Abstract
Clonal mosaicism for large chromosomal anomalies (duplications, deletions and uniparental disomy) was detected using SNP microarray data from over 50,000 subjects recruited for genome-wide association studies. This detection method requires a relatively high frequency of cells (>5–10%) with the same abnormal karyotype (presumably of clonal origin) in the presence of normal cells. The frequency of detectable clonal mosaicism in peripheral blood is low (<0.5%) from birth until 50 years of age, after which it rises rapidly to 2–3% in the elderly. Many of the mosaic anomalies are characteristic of those found in hematological cancers and identify common deleted regions that pinpoint the locations of genes previously associated with hematological cancers. Although only 3% of subjects with detectable clonal mosaicism had any record of hematological cancer prior to DNA sampling, those without a prior diagnosis have an estimated 10-fold higher risk of a subsequent hematological cancer (95% confidence interval = 6–18).
INTRODUCTION
Chromosomal mosaicism is the presence of different karyotypes in two or more cell lineages within an individual derived from a single zygote1,2. This karyotypic variation may arise early in development and involve both the soma and the germline or it may occur later and be restricted to one or more specific cell types. In cancer, chromosomal anomalies can initiate a neoplastic clone or arise during clonal evolution and serve as clonal markers3. Here we consider such clonal variation as a form of mosaicism, since the cancer cells may have acquired one or more chromosomal abnormalities, while other cells in the same tissue, or elsewhere in the body, retain the normal karyotype. Chromosomal mosaicism in humans has been well studied in embryos4,5, fetuses from spontaneous abortions6, children with birth defects or developmental delay7,8 and cancer patients9. However, little is known about the type, frequency and age distribution of acquired chromosomal anomalies in large samples from the general population9,10.
Data from genome-wide association studies now provide an opportunity to detect chromosomal variation in tens of thousands of people of all ages and to investigate the association of mosaicism with disease. Single nucleotide polymorphism (SNP) microarray data are used routinely to detect chromosomal anomalies (copy number variants (CNV) and uniparental disomy (UPD)) in clinical cytogenetic laboratories11,12 and to detect small CNVs in population studies13–15. However, the analytical methods used in population studies are not optimized for detecting large anomalies or mosaicism. Therefore, we developed an efficient method to identify and localize large (50 kb to whole-chromosome) anomalies and mosaicism within a single DNA sample. This method requires a relatively high frequency of cells (>5–10%) with the same abnormal karyotype (presumably of clonal origin) in the presence of normal cells. Therefore, we use the term ‘detectable clonal mosaicism’, rather than simply ‘chromosomal mosaicism’, to emphasize the observation of clones of cells with abnormal karyotype that occur at a frequency sufficient for detection using SNP microarray data.
DNA samples (primarily from peripheral blood) from over 50,000 people genotyped for the Gene-Environment Association Studies (GENEVA) consortium16 were analyzed to detect clonal mosaicism. The GENEVA studies include all ages from birth to old age, several major ethnic groups, and a variety of different health conditions, including healthy controls (Table 1, Supplementary Table 1, Supplementary Fig. 1). Here we characterize the types of chromosomal anomalies detected, show how the prevalence of detectable clonal mosaicism within blood cells increases with age, and examine the association between mosaic anomalies and hematological cancer.
Table 1.
Summary of GENEVA study characteristics.
| GENEVA study | Illumina arraya | Relatedness | Design | Ethnicityb | Mean Agec | % Male | Nd |
|---|---|---|---|---|---|---|---|
| Melanoma | Omni1M | mostly unrelatede | case-control | Eur | 52 | 58 | 2,947 |
| Lung Health | 660W | mostly unrelated | cohort | Eur | 54 | 63 | 4,087 |
| Cleft Lip/Palate | 610 | trios | case-parent trio | Eur & Asian | 33 | 52 | 6,860 |
| Addiction | 1M | mostly unrelated | case-control | Eur & AA | 39 | 46 | 2,790 |
| Lung Cancer | 550 | mostly unrelated | case-control | Eur | 66 | 72 | 5,518 |
| Blood clotting | Omni1M | sib pairs | population sample | Eur | 21 | 38 | 1,158 |
| Prostate cancer Japanese/Latino | 660W | mostly unrelated | case-control | Asian+ Hisp | 71 | 100 | 4,281 |
| Prostate cancer African-American | 1M | mostly unrelated | case-control | AA | 69 | 100 | 4,338 |
| Venous Thrombo-embolism | 660W | mostly unrelated | case-control | Eur | 55 | 49 | 2,591 |
| Birth weight Afro-Caribbean | 1M | mother-offspring duos | population sample | AC | 25 | 25 | 2,254 |
| Birth weight European | 610 | mother-offspring duos | population sample | Eur | 31 | 25 | 2,712 |
| Birth weight Hispanic | 1M | mother-offspring duos | population sample | Hisp | 29 | 21 | 1,419 |
| Dental caries | 610 | Families and singletons | population sample | Eur | 36 | 45 | 3,841 |
| Prematurity | 660W | mother-offspring duos | case-control | Eur | 30 | 26 | 3,725 |
| Glaucoma | 660W | mostly unrelated | case-control | Eur | 67 | 42 | 1,977 |
See Supplementary Table 1 for a full description of the array type.
Predominant ethnicity; most studies have small numbers of other ethnicities. Eur=European-ancestry, AA=African-American, Hisp=Hispanic, AC=Afro-Caribbean.
Mean age of participants older than 15 years. The Cleft, Birth Weight, Dental Caries and prematurity studies also have substantial numbers of infants and children less than 15 years old.
Total number of subjects with genotyped samples analyzed in this study.
“Unrelated” here means less relatedness than second degree relatives, based on estimation of identity-by-descent coefficients
RESULTS
Types of anomalies detected
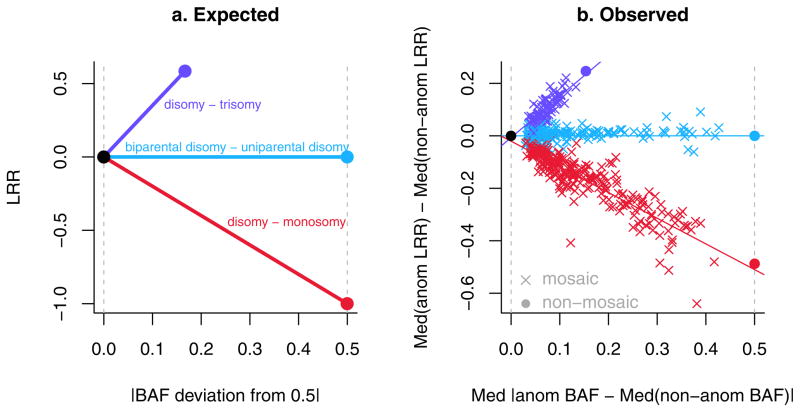
This report deals with autosomal anomalies, defined here as deviations from the normal biparental disomic state. Anomalies were detected using log R ratio (LRR) and B Allele Frequency (BAF)17. LRR is a measure of relative signal intensity (log2 of the ratio of observed to expected intensity, where the expectation is based on other samples). BAF is an estimate of the frequency of the B allele of a given SNP in the population of cells from which the DNA was extracted. In a normal cell, the B allele frequency at any locus is either 0 (AA), ½ (AB) or 1 (BB) and the expected LRR is 0. Both copy number changes and copy-neutral changes from biparental to uniparental disomy (UPD) result in changes in BAF, while copy number changes also affect LRR (Figures 1 and 2). Our detection method identifies both non-mosaic (constitutional) and clonal mosaic anomalies, which were distinguished subsequently using standards based on parent-offspring transmission in family studies and polymorphic CNVs in non-family studies. Three types of clonal mosaics were detected: mixtures of disomic and monosomic cells (deletions), mixtures of disomic and trisomic cells (duplications), and copy-neutral mixtures of biparental and acquired uniparental disomy (aUPD) (see examples in Figure 3 and Supplementary Figure 2). The aUPDs are primarily terminal segments, as expected for an origin through mitotic crossing over (Supplementary Fig 3), while some cases of whole-chromosome aUPD may be due to aneuploidy rescue (Supplementary Fig. 4).
Figure 1.
Expected values of B Allele Frequency (BAF) and Log R Ratio (LRR) for discrete copy number states. Mosaics have intermediate positions between these discrete states. Copy number is given above the horizontal lines in the LRR plot, while SNP genotypes are given in the BAF plot. M=maternal and P=paternal chromosome. States with M and P reversed are also possible. The scatter of points for copy number = 0 (homozygous deletion) represents background signal noise.
Figure 2.
Expected and observed values of B Allele Frequency (BAF) and Log R Ratio (LRR) metrics for clonal mosaic anomalies detected in GENEVA subjects. (a) Expected. (b) Observed (N=514). In (b), “med”=median, “anom”=within the anomaly, “nonanom”=non-anomalous autosomal regions of the same sample. The solid purple and red circles represent the mean values of non-mosaic anomalies and the solid black and cyan circles are theoretical.
Figure 3.
B Allele Frequency (BAF) and Log R Ratio (LRR) plots of four representative mosaic anomalies. Each pair of plots is for a different sample-chromosome combination and each point is a single SNP. Points in BAF plots are color-coded by genotype (red=AA, cyan=AB, purple-blue=BB, black=missing call). The vertical black lines indicate the breakpoint(s) of the anomaly. The vertical gray rectangle is the centromeric gap. Horizontal pink lines are drawn at 0, 1/3, 1/2, 2/3 and 1 in the BAF plots. The solid horizontal red line in each plot is the median value for non-anomalous regions of the autosomes. The horizontal dashed red line is the median value within the anomaly. (a) Mosaic acquired uniparental disomy for distal 12q is indicated by the split in the intermediate BAF band along with the lack of change in LRR. A non-mosaic uniparental isodisomy would have only two BAF bands (at 0 and 1). (b) Mosaic trisomy for chromosome 19 is indicated by a narrow split in the intermediate BAF band along with a small elevation of LRR. A non-mosaic trisomy would have a wider BAF split (at 1/3 and 2/3) and a larger elevation of LRR. (c) A mosaic deletion on 20q is indicated by a narrow split in the intermediate BAF band along with a small decrease in LRR. A non-mosaic heterozygous deletion would have no intermediate BAF bands and a larger decrease in LRR. (d) A mosaic deletion on 6q is indicated by a wide split in the intermediate BAF band along with a large decrease in LRR. The mosaic deletion in (d) has a greater proportion of cells containing the deletion than the one in (c). See Supplementary Figure 2 for additional examples.
Using a method optimized to detect large anomalies (50 kb to whole chromosome), we identified at least one non-mosaic anomaly (i.e. large CNV) in 75% of all subjects, at least one clonal mosaic anomaly in 0.80%, and both types in 0.69%. The median size of all anomalies detected is 150 kb (Supplementary Fig. 5) and the mean number per subject is 1.5, with a range of 0 to 13. There were 514 mosaic anomalies in 404 of 50,222 subjects analyzed.
The reproducibility (in 568 duplicate sample pairs) of all anomalies analyzed for mosaic status is 82% (with >80% overlap; see Methods and Supplementary Table 2 for details). For clonal mosaic anomalies in duplicate samples, the reproducibility is 15/22 = 68% and all discordant calls appear to be false negatives, based on examination of BAF/LRR plots. We also assessed the reproducibility of clonal mosaic anomaly calls in comparison with the results of Jacobs et al.18, who analyzed the same raw data for 5,510 subjects from the GENEVA Lung Cancer study. While both methods detected 83 mosaics, the GENEVA method described here detected an additional 28 mosaics (8 > 2 Mb) and the Jacobs method detected an additional 20 mosaics (all > 2 Mb). The overall reproducibility is 63% or, when considering only anomalies greater than 2 Mb (the size-detection limit of the Jacobs method), 75%. Both estimates are considerably greater than the 25–50% reproducibility across methods estimated for several common CNV-calling algorithms19. All of the discordant mosaic detections appear to be due to false negatives. The Jacobs method is more conservative with respect to size threshold (2 Mb), while our method is more conservative with respect to sample quality (but calling mosaics involving segments less than 2 Mb when sample quality is sufficient). Therefore, the false negative rate of both methods appears to be high and the prevalence of clonal mosaic anomalies detected here is likely to be underestimated. Mosaic detection is difficult when the fraction of abnormal cells is extreme, when the anomaly length is small or when sample quality is low (i.e. high BAF/LRR variability).
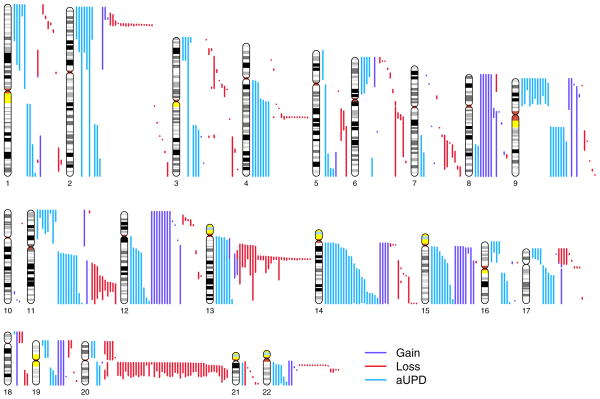
The clonal mosaic anomalies detected in GENEVA subjects were classified as 15.6% duplications, 50.4% deletions and 34.0% aUPDs. All three classes of mosaic anomalies are large (Figure 4 and Supplementary Fig. 6). Median lengths are 34.1 Mb for duplications, 3.8 Mb for deletions and 39.8 Mb for aUPD. Mosaic aneuploidies include +8, +9, +12, +14, +15, +18, +19, −21, and +22, while whole-chromosome mosaic UPDs include chromosomes 2, 3, 13, 14, and 15. Plots of the breakpoints of all mosaic anomalies are provided in Supplementary Figure 7 and genomic coordinates (along with other information) are provided in Supplementary Table 3.
Figure 4.
The lengths and chromosomal positions of the 514 clonal mosaic anomalies detected in GENEVA subjects. An ideogram of each autosome is shown with scaled and color-coded representations of each mosaic anomaly to the right.
There is a highly significant excess of subjects with multiple clonal mosaics, compared to the Poisson distribution expected if the anomalies occurred independently. The multiples are of two kinds: (a) ‘compound’ sets of anomalies adjacent to one another on a single chromosome, suggesting a single event or related mechanism of origin (e.g. Supplementary Figure 2g) and (b) non-adjacent sets. Among the 404 mosaic subjects, 64 had multiple mosaics of one or both types (while 2.6 were expected) and 55 had only non-adjacent sets (2.4 expected). The excess of multiple mosaics occurs for both CNVs and aUPD. The age of subjects with multiple anomalies is not significantly different than those with a single anomaly (p=0.99).
The frequency of detectable clonal mosaicism increases with age
The observed frequency of subjects with one or more clonal mosaic anomalies detected (‘mosaic status’) is shown in Figure 5 and Supplementary Table 4. It is low (< 0.5%) in subjects less than 50 years old, but increases thereafter to 2.7% in subjects over 80. The mosaic frequency is 0.2% in both the 0–14 (15/8535) and 15–29 year old group (16/6739), despite the fact that approximately half of the 0–14 year old subjects have a phenotypic abnormality (non-syndromic cleft lip/palate, prematurity or low birth weight). Excluding subjects less than 15 years old, in multiple logistic regression of mosaic status on age at DNA sampling, and adjusting for several covariates (study, sex, DNA source, and ethnicity), age is a highly significant predictor of mosaic status (p = 2 × 10−16, odds ratio=1.05, 95% confidence interval (CI)=1.04 – 1.07). Among the covariates, only study is significant (p=0.01) and a subsequent test of age-by-study interaction was not significant. It is notable that DNA source (92% from blood, 8% from saliva/buccal swabs) was not a significant predictor (p=0.45). When only blood samples are analyzed, the age effect estimate is the same (to three decimal places) and the p-value is only slightly higher (4 × 10−15). Copy-number mosaics and aUPD, when tested separately, each have a significant age effect and similar odds ratios (p-value for gain=0.01, loss=5 × 10−11, aUPD=6 × 10−8; OR (95% CI) for gain = 1.032 (1.005 – 1.061), loss = 1.057 (1.039 – 1.075), aUPD = 1.056 (1.035 – 1.077).
Figure 5.
The percentage of subjects having one or more mosaic anomalies within 5-year age bins. Vertical bars are 95% confidence intervals. For two cells with zero counts, the upper bar connects zero to the frequency with a lower 95% confidence interval of zero given the sample size. Expected leukemia values are given for reference and calculated using age- and sex-specific prevalence estimates (http://seer.cancer.gov).
This age effect is specific for mosaic anomalies. The same logistic regression performed with the non-mosaic anomalies did not have a significant age effect (p=0.11) and the sign of the regression coefficient estimate was reversed (Supplementary Figure 8). This result indicates that our classification method distinguishes effectively between acquired and constitutional anomalies.
To further explore the robustness of the age effect on clonal mosaicism, additional analyses were performed with each of the seven studies having more than 1,000 subjects over 50 years old (using both blood and saliva/buccal samples). Only the age effect was significant (p=8 × 10−16) in a combined logistic regression of mosaic status on study, sex, DNA source, ethnicity and smoking status (separately testing either ‘ever’ smoker or ‘never’ smoker). When only controls from these studies were analyzed together, the age effect remained highly significant (p=7 × 10−11). We also analyzed each study separately, with age and the case status specific to each study. A meta-analysis shows a highly significant effect of age (Figure 6), which is very robust to differences in both study and subject characteristics.
Figure 6.
Fixed-effects meta-analysis for effect of age at DNA sampling on mosaic status. Effect estimates are from logistic regression of mosaic status on age at DNA sampling, with adjustment for case status specific to each study. The summary estimate of the log odds ratio is 0.05 (95% CI=0.04 – 0.07) and the corresponding odds ratio is 1.06 (95% CI=1.04 – 1.07). Cochran’s Q test of heterogeneity has p-value=0.89. The sizes of the black boxes are proportional to the inverse of the squared standard error and the gray lines are 95% confidence intervals. The horizontal points of the diamond span the 95% confidence interval of the summary estimate. See Table 1 for study descriptions. AA = African American and JL = Japanese/Latino.
These cross-sectional analyses strongly suggest that most of the mosaic anomalies detectable by SNP microarrays appear late in life, because they arise more frequently and/or because they are more readily detected due to clonal expansion. This suggestion is supported by longitudinal observation in one GENEVA subject (the only subject sampled twice who had mosaicism in at least one sample). This subject was sampled at age 66 and again at age 72 (both with DNA from saliva). No mosaic anomalies were detected in the earlier sample, but the later sample contained 5 mosaic deletions, each on a different chromosome. Additional studies with subjects sampled at multiple ages are needed to evaluate the temporal origin and stability of mosaic anomalies.
In some GENEVA subjects, anomalies appear to have occurred early enough in development to be mosaic in both the soma and germline. In 35 parent-offspring pairs in which a mosaic anomaly was detected in the parent, there are three cases in which the offspring appears to be non-mosaic for the same anomaly (one deletion and two duplications), while there is no corresponding anomaly (mosaic or otherwise) in the remaining 32 offspring. Although this result suggests that a fairly large fraction of cases have mosaicism shared by the germline and soma, it may not be representative of the more frequent mosaics that occur in older subjects because parents in the family studies were sampled in their 20s and 30s (Table 1). The mosaics that appear in subjects less than 50 years of age may have different origins than those that appear later, when the frequency increases rapidly.
Mosaic anomalies characteristic of hematological cancers
The clonal mosaic anomalies detected in this study tend to cluster in location both within and among chromosome arms (Figure 4; Supplementary Fig. 7 and 9). Regions with multiple overlapping anomalies frequently coincide with regions of copy number change or aUPD characteristic of hematological cancers. Using the Mitelman "Recurrent Chromosome Aberrations in Cancer Database" (http://cgap.nci.nih.gov/Chromosomes/Mitelman), we found that 222 of 669 recurrent duplications and deletions found in hematological cancers have >80% overlap with at least one mosaic CNV in GENEVA subjects. Also, 77% of GENEVA mosaic CNVs have >80% overlap with the Mitelman aberrations and 48% overlap both cytological bands defining the limits of the aberration. The most common overlaps are 20q-, 13q-, 11q-, 17p-, 12+ and 8+.
Common deleted regions (CDR) of mosaic anomalies in different GENEVA subjects often pinpoint genes previously associated with the hematological cancers. The following examples are shown in Supplementary Figure 7: (1) On 13q, 31 deletions have a CDR of 299 kb, containing only one gene, DLEU7, which is thought to be a tumor suppressor20. In addition, 18 deletions on 13q cover RB1 and 24 cover MiR15a and MiR16-1. Deletions in this region (13q14) represent the most common cytogenetic abnormality in chronic lymphocytic leukemia (CLL)21, which is the most common leukemia in older adults (http://seer.cancer.gov). (2) On 4q, 14 deletions have a CDR of 214 kb containing only one gene, the TET2 oncogene, which is commonly deleted in myelodysplastic syndrome (MDS), myeloproliferative disorder (MPD) and acute myeloid leukemia (AML)22. (3) On 2p, 17 deletions have a CDR of 194 kb, which contains two genes, one of which is DNMT3A, recently found to be commonly mutated in AML-M523. (4) On 22q, 11 deletions have a CDR of 153 kb, which includes three genes, one of which is PRAME, which is frequently deleted in CLL24. (5) On 20q, 46 deletions have a CDR of 965 kb, containing 7 genes including L3MBTL1, which is a candidate tumor suppressor in del(20q12) myeloid disorders25.
Long (multi-megabase) segments of aUPD are frequently observed in cancers of many types26. In most cases, the UPD occurs on a terminal segment of one arm, consistent with origin by a single mitotic crossover, followed by outgrowth of one of the daughter cells. Acquired UPDs are frequently observed in hematological cancers such as MDS, MPD and AML and are associated with homozygosity of mutations in several tumor suppressors and oncogenes27,28. All autosomes (except chromosome 10) have at least one clonal mosaic aUPD in GENEVA subjects. Chromosomes 9 (with 24), 14 (with 21) and 11 (with 19) have the most aUPDs, which greatly exceed the expected number based on arm length (Supplementary Figure 9).
Despite the observation that many of the clonal mosaic anomalies observed here are characteristic of hematological cancer, the fraction of subjects with one or more mosaics who have a record of hematological cancer before DNA sampling is low. This fraction was estimated as 2.8% (95% CI=1.0 – 4.7%) in 291 mosaic subjects (with DNA from blood; from 13 GENEVA studies; using medical records, self-reported conditions and study exclusion criteria, as described in the Supplementary Note).
Hematological cancer incidence
We investigated whether detectable clonal mosaicism predisposes to incident hematological cancer after DNA sampling by using three GENEVA studies, which included cohorts with cancer diagnosis records both before and after DNA sampling. From the following studies, we analyzed 8,562 subjects who had DNA derived from blood and no record of hematological cancer prior to DNA sampling: (1) Glaucoma study, with subjects from the Nurses Health Study (NHS, N=363) and Health Professionals Follow-up Study (HPFS, N=285), (2) Lung Cancer study, with subjects from the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO, N= 1600) and (3) Prostate Cancer study, with subjects from the Multiethnic Cohort (MEC, N=6314). Among the 8,562 subjects analyzed for incident hematological cancer, 8,323 were non-mosaics with no events, 90 were non-mosaics with events, 134 were mosaics with no events, and 15 were mosaics with events (where ‘event’ is a hematological cancer diagnosis).
To test for an association between mosaic status and incident hematological cancer, we used a cause-specific Cox proportional hazards model to analyze time to a hematological cancer diagnosis from the date of DNA sampling, with right censoring at death and the endpoint of follow-up data. We performed a stratified analysis of the four cohorts, which included mosaic status and adjusted for age at DNA sampling, non-hematological cancer status (as a time-dependent covariate), ethnicity (two principal components) and sex (within the PLCO stratum). The hazard ratio estimate for mosaic status is 10.1 (95% CI=5.8 – 17.7) with a p-value of 3 × 10−10. A meta-analysis showed consistent results among cohorts and gave a very similar effect estimate (Supplementary Figure 10). These results estimate that the risk of hematological cancer is ten-fold higher for mosaic than for non-mosaic subjects.
Because both cancer and the clonal mosaic anomalies detected in this study increase with age, the adjustment for age at time of DNA sampling in the Cox regression model is critical. We modeled the age covariate as either a linear effect or as a non-linear effect (spline smoothing with 5 degrees of freedom) and found that the mosaic effect estimates and p-values are essentially identical.
Among the 15 mosaic subjects who had a hematological diagnosis after DNA sampling, four had myeloid leukemia, six had chronic lymphocytic leukemia, one had multiple myeloma, one had MDS, one had MPD and two had non-Hodgkin lymphoma. Thus, the 15 cases are about evenly divided between mature B-cell neoplasms and myeloid malignancies. Not surprisingly, the leukemias are over-represented among mosaic compared with non-mosaic subjects (p-value=0.005, Supplementary Tables 5 and 6). A variety of chromosomal anomalies were found in the mosaic subjects (Supplementary Table 7). Deletions covering the CDRs described above were found in several of these subjects: 13q- in five CLLs, 4q- in one chronic myelogenous leukemia (CML), 20q- in one multiple myeloma and one AML, and 22q- in one CLL. Five of the 15 mosaic subjects with incident hematological cancer had more than one mosaic anomaly, which is higher than in the remaining subjects within this set of cohort samples (25/134), although not significantly so (p=0.18).
Although the risk of incident hematological cancer is estimated as 10-fold higher for mosaic than for non-mosaic subjects (95% CI=5.8 – 17.7), it is important to note that the incidence rate in mosaics is low (10 year event rate of 0.143, 95% CI=0.065 – 0.214, Figure 7) and that only a small fraction of GENEVA mosaic subjects have a record of hematological cancer before DNA sampling (2.8%, 95% CI=1.0 – 4.7%). The period between first appearance of detectable clonal mosaicism and incidence of hematological cancer is of interest, but cannot be estimated from our data since mosaicism was present for an unknown period of time prior to DNA sampling. However, the median time of 3.5 years between DNA sampling and hematological cancer diagnosis provides a very rough minimum estimate (range 3.5 months to 10.7 years with N=15; see Figure 7).
Figure 7.
A Kaplan-Meier plot of the proportion of living subjects who remain free of hematological cancer as a function of time since the time of DNA sampling and determination of mosaic status. Estimates for mosaic (red) and non-mosaic (black) subjects are given separately (solid lines), each with their 95% confidence intervals (dashed lines). The vertical ticks represent censoring times. For the 15 mosaic subjects with incident cancer, the times between DNA sampling and diagnosis are 3.5, 6.1, 12.7, 18.8, 25.0, 36.9, 37.5, 42.9, 44.0, 46.2, 48.0, 60.4, 61.8, 91.1, and 130.5 months.
Non-hematological cancer
To investigate the relationship between mosaic status and non-hematological cancer, two types of analyses were done. First, in each of the three GENEVA case-control cancer studies (Lung Cancer, Prostate Cancer, Melanoma), we did logistic regression of mosaic status on case status and age at DNA sampling. Case status was not significant in any of the three studies or in a meta-analysis (one-tailed p=0.06). The estimated odds of having a clonal mosaic anomaly was higher among cancer cases than controls in the lung and prostate cancer studies, but lower in the melanoma study (Supplementary Fig. 11). Second, in the cohort studies (PLCO, HPFS, NHS and MEC), we did logistic regression of mosaic status on whether or not the subject had a non-hematological cancer prior to DNA sampling (excluding any hematological cancer cases). In these analyses the relationship is consistently positive, but small and not significant overall (one-tailed p=0.11, Supplementary Figure 12). In summary, the evidence hints at a positive relationship between mosaic status and non-hematological cancer, but lacks statistical significance. Therefore, further work is needed in larger sets of non-hematological cancer studies, including data on potential exposure, disease and treatment effects.
DISCUSSION
Here we have shown that the frequency of subjects with detectable clonal mosaicism for large chromosomal anomalies in peripheral blood is low (<0.5%) from birth until 50 years of age, after which it rises rapidly. This relationship between mosaicism and age is very robust to both study and subject characteristics. Among the covariates sex, ethnicity, smoking and disease status (exclusive of hematological cancer), none had a significant effect on mosaic status. The age effect in GENEVA subjects is consistent with a recent study showing that acquired differences in structural chromosome variants between members of monozygotic twin pairs (including clonal mosaic anomalies) are observed in pairs >55 years of age but not in younger pairs29. Nevertheless, longitudinal studies are required to rule out the possibility that a trend in environmental exposures across birth cohorts may contribute to the increase in mosaicism with age.
The observed increase in detectable clonal mosaicism late in life may be due to a change in the frequency with which chromosomal anomalies occur (i.e. increased somatic mutation rate) and/or their ability to form large clones (i.e. clonal expansion). Previous work has shown that the occurrence of chromosomal anomalies (rearrangements and aneuploidies) during cell division increases with age in cultured lymphocytes and fibroblasts30,31, that DNA damage accumulates with age in mouse hematopoietic stem cells32, and that mitotic recombination (leading to uniparental disomy) increases with replicative age in yeast33. This apparent increase in somatic mutation may result from age-related decline in genomic maintenance mechanisms (such as telomere attrition34). Clonal expansion of cells containing chromosomal anomalies could be due to either positive selection or to random changes in the frequencies of hematopoietic stem cell descendants. In principle, stem cell senescence and age-related decline in replicative function35 could result in a decrease in the effective population size of stem cells, leading to shifts in clonal composition analogous to random drift in small populations of individuals36. However, analyses of the clonal composition of blood cells, based on X-inactivation markers in healthy women, suggest stability over time and between lymphoid and myeloid lineages, even in the elderly37,38. Therefore, in most cases, positive selection may be required to establish clones of cells with chromosomal anomalies that are sufficiently large for detection with SNP microarrays. The potential for positive selection may increase with age as somatic mutations accumulate in genes that regulate cellular proliferation. For example, a highly proliferative clone may arise when a recessive tumor suppressor mutation becomes hemizygous in combination with a deletion, or homozygous due to aUPD. This suggestion is supported by the observation that acquired anomalies tend to cluster in certain genomic regions and that common deleted regions pinpoint genes previously associated with hematological cancer.
In the mosaics described in this study, the chromosomally abnormal cells constitute a significant fraction of white blood cells, since a minimum of 5–10% is required for detection by our method and many abnormal clones are substantially larger (Figure 2). The blood samples used for DNA extraction were not fractionated by white blood cell type. The abnormal blood cells within an individual may include multiple cell types if the anomaly arose in a multipotent hematopoietic stem cell that became predominant due to senescence or positive selection within the stem cell population. Alternatively, the abnormal cells may include a restricted set of cell types, particularly when the normal composition of blood (i.e. 60–70% of neutrophils and 20–40% of lymphocytes39) is altered by unregulated proliferation40.
There is a strong association between the clonal mosaic anomalies detected in our study and hematological cancer. We estimate the risk of acquiring a hematological cancer diagnosis as 10-fold higher for subjects with mosaic anomalies. This association is strongly supported by finding that many of the mosaic anomalies are characteristic of those found in hematological cancers. Nevertheless, the event numbers analyzed here are small and additional studies are needed across a broader diversity of cohorts to establish the clinical significance of these findings.
Notwithstanding the strong association with hematological cancer, we estimated that ~97% of subjects with clonal mosaic anomalies did not have a record of a hematological cancer prior to DNA sampling and the incidence rate is low (~ 14% over ten years in subjects who survive and are not lost to follow-up during this period). These results suggest that the clonal mosaicism observed in elderly subjects may be an asymptomatic condition with a predisposition to hematological cancer that is often not realized.
It is possible that many of the subjects with detectable clonal mosaicism in our study have monoclonal B-cell lymphocytosis (MBL), an asymptomatic condition with an estimated prevalence of 3–5% in the elderly. MBL is characterized by a clonal population of B lymphocytes with an immunophenotype similar to CLL or other B-cell malignancy41. Most, if not all, cases of CLL are preceded by MBL, but most cases of MBL do not progress to malignancy42,43. However, 85% of MBL detected in population screening studies have a B-cell count below 500/μl43, which is less than 10% of the normal white blood cell count. Since 10% is near the lower limit of detection for chromosomal mosaicism using our methods, the two types of clones may not be closely related. Nevertheless, further work on the relationship between B cell immunophenotypes and mosaic anomalies is warranted.
Although it appears that most of the clonal mosaicism observed in GENEVA subjects represents a non-malignant condition, further work is needed to evaluate the fraction of subjects who might have unrecorded malignant conditions such as MDS and MPD, or undiagnosed CLL. MDS and MPD were added to the Surveillance, Epidemiology, and End Results (SEER) cancer registries in 2001 and may still be under-recorded because they are often managed outside of the hospital setting44. Therefore, accurate prevalence data from widespread populations are not available, but local population estimates (0.1% MDS45 and 0.5% MPD46 in the elderly) are substantially less than the ~2.5% of GENEVA subjects with mosaic anomalies in the over 75 age.
This survey is the first large-scale study of acquired chromosomal anomalies in people of all ages and various states of health. Previously, the extent of chromosomal variation within developmentally normal individuals, in the absence of overt cancer, was largely unknown. The results presented here indicate that a significant fraction of blood cells in people without a prior history of hematological cancer may contain large chromosomal anomalies, including multi-megabase deletions, duplications and aUPD. The frequency of people with such clonal anomalies in a mosaic state is low up to about 50 years of age and then increases rapidly up to 2–3%. We find that these anomalies are associated with an approximately ten-fold higher risk of hematological cancer, but subjects with detectable clonal mosaicism may survive for years without having a hematological cancer diagnosis. Further work is needed to determine the stability of the mosaic state over time, to replicate and improve estimates of the predisposition to hematological cancer, and to identify anomalies associated with asymptomatic cancer precursor conditions. It also will be important to explore the health consequences of these anomalies for conditions other than cancer, such as immune system function.
METHODS
Study subjects, phenotypic data and genotyping
Subjects were recruited for 15 different studies belonging to the Gene Environment Association Studies (GENEVA) consortium16 (Table 1). Each study was approved by the institutional review board of the study investigator’s institution, and all subjects provided written informed consent for participation in the study. The Supplementary Note describes the phenotypes. Each study was genotyped on one of five different Illumina array types at the Center for Inherited Disease Research (CIDR), the Broad Institute Center for Genotyping and Analysis, or the University of Southern California (Supplementary Table 1). DNA samples were derived from blood (92%) or saliva/buccal swabs (8%). No lymphoblastoid cell line or whole-genome amplified samples were included in the analyses described here. Because cell lines may have artifactual mosaic anomalies47, mis-identification of DNA source is a concern. However, only the Addiction study had both cell line and non-cell line samples and the non-cell line samples analyzed here did not have an unusual frequency of mosaic anomalies. Genotypic data cleaning and calculation of BAF and LRR are described in the Supplementary Note. Sample sizes for analyses vary (as stated in Results) because a small proportion of the subjects are missing data for age at DNA sampling or other variables.
Anomaly detection and quality control
The method of anomaly detection is described in detail in the Supplementary Note and summarized here. Detection of anomalies (both mosaic and non-mosaic) was based on BAF and LRR metrics. The primary focus for detecting anomalies was BAF, because we wanted to identify copy-neutral events (mosaic UPD) and because BAF is much less noisy and prone to artifacts (such as GC waves48) than LRR. The main approach was to detect a split in the BAF intermediate band, which in normal (biparental disomic) samples is centered at 1/2 and corresponds to AB heterozygotes (Figure 1). In trisomic samples, this band splits into two components (AAB and ABB) at BAF= 1/3 and 2/3. In disomic-trisomic mosaics, the width of the split varies from zero to one third and LRR varies from zero to a theoretical value of log2(3/2). In disomic-monosomic mosaics, the width of the split varies from zero to one and LRR varies from 0 to a theoretical value of log2(1/2). In biparental-uniparental disomic mosaics, the width of the split varies from zero to one, while LRR remains constant at zero. These transitions are shown in Figure 2 as deviations from expected. In chromosomal regions containing heterozygous SNPs, use of BAF alone can detect duplications (both mosaic and non-mosaic), mosaic deletions, mosaic uniparental disomy and homozygous deletions. LRR is required to detect monosomic regions and duplications in regions lacking heterozygosity. Therefore, we implemented two separate but complementary methods, called ‘BAF’ and ‘LOH’ (the latter for LRR change detection in regions lacking heterozygosity). Anomalies detected by the BAF method were classified as mosaic or non-mosaic. Anomalies detected by the LOH method were used here only to define the BAF/LRR position of heterozygous deletions and not for mosaic detection. We did not attempt to identify non-mosaic segments of uniparental isodisomy, which have no heterozygosity and normal LRR.
In the BAF method, Circular Binary Segmentation (CBS)49 was used to detect change points in a metric modified from Itsara et al.15: sqrt(min(BAF,1-BAF,|BAF -median(BAF)|)) for SNPs called as missing or heterozygotes (i.e. excluding homozygotes). The use of missing calls allows detection of wide splits (e.g. Figure 3d). In the LOH method, CBS was applied to LRR values and combined with overlapping runs of homozygosity. By focusing on regions of homozygosity, we avoided a high false positive rate associated with a genome-wide search for changes in LRR. In both methods, the identification of anomalous segments involved establishing a non-anomalous baseline, choosing anomalous segments based on deviation from baseline, and applying quality control filters. Computations were done using the Bioconductor packages DNACopy and GWASTools. The latter was developed by our group; relevant functions are described in the Supplementary Note.
Quality control (QC) was done at the sample and anomaly level. Low quality samples (with high variance of BAF and/or LRR metrics or a high level of segmentation) were removed differentially for the two methods. Supplementary Table 1 shows the percentage of samples that passed QC for the BAF method (mean = 99.1%) and the LOH method (86.8%). In some studies, a high fraction of samples failed QC for LOH detection (maximum 47%), but the failure rates for BAF-detection (from which all mosaics were identified) are all low (maximum 8%). Anomaly-level QC involved several steps, including manual curation of all anomalies designated as mosaic and all other anomalies greater than 2 Mb in length. (see Supplementary Note). Manual curation involved evaluation of BAF/LRR plots, as shown in Figure 3 and Supplementary Fig. 2. Note that Supplementary Fig 2(m-t) shows a sample of eight of the smallest mosaic deletions. Features that distinguish mosaic from non-mosaic are described in the Figure 3 legend.
The reproducibility of anomaly detection was assessed using samples genotyped in duplicate (N = 568 pairs). For each sample pair, we defined a unit of observation as a contiguous chromosomal region containing an anomaly in one or both samples. Each unit is given a score equal to the length of the intersection divided by the length of the union of anomalies in that unit. A reproducibility measure was defined as the fraction of units with a score greater than either 0.30 or 0.80 (chosen for comparison with published CNV studies). We also calculated the average of the scores that were greater than zero. Supplementary Table 2 summarizes these quantities for each study. For BAF, the mean reproducibility measure was 90% with a 30% overlap threshold and 82% with 80% overlap. For LOH, the means were 71% (30% overlap) and 67% (80% overlap). The mean of scores greater than zero was 95% for BAF- and 96% for LOH-detected anomalies (30% threshold), indicating that when an anomaly is detected in both scans, the breakpoints are highly reproducible. These reproducibility estimates are higher than the 40–60% that is typical for detecting CNVs using Hidden Markov Models (HMM)19,50, perhaps in large part because we do not attempt to detect small anomalies (the 5th percentile of anomalies we detect is 35 kb). In our experience, standard methods of CNV detection, such as PennCNV51, tend to break up large anomalies into many segments and to miss large mosaics.
Identifying and classifying clonal mosaic anomalies
Clonal mosaic anomalies were identified in GENEVA family studies by using transmitted anomalies to characterize the bivariate BAF/LRR distribution expected for non-mosaic (constitutional) anomalies. For transmitted anomalies, this distribution is approximately bivariate normal within a study and we used this distribution to estimate a 95% prediction ellipse52, which defines an area likely to contain most of the constitutional anomalies (Supplementary Figure 13). Among the anomalies used to identify mosaics, the majority are 3N duplications. There is also a small cluster of 4N anomalies, but we did not attempt to detect 3N/4N mosaics. Anomalies outside of these two clusters contain mosaics and artifacts. The latter consist of false positives and anomalies with inaccurate breakpoints (which distorts the median BAF/LRR values). To distinguish between the mosaics and artifacts, we performed a manual review of BAF/LRR plots for all anomalies that fell outside of the 95% prediction ellipse and below the mean LRR for anomalies used to define the ellipse. The non-family studies were analyzed in a similar way, except that we replaced the class of transmitted anomalies with polymorphic CNVs. The latter were defined by hierarchical clustering to identify sets of anomalies with similar breakpoints. We then defined polymorphic sets as those with at least 4 members (but excluding sets with mean anomaly length greater than 10 Mb). We also included in the mosaic class three whole-autosome anomalies (12, 8, 22) that fell within the 3N ellipse, because constitutional trisomies for these chromosomes are not compatible with normal development1. Although we did not have access to biospecimens necessary for experimental validation of mosaics (i.e. live cells or those preserved for cytology), all anomalies classified as mosaics were manually reviewed and the BAF/LRR patterns that we observed are very similar to those reported by Peiffer17, Rodriguez-Santiago53 and Conlin7, who performed cytological validation for a variety of mosaic types.
Classification of clonal mosaic anomalies as duplication, deletion or aUPD was done using the median LRR and BAF deviations from non-anomalous segments (Figure 2b). Deviations from non-anomalous segments within the same sample were used to control for overall LRR variation among samples and for BAF asymmetry that occurs in some samples. Anomalies that are either terminal segments or whole chromosome and that have an LRR deviation within a ‘neutral zone’ (|LRR|<0.05) were classified as aUPD. This neutral zone was chosen because it includes nearly all of the wide splits (BAF deviation > 0.25) that have much smaller LRR deviations than expected for disomic/trisomic or disomic/monosomic transitions, while including very few interstitial anomalies (Supplementary Figure 3). All other anomalies (except for a few outliers) were classified as either duplications or deletions, depending on the sign of their LRR deviation. There is some ambiguity in classifying anomalies near the tip of the arrow, where the three transition zones intersect. This ambiguity is noted as ‘intensity.flag’ in Supplementary Table 3. Mixture proportions in mosaics can be estimated as position along the transitional line that connects the two constitutional states (Figure 2; see Supplementary Note).
All anomalies discussed in this paper are autosomal in the reference genome. Detection of X chromosome mosaics is complicated by the fact that LRR is a measure of the intensity of a sample relative to other samples. X chromosome LRR values (calculated in the standard way) are affected by the sex ratio in the sample set and are not comparable to those for the autosomes.
Statistical analysis
All statistical analyses were done in the R statistical package (http://www.R-project.org) using functions described in the Supplementary Note.
URLs
http://cgap.nci.nih.gov/Chromosomes/Mitelman, Mitelman, F., Johansson, B. & Mertens, F. (eds.). Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer, (2011).
http://seer.cancer.gov, SEER. US Estimated 33-Year L-D Prevalence Counts on 1/1/2008. (ed. Surveillance, E., and End Results (SEER) Program, National Cancer Institute, DCCPS, Surveillance Research Program, Statistical Research and Applications Branch, released April 2011, based on the November 2010 SEER data submission.) (2011).
http://www.R-project.org, The R Development Core Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL. (2006).
Supplementary Material
Acknowledgments
The GENEVA consortium thanks the subjects and the staff of all GENEVA studies for their important contributions. Support for the GENEVA genome-wide association studies was provided through the NIH Genes, Environment and Health Initiative (GEI). Some studies also received support from individual NIH Institutes. The grant numbers are: Melanoma (NCI R29CA70334, R01CA100264, P50CA093459); Lung Health (U01HG004738); Cleft lip/palate (NIDCR: U01DE018993, NIH contract: HHSN268200782096C); Addiction (U01HG004422, NIAAA: U10AA008401, NCI: P01CA089392, NIDA: R01DA013423, R01DA019963); Lung cancer (Z01CP010200); Blood clotting (R37 HL 039693); Prostate cancer (U01HG004726, NCI: CA63464, CA54281, CA1326792, RC2 CA148085); Venous thromboembolism (U01HG004735); Birth weight (U01HG004415); Dental Caries (NIDCR:U01DE018903 and R01DE014899, NIH CIDR contract: HHSN268200-782096C); Prematurity (U01HG004423); Glaucoma (U01HG004728, NEI: R01EY015473, NEI: R01EY015872); GENEVA Coordinating Center (U01 HG004446); Center for Inherited Disease Research (U01HG004438, HHSN268200782096C); Broad Center for Genotyping and Analysis (U01HG04424); Intramural Research Program of the NIH, National Library of Medicine; Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH. Dr. Pasquale was also supported by Physician Scientist award from Research to Prevent Blindness in NYC, an Ophthalmology Scholar Award from Harvard Medical School and from the Harvard Glaucoma Center of Excellence. Leila Zelnick was supported by T32 CA09168 from the National Cancer Institute. We thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. We thank Charles Laird and Gerald Marti for helpful comments on the manuscript, and Barbara Wakimoto and Daniel Gottschling for enlightening discussions. We also thank Kevin Jacobs for exchanging ideas and for working with us to estimate cross-method concordance of mosaic detection using the PLCO/GENEVA Lung Cancer study.
Footnotes
ACCESSION NUMBERS
The dbGaP accession numbers for the studies analyzed here are: phs000187.v1.p1, phs000335.v1.p1, phs000094.v1.p1, phs000092.v1.p1, phs000093.v2.p2, phs000304.v1.p1, phs000306.v2.p1, phs000289.v1.p1, phs000096.v3.p1, phs000096.v3.p2, phs000096.v3.p3, phs000095.v1.p1, phs000103.v1.p1, phs000308.v1.p1. See also Supplementary Table 1.
CONFLICTS OF INTEREST
Laura J. Bierut served as a consultant for Pfizer Inc. in 2008 and is an inventor on the patent “Markers for Addiction” (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
AUTHOR CONTRIBUTIONS
K.F.D, H.L., K.N.H and E.W.P. initiated the detection of chromosomal anomalies in GENEVA GWAS data. C.A.L. developed the automated methods of anomaly detection, with assistance from C.C.L., L.R.Z., C.P.M., V.E.S and A.N.M. C.C.L., C.A.L., K.R., L.R.Z., C.P.M., J.S., D.R.C., D.M.L, X.Z., S.C.N., S.M.G., M.P.C., J.I.U. and S.B. performed data analyses. C.A., Q.W., L.W., J.E.L., K.C.B., N.N.H., R.M., T.H.B., A.F.S., L.J.B., M.T.L., L.R.G., D.G., K.C.D., S.S.S., W.J.B., L.B.S., S.A.I., S.J.C., S.I.B., L.l.M., B.E.H., J.A.H., S.M.A., C.R., W.L.L., M.L.M., J.C.M., M.M., B.F., J.H.K., J.L.W., L.R.P., C.A.H. and N.C. contributed sample collections and phenotypic data. K.F.D., H.L., K.N.H., E.W.P, D.M., and A.C. performed genotyping. L.R.P., H.K., N.C., C.A.H., B.E.H. and K.R.M. provided data and interpretation for analysis of incident hematological cancer. C.C.L, C.A.L., L.R.Z., K.F.D, K.R., C.A., D.D., T.H.B., A.F.S., I.R., R.B.S., L.J.B., S.M.H., N.D.F., J.L., B.E.H., K.R.M., M.d.A., W.L.L, M.G.H., M.L.M., E.F., J.C.M., M.M, B.F., J.L.W., A.W., C.P.M., J.S., D.R.C., D.M.L., X.Z., J.I.U., S.B., S.C.N., S.M.G., P.H., G.P.J., A.N.M., C.C., V.E.S., H.L., K.N.H., E.W.P., D.M., A.C., N.S., T.M., L.R.P., C.A.H., N.C. and B.S.W. contributed ideas and advice during regular discussions of the project. C.C.L. coordinated the study. C.C.L. wrote the first draft of the paper, with guidance from a writing committee consisting of C.A.L., K.R., K.F.D., T.M., L.R.P., N.C. and B.S.W. All authors contributed to review and revision of the paper.
References
- 1.Miller OJ, Therman E. Human Chromosomes. Vol. 501. Springer-Verlag; 2001. [Google Scholar]
- 2.Strachan T, Read AP. Human Molecular Genetics. Wiley-Liss; New York: 1996. [Google Scholar]
- 3.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 4.Delhanty JD. Mechanisms of aneuploidy induction in human oogenesis and early embryogenesis. Cytogenet Genome Res. 2005;111:237–44. doi: 10.1159/000086894. [DOI] [PubMed] [Google Scholar]
- 5.Vanneste E, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–83. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 6.Hassold T. Mosaic trisomies in human spontaneous abortions. Hum Genet. 1982;61:31–5. doi: 10.1007/BF00291327. [DOI] [PubMed] [Google Scholar]
- 7.Conlin LK, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19:1263–75. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballif BC, et al. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A. 2006;140:2757–67. doi: 10.1002/ajmg.a.31539. [DOI] [PubMed] [Google Scholar]
- 9.Heim S, Mitelman F. Nonrandom chromosome abnormalities in cancer - an overview. In: Mitelman F, Heim S, editors. Cancer Cytogenetics. John Wiley & Sons, Inc; Hoboken, NJ: 2009. pp. 25–44. [Google Scholar]
- 10.Gardner RJM, Sutherland GR. Chromosome Abnormalities and Genetic Counseling. Oxford University Press; Oxford: 2004. [Google Scholar]
- 11.Maciejewski JP, Tiu RV, O’Keefe C. Application of array-based whole genome scanning technologies as a cytogenetic tool in haematological malignancies. Br J Haematol. 2009;146:479–88. doi: 10.1111/j.1365-2141.2009.07757.x. [DOI] [PubMed] [Google Scholar]
- 12.Dougherty MJ, et al. Implementation of high resolution single nucleotide polymorphism array analysis as a clinical test for patients with hematologic malignancies. Cancer Genet. 2011;204:26–38. doi: 10.1016/j.cancergencyto.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 13.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 14.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–12. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itsara A, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–61. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelis MC, et al. The Gene, Environment Association Studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol. 2010;34:364–72. doi: 10.1002/gepi.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peiffer DA, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–48. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs K, Yeager M, Zhou W, Wacholder S, Chanock S. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012 doi: 10.1038/ng.2270. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto D, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29:512–20. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pekarsky Y, Zanesi N, Croce CM. Molecular basis of CLL. Semin Cancer Biol. 2010;20:370–6. doi: 10.1016/j.semcancer.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohner H, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 22.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–15. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 24.Gunn SR, et al. Array CGH analysis of chronic lymphocytic leukemia reveals frequent cryptic monoallelic and biallelic deletions of chromosome 22q11 that include the PRAME gene. Leuk Res. 2009;33:1276–81. doi: 10.1016/j.leukres.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Gurvich N, et al. L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability. Proc Natl Acad Sci U S A. 2010;107:22552–7. doi: 10.1073/pnas.1017092108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuna M, Knuutila S, Mills GB. Uniparental disomy in cancer. Trends Mol Med. 2009;15:120–8. doi: 10.1016/j.molmed.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 27.O’Keefe C, McDevitt MA, Maciejewski JP. Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies. Blood. 2010;115:2731–9. doi: 10.1182/blood-2009-10-201848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghavan M, Gupta M, Molloy G, Chaplin T, Young BD. Mitotic recombination in haematological malignancy. Adv Enzyme Regul. 2010;50:96–103. doi: 10.1016/j.advenzreg.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 29.Forsberg LA, et al. Age-related somatic structural changes in the nuclear genome of human blood cells. Am J Hum Genet. 2012;90:217–28. doi: 10.1016/j.ajhg.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vorobtsova I, Semenov A, Timofeyeva N, Kanayeva A, Zvereva I. An investigation of the age-dependency of chromosome abnormalities in human populations exposed to low-dose ionising radiation. Mech Ageing Dev. 2001;122:1373–82. doi: 10.1016/s0047-6374(01)00275-5. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee AB, Thomas S. A longitudinal study of human age-related chromosomal analysis in skin fibroblasts. Exp Cell Res. 1997;235:161–9. doi: 10.1006/excr.1997.3673. [DOI] [PubMed] [Google Scholar]
- 32.Rossi DJ, et al. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell Cycle. 2007;6:2371–6. doi: 10.4161/cc.6.19.4759. [DOI] [PubMed] [Google Scholar]
- 33.Lindstrom DL, Leverich CK, Henderson KA, Gottschling DE. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002015. doi: 10.1371/journal.pgen.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–8. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–13. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 36.Crow JF, Kimura M. An Introduction to Population Genetics Theory. Vol. 591. Harper and Row; New York: 1970. [Google Scholar]
- 37.Prchal JT, et al. Clonal stability of blood cell lineages indicated by X-chromosomal transcriptional polymorphism. J Exp Med. 1996;183:561–7. doi: 10.1084/jem.183.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swierczek SI, et al. Hematopoiesis is not clonal in healthy elderly women. Blood. 2008;112:3186–93. doi: 10.1182/blood-2008-03-143925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischbach F, Dunning MB., III . A Manual of Laboratoy and Diagnostic Tests. Lippincott, Williams and Wilkins; Philadelphia: 1992. [Google Scholar]
- 40.Vandewoestyne ML, et al. Laser microdissection for the assessment of the clonal relationship between chronic lymphocytic leukemia/small lymphocytic lymphoma and proliferating B cells within lymph node pseudofollicles. Leukemia. 2011;25:883–8. doi: 10.1038/leu.2011.14. [DOI] [PubMed] [Google Scholar]
- 41.Marti GE, et al. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br J Haematol. 2005;130:325–32. doi: 10.1111/j.1365-2141.2005.05550.x. [DOI] [PubMed] [Google Scholar]
- 42.Landgren O, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–67. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shanafelt TD, Ghia P, Lanasa MC, Landgren O, Rawstron AC. Monoclonal B-cell lymphocytosis (MBL): biology, natural history and clinical management. Leukemia. 2010;24:512–20. doi: 10.1038/leu.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cogle CR, Craig BM, Rollison DE, List AF. Incidence of the myelodysplastic syndromes using a novel claims-based algorithm: high number of uncaptured cases by cancer registries. Blood. 2011;117:7121–5. doi: 10.1182/blood-2011-02-337964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neukirchen J, et al. Incidence and prevalence of myelodysplastic syndromes: data from the Dusseldorf MDS-registry. Leuk Res. 2011;35:1591–6. doi: 10.1016/j.leukres.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Ma X, Vanasse G, Cartmel B, Wang Y, Selinger HA. Prevalence of polycythemia vera and essential thrombocythemia. Am J Hematol. 2008;83:359–62. doi: 10.1002/ajh.21129. [DOI] [PubMed] [Google Scholar]
- 47.Simon-Sanchez J, et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 48.Diskin SJ, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 50.Lin P, et al. Copy number variation accuracy in genome-wide association studies. Hum Hered. 2011;71:141–7. doi: 10.1159/000324683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tracy ND, Young JC, Mason RL. Multivariate Control Charts for Individual Observations. Journal of Quality Technology. 1992;24:88–95. [Google Scholar]
- 53.Rodriguez-Santiago B, et al. Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. Am J Hum Genet. 2010;87:129–38. doi: 10.1016/j.ajhg.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.