Background: Topoisomerase IIIα (Top3α) and Blm dissolve Holliday junctions into non-crossover products.
Results: The Top3α C terminus binds to Blm and DNA substrates and is important in vivo.
Conclusion: The C-terminal domain of Top3α is required for dissolution and cellular functions.
Significance: The Top3α C terminus is an essential component of the dissolvasome complex.
Keywords: DNA Helicase, DNA Recombination, DNA Repair, DNA Topoisomerase, DNA Topology, Genomic Instability, Bloom Syndrome, Branch Migration, Drosophila Genetics, Holliday Junction
Abstract
Topoisomerase IIIα (Top3α) is an essential component of the double Holliday junction (dHJ) dissolvasome complex in metazoans, along with Blm and Rmi1/2. This important anti-recombinogenic function cannot be performed by Top3β, the other type IA topoisomerase present in metazoans. The two share a catalytic core but diverge in their tail regions. To understand this difference in function, we investigated the role of the unique C terminus of Top3α. The Drosophila C terminus contains an insert region not conserved among metazoans. This insert contributes an independent interaction with Blm, which may account for the absence of Rmi1 in Drosophila. Mutant Top3α lacking this insert maintains the ability to perform dHJ dissolution but only partially rescues a top3α null fly line, indicating an in vivo role for the insert. Truncation of the C terminus has a minimal effect on the type IA relaxation activity of Top3α; however, dHJ dissolution is greatly reduced. The Top3α C terminus was found to strongly interact with both Blm and DNA, which are critical to the dissolution reaction; these interactions are greatly reduced in the truncated enzyme. The truncation mutant also cannot rescue the viability of top3α null flies, indicating an essential in vivo role. Our data therefore suggest that the Top3α C terminus has an important role in dHJ dissolution (by providing an interaction interface for Blm and DNA) and an essential function in vivo.
Introduction
Maintaining genomic stability requires efficient repair of DNA breaks with minimal genetic changes. The complex of Bloom helicase (Blm) and topoisomerase IIIα (Top3α)2 has been shown to convergently migrate double Holliday junctions (dHJs), an intermediate of double-strand break repair, into solely non-crossover products (1, 2). The significance of this anti-recombinogenic function can be seen in the phenotype of Blm-deficient cells, which show a marked increase in sister chromatid exchanges (3) and loss of heterozygosity (4).
Blm and Top3α together are sufficient for dHJ dissolution (1, 2, 5). The two proteins are known to interact directly (6). In yeast, the ∼100 N-terminal amino acids of Sgs1, the yeast homolog of Blm, are required for interaction with Top3 (7, 8). The Top3α-interacting domain of human Blm has also been identified, although some ambiguity remains. Wu et al. (9) showed that both the N- and C-terminal ∼200 amino acids of human Blm can independently bind Top3α in a far Western analysis. However, Hu et al. (10) showed by co-immunoprecipitation that the N-terminal region is solely responsible for the interaction. Information on the region of Top3α that binds Blm is completely absent.
In addition to Blm and Top3α, the small structural proteins Rmi1 and Rmi2 were found to be stabilizing components of the complex in a variety of organisms (11–14). The deletion of either Rmi1 or Rmi2 recapitulates the Blm deficiency hallmark of increased sister chromatid exchanges (13, 15). However, Drosophila does not appear to possess any homologs of either structural protein (13).
In metazoans, there are a pair of type IA topoisomerases: Top3α and Top3β. Although both can perform relaxation on topologically constrained DNA with single-stranded regions (16–19), Top3β cannot substitute for Top3α in dHJ dissolution (2). Their in vivo effects are similarly distinct. Whereas Top3α is absolutely essential, Top3β mutants show only a modest reduction in life span (20–22), indicating that relaxation activity alone is insufficient for the full range of in vivo functions. Because Top3β has no functional interaction with Blm (23), the essential function of Top3α may be alongside Blm in the recombinational repair pathway.
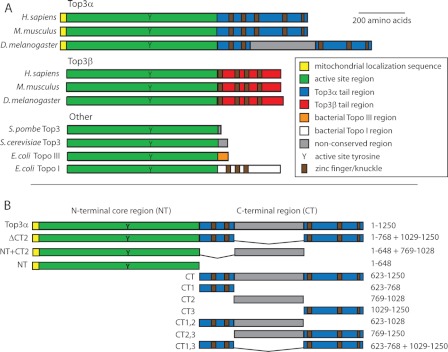
Top3α and Top3β share the type IA catalytic domain. The ∼600 N-terminal residues of Top3α and Top3β, including the catalytic core and active site tyrosine, are very similar, with 63% similarity and 43% identity between the human enzymes (57 and 35% in Drosophila, respectively). However, the proteins greatly diverge in length and sequence composition of their remaining C termini (see Fig. 1A), suggesting a potential region of protein-protein interaction specificity.
FIGURE 1.
A, alignment of the type IA topoisomerase family. The N-terminal region, including the active site tyrosine (Y), is conserved across all species (green). Among the metazoans, the two isoforms (Top3α and Top3β) are distinguished based on their distinct tail regions (blue and red). Although the tails loosely align across the metazoans, the Drosophila Top3α tail contains a unique insert (gray). Topo, topoisomerase. B, constructs of Drosophila Top3α used in this study. The boundaries were based on the alignment in A. Amino acids included are listed on the right. All constructs were made with an N-terminal GST tag and a C-terminal hexahistidine tag (not shown).
Here, we show that the unique C terminus of Drosophila Top3α is necessary for the interaction between Blm and Top3α, for double Holliday junction dissolution to occur efficiently, and to rescue the viability of top3α null flies. In addition to the conserved Top3α C terminus, the Drosophila enzyme contains an inserted region that can independently bind to Blm, perhaps compensating for the absence of the Rmi structural proteins. Top3α lacking this region can only partially rescue top3α null flies. The Top3α C terminus is also involved in binding to DNA substrates, substantially contributing to the binding activity. Together, these data reveal the essential role for the Top3α C terminus in recombinational repair.
EXPERIMENTAL PROCEDURES
Purification of Top3α, Top3αNT, Top3αΔCT2, and Top3αNT+CT2
Top3α, Top3αNT, Top3αΔCT2, and Top3αNT+CT2 were expressed and purified using the baculovirus system in Sf9 insect cells as described (22) with the following exceptions. Cells expressing the mutant proteins (Top3αNT, Top3αΔCT2, and Top3αNT+CT2) were resuspended in 20 mm Tris (pH 8.0), 20 mm KCl, 2 mm MgAc, 3 mm CaCl2, 25% glycerol, and 0.5% Nonidet P-40. After using a Dounce homogenizer, NaCl was added to 350 mm. The solution was incubated on ice for 30 min to precipitate DNA and then centrifuged at 20,000 × g for 20 min. The supernatant was loaded onto glutathione resin, and purification was continued as described above.
Purification of Top3αCT and C-terminal Domains
Top3αCT and C-terminal fragments were expressed in BL21(DE3)pLysS cells. Cells were grown at 37 °C for 2 h, induced with 1 mm isopropyl β-d-thiogalactopyranoside, and grown for 6 h at 30 °C. Cells were then pelleted and frozen. The following steps were performed at 4 °C, and all buffers contained 1:1000 protease inhibitor mixture (Sigma), 20 μg/ml leupeptin, and 5 mm 2-mercaptoethanol (immobilized metal affinity chromatography wash and elution buffers contained 2.5 mm 2-mercaptoethanol). Pellets were resuspended in 50 mm Tris (pH 7.9), 10 mm imidazole, 100 mm NaCl, and 10% glycerol, and the solution was sonicated to break open cells. The lysed cells were spun down at 10,000 × g for 25 min. The supernatant was then incubated with nickel metal affinity resin (Qiagen) for 1 h. The resin was washed with 10 volumes of 50 mm Tris (pH 7.9), 10 mm imidazole, 1 m NaCl, 10% glycerol, and 0.02% Triton X-100, followed by 50 mm Tris (pH 7.9), 10 mm imidazole, 150 mm NaCl, 10% glycerol, and 0.02% Triton X-100. Protein was eluted from the resin using 50 mm Tris (pH 7.9), 200 mm imidazole, 150 mm NaCl, 10% glycerol, and 0.02% Triton X-100 in 1-column volume aliquots. Peak fractions were determined by Bradford assay (Bio-Rad); pooled; diluted 5-fold with 50 mm Tris (pH 7.0), 150 mm NaCl, 10% glycerol, and 0.02% Triton X-100; and applied to glutathione-Sepharose resin (GE Healthcare). The glutathione resin was washed with 50 mm Tris (pH 7.0), 1 m NaCl, 10% glycerol, and 0.02% Triton X-100, followed by 50 mm Tris (pH 7.0), 150 mm NaCl, 10% glycerol, and 0.02% Triton X-100. Protein was eluted by overnight incubation with 50 mm Tris (pH 7.9), 150 mm NaCl, 10% glycerol, and 10 mm glutathione, followed by dialysis into 20 mm Tris (pH 7.5), 150 mm NaCl, and 50% glycerol.
Purification of Blm
Recombinant Blm was purified as by Weinert and Rio (24) with the following exceptions. All listed steps were performed at 4 °C, and all buffers contained 5 mm 2-mercaptoethanol, 1 mm benzamidine, and 0.1 mm PMSF. The nuclear pellet was resuspended in DNA precipitation buffer (15 mm Hepes-KOH (pH 7.6), 400 mm KCl, 5 mm MgCl2, 1 mm EDTA, and 1 mm EGTA) and gently homogenized. This solution was allowed to incubate for 30 min before centrifugation at 20,000 × g for 20 min. The supernatant was recovered, and 0.1 volume of saturated (NH4)2SO4 was added. Ammonium sulfate precipitation and storage were done as described above. Precipitates were resuspended in 10 ml of single-stranded DNA (ssDNA) wash buffer (20 mm Tris (pH 7.9), 200 mm NaCl, 10% glycerol, and 0.02% Triton X-100) and dialyzed twice into 2 liters of this buffer for 2 h. The dialyzed solution was then incubated with 2–3 ml of ssDNA-cellulose resin (United States Biochemical Corp.) for 30 min. The resin was washed with wash buffer for 10 min and then incubated with ssDNA elution buffer (20 mm Tris (pH 7.9), 1 m NaCl, 10% glycerol, and 0.02% Triton X-100) for 30 min. The eluate was dialyzed twice into 1 liter immunoprecipitation buffer (20 mm Tris (pH 7.5), 500 mm NaCl, 10% glycerol, and 0.02% Triton X-100) for 1 h and then incubated overnight with 100 μl of anti-Glu-Glu antibody resin (Covance). The resin was washed and eluted as described above. BSA was added to the eluate to a concentration of 0.25 mg/ml; the eluate was then dialyzed into storage buffer (40 mm Tris (pH 7.5), 150 mm NaCl, and 50% glycerol) for 3 h, flash-frozen, and stored at −80 °C.
Hyper-negatively Supercoiled Plasmid Substrate
Hyper-negatively supercoiled plasmid substrate was prepared as described previously (19). The plasmid pDHJS-AN+ (25) was used at an ethidium bromide/DNA ratio of 12 μg:100 ng.
Relaxation Activity Assay
Activity assays were similar to those performed in a previous study (22). Relaxation activity reactions contained 20 mm Hepes (pH 7.6), 1 mm MgCl2, 100 mm NaAc, 1 mm DTT, 0.1 mm EDTA, and 50 ng/μl DNA. Reactions proceeded at 37 °C for 30 min and were stopped by the addition of NaCl to 600 mm. After 5 min, loading dye and proteinase K (to 50 ng/μl) were added. After a 30-min incubation at 37 °C, samples were loaded onto a 1.5% Tris acetate/EDTA-agarose gel containing 0.5 ng/μl ethidium bromide and run overnight at 50 V. Gels were destained prior to imaging.
Creation of dHJ Substrate
The dHJ substrate was made as described by Plank and Hsieh (25) with the following exceptions. Cre reaction buffer contained 20 mm Tris, 300 mm LiCl, and 1 mm EDTA (pH 8.2). Instead of being electroeluted, Cre reaction products were gel-extracted; allowed to diffuse overnight into 10× buffer containing 50 mm Mops (pH 7.0), 10 mm EDTA (pH 8.0), and 750 mm NaCl; and recovered through a DEAE column (Qiagen).
dHJ Substrate Dissolution Assay
Dissolution of the dHJ substrate was performed as described by Plank et al. (2) with the following exceptions. Reactions were incubated at 30 °C for 30 min and contained 15 nm Top3α, 25 nm Top3αNT, or 20 nm Top3αΔCT2 and also 18 nm Blm.
Protein Binding Assay
Top3α and its derivatives were bound to glutathione-Sepharose resin. Equivalent amounts of bound protein were incubated with purified Blm for 30 min at 4 °C in 40 mm Tris (pH 7.5), 100 mm NaCl, 4 mm MgCl2, and 10% glycerol. The Top3αNT experiment included 5 μg/ml ethidium bromide. After incubation, the resin was washed three times with several volumes of the same buffer. Flow-through and resin samples were run on a 4–20% Criterion acrylamide gel (Bio-Rad), transferred to nitrocellulose, and blotted with mouse anti-Glu-Glu antibody. The blot was incubated with HRP-conjugated goat anti-mouse antibody and substrate (Thermo Scientific) and visualized with film (Eastman Kodak).
Drosophila Crosses
The Top3αNT and Top3αΔCT2-expressing transgenes were generated from the Top3α-YFP-expressing transgene (26) by excising the appropriate tail regions and adding back the nuclear localization sequence (amino acids 845–862) between the Top3α and YFP sequences. The transgenes were injected into fly larvae at the Duke Model System Genomics facility. These flies were mated with the homozygous lethal top3α54 line to create top3α54/CyO;NT/+ and top3α54/CyO;ΔCT2/+. Each line was mated inter se, and the progeny were phenotyped and counted to determine rescue efficiency.
DNA Binding Assay
Oligonucleotides were radiolabeled with [γ-32P]ATP (PerkinElmer Life Sciences) using T4 polynucleotide kinase (New England Biolabs). Oligonucleotides were purchased from IDT (supplemental Table S2). Where required, oligonucleotides were annealed at a 1:3 hot/cold ratio by temperature gradient in a thermocycler. Substrates were subsequently run on an 8% Tris borate/EDTA-polyacrylamide gel, excised, and electroeluted. Labeled DNA (0.25 nm) was incubated with 30 nm protein in 40 mm Tris (pH 7.5), 50 mm NaAc, 4 mm MgCl2, 1 mg/ml BSA, and 10 mm DTT for 30 min at 4 °C. Loading buffer was added, and samples were run on an 8% Tris borate/EDTA-polyacrylamide gel overnight at 4 °C. The gel was exposed to a PhosphorImager screen and scanned using a Storm scanner.
RESULTS
Top3α C Terminus Is Required for dHJ Dissolution
Because Top3α and Top3β have distinct cellular and biochemical functions, and the major structural difference resides in their C termini (Fig. 1A), we examined the properties of the Top3α C terminus. We constructed a series of C-terminal truncation mutants (Fig. 1B). The N-terminal 648 amino acid construct (Top3αNT) was purified and tested for relaxation activity. Top3αNT was active in relaxing a hyper-negatively supercoiled plasmid, with only a slight reduction in specific activity compared with the full-length enzyme (Fig. 2A).
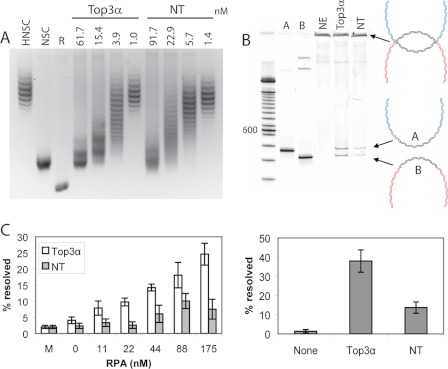
FIGURE 2.
A, full-length Top3α and Top3αNT display comparable relaxation activity. Top3α could relax a hyper-negatively supercoiled (HNSC) DNA to the negatively supercoiled (NSC) level but not to the fully relaxed (R) state. B, whereas full-length Top3α can support dissolution of the dHJ substrate, Top3αNT shows a significant reduction in dissolution activity. NE, no enzyme. Reaction products were digested with BamHI and run on an acrylamide gel. A diagram of the dHJ substrate and reaction products is shown to the right. Reactions are quantified in the graph below based on three independent experiments. C, stimulation by RPA is assisted by the Top3α C terminus. Dissolution reactions were conducted with varied amounts of RPA, and the amount of product was quantified from at least three independent experiments.
We then tested Top3αNT in the dHJ dissolution assay. The dHJ substrate contains two topologically constrained Holliday junctions separated by a homologous region of 165 bp (2). Top3α and Blm have previously been shown to dissolve the dHJ substrate into two separate double-stranded DNA circles (25). At comparable levels of relaxation activity, full-length Top3α showed clear dissolution products, whereas Top3αNT could only minimally support dHJ substrate dissolution, with a >2-fold reduction in dissolution activity (Fig. 2B).
Previously, it has been shown that replication protein A (RPA) specifically stimulates the dHJ dissolution reaction (2). We tested Top3αNT for stimulation of dissolution by RPA. Although stimulation of the dissolution reaction was seen for both full-length Top3α and Top3αNT, the stimulation increased proportionally to RPA concentration only when the full-length enzyme was used (Fig. 2C). Stimulation using Top3αNT was observed only at a higher level of RPA and did not continue to increase with RPA concentration.
Top3α C Terminus Is Involved in Binding Blm
Because dissolution requires coordination with Blm, we tested Top3αNT for the ability to bind Blm. Top3α and Top3αNT with N-terminal GST tags were bound to glutathione resin, incubated with purified Blm, and washed; bound Blm was visualized by Western blotting. Although bound Blm was clearly evident in the presence of full-length Top3α, Top3αNT showed only minimal binding (Fig. 3A). The clear difference between the two constructs suggests that the essential role of the Top3α C terminus may be to interact with Blm.
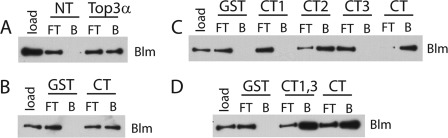
FIGURE 3.
Binding of purified Blm to various Top3α constructs. A, Blm binds to full-length Top3α but barely binds NT. B, Blm binds Top3αCT independently and does not bind a GST control. C, Blm cannot bind CT1 or CT3 alone but does bind CT2 independently. D, Blm can bind to covalently connected CT1 and CT3 (CT1,3). FT, flow-through; B, bound.
To test this hypothesis directly, Top3αCT was constructed and purified. Top3αCT was immobilized and incubated with purified Blm as described above. In contrast to GST alone, Top3αCT clearly showed retained Blm (Fig. 3B). Therefore, Top3αCT is able to bind Blm independently and presumably contributes this function to the full-length protein.
Drosophila Top3α C Terminus Contains Two Independent Blm-binding Domains
We made a series of C-terminal subdomain constructs to narrow down the binding region (Fig. 1B). When compared with the other metazoan enzymes, Drosophila Top3α contains a unique insert in its C terminus (Fig. 1A). The insert sequence is not homologous to any known protein and aligns only with the Top3α C termini of the genus Drosophila, among which it is conserved (supplemental Fig. S1). The first third of the insert is extremely glycine-rich (55%); the remainder is somewhat rich in lysine (8%) but contains no other outstanding features. This insert was used to define the boundaries for dividing the C terminus: the insert was designated CT2, the section prior to it was called CT1, and the section following it was termed CT3. Both individual (CT1, CT2, and CT3) and overlapping (CT1,2 and CT2,3) constructs were made (Fig. 1B). When tested individually, only the Drosophila unique region (CT2) bound Blm (Fig. 3C). This result was surprising given that Top3α in other species is known to interact with Blm. As expected, both CT1,2 and CT2,3 also interacted with Blm (supplemental Fig. S2A).
To determine whether CT2 was necessary or if the remaining tail was sufficient, we constructed CT1,3, which covalently connected regions 1 and 3. This CT1,3 construct was also able to bind to Blm independently (Fig. 3D). Therefore, the Drosophila Top3α enzyme appears to contain two regions capable of independently binding Blm: CT2, the insert present only in Drosophila, and CT1,3, which corresponds to the C terminus in other metazoans. To verify that the two domains (CT1,3 and CT2) can interact with Blm in the context of the enzyme, we added each back to the core domain, creating Top3αΔCT2 and Top3αNT+CT2. Both constructs were able to interact with Blm (supplemental Fig. S2B).
Drosophila Unique Region of Top3α C Terminus Is Not Required for dHJ Substrate Dissolution
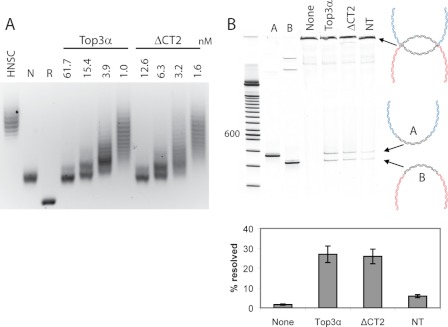
To investigate the role of CT2 in the context of dHJ dissolution, we assayed the Top3αΔCT2 construct. This enzyme is more comparable to human Top3α in length and composition. Similar to Top3αNT, Top3αΔCT2 was active in relaxation, with a specific activity comparable to that of the wild type (Fig. 4A). When added at equal activity, Top3αΔCT2 was able to support dHJ substrate dissolution similar to wild-type levels (Fig. 4B). Therefore, Drosophila Top3αΔCT2 is capable of recapitulating the in vitro activity of the full-length enzyme.
FIGURE 4.
A, full-length Top3α and Top3αΔCT2 show similar levels of activity in relaxing a hyper-negatively supercoiled (HNSC) DNA plasmid. Top3α could relax a hyper-negatively supercoiled (HNSC) DNA to the negatively supercoiled (NSC) level but not to the fully relaxed (R) state. B, Top3αΔCT2 is sufficient for dHJ substrate dissolution at wild-type levels. Dissolution reactions were ethanol-precipitated and digested with BamHI before being run on an acrylamide gel. A diagram of the dHJ substrate and reaction products is shown to right. Reactions from three independent experiments are quantified in the graph below.
Top3αCT Has Important in Vivo Role
Top3α is essential in flies but can be fully rescued by a transgene expressing a full-length Top3α-YFP construct (26). To test the in vivo role of the C terminus, a Top3αNT-YFP-expressing transgene was constructed. The Top3αNT-YFP construct failed to rescue the viability of the top3α null line (supplemental Table S1).
Although CT2 does not appear to be necessary in vitro, the region is highly conserved among Drosophila species (supplemental Fig. S1). To determine whether CT2 is necessary in vivo, a Top3αΔCT2-YFP-expressing transgene was constructed. A low percentage of rescue was seen with the Top3αΔCT2 construct (Table 1). Compared with control flies (top row), a single copy of the Top3αΔCT2-expressing transgene rescued only ∼5% of the homozygous top3α null flies, whereas two copies rescued ∼10%. This low level of rescue efficiency indicates that CT2 has critical in vivo functions, although the Top3αΔCT2 construct still retains partial functionality.
TABLE 1.
Top3αΔCT2 can partially rescue viability of top3α null flies
The top row lists flies with a wild-type copy, and the bottom row lists rescues. The number of flies is listed with the relative ratio in parentheses. The top3α null line is inviable (bottom left). In the presence of the mutant, ∼5% of flies with one copy and 10% with two copies are able to survive.
| +/+, 0 copies | ΔCT2/+, one copy | ΔCT2/ΔCT2, two copies | |
|---|---|---|---|
| top3α54/Top3α | 568 (1) | 1154 (2.0) | 556 (0.98) |
| top3α54/top3α54 | 0 (0) | 54 (0.10) | 56 (0.10) |
Top3α C Terminus Is Involved in Binding DNA
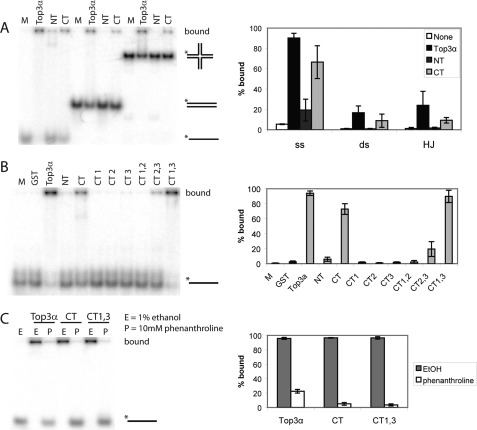
Dissolution requires a tripartite complex of Top3α, Blm, and DNA. To determine whether Top3αCT contributes to DNA interactions, the Top3α constructs were tested for DNA-binding activity using an EMSA with a series of radiolabeled DNA constructs (supplemental Table S2). Full-length Top3α, Top3αNT, and Top3αCT were incubated with single-stranded, double-stranded, and static-X (a non-migratable Holliday junction) DNAs as described previously (27). The full-length enzyme showed almost complete binding of all three substrate types (Fig. 5A). In contrast, Top3αNT showed very little binding above the background, with only the ssDNA showing a significant level of binding. This region of the protein contains type IA topoisomerase domains 1–4, which are known to interact with ssDNA (28). Top3αCT showed significant binding on all substrates. Although not equivalent to the full-length enzyme, Top3αCT contributed significantly to DNA binding. To test which subdomains are involved, the various Top3αCT constructs were tested on ssDNA. CT1,3 was the only subregion able to bind strongly (Fig. 5B), indicating that CT1 and CT3 work together.
FIGURE 5.
A, binding of Top3α, Top3αNT, and Top3αCT to single-stranded, double-stranded (ds), and static-X radiolabeled DNA substrates. The C terminus of Top3α provides the majority of the binding on all types of DNA. Top3α prefers ssDNA, which it requires for activity, but also binds the HJ structure. Data from five independent experiments are quantified in the graph. B, binding of Top3αCT subdomains to ssDNA. Quantified data from at least three independent experiments are shown in the graph. C, binding of Top3α, Top3αCT, and CT1,3 in the presence or absence of phenanthroline. Ethanol was used as a control. Binding was abrogated in the presence of phenanthroline. Quantified data from three independent experiments are shown in the graph. M, no protein added.
The Top3αCT region contains four putative zinc fingers and a zinc knuckle that could contribute to DNA binding (Fig. 1). To test for the importance of the zinc fingers in DNA binding, strong binders were incubated with either 1,10-phenanthroline, which is a zinc chelator, or ethanol, as the phenanthroline is dissolved in ethanol. Full-length Top3α, Top3αCT, and CT1,3 all showed substantially reduced binding in the presence of the zinc chelator (Fig. 5C), indicating that the zinc ion is essential for binding. Full-length Top3α retained some binding, presumably contributed from the active site region (N terminus), which does not contain zinc fingers and can independently bind ssDNA (Fig. 5A). Complete binding of full-length Top3α to ssDNA could be restored by washing away the phenanthroline and adding back zinc (supplemental Fig. S3). Taken together, these data indicate that Top3αCT and its zinc fingers are major contributors to DNA binding.
DISCUSSION
Despite possessing seemingly identical relaxation activity, Top3β cannot substitute for Top3α in vivo (20, 23) or in dHJ dissolution (2). The main structural difference between the two isoenzymes lies in their C termini. Therefore, we investigated the necessity and function of the Drosophila Top3α C terminus.
The relaxation activity of Top3αNT, which truncated the C terminus, was not significantly altered. However, dissolution of the dHJ substrate, a model recombination intermediate, was greatly reduced. Top3αNT was unable to interact with Blm directly, whereas Top3αCT could, indicating that Top3αCT provides a necessary platform for protein-protein interaction. Top3αCT is also involved in the stimulation of dissolution activity by RPA, as truncated enzyme did not display proportional stimulation. The stimulation by RPA could be related to the interaction with Blm, as Blm and RPA are known to interact directly (29), and Top3αCT provides the interaction with Blm.
The Drosophila Top3α C terminus contains an insert that is not homologous to the tails of other metazoans. This insert was used to divide the Drosophila Top3α C terminus for further study. The insert was termed CT2, and the divided pieces were called CT1 and CT3. CT1,3, which directly connects sections CT1 and CT3, is most homologous to the Top3α C-terminal domain of other metazoans. CT1,3 was capable of binding Blm, which is consistent with the notion that Top3α from other species can interact with Blm. However, CT2 was also able to bind Blm independently. This insert may be stabilizing the Top3α-Blm complex in place of Rmi1 and Rmi2, structural proteins found in the complex in other species that have so far not been found in Drosophila. In the species we studied, possession of CT2 consistently corresponds with a lack of Rmi proteins. Although CT1,3 and CT2 each had Blm-binding activity, CT1 and CT3 individually showed little activity in binding Blm or DNA. It is likely that CT1 and CT3 come together three-dimensionally to form a proper scaffold for binding. However, future studies are needed to determine the structural conformation of this region.
Drosophila Top3αΔCT2, which contains both the enzyme active site and the conserved CT1,3-binding region, was capable of performing dHJ dissolution at levels equal to the wild-type enzyme, indicating that CT2 is not required for dHJ dissolution in vitro. This mutant closely resembles the human and other metazoan Top3α enzymes, and therefore, the ability to perform dHJ dissolution is not unexpected. However, CT2 contains important in vivo functions, which are not apparent in our in vitro assays. When the Top3αΔCT2 mutant was inserted into flies, it was able to only partially rescue top3α null flies, and rescuing ability was correlated with the gene copy number of Top3αΔCT2. As Drosophila appears to lack the structural proteins Rmi1 and Rmi2, this insert may be contributing to stabilization of the Top3α-Blm complex.
Top3αCT also plays an extensive role in binding to DNA substrates. Although the catalytic core binds ssDNA as part of its activity, the majority of the observed ssDNA binding of the enzyme was due to Top3αCT. The tail region also contributed the majority of the binding for double-stranded and static-X substrates. Top3αCT is expected to bind DNA due to its putative zinc fingers, which align with those of other metazoans. When all zinc fingers were present, as in CT1,3, they were able to provide significant binding. The addition of a zinc chelator significantly reduced this binding activity, indicating that the protein is coordinating zinc ions to bind DNA substrates.
Recently, two models have been proposed for the molecular process of dHJ dissolution (30). The “unravel and unlink” model proposes a sequential reaction in which a region of ssDNA is first created between the junctions, followed by coordinated separation of the strands and renaturation. In the “HJ migration” model, the two enzymes are arranged in a complex such that Top3α is able to perform strand passage concomitant with the helicase migrating the junctions. Both models require concerted activity of the two enzymes. Our data reinforce the idea that the two proteins need to be coordinated for dissolution to occur. In the absence of interaction, only limited dissolution occurs. Because some limited dissolution is possible when RPA is present but Blm interaction is limited or missing (as with Top3αNT), this may indicate that the functions of the two enzymes may be separated, as in the unravel and unlink model. In this situation, Blm may be acting first to unwind the intervening strands, which are then held in the melted state by RPA until the topoisomerase can bind and act. This would explain why dHJ dissolution with Top3αNT is not above the background in the absence of RPA (Fig. 2C). It is also possible that the optimal situation (when full-length topoisomerase is present) is closer to the HJ migration model, with the unravel and unlink model occurring only under suboptimal situations. Further study will be needed to distinguish the exact mode of action of the complex.
The C terminus of Top3α appears to provide an important interface for protein-protein and protein-DNA interactions. These interactions are critical for the in vitro biochemical reactions, including dissolution of dHJs, as well as for the in vivo functions of the enzyme.
Supplementary Material
Acknowledgments
We thank Dr. Jianhong Wu for assistance with the Top3αΔCT2-YFP-expressing transgene, Dr. Donald Rio for the Drosophila Blm expression construct, and Dr. Paul Modrich for human RPA.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
- Top3α
- topoisomerase IIIα
- dHJ
- double Holliday junction
- NT
- N terminus
- CT
- C terminus
- ssDNA
- single-stranded DNA
- RPA
- replication protein A.
REFERENCES
- 1. Wu L., Hickson I. D. (2003) The Bloom syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 [DOI] [PubMed] [Google Scholar]
- 2. Plank J. L., Wu J., Hsieh T. S. (2006) Topoisomerase IIIα and Bloom helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc. Natl. Acad. Sci. U.S.A. 103, 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chaganti R. S., Schonberg S., German J. (1974) A manyfold increase in sister chromatid exchanges in Bloom syndrome lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 71, 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. LaRocque J. R., Stark J. M., Oh J., Bojilova E., Yusa K., Horie K., Takeda J., Jasin M. (2011) Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 108, 11971–11976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cejka P., Plank J. L., Bachrati C. Z., Hickson I. D., Kowalczykowski S. C. (2010) Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 17, 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu L., Hickson I. D. (2002) The Bloom syndrome helicase stimulates the activity of human topoisomerase IIIα. Nucleic Acids Res. 30, 4823–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bennett R. J., Noirot-Gros M. F., Wang J. C. (2000) Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275, 26898–26905 [DOI] [PubMed] [Google Scholar]
- 8. Fricke W. M., Kaliraman V., Brill S. J. (2001) Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 276, 8848–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu L., Davies S. L., North P. S., Goulaouic H., Riou J. F., Turley H., Gatter K. C., Hickson I. D. (2000) The Bloom syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275, 9636–9644 [DOI] [PubMed] [Google Scholar]
- 10. Hu P., Beresten S. F., van Brabant A. J., Ye T. Z., Pandolfi P. P., Johnson F. B., Guarente L., Ellis N. A. (2001) Evidence for BLM and topoisomerase IIIα interaction in genomic stability. Hum. Mol. Genet. 10, 1287–1298 [DOI] [PubMed] [Google Scholar]
- 11. Raynard S., Bussen W., Sung P. (2006) A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J. Biol. Chem. 281, 13861–13864 [DOI] [PubMed] [Google Scholar]
- 12. Wu L., Bachrati C. Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G. W., Hickson I. D. (2006) BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. U.S.A. 103, 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu D., Guo R., Sobeck A., Bachrati C. Z., Yang J., Enomoto T., Brown G. W., Hoatlin M. E., Hickson I. D., Wang W. (2008) RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 22, 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh T. R., Ali A. M., Busygina V., Raynard S., Fan Q., Du C. H., Andreassen P. R., Sung P., Meetei A. R. (2008) BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 22, 2856–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin J., Sobeck A., Xu C., Meetei A. R., Hoatlin M., Li L., Wang W. (2005) BLAP75, an essential component of Bloom syndrome protein complexes that maintain genome integrity. EMBO J. 24, 1465–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanai R., Caron P. R., Wang J. C. (1996) Human TOP3: a single-copy gene encoding DNA topoisomerase III. Proc. Natl. Acad. Sci. U.S.A. 93, 3653–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goulaouic H., Roulon T., Flamand O., Grondard L., Lavelle F., Riou J. F. (1999) Purification and characterization of human DNA topoisomerase IIIα. Nucleic Acids Res. 27, 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seki T., Seki M., Onodera R., Katada T., Enomoto T. (1998) Cloning of cDNA encoding a novel mouse DNA topoisomerase III (Topo IIIβ) possessing negatively supercoiled DNA relaxing activity, whose message is highly expressed in the testis. J. Biol. Chem. 273, 28553–28556 [DOI] [PubMed] [Google Scholar]
- 19. Wilson T. M., Chen A. D., Hsieh T. (2000) Cloning and characterization of Drosophila topoisomerase IIIβ. Relaxation of hyper-negatively supercoiled DNA. J. Biol. Chem. 275, 1533–1540 [DOI] [PubMed] [Google Scholar]
- 20. Li W., Wang J. C. (1998) Mammalian DNA topoisomerase IIIα is essential in early embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 95, 1010–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwan K. Y., Wang J. C. (2001) Mice lacking DNA topoisomerase IIIβ develop to maturity but show a reduced mean life span. Proc. Natl. Acad. Sci. U.S.A. 98, 5717–5721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Plank J. L., Chu S. H., Pohlhaus J. R., Wilson-Sali T., Hsieh T. S. (2005) Drosophila melanogaster topoisomerase IIIα preferentially relaxes a positively or negatively supercoiled bubble substrate and is essential during development. J. Biol. Chem. 280, 3564–3573 [DOI] [PubMed] [Google Scholar]
- 23. Seki M., Nakagawa T., Seki T., Kato G., Tada S., Takahashi Y., Yoshimura A., Kobayashi T., Aoki A., Otsuki M., Habermann F. A., Tanabe H., Ishii Y., Enomoto T. (2006) Bloom helicase and DNA topoisomerase IIIα are involved in the dissolution of sister chromatids. Mol. Cell. Biol. 26, 6299–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weinert B. T., Rio D. C. (2007) DNA strand displacement, strand annealing, and strand swapping by the Drosophila Bloom syndrome helicase. Nucleic Acids Res. 35, 1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Plank J. L., Hsieh T. S. (2006) A novel, topologically constrained DNA molecule containing a double Holliday junction: design, synthesis, and initial biochemical characterization. J. Biol. Chem. 281, 17510–17516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu J., Feng L., Hsieh T. S. (2010) Drosophila Topo IIIα is required for the maintenance of mitochondrial genome and male germ-line stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 6228–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Capp C., Wu J., Hsieh T. S. (2009) Drosophila RecQ4 has a 3′-5′ DNA helicase activity that is essential for viability. J. Biol. Chem. 284, 30845–30852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Changela A., DiGate R. J., Mondragón A. (2007) Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J. Mol. Biol. 368, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brosh R. M., Jr., Li J. L., Kenny M. K., Karow J. K., Cooper M. P., Kureekattil R. P., Hickson I. D., Bohr V. A. (2000) Replication protein A physically interacts with the Bloom syndrome protein and stimulates its helicase activity. J. Biol. Chem. 275, 23500–23508 [DOI] [PubMed] [Google Scholar]
- 30. Plank J., Hsieh T. S. (2009) Helicase-appended topoisomerases: new insight into the mechanism of directional strand transfer. J. Biol. Chem. 284, 30737–30741 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.