F, PTAA aggregates were located on the fibers in a highly periodic manner with distances of 42.2 ± 12.5 nm for population 1 and 36.3 ± 8.6 nm for population 2. Data are represented as the mean ± S.D. G, PTAA increases the resistance of mPrP(23–231) fibers to PK digestion. Aliquots of mPrP(23–231) fibers were incubated with 10 μg ml−1 PTAA prior to proteolysis with increasing concentrations of PK. The 1st lane contains nondigested mPrP(23–231) fibers. Scale bar, 100 nm.
We further analyzed the samples by negatively stained transmission electron microscopy (TEM) (Fig. 6B). Electron micrographs (EM) of nontreated control samples taken after 48 h showed long unbranched fibers, whereas fibers grown in the presence of PTAA (1–10 μg ml−1) appeared coated with globular and regularly spaced PTAA aggregates. Quantitative analysis revealed that PTAA forms two different populations of aggregates with average diameters of 43.9 ± 6.1 and 16.8 ± 2.5 nm when bound to the fibers compared with 25.6 ± 7.4 and 15.4 ± 2.7 nm for the free aggregates. Both populations bind to the fibers with a recognizable periodicity (Fig. 6, D–F), suggesting that PTAA interacts specifically with defined binding sites on the fibers. In addition, we partially observed coalescence of the fibers into more compact structures at these concentrations. Empty EM grids at high doses of PTAA confirmed the strong inhibitory effect of PTAA (Fig. 6B).
The presence of compact fiber conglomerates in the samples grown in the presence of PTAA at concentrations between 1 and 10 μg ml−1 prompted us to ask whether PTAA also interacts directly with existing mPrP(23–231) fibers. We therefore incubated preformed fibers with PTAA and analyzed their morphology by TEM. Similarly to the fibers grown in the presence of PTAA, fibers incubated with PTAA at concentrations between 1 and 10 μg ml−1 were associated with small aggregates of PTAA (Fig. 6, C and D). With increasing PTAA concentrations, we observed progressive coating and agglomeration of fibers (Fig. 6C).
We then asked whether PTAA directly interacts with native mPrP(23–231). Native mPrP(23–231) was serially diluted, and 0.2 μm PTAA was added to all samples. Fluorescence intensity and fluorescence polarization were measured (supplemental Fig. S11). PTAA was quenched by native mPrP(23–231), suggesting that the probe binds also native PrPC or a transition intermediate on-path toward aggregating.
To validate that the increased PK resistance of PrPSc in PTAA-treated RML6 brain homogenates and prion-infected slice cultures can be ascribed to a direct interaction of PTAA on PrPSc independently of the sample environment, we investigated whether PTAA also renders mPrP(23–231) fibers more stable to PK digestion. mPrP(23–231) fibers were incubated with 10 μg ml−1 PTAA prior to limited proteolysis with different PK concentrations and immunoblotting. Fig. 6G displays that PTAA directly acts on PrP as evidenced by a significant increase in PK resistance of recombinant mPrP(23–231) fibers. We conclude that PTAA prevents prion replication and accumulation by acting on both PrPC and PrPSc aggregates in a concentration-dependent manner.
DISCUSSION
We had previously found that LCPs interact with amyloids, including PrPSc. The fluorescent spectra emanating from PrPSc-complexed LCPs allowed for differentiation of prion strains, suggesting a remarkable degree of steric specificity (28). These observations prompted us to explore whether additional biophysical properties of prions, such as the stability of PrPSc aggregates and their infectiousness, may be affected by interactions with LCPs. The ultimate hope would be that these experiments might identify the polythiophene scaffold as a novel pharmacophore for the treatment of protein aggregation diseases.
Indeed, not only did LCP treatment of infectious brain homogenates significantly reduce their infectivity, but also prion-infected organotypic slice cultures accrued less infectivity when exposed to LCPs. Several polythiophenes with diverse charged side chains and backbone lengths were able to reduce infectivity when added to prion-containing brain homogenates. The antiprion activity of these compounds cannot be attributed to the charges of their side chains, because anionic, cationic, and zwitterionic compounds reduced prion infectivity to a similar extent. Also, neither the spacing between the side chains nor the rigidity of the molecules seemed to play a role for their antiprion activity. Based on theoretical considerations, the total avidity of LCPs for amyloids should increase with the number of low affinity binding sites and therefore with the length of the polymers. However, very long polymers may have chemical properties limiting their interactions. Hence, the activity of LCPs appears to be an intrinsic property of the polythiophene backbone itself.
The mode of action of LCPs may involve more than one mechanism. When used at low concentrations, the LCPs appear to render PrPSc more stable to PK digestion and to induce the formation of SDS-stable high molecular weight aggregates. Accordingly, PTAA transformed recombinant mPrP(23–231) fibers into more compact structures. At extremely high concentrations, PTAA and POMT, however, significantly decreased the resistance of PrPSc to PK. PTAA also caused a left shift in the conformational stability curve of PrPSc after exposure to increasing concentrations of the nonsurfactant-type chaotrope, GdnHCl. This concentration-dependent behavior of the LCPs resembles the mechanisms reported for several surfactants that were assumed to affect protein aggregation in a two-concentration regime by either interacting as single molecules or in equilibrium with small molecule aggregates (52) and were demonstrated to enhance or reduce the stability and detectability of PK-resistant PrPSc (18, 53, 54).
The concentration-dependent interaction of surfactants with aggregated proteins may induce conformational versatility. Low concentrations of SDS and lithium dodecyl sulfate were shown to act like negatively charged lipid interfaces favoring the formation of global hydrogen bonding, thereby facilitating the formation of β-sheet structure and e.g. promoting Aβ self-assembly (55–58), whereas micellar SDS inhibits aggregation by enhancing α-helical conformational states (59). A similar mode of action was reported for Congo red that induces β-sheet formation and self-propagation of monomeric Aβ (60, 61). LCPs might therefore act similarly to membrane mimetics that promote de novo PrP aggregation by converting PrP into compact aggregates that are more resistant to PK but less infectious.
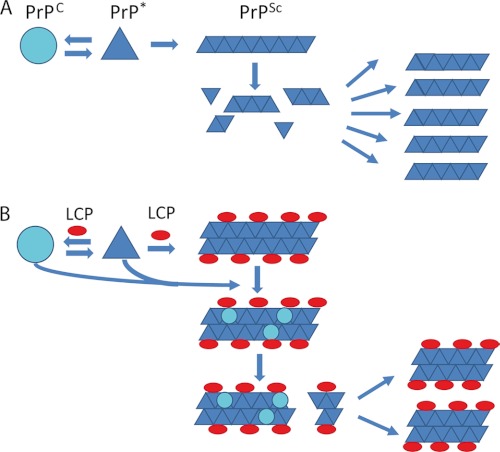
Accordingly, LCPs used at submicellar concentrations may prevent prion replication by hyperstabilization of the PrPSc conformation, resulting in reduced fiber frangibility, a critical determinant of prion infectivity (62, 63) that controls prion propagation by fragmentation of fibrils and elongation of these infectious seeds into new fibrils (64). The higher compactness of LCP-treated PrPSc aggregates may reduce the release of infectious particles, resulting in decreased prion infectivity. This scenario is supported by studies suggesting that large aggregates do not correlate well with high infectivity (65–67). Hence, LCPs can be protective by stabilizing fragile fibrils and perhaps even by incorporating pre-existing prions, and possibly also PrPC, into inert structures (Fig. 7).
FIGURE 7.
Model for the antiprion activity of the LCPs. A, in the prion model PrPC is in a reversible thermodynamic equilibrium with PrP*, which further aggregates into amyloid fibrils, PrPSc. When the fibrils reach a critical length, the fibril becomes more fragile, and fragmentation occurs. The newly formed ends of the fibril fragments are new nucleation sites for further fibril growth. B, antiprion activity of the LCPs seems to be based on interactions with PrPSc aggregates, possibly increasing their compactness. LCP-coated fibrils further embed pre-existing prions and even PrPC. The higher compactness of the PTAA-treated aggregates ultimately cause less fragmentation into infectious particles necessary for further prion replication.
The PK resistance of PrPSc is widely accepted as surrogate marker for prion infectivity and is often used for the screening of novel compounds for the treatment of prion diseases. However, we found a glaring dissociation between these two parameters. Along with previous observations (54, 68–74), these findings question the use of PK resistance as the sole screening parameter. Alternative methods, such as the MPA, appear to better reflect prion infectivity and may be better suited to screening for novel antiprion compounds.
Whereas the above data indicates that LCPs can interact with PrPSc, their antiprion effects on COCS suggest that LCPs may intervene at additional steps during the transition process of PrPC to PrPSc. In particular, ELISA measurements on COCS showed reduced levels of PrPC, suggesting that PTAA might deplete PrPC from the cell surface or modulate PrPC levels in prion trafficking. This may reduce the amount of PrPC accessible for the transition of PrPC into PrPSc. A similar mode of action was reported for other antiprion compounds (54, 75–81). This interpretation is supported by our finding of a direct interaction of PTAA with PrPC by performing an in vitro conversion assay with recombinant mPrP(23–231) in the presence of PTAA in a cell-free environment. PTAA strongly inhibits the formation of mPrP(23–231) fibers, supporting that PTAA directly interacted with PrPC by either stabilizing the spatial conformation of PrPC or protecting the binding site for the template PrPSc molecule. Both effects may prevent efficient transition of PrPC into PrPSc.
In conclusion, the interaction of the LCPs with PrPSc causes a structural transformation in a surfactant-like manner mostly independent of the molecular character of the LCPs. The treatment renders PrPSc more resistant to PK digestion without affecting proteolysis of PrPC and profoundly reduces prion infectivity. The data presented above link these effects to the hyperstabilization of PrPSc. Therefore, LCPs may represent a promising new class of antiprion compounds. The newly developed LCPs, pHTAA and pFTAA, were shown to cross the blood-brain barrier (35) and may represent promising candidates for further exploration and for in vivo studies.
Supplementary Material
Acknowledgments
We thank Dr. B. Seifert from the Institute of Biostatistics, University of Zurich, for performing the statistical analysis; Dr. Jens Sobek from the Functional Genomics Center Zurich for support with the Tecan Saphire 2 plate reader, and Rita Moos and Ahmet Varol for technical assistance.
This work was supported in part by grants from the European Union P-7 Health (project acronym LUPAS), the Swiss National Foundation, and the Novartis Research Foundation.

This article contains supplemental Figs. S1–S11 and Tables S1–S9.
- PrPSc
- scrapie isoform of PrPC
- PrP
- prion protein
- PrPC
- cellular isoform of PrP
- mPrP
- mouse PrP
- LCP
- luminescent conjugated polymer
- COCS
- cerebellar organotypic cultured slices
- RML6
- Rocky Mountain Laboratory strain, passage 6
- SCEPA
- scrapie cell end point assay
- MPA
- misfolded protein assay
- PTAA
- poly(thiophene-3-acetic acid)
- pFTAA
- pentamer formyl thiophene acetic acid (4′,3‴-biscarboxymethyl-[2,2′;5′,2″;5″,2‴;5‴,2‴-]quinquethiophene-5,5‴-dicarboxylic acid)
- pHTAA
- pentamer hydrogen thiophene acetic acid (3‴-(carboxymethyl-[2,2′;5′,2″;5″,2‴;5‴,2‴-]quinquethiophen-4′-yl)-acetic acid)
- POMT
- (poly(3-[5-amino-5-methoxycarboxyl-3-oxapentyl]-2,5-thiophenylene hydrochloride)
- POWT
- (poly(3-((S)-5-amino-5-carboxyl-3-oxapentyl)-2,5-thiophenylene) hydrochloride)
- PBAT
- (poly-((R)-1-carboxy-N,N,N-trimethyl-2-(2-(thiophen-3-yl)ethoxy)ethanaminium))
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GdnHCl
- guanidine HCl
- TEM
- transmission electron microscopy
- PPS
- pentosan polysulfate
- aq
- aqueous
- PK
- proteinase K
- DIV
- days in vitro.
REFERENCES
- 1. Mastrianni J. A. (2010) The genetics of prion diseases. Genet. Med. 12, 187–195 [DOI] [PubMed] [Google Scholar]
- 2. Edgeworth J. A., Gros N., Alden J., Joiner S., Wadsworth J. D., Linehan J., Brandner S., Jackson G. S., Weissmann C., Collinge J. (2010) Spontaneous generation of mammalian prions. Proc. Natl. Acad. Sci. U.S.A. 107, 14402–14406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weissmann C., Enari M., Klöhn P. C., Rossi D., Flechsig E. (2002) Transmission of prions. J. Infect. Dis. 186, S157–S165 [DOI] [PubMed] [Google Scholar]
- 4. Aguzzi A., Polymenidou M. (2004) Mammalian prion biology. One century of evolving concepts. Cell 116, 313–327 [DOI] [PubMed] [Google Scholar]
- 5. Haybaeck J., Heikenwalder M., Klevenz B., Schwarz P., Margalith I., Bridel C., Mertz K., Zirdum E., Petsch B., Fuchs T. J., Stitz L., Aguzzi A. (2011) Aerosols transmit prions to immunocompetent and immunodeficient mice. PLoS Pathog. 7, e1001257. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weissmann C. (2004) The state of the prion. Nat. Rev. Microbiol. 2, 861–871 [DOI] [PubMed] [Google Scholar]
- 8. Aguzzi A., Calella A. M. (2009) Prions. Protein aggregation and infectious diseases. Physiol. Rev. 89, 1105–1152 [DOI] [PubMed] [Google Scholar]
- 9. Aguzzi A. (2009) Cell biology. Beyond the prion principle. Nature 459, 924–925 [DOI] [PubMed] [Google Scholar]
- 10. Appleby B. S., Lyketsos C. G. (2011) Rapidly progressive dementias and the treatment of human prion diseases. Expert Opin. Pharmacother. 12, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korth C., May B. C., Cohen F. E., Prusiner S. B. (2001) Acridine and phenothiazine derivatives as pharmacotherapeutics for prion disease. Proc. Natl. Acad. Sci. U.S.A. 98, 9836–9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pocchiari M., Schmittinger S., Masullo C. (1987) Amphotericin B delays the incubation period of scrapie in intracerebrally inoculated hamsters. J. Gen. Virol. 68, 219–223 [DOI] [PubMed] [Google Scholar]
- 13. Collinge J., Gorham M., Hudson F., Kennedy A., Keogh G., Pal S., Rossor M., Rudge P., Siddique D., Spyer M., Thomas D., Walker S., Webb T., Wroe S., Darbyshire J. (2009) Safety and efficacy of quinacrine in human prion disease (PRION-1 study). A patient-preference trial. Lancet Neurol. 8, 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martínez-Lage J. F., Rábano A., Bermejo J., Martíez Pérez M., Guerrero M. C., Contreras M. A., Lunar A. (2005) Creutzfeldt-Jakob disease acquired via a dural graft. Failure of therapy with quinacrine and chlorpromazine. Surg. Neurol. 64, 542–545 [DOI] [PubMed] [Google Scholar]
- 15. Benito-León J. (2004) Combined quinacrine and chlorpromazine therapy in fatal familial insomnia. Clin. Neuropharmacol. 27, 201–203 [DOI] [PubMed] [Google Scholar]
- 16. Ingrosso L., Ladogana A., Pocchiari M. (1995) Congo red prolongs the incubation period in scrapie-infected hamsters. J. Virol. 69, 506–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tagliavini F., McArthur R. A., Canciani B., Giaccone G., Porro M., Bugiani M., Lievens P. M., Bugiani O., Peri E., Dall'Ara P., Rocchi M., Poli G., Forloni G., Bandiera T., Varasi M., Suarato A., Cassutti P., Cervini M. A., Lansen J., Salmona M., Post C. (1997) Effectiveness of anthracycline against experimental prion disease in Syrian hamsters. Science 276, 1119–1122 [DOI] [PubMed] [Google Scholar]
- 18. Caspi S., Halimi M., Yanai A., Sasson S. B., Taraboulos A., Gabizon R. (1998) The anti-prion activity of Congo red. Putative mechanism. J. Biol. Chem. 273, 3484–3489 [DOI] [PubMed] [Google Scholar]
- 19. Ouidja M. O., Petit E., Kerros M. E., Ikeda Y., Morin C., Carpentier G., Barritault D., Brugère-Picoux J., Deslys J. P., Adjou K., Papy-Garcia D. (2007) Structure-activity studies of heparan mimetic polyanions for anti-prion therapies. Biochem. Biophys. Res. Commun. 363, 95–100 [DOI] [PubMed] [Google Scholar]
- 20. Tsuboi Y., Doh-Ura K., Yamada T. (2009) Continuous intraventricular infusion of pentosan polysulfate. Clinical trial against prion diseases. Neuropathology 29, 632–636 [DOI] [PubMed] [Google Scholar]
- 21. Terada T., Tsuboi Y., Obi T., Doh-ura K., Murayama S., Kitamoto T., Yamada T., Mizoguchi K. (2010) Less protease-resistant PrP in a patient with sporadic CJD treated with intraventricular pentosan polysulfate. Acta Neurol. Scand. 121, 127–130 [DOI] [PubMed] [Google Scholar]
- 22. MacGregor I. R., Dawes J., Pepper D. S., Prowse C. V., Stocks J. (1985) Metabolism of sodium pentosan polysulfate in man measured by a new competitive binding assay for sulfated polysaccharides. Comparison with effects upon anticoagulant activity, lipolysis, and platelet α-granule proteins. Thromb. Haemost. 53, 411–414 [PubMed] [Google Scholar]
- 23. Supattapone S., Piro J. R., Rees J. R. (2009) Complex polyamines. Unique prion disaggregating compounds. CNS Neurol. Disord. Drug Targets 8, 323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nilsson K. P., Herland A., Hammarström P., Inganäs O. (2005) Conjugated polyelectrolytes. Conformation-sensitive optical probes for detection of amyloid fibril formation. Biochemistry 44, 3718–3724 [DOI] [PubMed] [Google Scholar]
- 25. Herland A., Nilsson K. P., Olsson J. D., Hammarström P., Konradsson P., Inganäs O. (2005) Synthesis of a regioregular zwitterionic conjugated oligoelectrolyte, usable as an optical probe for detection of amyloid fibril formation at acidic pH. J. Am. Chem. Soc. 127, 2317–2323 [DOI] [PubMed] [Google Scholar]
- 26. Nilsson K. P., Hammarström P., Ahlgren F., Herland A., Schnell E. A., Lindgren M., Westermark G. T., Inganäs O. (2006) Conjugated polyelectrolytes. Conformation-sensitive optical probes for staining and characterization of amyloid deposits. ChemBioChem. 7, 1096–1104 [DOI] [PubMed] [Google Scholar]
- 27. Nilsson K. P., Ikenberg K., Aslund A., Fransson S., Konradsson P., Röcken C., Moch H., Aguzzi A. (2010) Structural typing of systemic amyloidoses by luminescent-conjugated polymer spectroscopy. Am. J. Pathol. 176, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sigurdson C. J., Nilsson K. P., Hornemann S., Manco G., Polymenidou M., Schwarz P., Leclerc M., Hammarström P., Wüthrich K., Aguzzi A. (2007) Prion strain discrimination using luminescent conjugated polymers. Nat. Methods 4, 1023–1030 [DOI] [PubMed] [Google Scholar]
- 29. Gottlieb H. E., Kotlyar V., Nudelman A. (1997) NMR chemical shifts of common laboratory solvents as trace impurities. J. Org. Chem. 62, 7512–7515 [DOI] [PubMed] [Google Scholar]
- 30. Nilsson K. P., Aslund A., Berg I., Nyström S., Konradsson P., Herland A., Inganäs O., Stabo-Eeg F., Lindgren M., Westermark G. T., Lannfelt L., Nilsson L. N., Hammarström P. (2007) Imaging distinct conformational states of amyloid-β fibrils in Alzheimer's disease using novel luminescent probes. ACS Chem. Biol. 2, 553–560 [DOI] [PubMed] [Google Scholar]
- 31. Aslund A., Herland A., Hammarström P., Nilsson K. P., Jonsson B. H., Inganäs O., Konradsson P. (2007) Studies of luminescent conjugated polythiophene derivatives. Enhanced spectral discrimination of protein conformational states. Bioconjug. Chem. 18, 1860–1868 [DOI] [PubMed] [Google Scholar]
- 32. Ho H. A., Boissinot M., Bergeron M. G., Corbeil G., Doré K., Boudreau D., Leclerc M. (2002) Colorimetric and fluorometric detection of nucleic acids using cationic polythiophene derivatives. Angew. Chem. Int. Ed. Engl. 41, 1548–1551 [DOI] [PubMed] [Google Scholar]
- 33. Nilsson K. P., Rydberg J., Baltzer L., Inganäs O. (2003) Self-assembly of synthetic peptides control conformation and optical properties of a zwitterionic polythiophene derivative. Proc. Natl. Acad. Sci. U.S.A. 100, 10170–10174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nilsson K. P., Inganäs O. (2003) Chip and solution detection of DNA hybridization using a luminescent zwitterionic polythiophene derivative. Nat. Mater. 2, 419–424 [DOI] [PubMed] [Google Scholar]
- 35. Aslund A., Sigurdson C. J., Klingstedt T., Grathwohl S., Bolmont T., Dickstein D. L., Glimsdal E., Prokop S., Lindgren M., Konradsson P., Holtzman D. M., Hof P. R., Heppner F. L., Gandy S., Jucker M., Aguzzi A., Hammarström P., Nilsson K. P. (2009) Novel pentameric thiophene derivatives for in vitro and in vivo optical imaging of a plethora of protein aggregates in cerebral amyloidoses. ACS Chem. Biol. 4, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aslund A., Nilsson K. P., Konradsson P. (2009) Fluorescent oligo and poly-thiophenes and their utilization for recording biological events of diverse origin-when organic chemistry meets biology. J. Chem. Biol. 2, 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ding L., Jonforsen M., Roman L. S., Andersson M., Inganäs O. (2000) Synth. Met. 110, 133–140 [Google Scholar]
- 38. Falsig J., Aguzzi A. (2008) The prion organotypic slice culture assay, POSCA. Nat. Protoc. 3, 555–562 [DOI] [PubMed] [Google Scholar]
- 39. Falsig J., Julius C., Margalith I., Schwarz P., Heppner F. L., Aguzzi A. (2008) A versatile prion replication assay in organotypic brain slices. Nat. Neurosci. 11, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klöhn P. C., Stoltze L., Flechsig E., Enari M., Weissmann C. (2003) A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc. Natl. Acad. Sci. U.S.A. 100, 11666–11671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lau A. L., Yam A. Y., Michelitsch M. M., Wang X., Gao C., Goodson R. J., Shimizu R., Timoteo G., Hall J., Medina-Selby A., Coit D., McCoin C., Phelps B., Wu P., Hu C., Chien D., Peretz D. (2007) Characterization of prion protein (PrP)-derived peptides that discriminate full-length PrPSc from PrPC. Proc. Natl. Acad. Sci. U.S.A. 104, 11551–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Polymenidou M., Moos R., Scott M., Sigurdson C., Shi Y. Z., Yajima B., Hafner-Bratkovic I., Jerala R., Hornemann S., Wuthrich K., Bellon A., Vey M., Garen G., James M. N., Kav N., Aguzzi A. (2008) The POM monoclonals. A comprehensive set of antibodies to nonoverlapping prion protein epitopes. PLoS One 3, e3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zahn R., Liu A., Lührs T., Riek R., von Schroetter C., López García F., Billeter M., Calzolai L., Wider G., Wüthrich K. (2000) NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. U.S.A. 97, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lysek D. A., Wüthrich K. (2004) Prion protein interaction with the C-terminal SH3 domain of Grb2 studied using NMR and optical spectroscopy. Biochemistry 43, 10393–10399 [DOI] [PubMed] [Google Scholar]
- 45. Hornemann S., Christen B., von Schroetter C., Pérez D. R., Wüthrich K. (2009) Prion protein library of recombinant constructs for structural biology. FEBS J. 276, 2359–2367 [DOI] [PubMed] [Google Scholar]
- 46. Apetri A. C., Vanik D. L., Surewicz W. K. (2005) Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90–231. Biochemistry 44, 15880–15888 [DOI] [PubMed] [Google Scholar]
- 47. Almstedt K., Nyström S., Nilsson K. P., Hammarström P. (2009) Amyloid fibrils of human prion protein are spun and woven from morphologically disordered aggregates. Prion 3, 224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Polymenidou M., Prokop S., Jung H. H., Hewer E., Peretz D., Moos R., Tolnay M., Aguzzi A. (2011) Atypical prion protein conformation in familial prion disease with PRNP P105T mutation. Brain Pathol. 21, 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kranich J., Krautler N. J., Falsig J., Ballmer B., Li S., Hutter G., Schwarz P., Moos R., Julius C., Miele G., Aguzzi A. (2010) Engulfment of cerebral apoptotic bodies controls the course of prion disease in a mouse strain-dependent manner. J. Exp. Med. 207, 2271–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fischer M., Rülicke T., Raeber A., Sailer A., Moser M., Oesch B., Brandner S., Aguzzi A., Weissmann C. (1996) Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 15, 1255–1264 [PMC free article] [PubMed] [Google Scholar]
- 51. Peretz D., Scott M. R., Groth D., Williamson R. A., Burton D. R., Cohen F. E., Prusiner S. B. (2001) Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 10, 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Otzen D. E. (2010) Amyloid formation in surfactants and alcohols. Membrane mimetics or structural switchers? Curr. Protein Pept. Sci. 11, 355–371 [DOI] [PubMed] [Google Scholar]
- 53. Breyer J., Wemheuer W. M., Wrede A., Graham C., Benestad S. L., Brenig B., Richt J. A., Schulz-Schaeffer W. J. (2012) Detergents modify proteinase K resistance of PrP(Sc) in different transmissible spongiform encephalopathies (TSEs). Vet. Microbiol. 157, 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Milhavet O., Mangé A., Casanova D., Lehmann S. (2000) Effect of Congo red on wild-type and mutated prion proteins in cultured cells. J. Neurochem. 74, 222–230 [DOI] [PubMed] [Google Scholar]
- 55. Rangachari V., Reed D. K., Moore B. D., Rosenberry T. L. (2006) Secondary structure and interfacial aggregation of amyloid-β(1–40) on sodium dodecyl sulfate micelles. Biochemistry 45, 8639–8648 [DOI] [PubMed] [Google Scholar]
- 56. Tew D. J., Bottomley S. P., Smith D. P., Ciccotosto G. D., Babon J., Hinds M. G., Masters C. L., Cappai R., Barnham K. J. (2008) Stabilization of neurotoxic soluble β-sheet-rich conformations of the Alzheimer's disease amyloid-β peptide. Biophys. J. 94, 2752–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Terzi E., Hölzemann G., Seelig J. (1995) Self-association of β-amyloid peptide(1–40) in solution and binding to lipid membranes. J. Mol. Biol. 252, 633–642 [DOI] [PubMed] [Google Scholar]
- 58. Bokvist M., Lindström F., Watts A., Gröbner G. (2004) Two types of Alzheimer's β-amyloid(1–40) peptide membrane interactions. Aggregation preventing transmembrane anchoring versus accelerated surface fibril formation. J. Mol. Biol. 335, 1039–1049 [DOI] [PubMed] [Google Scholar]
- 59. Wahlström A., Hugonin L., Perálvarez-Marín A., Jarvet J., Gräslund A. (2008) Secondary structure conversions of Alzheimer Aβ(1–40) peptide induced by membrane-mimicking detergents. FEBS J. 275, 5117–5128 [DOI] [PubMed] [Google Scholar]
- 60. Lendel C., Bolognesi B., Wahlström A., Dobson C. M., Gräslund A. (2010) Detergent-like interaction of Congo red with the amyloid β peptide. Biochemistry 49, 1358–1360 [DOI] [PubMed] [Google Scholar]
- 61. Abelein A., Bolognesi B., Dobson C. M., Gräslund A., Lendel C. (2012) Hydrophobicity and conformational change as mechanistic determinants for nonspecific modulators of amyloid β self-assembly. Biochemistry 51, 126–137 [DOI] [PubMed] [Google Scholar]
- 62. Knowles T. P., Waudby C. A., Devlin G. L., Cohen S. I., Aguzzi A., Vendruscolo M., Terentjev E. M., Welland M. E., Dobson C. M. (2009) An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 [DOI] [PubMed] [Google Scholar]
- 63. Caughey B., Lansbury P. T. (2003) Protofibrils, pores, fibrils, and neurodegeneration. Separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26, 267–298 [DOI] [PubMed] [Google Scholar]
- 64. Wang Y. Q., Buell A. K., Wang X. Y., Welland M. E., Dobson C. M., Knowles T. P., Perrett S. (2011) Relationship between prion propensity and the rates of individual molecular steps of fibril assembly. J. Biol. Chem. 286, 12101–12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gabizon R., McKinley M. P., Prusiner S. B. (1987) Purified prion proteins and scrapie infectivity copartition into liposomes. Proc. Natl. Acad. Sci. U.S.A. 84, 4017–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Silveira J. R., Raymond G. J., Hughson A. G., Race R. E., Sim V. L., Hayes S. F., Caughey B. (2005) The most infectious prion protein particles. Nature 437, 257–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tixador P., Herzog L., Reine F., Jaumain E., Chapuis J., Le Dur A., Laude H., Béringue V. (2010) The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog. 6, e1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shaked G. M., Fridlander G., Meiner Z., Taraboulos A., Gabizon R. (1999) Protease-resistant and detergent-insoluble prion protein is not necessarily associated with prion infectivity. J. Biol. Chem. 274, 17981–17986 [DOI] [PubMed] [Google Scholar]
- 69. Lasmézas C. I., Deslys J. P., Robain O., Jaegly A., Beringue V., Peyrin J. M., Fournier J. G., Hauw J. J., Rossier J., Dormont D. (1997) Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science 275, 402–405 [DOI] [PubMed] [Google Scholar]
- 70. Sklaviadis T. K., Manuelidis L., Manuelidis E. E. (1989) Physical properties of the Creutzfeldt-Jakob disease agent. J. Virol. 63, 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hsiao K. K., Groth D., Scott M., Yang S. L., Serban H., Rapp D., Foster D., Torchia M., Dearmond S. J., Prusiner S. B. (1994) Serial transmission in rodents of neurodegeneration from transgenic mice expressing mutant prion protein. Proc. Natl. Acad. Sci. U.S.A. 91, 9126–9130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Piccardo P., Manson J. C., King D., Ghetti B., Barron R. M. (2007) Accumulation of prion protein in the brain that is not associated with transmissible disease. Proc. Natl. Acad. Sci. U.S.A. 104, 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Barron R. M., Campbell S. L., King D., Bellon A., Chapman K. E., Williamson R. A., Manson J. C. (2007) High titers of transmissible spongiform encephalopathy infectivity associated with extremely low levels of PrPSc in vivo. J. Biol. Chem. 282, 35878–35886 [DOI] [PubMed] [Google Scholar]
- 74. Chiesa R., Piccardo P., Quaglio E., Drisaldi B., Si-Hoe S. L., Takao M., Ghetti B., Harris D. A. (2003) Molecular distinction between pathogenic and infectious properties of the prion protein. J. Virol. 77, 7611–7622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shyng S. L., Lehmann S., Moulder K. L., Harris D. A. (1995) Sulfated glycans stimulate endocytosis of the cellular isoform of the prion protein, PrPC, in cultured cells. J. Biol. Chem. 270, 30221–30229 [DOI] [PubMed] [Google Scholar]
- 76. Enari M., Flechsig E., Weissmann C. (2001) Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc. Natl. Acad. Sci. U.S.A. 98, 9295–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Perrier V., Solassol J., Crozet C., Frobert Y., Mourton-Gilles C., Grassi J., Lehmann S. (2004) Anti-PrP antibodies block PrPSc replication in prion-infected cell cultures by accelerating PrPC degradation. J. Neurochem. 89, 454–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Peretz D., Williamson R. A., Kaneko K., Vergara J., Leclerc E., Schmitt-Ulms G., Mehlhorn I. R., Legname G., Wormald M. R., Rudd P. M., Dwek R. A., Burton D. R., Prusiner S. B. (2001) Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature 412, 739–743 [DOI] [PubMed] [Google Scholar]
- 79. Hijazi N., Shaked Y., Rosenmann H., Ben-Hur T., Gabizon R. (2003) Copper binding to PrPC may inhibit prion disease propagation. Brain Res. 993, 192–200 [DOI] [PubMed] [Google Scholar]
- 80. Pauly P. C., Harris D. A. (1998) Copper stimulates endocytosis of the prion protein. J. Biol. Chem. 273, 33107–33110 [DOI] [PubMed] [Google Scholar]
- 81. Brown L. R., Harris D. A. (2003) Copper and zinc cause delivery of the prion protein from the plasma membrane to a subset of early endosomes and the Golgi. J. Neurochem. 87, 353–363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.