Background: CdGAP is a Rac1/Cdc42 GTPase-activating protein (GAP) involved in the regulation of cell proliferation, migration, and invasion.
Results: A polybasic region (PBR) preceding the GAP domain binds to phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3).
Conclusion: The specific interaction between CdGAP and PI(3,4,5)P3 regulates CdGAP activity.
Significance: A PBR preceding the GAP domain is found in several RhoGAPs, suggesting an evolutionary conserved mechanism of regulation.
Keywords: Lipid-binding Protein, Phosphatidylinositol, Phosphatidylinositol 3-Kinase, Rac1, Rho GTPases, Signal Transduction, CdGAP, GTPase-activating Protein, Membrane Recruitment
Abstract
The Rho family of small GTPases are membrane-associated molecular switches involved in the control of a wide range of cellular activities, including cell migration, adhesion, and proliferation. Cdc42 GTPase-activating protein (CdGAP) is a phosphoprotein showing GAP activity toward Rac1 and Cdc42. CdGAP activity is regulated in an adhesion-dependent manner and more recently, we have identified CdGAP as a novel molecular target in signaling and an essential component in the synergistic interaction between TGFβ and Neu/ErbB-2 signaling pathways in breast cancer cells. In this study, we identified a small polybasic region (PBR) preceding the RhoGAP domain that mediates specific binding to negatively charged phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3). In vitro reconstitution of membrane vesicles loaded with prenylated Rac1 demonstrates that the PBR is required for full activation of CdGAP in the presence of PI(3,4,5)P3. In fibroblast cells, the expression of CdGAP protein mutants lacking an intact PBR shows a significant reduced ability of the protein mutants to induce cell rounding or to mediate negative effects on cell spreading. Furthermore, an intact PBR is required for CdGAP to inactivate Rac1 signaling into cells, whereas it is not essential in an in vitro context. Altogether, these studies reveal that specific interaction between negatively charged phospholipid PI(3,4,5)P3 and the stretch of polybasic residues preceding the RhoGAP domain regulates CdGAP activity in vivo and is required for its cellular functions.
Introduction
The Rho family of small GTPases, in particular RhoA, Rac1, and Cdc42, are key regulators of cytoskeleton remodeling and play important roles in many cellular functions, including cell polarization, cell migration and adhesion, apoptosis, membrane trafficking, and cell cycle progression (1–3). Rho GTPases are molecular switches that cycle between an active GTP- and an inactive GDP-bound state. This cycle is regulated by at least three families of regulatory proteins: guanine-nucleotide exchange factors (GEFs) (2) activate GTPases by promoting the release of bound GDP and replacement by GTP binding, guanine nucleotide-dissociation inhibitors (GDIs) bind and sequester the GDP-bound form in the cytoplasm, and GTPase-activating proteins (GAPs)5 stimulate the intrinsic GTPase activity, leading to inactivation of the GTPase (4, 5).
CdGAP (Cdc42 GTPase-activating protein) is a ubiquitously expressed RhoGAP protein with in vitro activity for Cdc42 and Rac1 (6, 7). CdGAP consists of an N-terminal GAP domain, a basic-rich (BR) central region, and a proline-rich domain (PRD) with an extended C-terminal region (8). CdGAP was identified as the first member of a subgroup of RhoGAP proteins, showing significant protein sequence homology and structural domain organization with the RhoGAP proteins GRIT (9–13), Noma-GAP (14, 15), and ARHGAP30 (16, 17). When overexpressed in various cell types, CdGAP induces a reduction in cell spreading and in lamellipodia formation (18–20). In U2OS osteoblast-like cells, CdGAP interacts with the paxillin-binding protein actopaxin and localizes to focal adhesions where it is activated following integrin engagement (19). CdGAP has also been implicated as an essential component in the synergistic interaction between TGFβ and Neu/ErbB2 signaling pathways in breast cancer cells. CdGAP is required for TGFβ and Neu/ErbB2-induced breast cancer cell motility and invasion, suggesting the possibility that CdGAP may act as a positive regulator of breast cancer (21). Furthermore, gain-of-function mutation in the human CdGAP/ARHGAP31 gene has recently been linked to a recognized developmental disorder, the Adams-Oliver syndrome (20).
RhoGAP proteins are regulated into cells by diverse molecular mechanisms, including protein-protein or lipid interactions, post-translational modifications, and proteolytic degradation (4, 5). Previous studies have unraveled at least two mechanisms of regulation of CdGAP activity. CdGAP is a substrate of ERK1/2 and GSK-3 protein kinases and phosphorylation of Thr-776 in the PRD negatively regulates its activity (8, 22). Furthermore, the association of CdGAP through a novel basic-rich motif with the SH3D domain of the endocytic scaffolding protein intersectin leads to inhibition of CdGAP activity (16, 18). To mediate their cellular effects, Rac1 and Cdc42 GTPases are post-translationally modified by the addition of a prenyl group to a conserved C-terminal cysteine residue (3). This modification serves to anchor the GTPases into membranes and is necessary for their activity (23–25). Accordingly, regulators of Rho GTPases need to be targeted to the membrane to exert their functions. This can be achieved either through protein-protein or lipid-protein interactions (26). Here, we found a small polybasic region (PBR) preceding the RhoGAP domain of CdGAP that mediates specific binding to phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3). In vitro reconstitution of membrane vesicles loaded with prenylated Rac1 shows that the PBR is required for full activation of CdGAP in the presence of PI(3,4,5)P3. In addition, the PBR is essential for CdGAP to regulate Rac1 in vivo and to mediate its GAP-dependent cellular effects.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies used in these experiments were as followed: rabbit anti-GST (Santa Cruz Biotechnology), rabbit anti-GFP (A6455, Molecular Probes, Burlington, ON), horseradish peroxidase (HRP)-labeled anti-rabbit IgG antibody (GE Healthcare, Piscataway, NJ), anti-Rac1 antibody (Temecula, CA), PDGF-BB (CalBiochem), and LY294002 (CalBiochem).
DNA Cloning
The plasmid pGEX2T-Rac1 was described previously (27). The plasmids pGEX-4T2-CdGAP(1–221), pGEX-4T2-CdGAP(17–221), pGEX-4T2-CdGAP(1–221)KQ, and pEGFPC1-related plasmids were constructed using standard cloning procedures (cloning details can be obtained upon request). The pGEX-BMX-PH construct was kindly provided by Dr. Jean-François Côté, Institut de Recherches Cliniques de Montréal, Montreal, Quebec (28). The R56AN169V double mutant construct was generated by taking advantage of the NheI restriction sites located 5′ to the EGFP gene in pEGFPC1-mCdGAP(1–820)R56A/N169V (19) (kindly provided by Dr. Chris Turner, SUNY, Syracuse, NY) and at 1814 bp in the coding sequence of M.m.CdGAP. The NheI fragment containing the R56AN169V amino acid substitutions was ligated with pEGFPC1-CdGAP(219–1425) digested with NheI to generate pEGFP-C1-CdGAP(1–1425)R56A/N169V. All plasmids were verified by sequencing.
Cell Culture and Transfection
COS-7 and HEK293 cells were cultured in Dulbecco's modified Eagle medium (DMEM, WISENT, Mississauga, ON) supplemented with 10% Fetal Bovine Serum (FBS, WISENT), 100 μg/ml streptomycin sulfate, and 100 units/ml penicillin G sodium salt (Invitrogen) in a 5% CO2 humidified environment at 37 °C. COS-7 cells were transfected with linear polyethylenimine MW 25,000 (PEI, Polysciences, Pennsylvania) using a 1:5 ratio (DNA:PEI). Media was replaced 3 h after transfection.
Purification of Recombinant Proteins from Escherichia coli
GST-Rac1 was produced and purified on glutathione-agarose beads as described previously (18). Active protein concentrations were determined by the filter binding assay using [3H]GTP as previously described (7). Production of GST-CdGAP proteins was achieved following the same protocol, except that proteins were induced with 0.4 mm isopropylthiogalactopyranoside (IPTG) for 16 h at 20 °C. Bacteria pellets were resuspended and incubated for 30 min at 4 °C in 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol (DTT) supplemented with 0.2 mg/ml lysozyme, 20 μg/ml DNase, 1 mm MgCl2, 1 mm PMSF, and Complete™ protease inhibitors (Roche) before sonication. Protein lysates obtained after centrifugation were incubated with glutathione-agarose beads (Sigma) for 2 h at 4 °C. The beads were washed five times in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT, and proteins were eluted with 20 mm glutathione in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT. Proteins were concentrated in 10 mm Tris-HCl pH 7.5, 2 mm MgCl2, 1 mm DTT with a Amicon Ultra-4 30K Centricon column concentrator (Sigma). Protein samples were aliquoted, snap-freezed in liquid nitrogen, and stored at −80 °C. Purity and concentration of the protein preparations were determined by Coomassie Blue-stained SDS-PAGE.
Lipid Overlay Assay
Membrane Lipid Strips™ (Echelon-Biosciences, Salt-Lake City, Utah) overlay assays were performed according to the manufacturer's recommendations. Briefly, membrane was blocked 1 h at room temperature with 3% fatty-acid free bovine serum albumin (BSA, Sigma) dissolved in TBS (10 mm Tris-HCl, 150 mm NaCl, pH 8.0) followed by incubation with GST-CdGAP(1–221) or GST-CdGAP(17–221) (10 nm) in TBS supplemented with 3% BSA at room temperature for 1 h. Membrane was washed three times for 10 min in TBS containing 0.1% Tween-20 before incubation with anti-GST (1:1000) and (HRP)-labeled anti-rabbit IgG (1:10000) antibodies. HRP activity was detected by Western Lightning Plus-ECL detection kit (Perkin Elmer).
Multilamellar Vesicles (MLVs) Preparation
Phosphatidylcholine (PC) and phosphatidylinositol-related lipids were all purchased from Avanti Polar Lipids (Alabaster, Alabama). PC and PI were bought as chloroform solubilized forms whereas PI(3)P, PI(4)P, PI(5)P, PI(4,5)P2, PI(3,4,5)P3 were obtained as ammonium salts and were dissolved at 1 mm in chloroform:methanol:water (80:18:2). Required amounts of lipids were mixed and dried under an argon stream. To generate MLVs, lipids were rehydrated in 20 mm Tris-HCl, pH 7.5, 25 mm NaCl and 4 mm EDTA for 1 h on ice, followed by vigorous vortexing to a final lipid concentration of 1 mm. MLVs were stored at 4 °C and used within 24 h.
Multilamellar Vesicle Flotation Assay
GST-tagged proteins used in the MLV flotation assay were first centrifuged at 436,000 × g for 15 min in an Optima™ TLX ultracentrifuge using a TLA-100 rotor (Beckman Coulter) to remove protein aggregates. A volume of 100 μl of the MLV solution (1 mm) was incubated with 25 μl of GST-tagged proteins (10 μm to 20 μm) and 25 μl of buffer (20 mm HEPES pH 7.2, 100 mm NaCl and 5 mm EDTA) for 30 min at 25 °C. A volume of 100 μl of 75% sucrose was added to the mix in order to bring sucrose concentration to 30%. This suspension was moved to a thickwall centrifugation tube and overlaid by 200 μl of 25% sucrose in buffer followed by 50 μl of sucrose-free buffer. Samples were centrifuged at 259,000 × g for 1 h with a TLS-55 rotor swinging-bucket rotor (Beckman Coulter). The bottom (250 μl) and top (100 μl) fractions were collected and 20% of each fraction analyzed by Western blotting. Densitometry was measured using Autoquant (Bio-Rad), and the bound/unbound ratio was compared with the reference value obtained for phosphatidylcholine MLVs.
Rac1 in Vitro Prenylation
E. coli-purified Rac1 is unprenylated and cannot bind to MLVs. Therefore, E. coli-purified Rac1 (1 μg) was prenylated in vitro in 50 mm HEPES pH 7.2, 150 mm KCl, 5 mm MgCl2, 1 mm DTT, 3 mm Nonidet P-40, supplemented with 2 mm geranylgeranyl pyrophosphate ammonium salt (Sigma-Aldrich), and 2 μg/μl recombinant rat Geranylgeranyltransferase I (Merck Biosciences, Darmstadt, Germany) in a volume of 25 μl following a 16-h incubation at 4 °C. Rac1-geranylgeranyl (Rac1GG) proteins were then purified as MLV-bound components. The reaction mix was divided in two fractions, and each was incubated with 20 μg of PC-based or PC:PI(3,4,5)P3 (9:1) MLVs in a buffer constituted of Tris-HCl pH 7.5, 25 mm NaCl and 4 mm EDTA in a final volume of 150 μl for 30 min at room temperature. MLVs were then purified as described above and resuspended in a final volume of 50 μl. Western blot analysis confirmed that ∼50% of Rac1 was co-purified with MLVs (data not shown).
In Vitro GAP Assay
For the in vitro GAP assays with GST-tagged CdGAP proteins (7), Rac1 (1 μg) was loaded with 10 μCi of [γ-32P]GTP (Perkin Elmer) in binding buffer (20 mm Tris-HCl, pH 7.5, 25 mm NaCl, 4 mm EDTA, 0.1 mm DTT) to a final volume of 20 μl at 30 °C for 10 min. Rac1GG bound to PC or PC:PI(3,4,5)P3 (9:1) MLVs (0.5 μg) was similarly loaded in a volume of 48 μl. The reactions were stopped by the addition of 20 mm MgCl2 and put on ice. [γ-32P]GTP-loaded Rac1 or Rac1GG (4.5 μl) (to give a final concentration of 0.5 μm) was added to 10.5 μl of the incubation buffer (20 mm Tris-HCl, pH 7.5, 0.1 mm DTT) supplemented with 1 mm GTP and 0.9 mg/ml BSA with or without GST-CdGAP(1–221), GST-CdGAP(17–221), or GST-CdGAP(1–221)KQ and incubated at 20 °C for 10 min. Fractions of 5 μl of the reaction mix were taken at 0 and 10 min and added to 1 ml of ice-cold wash buffer (50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2) before being filtered through pre-humidified nitrocellulose membranes (Millipore) to remove unbound GTP. Filters were rinsed with 6 ml of wash buffer and the amount of radioactivity was measured by scintillation counting. For in vitro GAP assays using GFP-tagged proteins, COS-7 cells were transiently transfected with pEGFPC1-CdGAP plasmids for 16 h before being lysed in lysis buffer (25 mm HEPES pH 7.5, 1% Nonidet P-40, 100 mm NaCl, 10 mm MgCl2, 5% glycerol, 5 mm Na3VO4, 50 mm NaF, 1 mm phenylmethylsulfonyl fluoride (PMSF), and Complete™ protease inhibitors). Supernatant was cleared by centrifugation (14,000 × g, 5 min), and incubated with 20 μl bead volume of protein-A-Sepharose (GE HealthCare) and 2 μl of anti-GFP antibodies for 2 h at 4 °C. Beads were then washed three times in lysis buffer followed by three washes in incubation buffer and were resuspended in a total volume of 100 μl of incubation buffer. Immmunoprecipitated GFP-tagged proteins were quantified using a Victor X3 2030 Multilabel Reader spectrofluorometer (PerkinElmer) with excitation and emission wavelength respectively of 485 nm and 535 nm. Approximately 0.1–0.5 μg of immunoprecipitated GFP-tagged proteins were used in each experiment. GAP assays were performed as described above.
Immunofluorescence Microscopy
For cell rounding experiments, COS-7 cells grown on glass coverslips were transfected with pEGFPC1-CdGAP expression vectors. After 40 h, cells were washed with PBS and fixed in 3.7% paraformaldehyde for 10 min and quenched in 0.1 m glycine for 5 min. Cells were permeabilized with 0.25% Triton X-100 for 5 min, washed and blocked with 0.2% BSA for 30 min followed by incubation with phalloidin-tetramethylrhodamine isothiocyanate (TRITC) (Sigma) for 15 min in blocking solution. Coverslips were mounted on glass slides using Prolong anti-fade mounting media (Invitrogen). For cell spreading experiments, COS-7 cells were transfected with plasmids for 16 h, trypsinized, and plated on glass coverslips pre-coated with fibronectin (Roche) in serum-free medium. After 1 h at 37 °C, cells were stimulated with 10 ng/ml PDGF for 15 and 60 min before fixation. For CdGAP membrane recruitment, serum-starved transfected COS-7 cells were treated with either DMSO or 50 μm LY294002 for 30 min before stimulation with 10 ng/ml PDGF. Cells were examined with an Olympus IX81 motorized inverted microscope using a 60× U PLAN S-APO oil objective lens. Images were recorded with a CoolSnap 4K (Photometrics) and analyzed with the Metamorph software (Molecular Devices). At least 50 cells per condition were analyzed in each of the three independent experiments.
Live Cell Time-lapse Imaging
Serum-starved transfected COS-7 cells were grown on glass bottom microwell dishes (MatTek) and treated with either DMSO or 50 μm LY294002 for 30 min before stimulation with 10 ng/ml PDGF. Cells were maintained in 5% CO2 and 37 °C environmental control system. Transfected cells were imaged every 20 s over periods of 10–20 min after PDGF stimulation. Z-stacks and time-lapse confocal series (4D) were captured by Zeiss LSM780 confocal microscope (Carl Zeiss, Inc) using a 63×/1.40 oil Plan-Apochromat. GFP was excited with 488 nm argon laser and detected with GaSaP detector. 4D Movie files were created using Image J software (NIH, Bethesda, MD).
CRIB Pull-down Assay
The CRIB domain of mouse PAK3 (amino acids 73–146) fused to glutathione S-transferase (GST-CRIB) was used to isolate GTP-bound Rac1 and was purified as previously described (29). HEK293 cells transfected with pEGFP-CdGAP expression vectors were lysed in buffer B (25 mm Hepes pH 7.5, 1% Nonidet P-40, 10 mm MgCl2, 100 mm NaCl, 5% glycerol, 5 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 50 mm sodium fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Equal amounts of proteins were incubated with glutathione-Sepharose beads coupled to 20 μg of GST-CRIB for 45 min at 4 °C. The beads were then washed twice with washing buffer (25 mm Hepes pH 7.5, 40 mm NaCl, 30 mm MgCl2, 0.5% Nonidet P-40, and 1 mm DTT) before being boiled in SDS sample buffer. The proteins were separated on a 12% SDS-PAGE, transferred to nitrocellulose and GTP-bound GTPases were visualized by immunoblotting usinganti-Rac1 antibody and ECL reagent detection kit (Perkin Elmer). The levels of GTP-bound proteins were assessed by densitometry and normalized to the total amount of GTPases detected in the total cell lysates.
Statistical Analysis
Statistical significance value (p values) was obtained by performing a two-sample unequal-variance Student's t test. Data are representative of at least three independent experiments.
RESULTS
A Polybasic Region (PBR) Preceding the RhoGAP domain Is Conserved within CdGAP Homologues
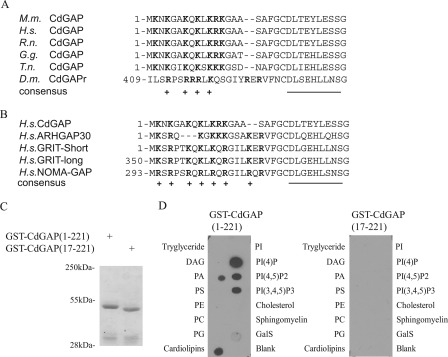
Protein sequence analysis of CdGAP vertebrate orthologues revealed 95% amino acid sequence identity for a cluster of 20 amino acids located at the N terminus of the GAP domain, which has a net charge of +7 (Fig. 1A). A similar region enriched in positively charged amino acids is also present in Drosophila melanogaster CdGAP (Fig. 1A) and in the closest homologues of CdGAP, namely ARHGAP30, GRIT, and NOMA-GAP (Fig. 1B). Binding of phospholipids to short basic sequences has been reported for a number of proteins, including the RhoGAP protein DLC1 (30). The high conservation of basic residues in CdGAP homologues strongly suggests that this PBR may serve a similar function for CdGAP.
FIGURE 1.
A conserved PBR in CdGAP binds to phospholipids. Amino acid sequence alignment of the PBR in CdGAP from different species (A) and closest homologs (B). Positively charged amino acids are highlighted in bold and marked (+), whereas amino acids forming the first α-helix of the GAP domain are underlined. C, Coomassie Blue-stained gel showing recombinant purified glutathione S-transferase (GST)-tagged CdGAP(1–221) and CdGAP(17–221). D, lipid overlay assay of GST fusion proteins showing selective binding of GST-CdGAP(1–221), but not GST-CdGAP(17–221), to nitrocellulose-bound phosphoinositides. M.m., Mus musculus; H.s., Homo Sapiens; R.n., Rattus Norvegicus; G.g., Gallus gallus, T.n., tetraodon nigroviridis, D.m., Drosophila melanogaster. DAG, diacylglycerol; PA, phosphatidic acid; PS, phosphatylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PI(4)P, phosphatidylinositol (4)-phosphate; PI(4,5)P2, phosphatidylinositol (4,5)-bisphosphate; PI(3,4,5)P3, phosphatidylinositol (3,4,5)-trisphosphate; GalS, 3-sulfogalactosylceramide.
CdGAP Interacts with Negatively Charged Phospholipids
To determine whether the PBR is able to bind negatively charged phospholipids, a lipid overlay assay was performed using purified GST fusion proteins of CdGAP encoding the N-terminal GAP domain (amino acids 1–221) or the GAP domain lacking the PBR (amino acids 17–221) (Fig. 1C). CdGAP(1–221) was found to strongly interact with phosphatidylinositol 4-phosphate (PI(4)P), phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2, phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3), and cardiolipins. It also bound to a lower extent to phosphatidic acid (PA) (Fig. 1D). Interestingly, CdGAP(17–221) lacking the PBR was unable to bind to the phospholipids (Fig. 1D). Therefore, the PBR preceding the RhoGAP domain is necessary to mediate in vitro binding to negatively charged phospholipids.
CdGAP Interacts Specifically with PI(3,4,5)P3 Incorporated into Membranes
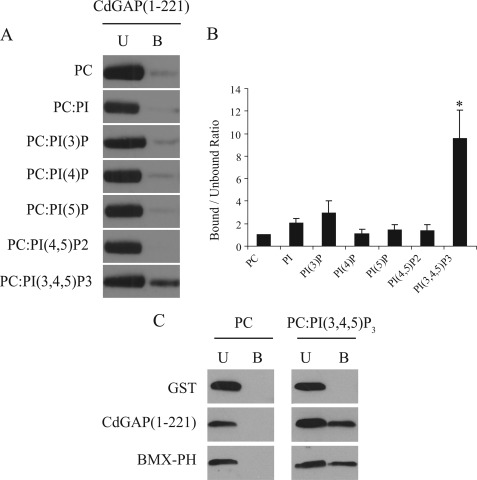
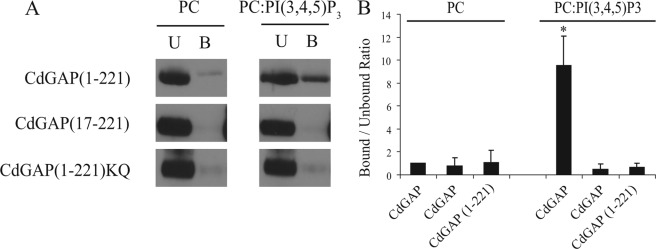
To investigate whether CdGAP interacts with specific phospholipids incorporated into membranes, we generated multilamellar vesicles (MLVs) using phosphatidylcholine (PC) with or without 10mol% of PI, PI(3)P, PI(4)P, PI(5)P, PI(4,5)P2, or PI(3,4,5)P3. In a MLV flotation assay, MLVs were incubated with GST-CdGAP(1–221) and MLV-bound proteins were separated from the unbound proteins by sucrose gradient centrifugation and quantified by Western blotting analysis (Fig. 2, A and B). We found a 9.5-fold increase association between CdGAP(1–221) and PI(3,4,5)P3-containing MLVs compared with the control PC alone (Fig. 2B) whereas the binding of CdGAP(1–221) to other phospholipids was negligible (Fig. 2, A and B). Furthermore, we found that binding of CdGAP(1–221) to PI(3,4,5)P3 was comparable to PI(3,4,5)P3-binding pleckstrin homology (PH) domain of BMX/Etk tyrosine kinase (28, 31) (Fig. 2C). These results show that CdGAP binds specifically to PI(3,4,5)P3-containing MLVs. To determine whether an intact PBR is required for its association with PI(3,4,5)P3, MLVs were incubated with GST-tagged CdGAP(1–221), CdGAP(17–221), or CdGAP(1–221)KQ, in which the seven positively charged residues have been substituted by glutamines. We observed that the removal of the PBR or the selective replacement of the basic residues by glutamines abolished the ability of CdGAP to associate with PI(3,4,5)P3-containing MLVs (Fig. 3, A and B). Together, these results show that an intact PBR is necessary to mediate the interaction of CdGAP with PI(3,4,5)P3-containing MLVs.
FIGURE 2.
CdGAP preferentially associates with phosphatidylinositol (3,4,5)-trisphosphate. A, MLVs composed of phosphatidylcholine or of a mix of 90 mol% phosphatidylcholine (PC) with 10 mol% of PI, phosphatidylinositol (3)-phosphate (PI(3)P), phosphatidylinositol (4)-phosphate (PI(4)P), phosphatidylinositol (5)-phosphate (PI(5)P), phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2), or phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3) were incubated with GST-CdGAP(1–221) in a MLV flotation assay. MLV-associated proteins were separated from unbound proteins by sucrose gradient centrifugation. Unbound and bound proteins were subjected to SDS-PAGE and analyzed by Western blotting with GST-specific antibodies (U, unbound; B, bound). B, quantitative analysis of blots shown in A, representing the bound-unbound ratios. The value obtained for PC MLVs was set to 1 and used as the reference point. Values represent an average of at least three independent experiments for each condition; error bars, S.E. *, p < 0.001 for the comparison to the reference point (PC MLVs). C, MLVs composed of PC or PC:PI(3,4,5)P3 were incubated with GST, GST-CdGAP(1–221), or GST-BMX-PH in MLV flotation assay.
FIGURE 3.
The PBR of CdGAP is required for PI(3,4,5)P3 interaction. A, MLVs composed of the indicated lipids were incubated with GST-CdGAP(1–221), CdGAP(17–221), or CdGAP(1–221)KQ. MLV-associated proteins were separated from unbound proteins by sucrose gradient centrifugation. Unbound and bound proteins were subjected to SDS-PAGE and analyzed by Western blotting with GST-specific antibodies (U, unbound; B, bound). B, quantitative analysis of blots shown in A, representing the bound/unbound ratios. The value obtained for GST-CdGAP(1–221) incubated with PC MLVs was set to 1 and used as the reference point. Values represent an average of at least three independent experiments for each condition; error bars, S.E. *, p < 0.001 for the comparison to the reference point (PC MLVs).
The GAP Activity of CdGAP Is Increased in the Presence of PI(3,4,5)P3-containing MLVs
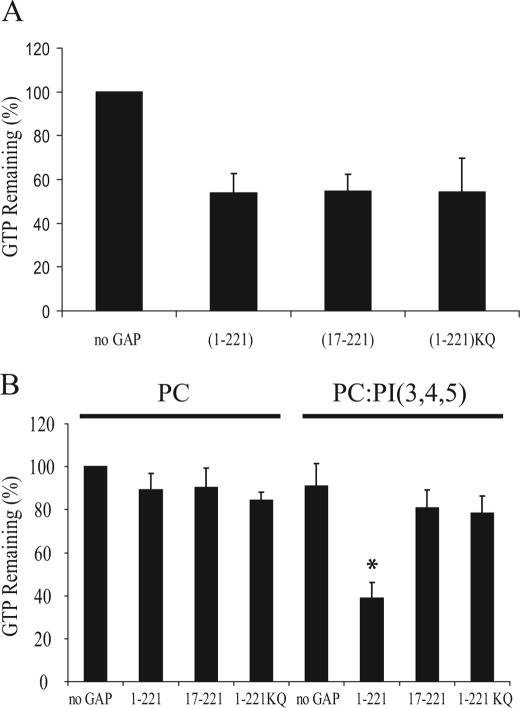
To examine the role of the PBR on CdGAP activity, we first determine the in vitro GAP activity of GST-CdGAP(1–221) and CdGAP protein mutants toward Rac1 in the absence of membranes. GTP-loaded Rac1 was incubated with equal amounts of GST-CdGAP(1–221), CdGAP(17–221), or (1–221)KQ, and as shown in Fig. 4A, the in vitro GAP activity was similar for all three proteins, demonstrating that the PBR does not influence the GAP activity in the absence of lipid binding. To be active, Rho GTPases are prenylated, which targets them to the membranes. Therefore, the in vivo stimulation of the intrinsic GTPase activity by GAPs should occur at the membranes. To assess the importance of PI(3,4,5)P3 interaction on CdGAP activity in the context of a membrane environment, we performed an in vitro GAP assay with geranylgeranylated Rac1 (Rac1GG) in the presence or absence of PI(3,4,5)P3-containing MLVs. Interestingly, we observed a 2.3-fold increase of CdGAP(1–221) activity in the presence of PI(3,4,5)P3-containing MLVs compared with PC alone-containing MLVs (Fig. 4B). The activity of the protein mutants CdGAP(17–221) or (1–221)KQ was similar to CdGAP(1–221) activity in the control PC-containing MLVs, however, in contrast to CdGAP(1–221), their activities were not affected by the presence of PI(3,4,5)P3 (Fig. 4B). Altogether, these results show that PI(3,4,5)P3 binding regulates CdGAP activity in vitro toward Rac1 in the specific context of membrane environment.
FIGURE 4.
The presence of PI(3,4,5)P3 on multilamellar vesicles loaded with prenylated Rac1 stimulates the GAP activity of CdGAP. A, [γ32P]GTP-loaded Rac1 was incubated with equal amounts of GST- CdGAP (1–221), CdGAP (17–221), or CdGAP (1–221)KQ. GTP hydrolysis was measured after 0 and 10 min incubation at 20 °C. The intrinsic Rac1 GTPase activity was set to 100% (no GAP). The graph shows a representative experiment performed in triplicate. Error bars, S.E. B, [γ32P]GTP-loaded geranylgeranylated Rac1 in the presence of MLVs was incubated with equal amounts of GST-CdGAP(1–221), CdGAP(17–221), or CdGAP(1–221)KQ. GTP hydrolysis was measured after 0 and 10 min incubation at 20 °C. The intrinsic Rac1 GTPase activity in the presence of control PC-containing MLVs was set to 100%. The graph shows a representative experiment performed in triplicate. Error bars, S.E. *, p < 0.001 for the comparison to the control (PC-containing MLVs).
The PBR Is Required for CdGAP-mediated Cellular Effects
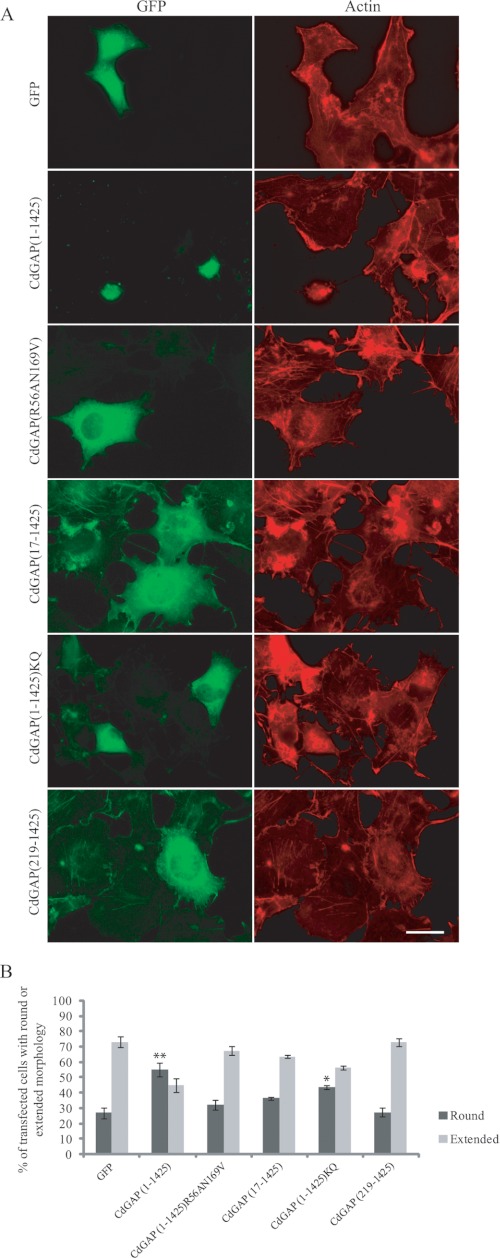
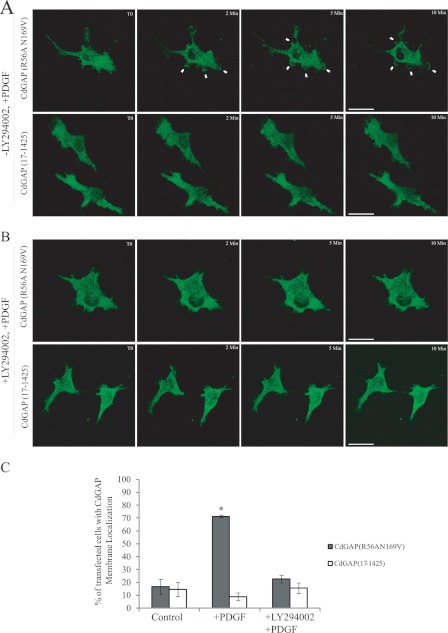
To study the role of the PBR in CdGAP cellular functions, we generated GFP-tagged CdGAP wild-type or mutant proteins. We have previously shown that overexpression of CdGAP in different cell types induces cell rounding in a GAP-dependent manner (6, 19). Thus, GFP-tagged CdGAP protein mutants were expressed in COS-7 cells and the percentage of transfected cells showing a rounded phenotype was determined (Fig. 5, A and B). Overexpression of CdGAP(1–1425) induced a cell rounding phenotype in 55% of transfected cells compared with 25% of control GFP-transfected cells (Fig. 5, A and B). As expected, CdGAP(219–1425) lacking the PBR and GAP domain or GAP-deficient CdGAP(R56AN169V) (19) were unable to induce cell rounding. Similarly, CdGAP(17–1425) lacking the PBR or CdGAP(1–1425)KQ were less efficient to induce cell rounding compared with CdGAP(1–1425). Therefore, the PBR is required for CdGAP to induce cell rounding. We next examined the translocation of CdGAP to plasma membrane in response to phosphatidylinositol 3-kinase (PI-3 kinase) activation. Since the expression of wild-type CdGAP inhibits PDGF-induced lamellipodia (7) and induces cell rounding in the majority of the cells (6, 19, 20), it was not possible to assess membrane translocation of the wild-type protein. Therefore, we examined membrane recruitment of the GAP-deficient CdGAP(R56AN169V) protein in PDGF-treated cells by time-lapse live imaging. CdGAP(R56AN169V) was translocated to the plasma membrane and membrane ruffles in COS-7 cells treated with PDGF to activate PI 3-kinase (Fig. 6, A and C; supplemental Movie S1). This membrane recruitment was blocked in the presence of the PI-3 kinase inhibitor LY294002 (Fig. 6, B and C). CdGAP(17–1425) lacking the PBR was not able to translocate to plasma membrane in cells treated with PDGF (Fig. 6, A and C; supplemental Movie S2). Together, these data show that the PBR is required for CdGAP to localize to the plasma membrane in response to PI-3 kinase activation.
FIGURE 5.
The PBR is required for CdGAP to mediate cell rounding in fibroblast cells. A, COS-7 cells transfected with GFP, GFP-CdGAP(1–1425) or protein mutants were fixed 36 h after transfection and stained for actin with phalloidin-TRITC. Scale bar, 5 μm. B, quantification analysis of A. Cells counted as round had a nucleus covering more than 50% of the cell surface. These values represent an average of three independent experiments, with at least 100 cells counted per condition and per experiment; error bars, S.E. **, p < 0.001; *, p < 0.05 for the comparison to the control (GFP).
FIGURE 6.
PI 3-kinase activation causes CdGAP plasma membrane recruitment. A and B, time-lapse series of COS-7 cells expressing GFP-CdGAP(R56AN169V) or GFP-CdGAP(17–1425). Serum-starved transfected COS-7 cells were treated with either DMSO or 50 μm LY294002 for 30 min before stimulation with 10 ng/ml PDGF. Scale bar, 5 μm. White arrowheads show membrane ruffles. C, quantification analysis of fixed cells, these values represent an average of three independent experiments, with at least 50 cells counted per condition and per experiment; error bars, S.E. *, p < 0.01 for the comparison to the control.
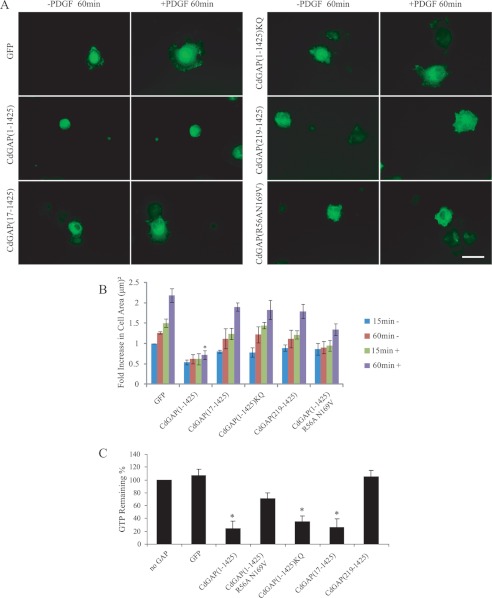
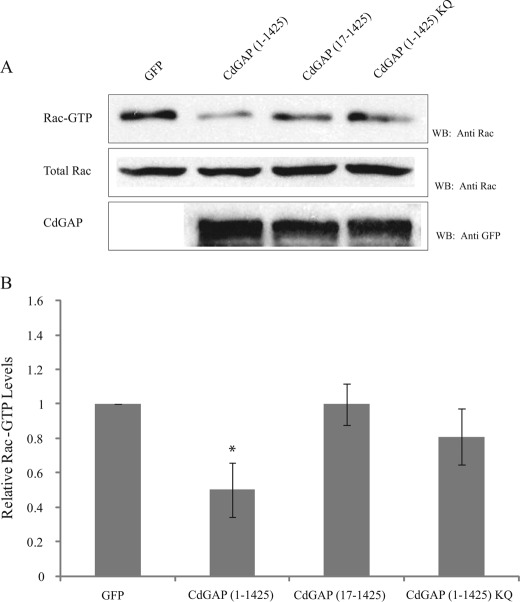
CdGAP-expressing cells have previously been shown to spread on extracellular matrix at a lower rate than control cells (19). Furthermore, the GAP activity of CdGAP is regulated in an adhesion-dependent manner (19). Thus, we assessed the role of CdGAP protein mutants in cell spreading experiments by quantifying the ability of CdGAP-expressing cells to adhere and spread on fibronectin in response to PDGF stimulation that activate PI-3 kinase. CdGAP(1–1425)-expressing cells treated with PDGF were able to attach to the matrix but were inefficient at spreading compared with the GFP-expressing control cells (Fig. 7, A and B). CdGAP(219–1425) lacking the PBR and GAP domain was able to induce cell spreading on fibronectin, with a 2.5-fold increase in cell area compared with CdGAP(1–1425)-expressing cells after 60 min of PDGF stimulation (Fig. 7, A and B). GAP-deficient CdGAP(R56AN169V) or the PBR-defective mutant proteins were more efficient than CdGAP(1–1425) to induce cell spreading, with a 2–2.7-fold increase in cell area after a 60-min PDGF stimulation (Fig. 7, A and B). To assess whether these effects were directly linked to the intrinsic GTPase-stimulating activity of these proteins or could be mediated by the proper localization of CdGAP proteins to membranes, GFP-tagged CdGAP proteins expressed in COS-7 cells were immunoprecipitated, and equal amounts of CdGAP proteins were used in an in vitro GAP assay using recombinant Rac1 as a substrate (Fig. 7C). CdGAP(1–1425) and the PBR-defective CdGAP protein mutants showed a strong in vitro GAP activity toward Rac1. These results correlate well with the in vitro GAP assays using CdGAP(17–221) or (1–221)KQ toward Rac1 in the absence of membranes (Fig. 4A). However, when we examined the in vivo GAP activity of PBR-deficient CdGAP proteins toward endogenous Rac1 in HEK293 cells, we found a significant decrease in Rac1-bound GTP levels in the presence of CdGAP(1–1425) expression but not with the PBR-deficient CdGAP proteins (Fig. 8). Taken together, these data demonstrate that the PBR is not required for the intrinsic GTPase-stimulating activity of CdGAP, but is necessary for the membrane recruitment of CdGAP to mediate its GAP-dependent cellular effects.
FIGURE 7.
The PBR is required for CdGAP to mediate its effects on cell spreading in response to PDGF stimulation. A, COS-7 cells transfected with GFP, GFP-CdGAP(1–1425) or protein mutants were serum-starved overnight trypsinized, and resuspended 16 h post-transfection. Cells plated on fibronectin-coated coverslips were stimulated with PDGF for 60 min before fixation. Scale bar, 5 μm. B, images were analyzed using Metamorph, and the area covered by each cell was measured throughout the experiment, these values represent an average of three independent experiments, with at least 50 cells counted per condition and per experiment error bars, S.E. *, p < 0.01 for the comparison to control. C, GFP-tagged CdGAP proteins isolated by immunoprecipitation from COS-7 cell protein lysates using an anti-GFP antibody were used in an in vitro GAP assay with [γ-32P]GTP-loaded Rac1. GTP hydrolysis was measured after 0 and 10 min incubation at 20 °C. The intrinsic Rac1 GTPase activity was set to 100% (no GAP). The graph shows a representative experiment performed in triplicate; error bars, S.E. *, p < 0.01 for the comparison to the control.
FIGURE 8.
The PBR is required for the in vivo GAP activity of CdGAP. A, protein lysates from HEK293 cells expressing GFP, GFP-CdGAP (1–1425) or protein mutants were incubated with GST-CRIB coupled to glutathione-agarose beads. GTP-bound Rac1 was detected by Western blotting using anti-Rac1 antibody. Total cell lysates were probed for Rac1 expression and GFP-tagged CdGAP proteins. B, quantification of the relative Rac1-GTP levels was determined by densitometry. Values represent the average of at least four independent experiments; error bars, S.E. *, p < 0.001 for the comparison to the control.
DISCUSSION
In this study, we demonstrate that a polybasic region preceding the GAP domain at the N terminus of CdGAP is required to mediate specific binding to the negatively charged phospholipids PI(3,4,5)P3. This lipid interaction is essential for the in vivo GAP activity and the cellular functions of CdGAP. The fact that Rho GTPases are anchored into membranes to exert their cellular functions suggests the need for effector and regulatory proteins to be also targeted at membranes, in close proximity to their specific targets. Indeed, many effector proteins, including WASP, PAKs, and Cdc42-effector proteins (CEP) comprise a region enriched in basic residues preceding their Cdc42/Rac-interactive domain (CRIB) and involved in lipid binding (32, 33). Each RhoGEF of the Dbl family of proteins includes a PH domain, which associates with low affinity to phosphatidylinositol groups (34). Some RhoGAP proteins also contain putative lipid-binding domains such as PH, C1, C2, Sec14, PX, and BAR domains although lipid interaction remains to be demonstrated for most of these proteins (5). The PH domains of the BMX/Etk (28, 31) and AKT protein kinases (35, 36) interact with high affinity with PI(3,4,5)P3 and indeed, we also found comparable interactions between PI(3,4,5)P3 binding to CdGAP(1–221) or to the PH domain of BMX.
Two previous studies on p190RhoGAP and DLC1 have reported lipid interaction through a PBR, which appeared to be essential for their GAP activity and cellular functions (30, 37, 38). In each of them, the PBR is preceding the GAP domain, at a site that fits with a model in which the GAP domain could simultaneously bind a GTPase and associate with the membrane through the PBR (39). The specificity and activity of p190RhoGAP have previously been shown to be modulated in vitro by the presence of phosphatidylserine (PS), PI, and PI(4,5)P2 (38). Although the PBR in p190RhoGAP was shown to be required for p190RhoGAP association with PS only, there is a possibility that it could also serve as a PI and/or PI(4,5)P2-binding site (37). In the case of DLC1, lipid interaction with the PBR was addressed in an experiment using immobilized lipids and demonstrated a strong preference for PI(4,5)P2 (30). For CdGAP, a similar experiment using a lipid overlay assay showed a strong association of the PBR with PI(4)P, PI(4,5)P2, PI(3,4,5)P3, and cardiolipins. However, when these lipids were inserted into vesicular membranes, only PI(3,4,5)P3 specifically bound to CdGAP. To our knowledge, this is the first example of a RhoGAP binding specifically to PI(3,4,5)P3 through a polybasic amino acid cluster.
The relative lipid binding specificity of the PBR from CdGAP, DLC1, and p190RhoGAP proteins cannot be strictly explained by electrostatic forces between the positively charged PBR and association with the negatively charged head group of the lipid. The fact that PI(3,4,5)P3, with a net charge of −3, is not binding to DLC1 with more affinity than PI(4,5)P2 and that CdGAP does not show more affinity toward PI(4,5)P2 than it does for PI(3)P, PI(4)P, or PI(5)P suggest that the PBRs adopt a three-dimensional structure required for lipid binding specificity. The conserved hydrophobic residues in the PBR have been proposed to induce the formation of an amphipathic α-helix upon membrane association, as in ARF small GTPases and ARFGAPs (40, 41). The relative conservation of leucine and phenylalanine residues at positions 10 and 19 in CdGAP suggests that these residues could behave in a similar way, forcing the positioning of positively charged side chains within the PBR after their insertion into a membrane. Alternatively, the nearby globular GAP domain could stabilize the PBR in a conformation allowing particular positioning of its positively charged side chains.
To date, most of the GAP assays were performed in an in vitro context that did not take into account the membrane environment. Although their composition does not completely reflect the in vivo nature of cellular membranes, PC-constituted MLVs used with prenylated Rho GTPases represent a good alternative to study the effect of different lipids on the activity of GTPases in the presence of their regulators. Similar to DLC1 activity regulated by PIP(4,5)P2, the GAP activity of CdGAP was activated in the presence of PIP(3,4,5)P3 on MLVs. Furthermore, the PBR binding to PI(3,4,5)P3 is required for CdGAP to be translocated at the plasma membrane in a PI-3 kinase-dependent manner and to mediate its GAP-dependent cellular functions. Cells expressing CdGAP mutant proteins lacking the PBR or containing a PBR with lysine-to-glutamine substitutions produced a reduced number of rounded cells, which correlates well with the altered in vivo GAP activity of the protein mutants. In addition, the expression of CdGAP(17–1425) or CdGAP(1–1425)KQ mutant proteins increased the ability of COS-7 cells to spread on fibronectin similar to the GAP-inactivated mutant protein. Taken together, these results show the importance of an intact PBR for CdGAP cellular functions. Considering that the intrinsic GTPase-stimulating activity of CdGAP is not altered by the removal of the PBR or the substitution of basic residues to glutamine, we suggest that binding to PI(3,4,5)P3 is responsible to recruit CdGAP to the membranes where it can interact with GTPases and exert its regulatory function.
The specificity of CdGAP for PI(3,4,5)P3 suggests that its GAP activity may be restricted at sites of PI(3,4,5)P3 production. Cellular abundance of PI(3,4,5)P3 is estimated to be under 0.15% of all PI products whereas PI(4)P and PI(4,5)P2 together represent around 5% of these lipids (33). The low concentration of PI(3,4,5)P3 suggests that it may be the clutch controlling CdGAP activity at specific cellular localization. CdGAP has been shown to bind to actopaxin, which promotes its localization to focal adhesion complexes and its activation following integrin engagement (19). Considering that integrins activate PI3K to produce PI(3,4,5)P3, it is tempting to propose that integrin activation of CdGAP could be partly mediated by local production of PI(3,4,5)P3.
Analysis of the amino acid sequences preceding the GAP domain of mammalian RhoGAPs suggests a high prevalence of PBRs in RhoGAPs. Alternatively, RhoGAP proteins lacking a PBR are found to contain known lipid binding motifs such as PH, C1, C2, or sec14 domains, preceding the GAP domain (5). This suggests the interesting possibility that, similar to CdGAP, DLC1, and p190RhoGAP, PBRs may serve a similar function in these RhoGAP proteins. Therefore, the observation that a PBR preceding the GAP domain is found in several RhoGAP proteins suggests that a PBR-GAP domain may be seen as an evolutionary conserved mechanism of regulation of RhoGAP proteins.
Supplementary Material
Acknowledgments
We thank the MUHC-RI Imaging facility and Drs. Stéphane Laporte and Min Fu for expert help with time-lapse live cell imaging experiments. We would also like to thank Vilayphone Luangrath for technical assistance.
This work was supported by the Canadian Institutes for Health Research Grant MOP-84449 and the Canada Foundation for Innovation-Leaders Opportunity Fund.

This article contains supplemental Movies S1 and S2.
- GAP
- GTPase-activating proteins
- CdGAP
- Cdc42 GTPase-activating protein
- C1
- phorbol esters/diacylglycerol-binding domain
- C2
- Ca2+-dependent lipid binding
- MLVs
- multilamellar vesicles
- PBR
- polybasic region
- PH
- Pleckstrin homology
- PI
- phosphatidylinositol
- PI3K
- phosphatidylinositol 3-kinase
- PRD
- proline-rich domain
- PX
- Phox domain
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Heasman S. J., Ridley A. J. (2008) Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701 [DOI] [PubMed] [Google Scholar]
- 2. Jaffe A. B., Hall A. (2005) Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 3. Wennerberg K., Der C. J. (2004) Rho-family GTPases: it's not only Rac and Rho (and I like it). J. Cell Sci. 117, 1301–1312 [DOI] [PubMed] [Google Scholar]
- 4. Bernards A., Settleman J. (2004) GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14, 377–385 [DOI] [PubMed] [Google Scholar]
- 5. Tcherkezian J., Lamarche-Vane N. (2007) Current knowledge of the large RhoGAP family of proteins. Biol. Cell 99, 67–86 [DOI] [PubMed] [Google Scholar]
- 6. Tcherkezian J., Triki I., Stenne R., Danek E. I., Lamarche-Vane N. (2006) The human orthologue of CdGAP is a phosphoprotein and a GTPase-activating protein for Cdc42 and Rac1 but not RhoA. Biol. Cell 98, 445–456 [DOI] [PubMed] [Google Scholar]
- 7. Lamarche-Vane N., Hall A. (1998) CdGAP, a novel proline-rich GTPase-activating protein for Cdc42 and Rac. J. Biol. Chem. 273, 29172–29177 [DOI] [PubMed] [Google Scholar]
- 8. Tcherkezian J., Danek E. I., Jenna S., Triki I., Lamarche-Vane N. (2005) Extracellular signal-regulated kinase 1 interacts with and phosphorylates CdGAP at an important regulatory site. Mol. Cell. Biol. 25, 6314–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakazawa T., Watabe A. M., Tezuka T., Yoshida Y., Yokoyama K., Umemori H., Inoue A., Okabe S., Manabe T., Yamamoto T. (2003) p250GAP, a novel brain-enriched GTPase-activating protein for Rho family GTPases, is involved in the N-methyl-d-aspartate receptor signaling. Mol. Biol. Cell 14, 2921–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao C., Ma H., Bossy-Wetzel E., Lipton S. A., Zhang Z., Feng G. S. (2003) GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J. Biol. Chem. 278, 34641–34653 [DOI] [PubMed] [Google Scholar]
- 11. Moon S. Y., Zang H., Zheng Y. (2003) Characterization of a brain-specific Rho GTPase-activating protein, p200RhoGAP. J. Biol. Chem. 278, 4151–4159 [DOI] [PubMed] [Google Scholar]
- 12. Okabe T., Nakamura T., Nishimura Y. N., Kohu K., Ohwada S., Morishita Y., Akiyama T. (2003) RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the β-catenin-N-cadherin and N-methyl-d-aspartate receptor signaling. J. Biol. Chem. 278, 9920–9927 [DOI] [PubMed] [Google Scholar]
- 13. Nakamura T., Hayashi T., Nasu-Nishimura Y., Sakaue F., Morishita Y., Okabe T., Ohwada S., Matsuura K., Akiyama T. (2008) PX-RICS mediates ER-to-Golgi transport of the N-cadherin/β-catenin complex. Genes Dev. 22, 1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rosário M., Franke R., Bednarski C., Birchmeier W. (2007) The neurite outgrowth multiadaptor RhoGAP, NOMA-GAP, regulates neurite extension through SHP2 and Cdc42. J. Cell Biol. 178, 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chiang S. H., Hwang J., Legendre M., Zhang M., Kimura A., Saltiel A. R. (2003) TCGAP, a multidomain Rho GTPase-activating protein involved in insulin-stimulated glucose transport. EMBO J. 22, 2679–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Primeau M., Ben Djoudi Ouadda A., Lamarche-Vane N. (2011) Cdc42 GTPase-activating protein (CdGAP) interacts with the SH3D domain of Intersectin through a novel basic-rich motif. FEBS Lett. 585, 847–853 [DOI] [PubMed] [Google Scholar]
- 17. Naji L., Pacholsky D., Aspenström P. (2011) ARHGAP30 is a Wrch-1-interacting protein involved in actin dynamics and cell adhesion. Biochem. Biophys. Res. Commun. 409, 96–102 [DOI] [PubMed] [Google Scholar]
- 18. Jenna S., Hussain N. K., Danek E. I., Triki I., Wasiak S., McPherson P. S., Lamarche-Vane N. (2002) The activity of the GTPase-activating protein CdGAP is regulated by the endocytic protein intersectin. J. Biol. Chem. 277, 6366–6373 [DOI] [PubMed] [Google Scholar]
- 19. LaLonde D. P., Grubinger M., Lamarche-Vane N., Turner C. E. (2006) CdGAP associates with actopaxin to regulate integrin-dependent changes in cell morphology and motility. Curr. Biol. 16, 1375–1385 [DOI] [PubMed] [Google Scholar]
- 20. Southgate L., Machado R. D., Snape K. M., Primeau M., Dafou D., Ruddy D. M., Branney P. A., Fisher M., Lee G. J., Simpson M. A., He Y., Bradshaw T. Y., Blaumeiser B., Winship W. S., Reardon W., Maher E. R., FitzPatrick D. R., Wuyts W., Zenker M., Lamarche-Vane N., Trembath R. C. (2011) Gain-of-function mutations of ARHGAP31, a Cdc42/Rac1 GTPase regulator, cause syndromic cutis aplasia and limb anomalies. Am. J. Hum. Genet. 88, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He Y., Northey J. J., Primeau M., Machado R. D., Trembath R., Siegel P. M., Lamarche-Vane N. (2011) CdGAP is required for transforming growth factor β- and Neu/ErbB-2-induced breast cancer cell motility and invasion. Oncogene 30, 1032–1045 [DOI] [PubMed] [Google Scholar]
- 22. Danek E. I., Tcherkezian J., Triki I., Meriane M., Lamarche-Vane N. (2007) Glycogen synthase kinase-3 phosphorylates CdGAP at a consensus ERK 1 regulatory site. J. Biol. Chem. 282, 3624–3631 [DOI] [PubMed] [Google Scholar]
- 23. Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fivaz M., Meyer T. (2003) Specific localization and timing in neuronal signal transduction mediated by protein-lipid interactions. Neuron 40, 319–330 [DOI] [PubMed] [Google Scholar]
- 25. Wheeler A. P., Ridley A. J. (2004) Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp. Cell Res. 301, 43–49 [DOI] [PubMed] [Google Scholar]
- 26. Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. (2002) A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lamarche N., Tapon N., Stowers L., Burbelo P. D., Aspenström P., Bridges T., Chant J., Hall A. (1996) Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell 87, 519–529 [DOI] [PubMed] [Google Scholar]
- 28. Côté J. F., Motoyama A. B., Bush J. A., Vuori K. (2005) A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signaling. Nat. Cell Biol. 7, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li X., Saint-Cyr-Proulx E., Aktories K., Lamarche-Vane N. (2002) Rac1 and Cdc42 but not RhoA or Rho kinase activities are required for neurite outgrowth induced by the Netrin-1 receptor DCC (deleted in colorectal cancer) in N1E-115 neuroblastoma cells. J. Biol. Chem. 277, 15207–15214 [DOI] [PubMed] [Google Scholar]
- 30. Erlmann P., Schmid S., Horenkamp F. A., Geyer M., Pomorski T. G., Olayioye M. A. (2009) DLC1 activation requires lipid interaction through a polybasic region preceding the RhoGAP domain. Mol. Biol. Cell 20, 4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Várnai P., Rother K. I., Balla T. (1999) Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton's tyrosine kinase pleckstrin homology domain visualized in single living cells. J. Biol. Chem. 274, 10983–10989 [DOI] [PubMed] [Google Scholar]
- 32. Knaus U. G., Wang Y., Reilly A. M., Warnock D., Jackson J. H. (1998) Structural requirements for PAK activation by Rac GTPases. J. Biol. Chem. 273, 21512–21518 [DOI] [PubMed] [Google Scholar]
- 33. Papayannopoulos V., Co C., Prehoda K. E., Snapper S., Taunton J., Lim W. A. (2005) A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol. Cell 17, 181–191 [DOI] [PubMed] [Google Scholar]
- 34. Zheng Y. (2001) Dbl family guanine nucleotide exchange factors. Trends Biochem. Sci. 26, 724–732 [DOI] [PubMed] [Google Scholar]
- 35. Andjelkovi M., Alessi D. R., Meier R., Fernandez A., Lamb N. J., Frech M., Cron P., Cohen P., Lucocq J. M., Hemmings B. A. (1997) Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 272, 31515–31524 [DOI] [PubMed] [Google Scholar]
- 36. Currie R. A., Walker K. S., Gray A., Deak M., Casamayor A., Downes C. P., Cohen P., Alessi D. R., Lucocq J. (1999) Role of phosphatidylinositol 3,4,5-trisphosphate in regulating the activity and localization of 3-phosphoinositide-dependent protein kinase-1. Biochem. J. 337, 575–583 [PMC free article] [PubMed] [Google Scholar]
- 37. Lévay M., Settleman J., Ligeti E. (2009) Regulation of the substrate preference of p190RhoGAP by protein kinase C-mediated phosphorylation of a phospholipid binding site. Biochemistry 48, 8615–8623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ligeti E., Dagher M. C., Hernandez S. E., Koleske A. J., Settleman J. (2004) Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J. Biol. Chem. 279, 5055–5058 [DOI] [PubMed] [Google Scholar]
- 39. Ligeti E., Settleman J. (2006) Regulation of RhoGAP specificity by phospholipids and prenylation. Methods Enzymol. 406, 104–117 [DOI] [PubMed] [Google Scholar]
- 40. Bigay J., Casella J. F., Drin G., Mesmin B., Antonny B. (2005) ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 24, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kliouchnikov L., Bigay J., Mesmin B., Parnis A., Rawet M., Goldfeder N., Antonny B., Cassel D. (2009) Discrete determinants in ArfGAP2/3 conferring Golgi localization and regulation by the COPI coat. Mol. Biol. Cell 20, 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.