Background: TMEM106B, a major risk factor for FTLD, is a protein of unknown function and cellular properties.
Results: TMEM106B is a glycosylated type 2 membrane protein that localizes to late endosomes/lysosomes.
Conclusion: The cellular properties of TMEM106B suggest a function in protein turnover in endosomes/lysosomes.
Significance: These findings provide the biochemical and cell biological basis for elucidating the pathological role of TMEM106B in FTLD.
Keywords: Lysosomal Glycoproteins, Lysosomes, Membrane Proteins, Neurodegeneration, Neurodegenerative Diseases, Frontotemporal Lobar Degeneration (FTLD), Progranulin (GRN), TAR DNA-binding protein (TDP-43), TMEM106B
Abstract
TMEM106B was identified as a major risk factor in a genome-wide association study for frontotemporal lobar degeneration (FTLD) with TAR DNA-binding protein (TDP)-43 pathology. The most significant association of TMEM106B single nucleotide polymorphisms with risk of FTLD-TDP was observed in patients with progranulin (GRN) mutations. Subsequent studies suggested an inverse correlation between TMEM106B expression and GRN levels in patient serum. However, in this study, this was not confirmed as we failed to detect a significant alteration of GRN levels upon knockdown or exogenous expression of TMEM106B in heterologous cells. To provide a basis for understanding TMEM106B function in health and disease, we investigated the membrane orientation and subcellular localization of this completely uncharacterized protein. By differential membrane extraction and sequential mutagenesis of potential N-glycosylation sites, we identified TMEM106B as a type 2 integral membrane protein with a highly glycosylated luminal domain. Glycosylation is partially required for the transport of TMEM106B beyond the endoplasmic reticulum to late cellular compartments. Endogenous as well as overexpressed TMEM106B localizes to late endosomes and lysosomes. Interestingly, the inhibition of vacuolar H+-ATPases significantly increased the levels of TMEM106B, a finding that may provide an unexpected biochemical link to GRN, because this protein is also strongly increased under the same conditions. Our findings provide a biochemical and cell biological basis for the understanding of the pathological role of TMEM106B in FTLD, an incurable neurodegenerative disorder.
Introduction
FTLD4 is a progressive fatal neurodegenerative disorder and the second most prevalent form of dementia in people under the age of 60 years after Alzheimer disease (1). FTLD patients present with changes in personality and behavior frequently accompanied by progressive nonfluent aphasia. Some patients develop symptoms of parkinsonism or amyotrophic lateral sclerosis (2). There are two major pathological subtypes of FTLD (3). About 40% of FTLD patients are pathologically characterized by inclusions of hyperphosphorylated Tau (FTLD-Tau). The more common FTLD variant is characterized by TDP-43 inclusions (FTLD-TDP) (4–6). In rare cases of FTLD, cytoplasmic deposits of the fused in sarcoma protein are observed (7, 8). In addition, very recently ubiquilin 2 (UBQLN2) has also been shown to be deposited in patients with amyotrophic lateral sclerosis and dementia (9).
Although rare FTLD-TDP-causing mutations were identified in the valosin-containing protein (VCP) (10, 11) and the charged multivesicular body protein 2B (CHMP2B) (12), many more mutations are located in the GRN gene. Autosomal dominant mutations in the GRN gene, which have been identified by genetic linkage studies and/or mutation screenings, account for 20% of familial FTLD-TDP cases (13–16). Of the mutations reported to date, most are loss-of-function mutations leading to GRN haploinsufficiency (5, 15), which results in a severe reduction of GRN levels in tissues and biological fluids of patients (17–20). Additionally, missense mutations (21–23) might lead to folding defects, aberrant processing (24), or cytoplasmic missorting and degradation of GRN (25, 26) and thereby result in reduced secretion (20, 26). Because GRN mutations are not fully penetrant, carriers of identical mutations show a high variability in age of onset and pathological presentation. Thus, additional genetic factors or environmental influences were postulated to play a role in the manifestation of the disease (27). Consistent with that hypothesis, the first genome-wide association study in patients with FTLD-TDP inclusions identified three single nucleotide polymorphisms at the TMEM106B gene locus on chromosome 7p21.3 as a risk factor (28). TMEM106B variants specifically increase the risk for FTLD-TDP in patients with mutations in the GRN (28). Although one study could not confirm these findings (29), multiple replication studies reproduced the genome-wide association study (30–32) stressing the importance of TMEM106B as a risk factor for FTLD. Van Deerlin et al. (28) demonstrated a more than 2.5-fold increase of TMEM106B mRNA expression in cases of FTLD-TDP compared with healthy controls. Moreover, disease-associated TMEM106B variants apparently reduce GRN in plasma (30, 31) and thus decrease the age at disease onset of GRN mutation carriers (30, 31). However, these results are still under debate (33) and could not be confirmed by others (32). So far, our knowledge of the cell biological properties of TMEM106B is far too limited to allow any suggestions of how TMEM106B could affect TDP-43 pathology in a GRN-dependent manner. We therefore investigated membrane orientation and subcellular localization of TMEM106B. In addition, we examined whether TMEM106B expression is affected by inhibition of vacuolar H+-ATPases, which is known to increase GRN expression levels (34). Finally, we investigated whether TMEM106B expression influences GRN levels in cell culture.
EXPERIMENTAL PROCEDURES
cDNA Constructs
Human TMEM106B cDNA (clone IRATp970G1031D) was obtained from Source BioScience LifeSciences (Nottingham, UK). TMEM106B wild type (WT) cDNA was amplified by PCR and subcloned into the BamHI and XhoI restriction sites of the pcDNA 3.1/Hygro(+) or the pcDNATM4/TO expression vector (Invitrogen). The HA tag was introduced by a 5′- or 3′-primer. TMEM106B point mutations N1–5 (N1, N145S; N2, N151S; N3, N164S; N4, N183S; N5, N256S) were introduced by site-directed mutagenesis (Stratagene, La Jolla, CA) according to the manufacturer's instructions and verified by DNA sequencing.
Cell Culture and Transfection
Human cervical carcinoma (HeLa) cells, human embryonic kidney (HEK 293T) cells, and the T-RExTM 293 cell line (Invitrogen) for tetracycline-inducible expression were cultured in Dulbecco's modified Eagle's medium (DMEM) with Glutamax I (Invitrogen) supplemented with 10% (v/v) fetal calf serum (Invitrogen) and penicillin/streptomycin (PAA Laboratories, Pasching, Austria). Human neuroblastoma cells (SH-SY5Y) were cultured in Dulbecco's modified Eagle's medium: nutrient mixture F-12 (DMEM/F-12) supplemented with 15% (v/v) fetal calf serum and penicillin/streptomycin. Transient transfection of cells was carried out using either LipofectamineTM 2000 (Invitrogen) or FuGENE® HD transfection reagent (Roche Applied Science) according to the manufacturers' protocols. Stable cell lines were obtained through transfection of TMEM106B pcDNATM4/TO constructs (N-terminally HA-tagged) into the T-RExTM 293 cell line. For stable TMEM106B-expressing cell lines, transfected cells were selected with 400 ng/μl ZeocinTM (Invitrogen), and single cell clones were picked. To induce TMEM106B expression, stable cell clones were treated with 0.2 μg/ml tetracycline (Sigma) for 12–24 h.
siRNA-mediated Knockdown of TMEM106B
TMEM106B knockdown in HEK 293T and SH-SY5Y cells was achieved by using a pool of pre-designed siRNAs (D-020307-17, D-020307-04, D-020307-03, and D-020307-02; Thermo Fisher Scientific, Waltham, MA). Nontargeting siRNA pool, negative control 1 (D-001210-01-20; Thermo Fisher Scientific, Waltham, MA), was used to assess unspecific effects of siRNA delivery. Cells were reversely transfected with siRNA (10 nm) and LipofectamineTM RNAiMAX (Invitrogen) according to the manufacturer's instruction and analyzed 72 h post-transfection.
Antibodies
The following antibodies were used for immunoblotting: rabbit polyclonal anti-human GRN antibody (Invitrogen; 1:700), mouse monoclonal anti-β-actin antibody (Sigma; 1:2000), rabbit polyclonal anti-calnexin antibody (StressGen, San Diego, CA; 1:2000), mouse monoclonal anti-14-3-3-β antibody (Santa Cruz Biotechnology, Santa Cruz, CA; 1:200), polyclonal anti-HA antibody (Sigma; 1:250), a horseradish peroxidase-conjugated rat monoclonal anti-HA antibody 3F10 (Roche Applied Science; 1:1000), and a generated rat monoclonal anti-TMEM106B antibody directed against the N terminus (1:10). Secondary antibodies were horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit IgG (Promega, Madison, WI; 1:10,000). The following antibodies were used for immunocytochemistry: mouse monoclonal anti-Grp78 (BiP) antibody (StressGen, San Diego; 1:200); mouse monoclonal anti-giantin antibody (Alexis, Lörrach, Germany; 1:600); a rat monoclonal anti-HA antibody 3F10 (Roche Applied Science; 1:200); and a rabbit polyclonal anti-TMEM106B antibody raised against rat TMEM106B (amino acids 1–91). The mouse monoclonal anti-LAMP1 antibody H4A3 (1:200) and the mouse monoclonal anti-LAMP2 antibody H4B4 (1:200) were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health, and maintained by the Dept. of Biology, University of Iowa (Iowa City, IA). Secondary antibodies were Alexa 555-, Alexa 488-, and Alexa 647-conjugated goat anti-mouse, anti-rabbit, or anti-rat IgG (Invitrogen; 1:500).
Immunocytochemistry
Transfected HeLa cells, transfected HEK 293T cells, or induced T-RExTM 293 cells stably expressing HA-TMEM106B were grown on poly-l-lysine-coated coverslips, and immunocytochemistry was performed as described before (34). The coverslips were mounted on glass slides using ProLong® Gold antifade reagent (Invitrogen). Images were obtained on a Zeiss confocal laser scanning microscope (LSM 510 META) using oil immersion 1×00/1.4 and ×60/1.4 objectives and the LSM software Version 3.5 (Carl Zeiss MicroImaging, Göttingen, Germany).
Preparation of Conditioned Media, Cell Lysates, and Immunoblotting
Conditioned media and cell lysates were prepared and analyzed as described (34). For the separation of TMEM106B by PAGE, 4 m urea SDS gels and a urea sample buffer without β-mercaptoethanol were used. Signals on immunoblots were visualized by horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (GE Healthcare), which was detected by a Luminescent Image Analyzer LAS-4000 (Fujifilm Life Science, Tokyo, Japan). Quantification was performed with the MultiGauge Version 3.0 software.
Membrane Preparation
Cells were scraped and pelleted as described (34). For swelling, the cells were incubated on ice for 30 min in hypotonic buffer supplemented with a protease inhibitor mixture (Sigma). The cell suspension was needled and subsequently centrifuged for 10 min at 4 °C and at 1500 × g. The resulting supernatant was ultracentrifuged for 1 h at 4 °C and 100,000 × g. The pellet contained all membrane proteins, and the supernatant contained the cytosolic proteins. To further distinguish between integral membrane proteins and membrane-associated proteins, a carbonate extraction was performed as described (35). Briefly, membranes were incubated on ice for 30 min in carbonate buffer supplemented with a protease inhibitor mixture (Sigma). Then, the sample was ultracentrifuged as described above. The resulting pellet contained the integral membrane proteins, and the supernatant contained the membrane-associated proteins.
Deglycosylation with N-Glycosidase F and Endoglycosidase H
For N-glycosidase F and endoglycosidase H treatment, either total cell lysates (30 μg of protein) or immunoprecipitated TMEM106B (polyclonal anti-HA antibody; 1:250; Sigma) was used as indicated. Immunoprecipitated TMEM106B was eluted from protein A-Sepharose as described (36). Deglycosylation was performed using 1 unit of N-glycosidase F (Roche Applied Science) in N-glycosidase F buffer (100 mm sodium phosphate, pH 8, 25 mm EDTA, 0.1% Triton X-100, 0.1% β-mercaptoethanol, 0.1% SDS, protease inhibitor mixture) or 5 milliunits endoglycosidase H (Roche Applied Science) in the appropriate buffer (200 mm sodium citrate, pH 5.8, 0.1% β-mercaptoethanol, 0.1% SDS, protease inhibitor mixture). Samples and controls without enzyme were incubated for 16 h at 37 °C. The samples were then mixed with urea loading buffer and separated on urea SDS gels.
Drug Treatment
To inhibit N-glycosylation, cells were treated with 10 μg/ml tunicamycin, and transcription was inhibited by 1 μm actinomycin D and translation by 20 μg/ml cycloheximide. Lysosomal degradation was inhibited by 10 μm leupeptin or a mixture of 10 μm leupeptin, 10 μm E64, and 5 μm antipain. Proteasomal degradation was inhibited by 1 μm epoxomicin (all Sigma). To inhibit vacuolar H+-ATPases, cells were treated with 30 nm bafilomycin A1 (BafA1) (Merck). All treatments were carried out for 16 h at 37 °C.
Quantifying mRNA with Real Time RT-PCR
For qRT-PCR, total RNA preparation and reverse transcription were performed as described (34). qRT-PCRs were carried out on a 7500 Fast Real Time PCR system (Applied Biosystems, Carlsbad, CA) with TaqMan technology using human TMEM106B (Hs00998849; exon boundary 7–8), human GRN (Hs00173570; exon boundary 1–2), and human GAPDH (4326317E) primer sets (Applied Biosystems, Carlsbad, CA). Each sample was analyzed in triplicate, and levels of TMEM106B cDNA were normalized to GAPDH cDNA according to the ΔΔCt method using the equation 2−(CtTMEM106B − CtGAPDH)treatment − (CtTMEM106B − CtGAPDH)control.
Enzyme-linked Immunosorbent Assay (ELISA) for Human GRN
Secreted GRN in conditioned media was quantified in a sandwich ELISA as described before (34).
Statistics
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA). If more than two groups were compared, a one-way ANOVA followed by the Dunnett's test or the Tukey's test was applied. If only two groups were compared, an unpaired Student's t test was used with a significance level α of 0.05.
RESULTS
Membrane Association and Glycosylation of TMEM106B
Because nothing is known about the cellular properties of TMEM106B, we first generated a tetracycline-inducible T-RExTM 293 cell line stably expressing N-terminally HA-tagged WT TMEM106B (HA-TMEM106B), and we conducted a detailed study to determine a potential membrane association and orientation of TMEM106B. Bioinformatic prediction programs like TMHMM 2.0 and TMpred suggest that TMEM106B is a membrane protein that lacks an N-terminal signal peptide and may contain either one or two transmembrane domains. To prove whether TMEM106B is membrane-associated or membrane-inserted, as predicted, we extracted membranes with carbonate (35). HA-tagged TMEM106B was detected as an ∼56-kDa membrane protein, and no TMEM106B was obtained in the cytosolic fraction (Fig. 1A). Upon carbonate extraction of the membrane fraction, no TMEM106B was observed in the supernatant, whereas TMEM106B was quantitatively recovered within the membrane pellet (i.e. carbonate pellet) (Fig. 1A). These findings therefore demonstrate that TMEM106B is an integral membrane protein. A typical signature of numerous membrane proteins is their post-translational glycosylation, which occurs during the transport through the secretory pathway. TMEM106B contains five putative consensus sequence motifs for N-glycosylation (Asn-Xaa-(Ser/Thr)), but no potential motif for O-glycosylation. To determine whether TMEM106B is a glycoprotein, lysates of T-RExTM 293 cells stably expressing WT TMEM106B were treated with N-glycosidase F to remove all N-linked glycans. Treatment with N-glycosidase F resulted in a substantial molecular weight shift demonstrating that TMEM106B is expressed as a glycoprotein (Fig. 1B). However, deglycosylated TMEM106B still migrates above its predicted molecular mass of 31 kDa and appears as a double band upon deglycosylation, which suggests additional modifications (Fig. 1B). Next, we analyzed whether the N-linked glycans are of complex type indicating trafficking of TMEM106B beyond early Golgi compartments. T-RExTM 293 cells expressing WT TMEM106B were treated with endoglycosidase H, which removes N-linked mannose-rich but not fully processed complex glycans. Indeed, TMEM106B is partially resistant to treatment with endoglycosidase H (Fig. 1B), suggesting transport to late secretory compartments.
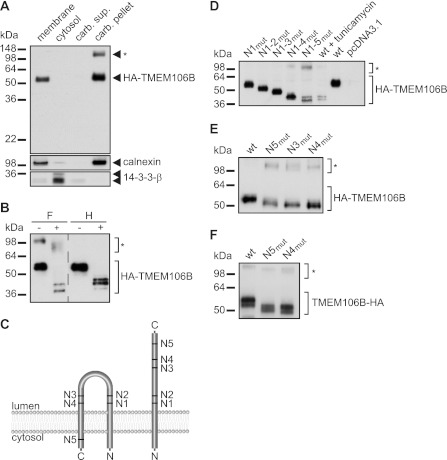
FIGURE 1.
TMEM106B is a highly glycosylated type 2 transmembrane protein. A, cellular fractionation of tetracycline-induced T-RExTM 293 cells stably expressing N-terminally HA-tagged WT TMEM106B (HA-TMEM106B). Total membranes, cytosol and carbonate-extracted membranes (carbonate (carb.) pellet and supernatant (sup.)), were analyzed for TMEM106B by immunoblotting with an anti-HA antibody. Calnexin and 14-3-3-β are controls for integral membrane proteins and cytosolic proteins, respectively. * indicates putative TMEM106B aggregates. B, deglycosylation of TMEM106B. Lysates of T-RExTM 293 cells stably expressing HA-TMEM106B were treated with N-glycosidase F (F) or endoglycosidase H (H) for 16 h. Controls were incubated in the appropriate buffer without the enzyme. TMEM106B was detected by immunoblotting with an anti-HA antibody. * indicates putative TMEM106B aggregates. C, model of the two probable topologies of TMEM106B. N1–N5 indicate the five potential glycosylation motifs within TMEM106B. D, determination of the N-glycosylation sites of TMEM106B. The critical Asn residues of the five potential N-glycosylation motifs of HA-TMEM106B were sequentially mutated to Ser, so that each mutant contains an additionally deleted glycosylation site (N1mut to N1–5mut). The mutated HA-TMEM106B cDNA constructs were transiently transfected into HEK 293T cells. As a control for the migration behavior of unglycosylated and fully glycosylated TMEM106B, cells transfected with WT HA-TMEM106B were treated with 10 μg/ml tunicamycin or solvent for 16 h. HA-TMEM106B variants were detected by immunoblotting with an anti-HA antibody. * indicates putative TMEM106B aggregates. Note that each additionally introduced Asn-to-Ser mutation results in a further molecular weight shift indicating that all potential glycosylation sites are utilized. Note that preventing TMEM106B glycosylation, either by mutated glycosylation sites or by inhibitor treatment, leads to a reduced expression level probably due to impaired folding and therefore increased degradation. E and F, verification of the glycosylation site N5. Mutations of the single glycosylation sites N3, N4, and N5 were generated (N3mut, N4mut, and N5mut) in N-terminally HA-tagged TMEM106B (HA-TMEM106B) (E) and of N4 and N5 (N4mut and N5mut) in C-terminally tagged TMEM106B (TMEM106B-HA) (F). The indicated constructs were transiently transfected in HEK 293T cells. Lysates were analyzed for TMEM106B expression by immunoblotting with an anti-HA antibody. * indicates putative TMEM106B aggregates. Note that the HA tag does not interfere with the glycosylation at the N5 site.
TMEM106B Is a Type 2 Transmembrane Protein
After demonstrating that TMEM106B is a glycosylated integral membrane protein, we investigated its membrane orientation. On the basis of the prediction of one or two potential transmembrane domains, the lack of an N-terminal signal sequence and the modification by glycosylation, we assumed that TMEM106B could adopt two distinct topologies (Fig. 1C). If only the N-terminal hydrophobic domain was utilized for membrane insertion, TMEM106B would adopt a type 2 orientation, whereas if both potential transmembrane domains were used, TMEM106B would be inserted as a hairpin protein with a large luminal loop and with its N and C termini located in the cytosol (Fig. 1C). To discriminate between both possibilities, we made use of the predicted N-glycosylation sites (N1–5) within the primary sequence of TMEM106B (Fig. 1C). We sequentially inactivated all five predicted sites by mutagenesis of the critical Asn residues to Ser. If these sites were used for glycosylation in vivo, a molecular weight shift of the TMEM106B variant should be observed upon mutagenesis. Moreover, each molecular weight shift would be indicative for a luminal positioning of this domain. Indeed, sequential mutagenesis of glycosylation sites N1–5 resulted in a corresponding stepwise reduction of the molecular weight of TMEM106B (Fig. 1D). Importantly, mutagenesis of all glycosylation sites, including site N5 (N1–5mut), additionally reduced the molecular weight of TMEM106B (Fig. 1D). Furthermore, mutant TMEM106B lacking all five glycosylation sites migrates at the same molecular weight as nonglycosylated WT TMEM106B (Fig. 1D). To verify that glycosylation site N5 is indeed utilized in living cells and thus facing luminal compartments, we compared the molecular weight of TMEM106B containing only a single mutation of glycosylation site N5 (N5mut) with that of WT TMEM106B and other single site mutants (N3mut and N4mut). Indeed, the single mutation of glycosylation site N5 resulted in a lowering of the molecular weight of TMEM106B similar to single mutations of site N3 or N4 (Fig. 1E). To prove that the N-terminal HA tag does not interfere with the membrane orientation of TMEM106B, we additionally analyzed a C-terminally tagged variant of TMEM106B. Mutation of glycosylation site N5 of the C-terminally tagged TMEM106B resulted in a lowering of the molecular weight similar to N-terminally HA-tagged TMEM106B variants (Fig. 1F). This proves that glycosylation site N5, which is located C-terminal to the predicted second transmembrane domain (see Fig. 1C), is indeed used for glycosylation and must therefore be located within the cellular lumen. Thus, TMEM106B is an integral type 2 membrane protein (Fig. 1C, right model).
Subcellular Localization of TMEM106B
After demonstrating that TMEM106B is a complex glycosylated integral type 2 membrane protein, we investigated its subcellular localization in T-RExTM 293 cells stably expressing HA-TMEM106B. Cells were subjected to immunofluorescence to visualize TMEM106B and a variety of marker proteins for subcellular compartments, including BiP (ER), giantin (Golgi), and LAMP1 (late endosomes and lysosomes). Stably expressed HA-TMEM106B colocalized predominantly with the endosomal/lysosomal marker protein LAMP1 (Fig. 2A). The predominant localization of TMEM106B in late endosomal/lysosomal compartments was further confirmed by costaining with transiently expressed GFP-Rab7 (37), a late endosomal/lysosomal marker protein (supplemental Fig. 1).
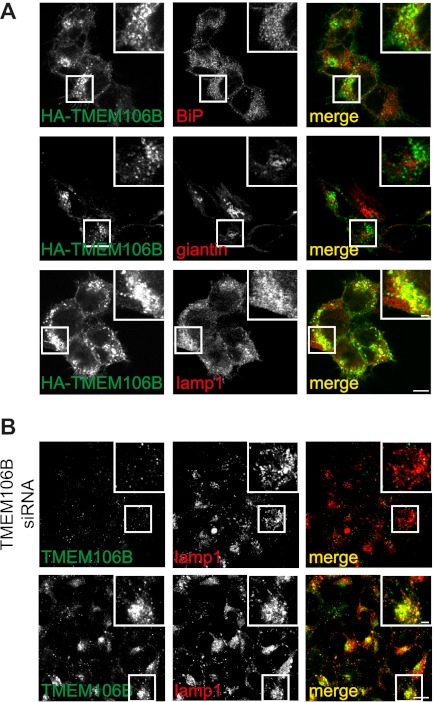
FIGURE 2.
TMEM106B is located in the secretory pathway, mainly in endosomes/lysosomes. A, immunocytochemistry of T-RExTM 293 cells stably expressing TMEM106B. A T-RExTM 293 cell clone stably expressing WT HA-TMEM106B at a low level was grown on coated coverslips, and TMEM106B expression was induced with 0.2 μg/ml tetracycline. 16 h after tetracycline induction, cells were stained with an anti-HA antibody for TMEM106B (green) and costained with the cell marker antibodies BiP (ER, red), giantin (Golgi, red) and LAMP1 (lysosomes, red). Scale bars represent 10 and 2.5 μm (inset). B, immunocytochemistry of endogenous TMEM106B. HEK 293T cells were grown on coverslips. Cells were subsequently stained for TMEM106B (green) and costained with the lysosomal marker antibody against LAMP1 (red). Additionally, TMEM106B was knocked down by siRNA transfection to verify the specificity of the antibody directed to TMEM106B. Scale bars, 10 μm and 2.5 μm (inset). Note that endogenous TMEM106B similar to stably expressed WT HA-TMEM106B colocalizes almost exclusively with lysosomes.
To prove that endogenous TMEM106B also localizes to late endosomes/lysosomes, we generated a polyclonal antibody to TMEM106B. In untransfected cells, this antibody detects TMEM106B predominantly in LAMP1-positive compartments (Fig. 2B). The endogenous late endosomal/lysosomal staining was specific as it could be completely abolished by siRNA-mediated knockdown of TMEM106B (Fig. 2B).
Cellular Transport of TMEM106B Requires Complex Glycosylation
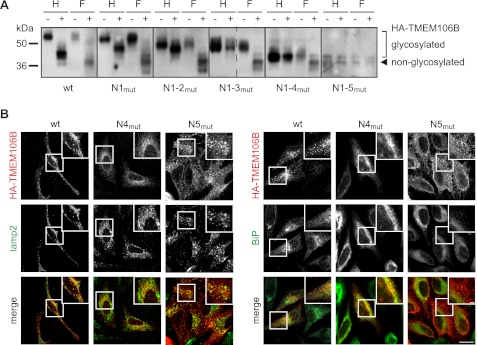
To investigate whether glycosylation affects the cellular transport of TMEM106B to late secretory compartments, we first examined which glycosylation sites contain complex glycans and consequently become endoglycosidase H-resistant. To do so, the glycosylation site mutants described above (Fig. 1D) were analyzed for complex glycosylation by N-glycosidase F and endoglycosidase H treatment (Fig. 3A). Combined mutation of the glycosylation sites N1, N2, and N3 (N1–3mut) still results in an endoglycosidase H-resistant TMEM106B variant indicating that complex glycosylation occurs for the remaining glycosylation sites N4 and N5 (Fig. 3A). Mutation of the noncomplex glycosylation sites (N1–3mut) has no influence on the localization of TMEM106B (supplemental Fig. 2). In contrast, abolishing glycosylation completely by mutation of all glycosylation sites (N1–5mut) results in an accumulation of TMEM106B in the ER and an impaired transport to late endosomal/lysosomal compartments (supplemental Fig. 2). To address the question whether complex glycosylation at site N4 and/or N5 influences lysosomal targeting, we analyzed the cellular localization of the individual complex glycosylation site mutants of TMEM106B (N4mut and N5mut) in HeLa cells transiently transfected with N-terminally HA-tagged TMEM106B constructs. Abolishing the glycosylation at the N4 site impairs forward transport to late endosomes/lysosomes and leads to an accumulation of TMEM106B N4mut within the ER (Fig. 3B, right panel), whereas the N5 glycosylation site mutant (TMEM106B N5mut) is not retained within the ER (Fig. 3B, right panel). Instead, this variant shows a strong staining at lamellipodia (Fig. 3B) consistent with a prominent cell surface localization. Thus, glycosylation at the N4 site appears to be required for anterograde trafficking, whereas complex glycosylation at the N5 site appears to influence direct sorting of TMEM106B to endosomes.
FIGURE 3.
Glycosylation of TMEM106B influences its localization. A, deglycosylation of HA-TMEM106B mutants containing sequentially deleted glycosylation motifs (N1mut to N1–5mut). HA-TMEM106B cDNA constructs (N1mut to N1–5mut) as well as WT HA-TMEM106B were transiently transfected into HEK 293T cells. 24 h after transfection, cell lysates were subjected to immunoprecipitation and treatment with N-glycosidase F (F) and endoglycosidase H (H) for 16 h. Controls were incubated in the appropriate buffer without the enzyme. TMEM106B was detected by immunoblotting with an anti-HA antibody. B, immunocytochemistry of HeLa cells transiently transfected with WT, N4mut, and N5mut HA-TMEM106B constructs. TMEM106B was stained with an anti-HA antibody (red) and costained with antibodies directed to LAMP2 (lysosomes, green) and BiP (ER, green). Scale bars, 10 and 2.5 μm (inset). Note that N4mut (N183S) is retained in the ER, whereas N5mut (N256S) shows lysosomal and strong lamellipodial localization.
TMEM106B Expression Does Not Influence GRN Levels
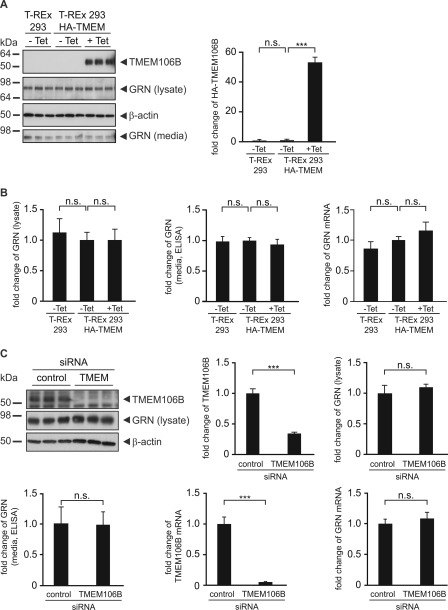
Because TMEM106B is associated with the risk for FTLD-TDP specifically in patients with GRN mutations (28) and an inverse correlation has been shown between increased TMEM106B mRNA levels (28) and reduced GRN in the plasma of patients with disease-associated TMEM106B variants (30, 31), we analyzed whether exogenous expression or siRNA-mediated knockdown of TMEM106B affects GRN levels in cell culture. Tetracycline induction of WT TMEM106B in stably transfected T-RExTM 293 cell lines did not cause a change of GRN protein levels in cell lysates or media (Fig. 4, A and B) nor of GRN mRNA levels (Fig. 4B). In addition, transient expression of TMEM106B in SH-SY5Y cells, a human neuroblastoma cell line, did not significantly affect GRN levels in cell lysates (supplemental Fig. 3A). Because exogenous expression of TMEM106B did not affect GRN levels, we examined whether reduction of TMEM106B had an effect on GRN protein levels and GRN mRNA. Although siRNA-mediated TMEM106B knockdown was very efficient, no change of GRN protein and GRN mRNA levels could be observed in HEK 293T cells (Fig. 4C) and SH-SY5Y cells (supplemental Fig. 3B).
FIGURE 4.
TMEM106B expression does not affect GRN levels. A, GRN levels in T-RExTM 293 cells stably expressing WT HA-TMEM106B. T-RExTM 293 HA-TMEM106B cells were not tetracycline-induced (−Tet) or induced (+Tet) with 0.2 μg/ml tetracycline for 16 h. T-RExTM 293 cells were used as control. Conditioned media and lysates were analyzed for TMEM106B and GRN expression by immunoblotting. β-Actin serves as a loading control. The bar graph represents the quantification of cell lysates for TMEM106B expression by measuring the chemiluminescence on the immunoblot. Data are shown as fold change normalized to noninduced cells, means ± S.D. (n = 3) are depicted (***, p < 0.001 and n.s. (not significant) by one-way ANOVA post hoc Dunnett's test). B, bar graphs represent GRN levels in lysates quantified from the immunoblot and media quantified by ELISA. GRN mRNA levels were quantified by qRT-PCR. Data are shown as fold change in GRN levels after TMEM106B induction, means ± S.D. (n = 3) are depicted (n.s. by one-way ANOVA post hoc Dunnett's test). C, knockdown of TMEM106B does not affect GRN levels. TMEM106B knockdown in HEK 293T cells was achieved by the transfection of a TMEM106B siRNA pool. Nontargeting siRNA was transfected as a control. 72 h after the transfection, conditioned media of the last 16 h of siRNA transfection were collected, and cell lysates and total RNA were prepared. Subsequently, lysates were analyzed for endogenous TMEM106B and GRN expression by immunoblotting. β-Actin serves as a loading control. The bar graphs represent the quantification of cell lysates for TMEM106B and GRN expression by measuring the chemiluminescence on the immunoblot. Conditioned media were analyzed for GRN levels by ELISA. mRNA levels of TMEM106B and GRN were analyzed by qRT-PCR and normalized to control siRNA transfected cells. All quantifications are shown as fold change, means ± S.D. (n = 3) are depicted (***, p < 0.001 and n.s. by unpaired Student's t test).
Protein Levels of TMEM106B Are Increased upon Inhibition of H+-ATPases
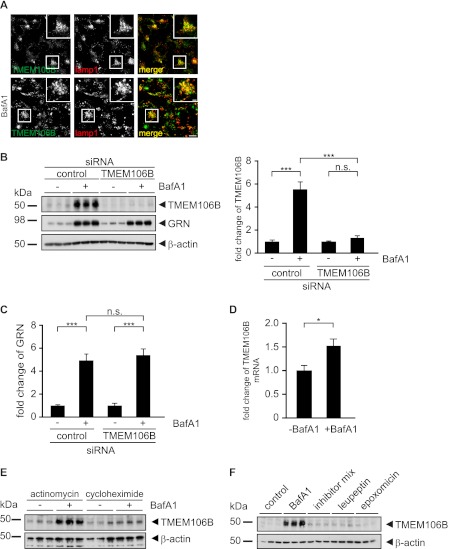
We have previously shown that GRN expression is significantly increased upon inhibition of vacuolar H+-ATPases by BafA1 by a post-transcriptional mechanism (34). Because TMEM106B is located in BafA1-sensitive compartments, we investigated whether TMEM106B levels are affected by inhibiting vacuolar H+-ATPases. Upon inhibition of H+-ATPases by BafA1, endogenous TMEM106B accumulated within LAMP1-positive late endosomes/lysosomes (Fig. 5A). This finding suggests that BafA1 may affect the expression or degradation of TMEM106B. Indeed, endogenous TMEM106B protein levels significantly increased upon inhibition of vacuolar H+-ATPases by BafA1 (Fig. 5B). Detection of endogenous TMEM106B is specific because its immunoreactive signal is almost completely abolished upon siRNA-mediated TMEM106B knockdown (Fig. 5B). Because we could not confirm a direct influence of TMEM106B expression on GRN levels, we next analyzed whether TMEM106B is required for enhanced GRN expression upon inhibition of vacuolar H+-ATPase. Therefore, we examined the increase of GRN levels upon BafA1 treatment after knockdown or overexpression of TMEM106B. However, neither TMEM106B knockdown (Fig. 5, B and C) nor exogenous expression (supplemental Fig. 4, A and B) affected the BafA1-mediated increase in GRN expression. Thus, TMEM106B is not involved in regulating pH-sensitive GRN expression. Next, we investigated whether the BafA1-mediated increase in TMEM106B protein levels occurs by a post-transcriptional mechanism similar to that of GRN (34). Therefore, we examined whether the increase of TMEM106B protein levels is dependent on transcription. Although the mRNA of TMEM106B showed a minor 1.5-fold increase (Fig. 5D), inhibition of transcription by actinomycin did not affect the massive elevation of TMEM106B protein levels caused by BafA1 treatment (Fig. 5E). In contrast, inhibition of translation by cycloheximide strongly reduced the BafA1 effect on TMEM106B levels (Fig. 5E). Therefore, stabilization of TMEM106B protein alone cannot account for the elevated protein levels after BafA1 treatment. This is further supported by the finding that inhibition of lysosomal degradation does not result in an increase in TMEM106B protein levels to the same extent as BafA1 treatment (Fig. 5F). Taken together, these data suggest a predominant post-transcriptional mechanism involved in the BafA1-mediated increase in TMEM106B expression.
FIGURE 5.
Endogenous TMEM106B accumulates in lysosomes upon BafA1 treatment. A, subcellular localization of endogenous TMEM106B after BafA1 treatment. HEK 293T cells were grown on coverslips and treated with/without 30 nm BafA1 for 16 h. Cells were subsequently stained for TMEM106B (green) and costained with the lysosomal marker antibody against LAMP1 (red). Scale bars, 10 and 2.5 μm (inset). B, effects of BafA1 treatment on endogenous TMEM106B expression levels. HEK 293T cells were transfected with a control or a TMEM106B siRNA pool and treated with/without 30 nm BafA1 for 16 h. Cell lysates were prepared 72 h after the siRNA transfection and analyzed by immunoblotting for TMEM106B and GRN expression and for β-actin to verify equal loading. TMEM106B increase after BafA1 treatment was quantified (right panel) in cell lysates normalized to untreated cells and depicted as means ± S.D. (n = 3) (***, p < 0.001 and by one-way ANOVA post hoc Tukey's test). n.s. (not significant). C, BafA1-mediated GRN increase is not affected by TMEM106B knockdown. GRN increase after BafA1 treatment with/without TMEM106B knockdown was quantified from the immunoblot (B), normalized to untreated cells, and depicted as means ± S.D. (n = 3) (***, p < 0.001 and n.s. by one-way ANOVA post hoc Tukey's test). D, mRNA levels of TMEM106B upon BafA1 treatment. TMEM106B mRNA levels of control and BafA1-treated cells were quantified by qRT-PCR and normalized to control cells (means ± S.D. (n = 3); *, p < 0.05, by unpaired Student's test). E, HEK 293T were treated with actinomycin or cycloheximide during treatment with/without BafA1. Cell lysates were analyzed by immunoblotting for TMEM106B and for β-actin to verify equal loading. F, HEK 293T were treated with BafA1, a mixture of lysosomal protease inhibitors (leupeptin, E64, and antipain), leupeptin, and epoxomicin. Cell lysates were analyzed by immunoblotting for TMEM106B and for β-actin to verify equal loading.
DISCUSSION
Major progress has been made in the understanding of the overlapping pathology and clinical symptoms of FTLD and amyotrophic lateral sclerosis specifically by the identification of the genetic components as well as the protein deposits involved (38). Although many novel FTLD and amyotrophic lateral sclerosis associated genes have now been discovered, understanding the cellular consequences of the disease-associated mutations lags far behind. This is particularly obvious for TMEM106B, which is apparently a risk factor for FTLD-TDP (28) probably even to a similar extent as ApoE for Alzheimer disease.
However, for TMEM106B not even the most basic information about its membrane orientation and its subcellular localization was available. We therefore performed a comprehensive protein biochemical and cell biological analysis of this uncharacterized protein. We first demonstrated that TMEM106B is an integral membrane protein using sequential membrane extraction protocols. Furthermore, five asparagine residues were identified as N-glycosylation sites. Making use of these N-glycosylation sites, we could unequivocally show that TMEM106B adopts a type 2 membrane orientation with its large C-terminal domain within the lumen and the smaller N-terminal domain within the cytosol. Specifically, the usage of the most C-terminal N-glycosylation site at amino acid Asn256 (N5) excludes a potential hairpin structure with both the N and the C termini located within the cytosol. If the latter was the case, mutation of this site would not affect the apparent molecular weight of the native protein. However, upon inactivation of this single glycosylation site, we observed a molecular weight shift indicative of a luminal glycosylation. The type 2 orientation was independent of tags fused to either the N or the C terminus. In both cases, inactivation of the N5 site resulted in an equal reduction of the molecular weight. Next, we investigated whether TMEM106B is transported through the secretory pathway and where it accumulates in the cell. Immunohistochemistry and a partial endoglycosidase H resistance of TMEM106B are consistent with a transport into late secretory compartments. Importantly, the endosomal/lysosomal transport of TMEM106B was not due to ectopic expression because similar findings were also made for endogenous TMEM106B. Thus, TMEM106B occurs as a type 2-oriented membrane protein in vivo, which is targeted through the secretory pathway to late endosomes and lysosomes.
The lysosomal targeting signal of TMEM106B is currently not known. However, TMEM106B may reach late endosomes/lysosomes via sorting from the trans-Golgi network, which requires complex glycosylation and/or internalization of plasma membrane-targeted TMEM106B. In line with such sorting mechanisms, TMEM106B lacking the N5 glycosylation site is preferentially targeted to the plasma membrane, but it still reaches late endosomes/lysosomes, probably due to cell surface reinternalization. Indeed, lysosomal membrane proteins often use both pathways. Therefore, the indirect pathway may function as a backup pathway because the direct targeting mechanism can be saturated due to a limited expression of coat or adaptor proteins (reviewed in Refs. 39, 40).
The localization of TMEM106B in late endosomes and lysosomes may provide an unexpected link to GRN. We have previously shown that the inhibition of vacuolar H+-ATPases by BafA1 causes a significant increase of GRN within cell lysates and conditioned media (34). In fact, we provided evidence that Food and Drug Administration-approved inhibitors of lysosomal acidification such as chloroquine exert a similar activity, which could even be of therapeutic relevance for patients suffering from GRN haploinsufficiency. Mechanistically, the increase of GRN levels due to inhibition of vacuolar H+-ATPases largely occurs by a post-transcriptional regulation (34). Surprisingly, we now found that the protein levels of TMEM106B are also significantly increased upon BafA1 treatment most likely by a predominant post-transcriptional mechanism. However, it remains to be shown whether and how the BafA1-mediated increase of TMEM106B and GRN is functionally connected. Interestingly, whereas BafA1 leads to increased GRN levels in early secretory compartments, its effect on TMEM106B appears to be restricted to the endosomal/lysosomal compartment because a strong increase of endosomal/lysosomal localization of TMEM106B is observed upon inhibition of vacuolar H+-ATPases. Together with our finding that overexpression as well as knockdown of TMEM106B fails to influence GRN levels, the BafA1-mediated increase of both GRN and TMEM106B demonstrates that there is not necessarily a negative correlation between TMEM106B and GRN expression as suggested previously (30, 31).
Interestingly, the localization of TMEM106B in BafA1-sensitive late endosomes/lysosomes links TMEM106B to a cellular compartment, which has been implicated previously in familial FTLD-TDP. It has been shown that rare mutations in CHMP2B are causative for FTLD-TDP (12). CHMP2B is a component of the ESCRT-III complex, which is involved in budding and fission of cellular membranes. Moreover, FTLD-associated mutations in CHMP2B result in impaired endocytosis (12, 41) and maturation of dendritic spines (42). One may speculate that the CHMP2B mutations prevent the recruitment of accessory proteins, like Rab7, required for membrane fusion (43). Interestingly, TMEM106B preferentially localizes to Rab7-positive compartments. In addition, one may also speculate that TMEM106B affects sortilin-facilitated endocytosis of GRN and its subsequent transport to late endosomes and lysosomes (44). However, further work is required to address the question of whether TMEM106B is directly required for assisting neuronal autophagy similar to CHMP2B (45–47) or whether TMEM106B affects the sortilin-facilitated endocytosis and subsequent transport of GRN to lysosomes (44). Finally, the localization of TMEM106B within endosomes and lysosomes as well as its increased protein levels upon inhibition of H+-ATPases may be in line with the accumulating evidence for lysosomal dysfunction in neurodegenerative diseases, including Alzheimer disease and FTLD (48–52).
Supplementary Material
Acknowledgments
We thank Marino Zerial for providing the EGFP-Rab constructs and Dorothee Dormann for the GFP-LC3 construct.
This work was supported in part by Sonderforschungsbereich Molecular Mechanisms of Neurodegeneration Grant SFB 596, the Competence Network for Neurodegenerative Diseases of the Bundesministerium für Bildung und Forschung, and a fellowship from the Hans and Ilse Breuer Foundation (to C. M. L.).

This article contains supplemental Figs. S1–S4 and additional references.
This article was selected as a Paper of the Week.
- FTLD
- frontotemporal lobar degeneration
- GRN
- progranulin
- ER
- endoplasmic reticulum
- qRT
- quantitative RT
- ANOVA
- analysis of variance.
REFERENCES
- 1. Graff-Radford N. R., Woodruff B. K. (2007) Frontotemporal dementia. Semin. Neurol. 27, 48–57 [DOI] [PubMed] [Google Scholar]
- 2. Neary D., Snowden J. S., Gustafson L., Passant U., Stuss D., Black S., Freedman M., Kertesz A., Robert P. H., Albert M., Boone K., Miller B. L., Cummings J., Benson D. F. (1998) Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology 51, 1546–1554 [DOI] [PubMed] [Google Scholar]
- 3. Mackenzie I. R., Neumann M., Bigio E. H., Cairns N. J., Alafuzoff I., Kril J., Kovacs G. G., Ghetti B., Halliday G., Holm I. E., Ince P. G., Kamphorst W., Revesz T., Rozemuller A. J., Kumar-Singh S., Akiyama H., Baborie A., Spina S., Dickson D. W., Trojanowski J. Q., Mann D. M. (2010) Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 119, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cairns N. J., Bigio E. H., Mackenzie I. R., Neumann M., Lee V. M., Hatanpaa K. J., White C. L., 3rd, Schneider J. A., Grinberg L. T., Halliday G., Duyckaerts C., Lowe J. S., Holm I. E., Tolnay M., Okamoto K., Yokoo H., Murayama S., Woulfe J., Munoz D. G., Dickson D. W., Ince P. G., Trojanowski J. Q., Mann D. M. (2007) Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 114, 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cruts M., Van Broeckhoven C. (2008) Loss of progranulin function in frontotemporal lobar degeneration. Trends Genet. 24, 186–194 [DOI] [PubMed] [Google Scholar]
- 6. Mackenzie I. R., Rademakers R. (2007) The molecular genetics and neuropathology of frontotemporal lobar degeneration: recent developments. Neurogenetics 8, 237–248 [DOI] [PubMed] [Google Scholar]
- 7. Neumann M., Rademakers R., Roeber S., Baker M., Kretzschmar H. A., Mackenzie I. R. (2009) A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132, 2922–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Urwin H., Josephs K. A., Rohrer J. D., Mackenzie I. R., Neumann M., Authier A., Seelaar H., Van Swieten J. C., Brown J. M., Johannsen P., Nielsen J. E., Holm I. E., FReJA Consortium, Dickson D. W., Rademakers R., Graff-Radford N. R., Parisi J. E., Petersen R. C., Hatanpaa K. J., White C. L., 3rd, Weiner M. F., Geser F., Van Deerlin V. M., Trojanowski J. Q., Miller B. L., Seeley W. W., van der Zee J., Kumar-Singh S., Engelborghs S., De Deyn P. P., Van Broeckhoven C., Bigio E. H., Deng H. X., Halliday G. M., Kril J. J., Munoz D. G., Mann D. M., Pickering-Brown S. M., Doodeman V., Adamson G., Ghazi-Noori S., Fisher E. M., Holton J. L., Revesz T., Rossor M. N., Collinge J., Mead S., Isaacs A. M. (2010) FUS pathology defines the majority of tau- and TDP-43-negative frontotemporal lobar degeneration. Acta Neuropathol. 120, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deng H. X., Chen W., Hong S. T., Boycott K. M., Gorrie G. H., Siddique N., Yang Y., Fecto F., Shi Y., Zhai H., Jiang H., Hirano M., Rampersaud E., Jansen G. H., Donkervoort S., Bigio E. H., Brooks B. R., Ajroud K., Sufit R. L., Haines J. L., Mugnaini E., Pericak-Vance M. A., Siddique T. (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Watts G. D., Wymer J., Kovach M. J., Mehta S. G., Mumm S., Darvish D., Pestronk A., Whyte M. P., Kimonis V. E. (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 36, 377–381 [DOI] [PubMed] [Google Scholar]
- 11. Watts G. D., Thomasova D., Ramdeen S. K., Fulchiero E. C., Mehta S. G., Drachman D. A., Weihl C. C., Jamrozik Z., Kwiecinski H., Kaminska A., Kimonis V. E. (2007) Novel VCP mutations in inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Clin. Genet. 72, 420–426 [DOI] [PubMed] [Google Scholar]
- 12. Skibinski G., Parkinson N. J., Brown J. M., Chakrabarti L., Lloyd S. L., Hummerich H., Nielsen J. E., Hodges J. R., Spillantini M. G., Thusgaard T., Brandner S., Brun A., Rossor M. N., Gade A., Johannsen P., Sørensen S. A., Gydesen S., Fisher E. M., Collinge J. (2005) Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 37, 806–808 [DOI] [PubMed] [Google Scholar]
- 13. Baker M., Mackenzie I. R., Pickering-Brown S. M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A. D., Rollinson S., Cannon A., Dwosh E., Neary D., Melquist S., Richardson A., Dickson D., Berger Z., Eriksen J., Robinson T., Zehr C., Dickey C. A., Crook R., McGowan E., Mann D., Boeve B., Feldman H., Hutton M. (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 [DOI] [PubMed] [Google Scholar]
- 14. Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J. J., van Duijn C., Peeters K., Sciot R., Santens P., De Pooter T., Mattheijssens M., Van den Broeck M., Cuijt I., Vennekens K., De Deyn P. P., Kumar-Singh S., Van Broeckhoven C. (2006) Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442, 920–924 [DOI] [PubMed] [Google Scholar]
- 15. Gijselinck I., Van Broeckhoven C., Cruts M. (2008) Granulin mutations associated with frontotemporal lobar degeneration and related disorders: an update. Hum. Mutat. 29, 1373–1386 [DOI] [PubMed] [Google Scholar]
- 16. Gass J., Cannon A., Mackenzie I. R., Boeve B., Baker M., Adamson J., Crook R., Melquist S., Kuntz K., Petersen R., Josephs K., Pickering-Brown S. M., Graff-Radford N., Uitti R., Dickson D., Wszolek Z., Gonzalez J., Beach T. G., Bigio E., Johnson N., Weintraub S., Mesulam M., White C. L., 3rd, Woodruff B., Caselli R., Hsiung G. Y., Feldman H., Knopman D., Hutton M., Rademakers R. (2006) Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum. Mol. Genet. 15, 2988–3001 [DOI] [PubMed] [Google Scholar]
- 17. Bird T. D. (2009) Progranulin plasma levels in the diagnosis of frontotemporal dementia. Brain 132, 568–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Finch N., Baker M., Crook R., Swanson K., Kuntz K., Surtees R., Bisceglio G., Rovelet-Lecrux A., Boeve B., Petersen R. C., Dickson D. W., Younkin S. G., Deramecourt V., Crook J., Graff-Radford N. R., Rademakers R. (2009) Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain 132, 583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghidoni R., Benussi L., Glionna M., Franzoni M., Binetti G. (2008) Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology 71, 1235–1239 [DOI] [PubMed] [Google Scholar]
- 20. Sleegers K., Brouwers N., Van Damme P., Engelborghs S., Gijselinck I., van der Zee J., Peeters K., Mattheijssens M., Cruts M., Vandenberghe R., De Deyn P. P., Robberecht W., Van Broeckhoven C. (2009) Serum biomarker for progranulin-associated frontotemporal lobar degeneration. Ann. Neurol. 65, 603–609 [DOI] [PubMed] [Google Scholar]
- 21. Brouwers N., Sleegers K., Engelborghs S., Maurer-Stroh S., Gijselinck I., van der Zee J., Pickut B. A., Van den Broeck M., Mattheijssens M., Peeters K., Schymkowitz J., Rousseau F., Martin J. J., Cruts M., De Deyn P. P., Van Broeckhoven C. (2008) Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology 71, 656–664 [DOI] [PubMed] [Google Scholar]
- 22. Schymick J. C., Yang Y., Andersen P. M., Vonsattel J. P., Greenway M., Momeni P., Elder J., Chiò A., Restagno G., Robberecht W., Dahlberg C., Mukherjee O., Goate A., Graff-Radford N., Caselli R. J., Hutton M., Gass J., Cannon A., Rademakers R., Singleton A. B., Hardiman O., Rothstein J., Hardy J., Traynor B. J. (2007) Progranulin mutations and amyotrophic lateral sclerosis or amyotrophic lateral sclerosis-frontotemporal dementia phenotypes. J. Neurol. Neurosurg. Psychiatry 78, 754–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Zee J., Le Ber I., Maurer-Stroh S., Engelborghs S., Gijselinck I., Camuzat A., Brouwers N., Vandenberghe R., Sleegers K., Hannequin D., Dermaut B., Schymkowitz J., Campion D., Santens P., Martin J. J., Lacomblez L., De Pooter T., Peeters K., Mattheijssens M., Vercelletto M., Van den Broeck M., Cruts M., De Deyn P. P., Rousseau F., Brice A., Van Broeckhoven C. (2007) Mutations other than null mutations producing a pathogenic loss of progranulin in frontotemporal dementia. Hum. Mutat. 28, 416. [DOI] [PubMed] [Google Scholar]
- 24. Wang J., Van Damme P., Cruchaga C., Gitcho M. A., Vidal J. M., Seijo-Martínez M., Wang L., Wu J. Y., Robberecht W., Goate A. (2010) Pathogenic cysteine mutations affect progranulin function and production of mature granulins. J. Neurochem. 112, 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mukherjee O., Wang J., Gitcho M., Chakraverty S., Taylor-Reinwald L., Shears S., Kauwe J. S., Norton J., Levitch D., Bigio E. H., Hatanpaa K. J., White C. L., Morris J. C., Cairns N. J., Goate A. (2008) Molecular characterization of novel progranulin (GRN) mutations in frontotemporal dementia. Hum. Mutat. 29, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shankaran S. S., Capell A., Hruscha A. T., Fellerer K., Neumann M., Schmid B., Haass C. (2008) Missense mutations in the progranulin gene linked to frontotemporal lobar degeneration with ubiquitin-immunoreactive inclusions reduce progranulin production and secretion. J. Biol. Chem. 283, 1744–1753 [DOI] [PubMed] [Google Scholar]
- 27. Van Deerlin V. M., Wood E. M., Moore P., Yuan W., Forman M. S., Clark C. M., Neumann M., Kwong L. K., Trojanowski J. Q., Lee V. M., Grossman M. (2007) Clinical, genetic, and pathologic characteristics of patients with frontotemporal dementia and progranulin mutations. Arch. Neurol. 64, 1148–1153 [DOI] [PubMed] [Google Scholar]
- 28. Van Deerlin V. M., Sleiman P. M., Martinez-Lage M., Chen-Plotkin A., Wang L. S., Graff-Radford N. R., Dickson D. W., Rademakers R., Boeve B. F., Grossman M., Arnold S. E., Mann D. M., Pickering-Brown S. M., Seelaar H., Heutink P., van Swieten J. C., Murrell J. R., Ghetti B., Spina S., Grafman J., Hodges J., Spillantini M. G., Gilman S., Lieberman A. P., Kaye J. A., Woltjer R. L., Bigio E. H., Mesulam M., Al-Sarraj S., Troakes C., Rosenberg R. N., White C. L., 3rd, Ferrer I., Lladó A., Neumann M., Kretzschmar H. A., Hulette C. M., Welsh-Bohmer K. A., Miller B. L., Alzualde A., Lopez de Munain A., McKee A. C., Gearing M., Levey A. I., Lah J. J., Hardy J., Rohrer J. D., Lashley T., Mackenzie I. R., Feldman H. H., Hamilton R. L., Dekosky S. T., van der Zee J., Kumar-Singh S., Van Broeckhoven C., Mayeux R., Vonsattel J. P., Troncoso J. C., Kril J. J., Kwok J. B., Halliday G. M., Bird T. D., Ince P. G., Shaw P. J., Cairns N. J., Morris J. C., McLean C. A., DeCarli C., Ellis W. G., Freeman S. H., Frosch M. P., Growdon J. H., Perl D. P., Sano M., Bennett D. A., Schneider J. A., Beach T. G., Reiman E. M., Woodruff B. K., Cummings J., Vinters H. V., Miller C. A., Chui H. C., Alafuzoff I., Hartikainen P., Seilhean D., Galasko D., Masliah E., Cotman C. W., Tuñón M. T., Martínez M. C., Munoz D. G., Carroll S. L., Marson D., Riederer P. F., Bogdanovic N., Schellenberg G. D., Hakonarson H., Trojanowski J. Q., Lee V. M. (2010) Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 42, 234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rollinson S., Mead S., Snowden J., Richardson A., Rohrer J., Halliwell N., Usher S., Neary D., Mann D., Hardy J., Pickering-Brown S. (2011) Frontotemporal lobar degeneration genome wide association study replication confirms a risk locus shared with amyotrophic lateral sclerosis. Neurobiol. Aging 32, 758 e751–757 [DOI] [PubMed] [Google Scholar]
- 30. Cruchaga C., Graff C., Chiang H. H., Wang J., Hinrichs A. L., Spiegel N., Bertelsen S., Mayo K., Norton J. B., Morris J. C., Goate A. (2011) Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch. Neurol. 68, 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Finch N., Carrasquillo M. M., Baker M., Rutherford N. J., Coppola G., Dejesus-Hernandez M., Crook R., Hunter T., Ghidoni R., Benussi L., Crook J., Finger E., Hantanpaa K. J., Karydas A. M., Sengdy P., Gonzalez J., Seeley W. W., Johnson N., Beach T. G., Mesulam M., Forloni G., Kertesz A., Knopman D. S., Uitti R., White C. L., 3rd, Caselli R., Lippa C., Bigio E. H., Wszolek Z. K., Binetti G., Mackenzie I. R., Miller B. L., Boeve B. F., Younkin S. G., Dickson D. W., Petersen R. C., Graff-Radford N. R., Geschwind D. H., Rademakers R. (2011) TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 76, 467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Zee J., Van Langenhove T., Kleinberger G., Sleegers K., Engelborghs S., Vandenberghe R., Santens P., Van den Broeck M., Joris G., Brys J., Mattheijssens M., Peeters K., Cras P., De Deyn P. P., Cruts M., Van Broeckhoven C. (2011) TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain 134, 808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Zee J., Van Broeckhoven C. (2011) TMEM106B a Novel Risk Factor for Frontotemporal Lobar Degeneration. J. Mol. Neurosci., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Capell A., Liebscher S., Fellerer K., Brouwers N., Willem M., Lammich S., Gijselinck I., Bittner T., Carlson A. M., Sasse F., Kunze B., Steinmetz H., Jansen R., Dormann D., Sleegers K., Cruts M., Herms J., Van Broeckhoven C., Haass C. (2011) Rescue of progranulin deficiency associated with frontotemporal lobar degeneration by alkalizing reagents and inhibition of vacuolar ATPase. J. Neurosci. 31, 1885–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonifacino J. S., Dell'Angelica E. C., Springer T. A. (2001) Immunoprecipitation. in Curr. Protoc. Protein Sci. Chapter 9, Unit 9.8 [DOI] [PubMed] [Google Scholar]
- 37. Rink J., Ghigo E., Kalaidzidis Y., Zerial M. (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 [DOI] [PubMed] [Google Scholar]
- 38. Da Cruz S., Cleveland D. W. (2011) Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 21, 904–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Braulke T., Bonifacino J. S. (2009) Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793, 605–614 [DOI] [PubMed] [Google Scholar]
- 40. Saftig P., Klumperman J. (2009) Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 [DOI] [PubMed] [Google Scholar]
- 41. van der Zee J., Urwin H., Engelborghs S., Bruyland M., Vandenberghe R., Dermaut B., De Pooter T., Peeters K., Santens P., De Deyn P. P., Fisher E. M., Collinge J., Isaacs A. M., Van Broeckhoven C. (2008) CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum. Mol. Genet. 17, 313–322 [DOI] [PubMed] [Google Scholar]
- 42. Belly A., Bodon G., Blot B., Bouron A., Sadoul R., Goldberg Y. (2010) CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J. Cell Sci. 123, 2943–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Urwin H., Authier A., Nielsen J. E., Metcalf D., Powell C., Froud K., Malcolm D. S., Holm I., Johannsen P., Brown J., Fisher E. M., van der Zee J., Bruyland M., FReJA Consortium, Van Broeckhoven C., Collinge J., Brandner S., Futter C., Isaacs A. M. (2010) Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet. 19, 2228–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu F., Padukkavidana T., Vægter C. B., Brady O. A., Zheng Y., Mackenzie I. R., Feldman H. H., Nykjaer A., Strittmatter S. M. (2010) Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 68, 654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E. M., Isaacs A., Brech A., Stenmark H., Simonsen A. (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179, 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee J. A., Gao F. B. (2008) Roles of ESCRT in autophagy-associated neurodegeneration. Autophagy 4, 230–232 [DOI] [PubMed] [Google Scholar]
- 47. Rusten T. E., Simonsen A. (2008) ESCRT functions in autophagy and associated disease. Cell Cycle 7, 1166–1172 [DOI] [PubMed] [Google Scholar]
- 48. Bahr B. A., Bendiske J. (2002) The neuropathogenic contributions of lysosomal dysfunction. J. Neurochem. 83, 481–489 [DOI] [PubMed] [Google Scholar]
- 49. Lee J. H., Yu W. H., Kumar A., Lee S., Mohan P. S., Peterhoff C. M., Wolfe D. M., Martinez-Vicente M., Massey A. C., Sovak G., Uchiyama Y., Westaway D., Cuervo A. M., Nixon R. A. (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee S., Sato Y., Nixon R. A. (2011) Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J. Neurosci. 31, 7817–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nixon R. A., Yang D. S. (2011) Autophagy failure in Alzheimer's disease-locating the primary defect. Neurobiol. Dis. 43, 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rubinsztein D. C. (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.