Abstract
Objective
We have conducted a GWAS in a Caucasian cohort of juvenile idiopathic arthritis (JIA) patients and have previously published findings limited to autoimmune loci shared with other diseases. The goal of this study was to identify novel JIA predisposing loci using genome-wide approaches.
Methods
The Discovery cohort consisted of Caucasian JIA cases (814) and local controls (658) genotyped on the Affymetrix SNP 6.0 Array along with 2400 out-of-study controls. A replication study consisted of 10 SNPs genotyped in 1744 cases and 7010 controls from the US and Europe.
Results
Analysis within the Discovery cohort provided evidence of associations at 3q13 within C3orf1 and near CD80 (rs4688011, OR=1.37, P=1.88×10−6), and 10q21 near the gene JMJD1C [rs6479891, odds ratio (OR) =1.59, P=6.1×10−8; rs12411988, OR=1.57, P=1.16×10−7 and rs10995450, OR = 1.31, P=6.74×10−5]. Meta-analysis continued to provide evidence for association for these 4 SNPs (rs4688011, P=3.6×10−7, rs6479891, P=4.33×10−5; rs12411988, P=2.71×10−5; and rs10995450, 5.39×10−5;). Gene expression data from 68 JIA cases and 23 local controls showed cis eQTL associations for C3orf1 SNP rs4688011 (P=0.024 or P=0.034, depending on probe set) and the JMJD1C SNPs rs6479891 and rs12411988 (P=0.01 and P=0.008, respectively). A variance component liability model estimated that common SNP variation accounts for ~1/3 of JIA susceptibility.
Conclusions
Genetic association results and correlated gene expression findings provide evidence of association at 3q13 and 10q21 for JIA and offer novel genes as plausible candidates in disease pathology.
Introduction
Juvenile idiopathic arthritis (JIA) is a debilitating complex genetic disorder characterized by inflammation of the joints and other tissues and shares histopathological features with other autoimmune diseases. Clinically, the International League of Associations for Rheumatology (ILAR) classification includes seven JIA subtypes (1). Overall there are about 50,000 children with JIA in the USA, approximately 1 per 1000 births (2), which is about the same incidence as juvenile diabetes. While JIA is relatively uncommon compared to some adult onset disorders, it may have a stronger genetic contribution since children with JIA have had less time for environment and behavior to influence disease risk relative to adults. Most subtypes of JIA are more prevalent in females. In addition, individuals from JIA families have increased risk for other autoimmune diseases (3). Unlike many other autoimmune diseases, JIA is more common in children of European ancestry, and the distribution of JIA subtypes does differ significantly across ethnic groups (4).
Two of the JIA subtypes, oligoarticular (which includes both persistent and extended forms) and IgM rheumatoid factor negative polyarticular (polyRFneg) disease make up the majority of patients. Oligoarticular disease is the most common subtype, occurs particularly in younger children and has an average age at onset of approximately five years. This subtype is generally associated with anti-nuclear antibodies (ANA) and is characterized by involvement of four or fewer joints in the first 6 months of disease. The involvement of additional joints (> 4 joints in total) over time distinguishes extended oligoarticular from persistent oligoarticular disease. There is no adult disease equivalent for oligoarticular JIA (5). Patients who have more than 4 affected joints within the first 6 months of disease are referred to as polyarticular and further are classified based on the absence or presence of IgM rheumatoid factor (RF) as separate JIA subtypes. Other subtypes of JIA include systemic JIA, enthesitis-related arthritis, juvenile psoriatic arthritis and undifferentiated JIA. This report is focused on oligoarticular and polyRFneg disease in order to maximize both homogeneity and sample size in the study cohorts.
There is convincing evidence of a strong genetic component to the risk of JIA, based on evidence inferred from twin and affected sib-pair studies, with an estimated sibling recurrence risk (λs) equal to 15 (6). Like other autoimmune diseases, genetic variation within the Human Leukocyte Antigen (HLA) region defines the strongest known genetic risk factors for JIA. Both HLA class I and class II haplotypes associated with JIA risk are distinct from those associated with rheumatoid arthritis (7) and differ among JIA subtypes (8). It is estimated that the HLA-DR region accounts for 17% of the sibling recurrence risk for JIA, with other Major Histocompatibility Complex (MHC) regions contributing additional risk (9).
The two published JIA genome-wide association studies (GWAS) (10, 11) which identified TRAF1-C5 and VTCN1 respectively, included only modest numbers of cases or markers assayed and therefore were limited in statistical power. Polymorphisms implicated in other autoimmune diseases have been associated with JIA by us and by others (12-14). From our GWAS dataset, we have previously reported SNPs representing PTPN22, PTPN2, IL2RA, TNFAIP3, COG6, ADAD1-IL2-IL21 and STAT4 loci (14), but these findings still only explain a portion of JIA susceptibility. To identify additional novel JIA-predisposing loci, we continued our analysis of the largest genome-wide dataset to date. A two-stage design was used that included a first stage “Discovery cohort”, which was genotyped using the Affymetrix Genome-Wide SNP Array 6.0 (Affymetrix Inc., Foster City, CA), and a second stage where ten SNPs with strong statistical support were genotyped in replication samples. Gene expression data available for a subset of the Discovery cohort (15) allowed an integrated analysis for relevant expression quantitative trait loci (eQTL) to improve our ability to discover genetic risk factors for complex traits.
Materials and Methods
Discovery cohort
The 814 JIA cases and 658 local controls of self-reported non-Hispanic European American (EA) ancestry genotyped for this GWAS have been previously described (15). The cases were limited to the two most common subtypes of ILAR-defined JIA, polyRFneg and oligoarticular JIA (both persistent and extended). Of the 814 cases, 113 (14%) were from multiplex pedigrees. In each pedigree, one polyRFneg or oligoarticular JIA case was randomly selected for genotyping. The local control cohort included healthy children without known major health conditions recruited from the geographical area served by Cincinnati Children’s Hospital Medical Center (CCHMC). To increase statistical power, genotype data from 2400 “out-of-study” controls from the Molecular Genetics of Schizophrenia non-GAIN samples available from dbGaP were combined with the local controls. This yielded a ratio of ~1:3.8 comparing cases to controls.
Replication cohort
To attempt to replicate associations in the initial cohort, five independent JIA case (persistent oligoarticular, extended oligoarticular or polyRFneg) and control sample collections of self-reported Caucasian ethnicity were genotyped (Table S1). The “Texas”, “Utah” and “German” samples have been previously described (15). The “UK samples” comprised 755 persistent oligoarticular, extended oligoarticular or polyRFneg JIA cases from three sources: 1) The British Society for Paediatric and Adolescent Rheumatology (BSPAR) National Repository of JIA; 2) UK Caucasian patients with long-standing JIA (34); and 3) a five-center prospective inception cohort collected as part of the Childhood Arthritis Prospective Study (CAPS) (35). Control genotype data was extracted from the Wellcome Trust Case Control Consortium 2 (WTCCC2) European Genome-phenome Archive website (http://www.ebi.ac.uk/ega/page.php). The “Delaware samples” were collected at the Nemours/Alfred I. DuPont Hospital for Children and include 24 JIA cases. This study was approved by the Institutional Review Board of CCHMC and collaborating centers.
Genotyping
For the initial phase, genotyping was done at the Affymetrix Service Center using the Genome-Wide Human SNP Array 6.0. To avoid technical artifacts, samples were arranged in batches that each included cases, controls, both genders and all disease subtypes. The Birdseed (version 2) calling algorithm (Affymetrix, Inc.) yielded an overall call rate of 98.97%. The genotypic data were used to test for cryptic relatedness (duplicates and first degree relatives), autosomal heterozygosity outliers (|Fst statistic| > 0.07), and plate effects. Genotype calling cluster plots were examined by batch for all SNPs reported. SNPs met criteria of having less than 5% missing genotype calls, no evidence of differential missingness between cases and controls (P>0.05), no evidence of departure from expectation in Hardy-Weinberg equilibrium (HWE) proportions (cases P>0.0001; controls P>0.01), and MAF>0.05 in cases and controls. Individual SNPs that violated HWE or had MAF<0.05 and showed strong evidence of association were examined individually in context to flanking markers.
TaqMan® SNP genotyping (Applied Biosystems, Foster City, CA) was done for the Texas, Delaware, Utah and German replication cohorts using either pre-designed or custom assays. Genotyping was performed according to Applied Biosystems’ recommendations using as starting material 16 ng of genomic DNA. Amplification was accomplished using a 384-well format PTC-200 (MJ Research, Hercules, CA) in total volume of 5 μl. PCR conditions were 95°C for 10 min, followed by 50 cycles at 95°C for 15 seconds, and 60°C for 1 min. Following amplification, products were analyzed on an Applied Biosystems 7300 Real-Time PCR System. SNPs were genotyped in UK JIA cases and controls using the Sequenom iPlex MassARRAY platform according to manufacturer’s instructions (Sequenom, San Diego, California, USA, http://www.sequenom.com, accessed 9 July 2010). A 90% sample quality control rate and 90% SNP genotyping success rate was imposed on the analysis. Due to technical genotyping reasons, the UK replication cohort was genotyped for rs10995447 rather than rs10995450 (r2=0.56).
Statistical Methods
Admixture and SNP statistical quality control
To account for potential population substructure, a principal component (PC) analysis was computed using SNPs that passed quality control and were not in genomic regions with long range LD (16). Case-control association analyses were computed adjusting for two PCs that minimized the inflation factor, such that adding additional or other PCs did not further reduce the inflation factor. Replication samples were not genotyped for admixture analysis.
Association Analysis
Four tests of genotypic association were computed using the program SNPGWA (www.phs.wfubmc.edu): dominant, additive and recessive models, and lack-of-fit to an additive model. The additive and recessive models required at least 10 and 30 homozygotes for the minor allele, respectively. The genetic model and odds ratios were defined relative to the minor allele. The primary inference for this study was based on the additive genetic model unless the lack-of-fit to an additive model was statistically significant (P-value<0.05). If the lack-of-fit test was significant, then the minimum P-value from the dominant, additive or recessive models was reported.
Replication Study Analysis
The 10 SNPs taken forward in the replication study represent novel regions not previously reported for JIA (15); in the replication study a few SNPs associated with JIA were chosen that were in linkage disequilibrium with each other. In all cases, SNP intensity plots showed robust genotype calling and out-of-study and local controls were consistent in allele frequency. The replication genotypes were tested for association by meta-analysis using the weighted inverse normal method, where the weights were the square root of the sample size. Here, the 24 Delaware JIA cases were combined with the large UK collection due to a lack of regional controls. Finally, a meta-analysis of the Discovery and Replication cohorts was also computed both weighted and unweighted. For the former, the weighted inverse normal method was used and weighted by the square root of the sample size. Since rs10995447 was genotyped instead of rs10995450 in the UK replication cohort, the meta-analysis of rs109995447 combined the evidence of association only from the Discovery and UK cohorts.
eQTL analysis
Gene expression values derived from peripheral blood mononuclear cells (68 JIA cases and 23 healthy controls) were determined using the Affymetrix U133 plus 2.0 GeneChip® as previously reported (16, 31). To test for an association between each SNP in the GWAS and expression level, analysis of covariance was computed using the natural log of expression levels as the outcome and the two PCs as covariates. Due to the small sample size, only dominant and additive genetic models were computed. To assess functional impact, in silico examination of all Table 1 SNPs (or proxy SNPs, r2>0.5, HapMap CEU) was also completed using gene expression data derived from lymphoblastoid cell lines (LCL) for 378 asthmatic children (www.sph.umich.edu/csg/liang/imputation/) (36). Cis associations (P<0.05) are reported. Affymetrix probeset annotations were confirmed by comparing consensus sequences in Refseq databases.
Table 1. Genome-wide Association Results in the JIA Discovery Cohort*.
| Sample size (N) | MAF | Genotype freq (AA/AB/BB) | Association Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| SNP | Gene | Chr | Mb | MA | JIA | Ctrl | JIA | Ctrl | JIA | Ctrl | P-value | OR[95%CI] |
| rs6766899 | CDGAP | 3 | 120.59 | T | 806 | 3043 | 0.26 | 0.22 | 0.54/0.41/0.06 | 0.62/0.34/0.05 | 9.82E-05 | 1.37[1.17-1.60] |
| rs4688011 | C3orf1 | 3 | 120.71 | T | 814 | 3046 | 0.24 | 0.19 | 0.59/0.35/0.07 | 0.66/0.3/0.03 | 1.88E-06 | 1.37[1.21-1.57] |
| rs13139573 | IL15 | 4 | 142.84 | T | 813 | 3058 | 0.44 | 0.48 | 0.33/0.47/0.2 | 0.26/0.52/0.22 | 2.44E-04 | 0.73[0.62-0.86] |
| rs4254850 | IL15 | 4 | 142.91 | G | 811 | 3058 | 0.43 | 0.47 | 0.34/0.46/0.19 | 0.27/0.51/0.22 | 7.84E-05 | 0.72[0.61-0.85] |
| rs10995447 | NRBF2-EGR2 | 10 | 64.55 | T | 813 | 3058 | 0.22 | 0.18 | 0.6/0.36/0.04 | 0.67/0.29/0.04 | 1.09E-04 | 1.37[1.17-1.61] |
| rs6479891 | JMJD1C | 10 | 64.68 | T | 814 | 3049 | 0.19 | 0.13 | 0.66/0.31/0.03 | 0.75/0.23/0.02 | 6.10E-08 | 1.59[1.34-1.87] |
| rs10761747 | JMJD1C | 10 | 64.79 | G | 812 | 3056 | 0.26 | 0.21 | 0.55/0.38/0.07 | 0.63/0.32/0.05 | 7.44E-05 | 1.29[1.14-1.46] |
| rs12411988 | REEP3 | 10 | 64.99 | C | 814 | 3058 | 0.18 | 0.13 | 0.66/0.31/0.02 | 0.76/0.22/0.02 | 1.16E-07 | 1.57[1.33-1.86] |
| rs12719740 | IGF1R-FAM169B | 15 | 96.89 | T | 812 | 3058 | 0.23 | 0.17 | 0.6/0.35/0.05 | 0.69/0.28/0.03 | 6.80E-08 | 1.45[1.27-1.66] |
| rs9302588 | CHD9-TOX3 | 16 | 51.55 | T | 814 | 3058 | 0.23 | 0.18 | 0.59/0.36/0.05 | 0.67/0.29/0.04 | 8.13E-06 | 1.44[1.23-1.68] |
Chr=chromosome; Mb=megabase; MA=minor allele; MAF=minor allele frequency; Ctrl=controls; OR=odds ratio; CI=95% confidence interval. The additive model is presented unless the test for lack of fit to an additive model was significant (P < 0.05) in the Discovery cohort, as for rs6766899, rs13139573, rs4254850, rs10995447, rs6479891, rs12411988 and rs9302588 (all dominant). Positions are from NCBI build 36 throughout.
Estimation of genetic variance
The cumulative variance explained by common SNP variation was estimated using a variance component model and Restricted Maximum Likelihood estimation as implemented in the GCTA software package (27). The variance component models adjusted for gender and the two PCs for population structure, with separate variance components for each chromosome and one for the extended MHC region extending from HIST1H2AA (telomeric end) to RPL12P1 (centromeric end). Estimates using Yang’s correction factor (c=0 from formula 9) for imperfect LD with causal variants were nearly identical (not shown). The estimates employ SNPs within the GWAS that had <1% missing (555,355 SNPs). In addition, individuals were excluded using a relatedness threshold of 0.025 consistent with Yang et al. (27). The stringent relatedness criteria resulted in dropping 82 individuals from the within study analysis and 219 individuals when out-of-study controls were included in the analysis. To note, results were comparable using GWAS data with and without these additional exclusions.
Results
The Discovery cohort included 814 JIA cases, 658 local controls of self-reported non-Hispanic, European-American ancestry and 2400 “out-of-study” controls from the Molecular Genetics of Schizophrenia non-GAIN samples available from dbGAP. The demographic details are provided in Supplemental Table 1. The cases were classified based on the ILAR revised criteria for JIA (17). To control for potential population substructure, two principal components were identified and included in the logistic regression model as covariates. The genome-wide inflation factor was 1.04 and there was little evidence of systematic departure from expectation as shown in the Q-Q plot (Figure S1). A total of 561,137 SNPs had MAF>0.05, no differential missingness between cases and controls, < 5% missing data and no evidence of HWE departures (P<0.01 in controls, P<0.0001 in cases). P-values of ≤ 5 × 10−8 were considered significant for genome-wide testing.
The results of the principal component adjusted analysis identified both HLA and non-HLA associations with JIA (Figure 1A). The strongest associations were within the MHC region and include HLA Class I and Class II loci (Figure 1B) as well as non-HLA loci robust to linkage disequilibrium (manuscript in preparation). The strongest SNP associations outside the MHC region included novel loci as well as loci implicated in other autoimmune diseases. We have previously reported the overlap with autoimmune disease susceptibility loci in this dataset (14). The shared autoimmune loci are labeled in blue on the Manhattan plot (Figure 1A). To extend association findings for JIA, ten SNPs, representing five regions not yet reported in JIA, were selected for replication testing in independent samples. These SNPs and the association results for the Discovery cohort are listed in Table 1 and represent chromosomal regions 3q13, 4q31, 10q21, 15q26 and 16q12.
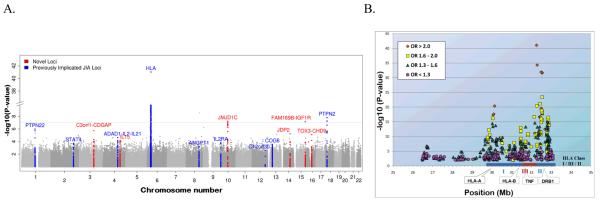
Figure 1.
A. JIA genome-wide association results (−log10[p]) plotted on a genomic scale (Manhattan plot). The horizontal line represents a p-value of 5×10−8. This conservative threshold for genome-wide significance ignores linkage disequilibrium in this Caucasian cohort. B. Association results for the extended MHC region (chromosome 6, 26-34 Mb). SNPs with P < 0.01 are represented and color-coded by odds ratio (OR) strata. SNPs of interest include (i) rs1035798 (position 32,259,200 bp, gene AGER; OR=2.32, P=9.5 × 10-42); (ii) rs2071286 (position 32,287,874 bp, gene NOTCH4; OR=2.21, P=5.0 × 10-35); (iii) rs10947261 (position 32,481,210 bp, gene BTNL2; OR=2.66, P=1.1 × 10-32); (iv) rs9268858 (position 32,537,736 bp, 17 kb from HLA-DRA; OR=2.70, P=1.9 × 10-32); (v) rs9268853 (position 32,537,621 bp, 17 kb from HLA-DRA; OR=2.70, P=2.3 × 10-32); (vi) rs9366752 (position 30,132,656 Mb, 2 kb from HLA-H; OR=2.14, P=1.9 × 10-21).
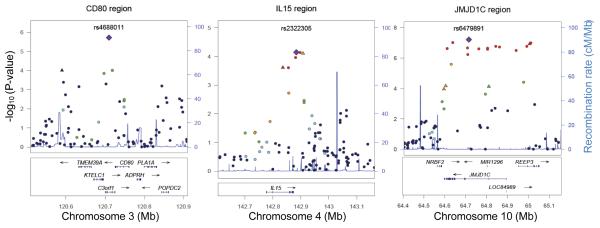
Considering the genome-wide association analysis in the Discovery cohort, evidence for association with JIA was found for the chromosome 3q13 region which includes the genes CD80, KTELC1 and C3orf1. Visual resolution of the association pattern across this region is provided in Figure 2 and indicates that the signal maps to a single linkage disequilibrium block. Replication was attempted for rs4688011 and rs6766899 based on the Discovery cohort statistical findings (OR=1.37, P=1.9×10−6 and OR=1.37, P=9.8×10−5, respectively). Association with JIA was consistent in the Replication cohort for rs4688011 (OR=1.18, P=0.0033) and was the strongest of all markers tested in meta-analyses (OR=1.23, P=3.6×10−7, Table 2). Due to the disparate relative sample sizes of the cohorts, the weighted meta-analysis tends to be dominated by the UK cohort. Weighting each cohort equally modestly increased the evidence of association for both of these loci (Table 2).
Figure 2.
Regional plots of JIA loci. Genotyped and imputed SNPs are plotted with their Discovery P-values (−log10[p]) as a function of genomic position (assembly hg18) within a 400 kb region surrounding the most significant SNP (purple diamond). Recombination rates from the HapMap Phase II CEU are plotted in blue to reflect the regional linkage disequilibrium structure. In each region the index SNP is represented by a large purple diamond, and the color of all other SNPs (circles) indicates LD with the index SNP based on pairwise r2 values from HapMap CEU (red, r2 > 0.8; orange, r2 = 0.6-0.8; green, r2 = 0.4-0.6; light blue, r2 = 0.2-0.4; dark blue, r2 < 0.2). SNPs chosen for replication are represented by a triangle. Known human genes in the UCSC Genome Browser are indicated at the bottom of the plot.
Table 2. Replication of JIA association findings*.
| Meta-Analysis of Replication Cohorts | Meta-Analysis Combined Discovery and Replication Cohorts |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||||
| Sample size (N) |
MAF | MAF | |||||||||||||
|
| |||||||||||||||
| SNP | Gene | Chr | Mb | MA | JIA | Ctrl | JIA | Ctrl | P- value |
OR[95%CI] | JIA | Ctrl | Weighted P-value |
OR[95%CI] | Unweighted P-value |
|
|
|
||||||||||||||
| rs6766899 | CDGAP | 3 | 120.59 | T | 1726 | 6997 | 0.24 | 0.22 | 0.2260 | 1.08[0.96-1.21] | 0.24 | 0.22 | 1.55E-03 | 1.15[1.05-1.26] | 2.60E-04 |
| rs4688011 | C3orf1 | 3 | 120.71 | T | 1722 | 6982 | 0.21 | 0.19 | 0.0033 | 1.18[1.06-1.3] | 0.22 | 0.19 | 3.60E-07 | 1.23[1.13-1.33] | 1.23E-07 |
| rs13139573 | IL15 | 4 | 142.84 | T | 1702 | 6985 | 0.45 | 0.46 | 0.0222 | 0.87[0.77-0.99] | 0.45 | 0.47 | 8.18E-05 | 0.83[0.75-0.92] | 7.11E-05 |
| rs4254850 | IL15 | 4 | 142.91 | G | 969 | 1573 | 0.46 | 0.47 | 0.5001 | 1.39[0.82-2.34] | 0.44 | 0.46 | 9.93E-02 | 1.16[0.79-1.69] | 4.37E-04 |
| rs10995447 |
NRBF2- EGR2 |
10 | 64.55 | T | 969 | 1607 | 0.18 | 0.18 | 0.6480 | 0.88[0.58-1.35] | 0.20 | 0.18 | 7.49E-02 | 1.00[0.73-1.36] | 7.46E-04 |
| rs10995450 |
NRBF2- EGR2 |
10 | 64.56 | T | 754 | 5380 | 0.20 | 0.18 | 0.0465 | 1.14[1.00-1.31] | 0.21 | 0.18 | 5.39E-05 | 1.21[1.10-1.34] | 6.69E-05 |
| rs6479891 | JMJD1C | 10 | 64.68 | T | 1728 | 6982 | 0.15 | 0.14 | 0.1920 | 1.06[0.93-1.21] | 0.16 | 0.14 | 4.33E-05 | 1.19[1.07-1.32] | 2.26E-07 |
| rs10761747 | JMJD1C | 10 | 64.79 | G | 1719 | 6987 | 0.22 | 0.21 | 0.1430 | 1.05[0.95-1.16] | 0.23 | 0.21 | 6.35E-04 | 1.11[1.02-1.20] | 1.32E-04 |
| rs12411988 | REEP3 | 10 | 64.99 | C | 1701 | 6978 | 0.14 | 0.13 | 0.1320 | 1.06[0.93-1.21] | 0.15 | 0.13 | 2.71E-05 | 1.18[1.06-1.31] | 2.91E-07 |
| rs12719740 |
IGF1R- FAM169B |
15 | 96.89 | T | 1725 | 6974 | 0.19 | 0.19 | 0.5670 | 1.05[0.95-1.17] | 0.20 | 0.18 | 5.19E-04 | 1.15[1.06-1.25] | 6.97E-07 |
| rs9302588 |
CHD9- TOX3 |
16 | 51.55 | T | 1728 | 6957 | 0.21 | 0.20 | 0.3700 | 1.03[0.92-1.16] | 0.21 | 0.19 | 1.27E-03 | 1.13[1.03-1.25] | 4.13E-05 |
|
|
|
||||||||||||||
Abbreviations and genetic models are the same as in Table 1. Replication results are from a meta- analysis of cohorts from Delaware, Germany, Texas, UK and Utah. For technical reasons in the UK samples, rs10995450 was genotyped as a proxy for rs10995447 (r2=0.55 and p-value = 6.74E-05; OR 1.31[1.15-1.50] in the Discovery cohort). The meta-analyses for the Replication Cohort and the combined Discovery and Replication Cohorts were computed using the weighted inverse normal method. The combined cohorts were also computed with equal weighting of the cohorts (unweighted).
Compelling evidence for genetic association was also found at 10q21, a region that has not been reported for any other autoimmune disease. Specifically, rs6479891 (OR=1.59, P=6.1×10−8) is located within the jumonji domain containing 1C (JMJD1C) gene and represents the single strongest finding outside the MHC region for the Discovery cohort. Analysis of the Replication cohort for SNPs in the 10q21.3 region revealed odds ratios that were consistent in direction, but of smaller magnitude and less significance (Table 2) than the Discovery cohort. Notably, only one of the 5 SNPs, rs10995450, reached P<0.05 levels of statistical significance in the Replication cohort and noting that only the Discovery and UK cohorts were genotyped for this SNP. Weighting each cohort equally increased the evidence for association for all but rs10995450, which had comparable evidence under either approach.
An eQTL analysis was performed to assess the potential impact of the polymorphisms on expression levels of neighboring genes using two resources: 1) a dataset comprised of 68 JIA cases and 23 healthy controls from the GWAS and 2) an online dataset with GWAS and expression analysis from lymphoblastoid cells lines (LCL) collected from individuals with asthma (18).
The SNPs that showed association in 3q13 were significantly associated with expression levels in the eQTL analyses (Table 3). Specifically, in the JIA/control dataset, both rs4688011 in C3orf1 and rs6766899 in CDGAP were associated with the TMEM39A probe set 222690_s_at (P=0.024 and P=0.0097, respectively). The order of genes on chromosome 3, and illustrated in Figure 2, depicts TMEM39A as located between CDGAP and C3orf1. Interestingly, rs6766899 genotypes correlated with expression levels for CD80 (P=0.065, 1554519_at) as well. Considering the publically available online LCL dataset relative to our chromosome 3 results, we found that rs4447803, which is in LD with rs4688011 (r2=0.54), was strongly associated with KTELC1 expression (P=5.7×10−6) with differences in expression levels comparable in magnitude to reports of KTELC1 expression in celiac disease (19). (Note: for Celiac disease, the associated SNP is rs1599796, which is in almost complete LD with rs4688011 (r2 = 0.95 in CEU)). Neither C3orf1 nor CD80 expression data was available in the online LCL dataset.
Table 3. Significant findings for eQTL analysis considering regions supported by genetic association*.
| cis eQTL | Normalized expression values based on genotype |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Chr | SNP (locus) | Mb | Expression Probe set |
Distance (kb) |
Gene | P-value | MA | 1 | 2 |
| 3 | rs6766899 (CDGAP) |
120.59 | 222690_s_at | 42.45 | TMEM39 A |
0.0097 | T | 1.01 | 0.92 |
| 240232_at | 110.12 | C3orf1 | 0.031 | 1.08 | 0.83 | ||||
| 1554519_at | 135.89 | CD80 | 0.065 | 1.00 | 0.99 | ||||
| 238505_at | 191.27 | ADPRH | 0.0364 | 1.17 | 0.97 | ||||
|
| |||||||||
| rs4688011 (C3orf1) |
120.71 | 222690_s_at | 44.78 | TMEM39 A |
0.024 | T | 1.04 | 0.98 | |
| 228042_at | 71.28 | ADPRH | 0.0339 | 1.01 | 0.96 | ||||
| rs4447803 (C3orf1) |
120.72 | 218587_s_at | 22.42 | KTELC1 | §5.7E-06 | A | NA | NA | |
|
| |||||||||
| 10 | rs10995447 | 64.55 | 230007_at | 47.47 | JMJD1C | 0.0318 | T | 0.99 | 1.02 |
| rs10995450 (NRBF2- EGR2) |
64.56 | 223650_s_at | 4.10 | NRBF2 | 0.0303 | T | 1.13 | 0.97 | |
| 241391_at | 38.08 | JMJD1C | §0.00058 | NA | NA | ||||
|
| |||||||||
| rs6479891 (JMJD1C) |
64.68 | 223650_s_at | 91.67 | NRBF2 | 0.0109 | T | 1.12 | 1.00 | |
| 230007_at | 0 | JMJD1C | 0.042 | 1.05 | 1.04 | ||||
|
| |||||||||
| rs10761747 (JMJD1C) |
64.79 | 223650_s_at | 193.37 | NRBF2 | 0.0306 | G | 1.12 | 0.99 | |
| 1556622_s_at | 0 | JMJD1C | 0.0442 | 1.03 | 0.90 | ||||
| 241391_at | 0 | JMJD1C | §1.6E-06 | NA | NA | ||||
|
| |||||||||
| rs12411988 (REEP3) |
64.99 | 230007_at | 89.68 | JMJD1C | 0.008 | C | 1.04 | 1.04 | |
Principal component-adjusted p-values were used for the local dataset. Lymphoblastoid cell line data available at http://www.sph.umich.edu/csg/liang/imputation/ provide additional support for functional relationships between genotype and gene expression levels (p-values from this dataset are denoted by “§”. rs4447803 is a SNP proxy (r2= 0.54) for rs4688011. For eQTL analysis, the expression values were log-transformed. The distance from SNP to nearest end of gene is from NCBI build 36. MA = minor allele. “1” and “2” refer to number of copies of the genotyped minor allele. The geometric mean of normalized expression values were computed for genotypes 0, 1 and 2 and the results for “1” and “2” are presented as a ratio to the “0” category (homozygous major allele). NA=data not available.
Similarly, the SNPs that showed association with JIA in 10q21 were associated with certain expression levels (Table 3). Specifically, rs12411988 and rs6479891 were associated with expression levels measured by the JMJD1C probe set 230007_at (P=0.008 and P=0.042, respectively; Table 3) and other JIA-associated SNPs in the region were also associated with either JMJD1C or NRBF2 expression. These included SNPs in modest linkage disequilibrium (LD) with rs6479891, such as rs10761747 and rs10995450 (r2=0.48 and 0.57, respectively). This is notable since in the online LCL dataset, both SNPs were associated with differential expression for the JMJD1C probe set 241391_at (P=1.6×10−6 and P=5.8×10−4, respectively) (18). In total, there were eight SNPs from the JMJD1C region in the online dataset that were highly associated with expression of this gene. These SNPs were in high LD (r2>0.77) with JIA associated SNPs and located within or near JMJD1C which encompasses a region of over 150 kb (Figure 2). Furthermore, one of the SNPs that relates to gene expression values (rs10761725) codes for a conservative substitution (Ser to Thr) in the JMJD1C protein, but is not available in the Affymetrix SNP dataset.
A third region of interest identified by genome-wide analysis was located at 4q31 and includes the gene encoding interleukin 15 (IL15). One SNP, rs4254850 showed evidence of association (OR=0.72; P=7.8×10−5), but not replication, whereas a second nearby SNP in high LD with rs425850 with more modest results in the Discovery cohort was supported by modest evidence of association in the Replication cohort (rs13139573, r2=0.84, Discovery cohort P=2.4×10−4; Replication cohort P=0.02). The regional association plot (Figure 2) highlights the statistical strength of the association in this region and the localization of the signal to the IL15 gene. No correlation with expression levels was found for IL15 loci in the eQTL analysis.
Replication was also attempted for two additional loci identified in the genome-wide analysis. An association with JIA was found for rs9302588 in the 16q12 region (OR=1.44, P=8.1×10−6, Discovery Cohort), but this finding lacked evidence for association in the Replication cohort. Similarly, rs12719740 on 15q26 was associated with JIA in the Discovery cohort (OR=1.45, P=6.8×10−8) but not supported by replication.
While the genetic architecture of JIA susceptibility has been informed by this and other studies, it still remains unclear how much common genetic variation accounts for the risk to JIA, as opposed to rare variants, epigenetics and gene-gene as well as gene-environment interactions. To address this, we used this GWAS data to estimate the fraction of the JIA risk that could be attributed to common SNP variation (minor allele frequency > 0.05) using a variance component threshold liability model (27) assuming a disease prevalence ranging from 25 to 140 per 100,000 (28). We found that common SNP variation accounts for an estimated one third of JIA susceptibility. This result holds even without the out-of study controls (Table 4). When partitioning this estimate into the individual chromosomes and extended MHC region, the extended MHC accounted for about ~8% while the remainder of the genome accounted for ~20% of the variation in JIA susceptibility.
Table 4.
Estimates of phenotypic variance in JIA susceptibility explained by genome-wide single nucleotide polymorphisms among unrelated individuals using a threshold model and the restricted maximum likelihood method*.
| Estimates of Explained Variance on Liability Scale (±SE) |
||||||
|---|---|---|---|---|---|---|
| Combined(Within and Out-of-Study Controls) |
Within Study Data Only |
|||||
|
|
||||||
| Prevalence of JIA |
Entire Genome |
xMHC Region |
Non- xMHC |
Entire Genome |
xMHC Region |
Non- xMHC |
| 25 per 100,000 | 0.30±0.04 | 0.08±0.01 | 0.18±0.04 | 0.29±0.03 | 0.05±0.01 | 0.19±0.07 |
| 40 per 100,000 | 0.32±0.04 | 0.08±0.01 | 0.19±0.04 | 0.31±0.03 | 0.06±0.01 | 0.21±0.07 |
| 50 per 100,000 | 0.33±0.04 | 0.09±0.01 | 0.20±0.04 | 0.32±0.03 | 0.06±0.01 | 0.21±0.07 |
| 80 per 100,000 | 0.35±0.04 | 0.09±0.01 | 0.21±0.04 | 0.34±0.03 | 0.06±0.01 | 0.23±0.08 |
| 140 per 100,000 | 0.38±0.04 | 0.10±0.01 | 0.23±0.05 | 0.38±0.03 | 0.07±0.01 | 0.25±0.08 |
Estimates are provided ± standard error. Single nucleotide polymorphisms used had less than 1% missing data, no evidence of differential missingness between cases and controls (P>0.05), and no evidence of departure from Hardy Weinberg equilibrium proportions (controls P>0.01, cases P>1×10−6). The extended Major Histocompatibility Complex (xMHC) is defined as the region extending from HIST1H2AA (telomeric end) to RPL12P1 (centromeric end),.
Discussion
This work represents, to our knowledge, the largest GWAS of JIA cases to date and focuses on the two most common subtypes, oligoarticular and polyRFneg JIA subtypes. There was no evidence that the associations varied between these two subtypes for the SNPs included in the replication studies. Sufficient numbers of samples are not yet available to allow JIA subtypes to be considered independently. Oligoarticular and polyRFneg JIA subtypes have overlapping HLA associations, share a female gender bias, and are distinguished primarily based on the number of joints involved. Thus, a priori they are the most logical subtypes to combine to maximize the power to detect common JIA susceptibility loci and minimize heterogeneity.
We report novel JIA-associated loci and supporting eQTL results that extend the JIA associations beyond those previously reported (PTPN2, PTPN22, IL2RA, ADAD1-IL2-IL21, ANGPT1, COG6, C12orf30 and STAT4). The eQTL analyses are reinforced by two datasets which include a subgroup of the same patients and controls used in our GWAS as well as a publically available dataset that measures gene expression in lymphoblastoid cell lines. The strongest replicated evidence for association with JIA was found at the chromosome 3q13 region which includes CD80, a costimulatory molecule necessary for T cell activation, and KTELC1, an O-glucosyltransferase that modifies the Notch receptor in Drosophila (29). Notch signaling is important at several stages of T cell development and differentiation and has been proposed as a target for selective therapy to treat autoimmune disorders. Consistent with a theme of overlapping association findings in autoimmune disease are reports of genetic association in this region for celiac disease that can be related to gene expression findings (19).
Evidence for association within jumonji domain containing 1C (JMJD1C) was observed in the Discovery cohort and although consistent in direction, a less significant effect was detected in the Replication cohort. JMJD1C encodes a hormone-dependent transcription factor that regulates expression of a variety of target genes. It includes a jumonji-domain that functions by removing methyl marks on histones that are associated with gene regulation (30). JMJD1C expression has been reported in multiple immune cells, including B cells as well as CD4+ and CD8+ T cells (www.biogps.gnf.org). It is noteworthy that a gene expression signature (50 expression probe sets) that identified a subset of JIA patients characterized with a disease course of chronically active arthritis included probe sets specific for both JMJD1C and the previously reported PTPN2 loci (14, 31). This overlap of findings cannot be explained by chance and suggests a functional relationship between genetic differences and gene expression findings in processes related to disease pathogenesis. This region merits further study not only to validate the genetic association, but also to explore the patterns of histone methylation that may relate to disease.
There are additional SNPs-JIA associations that have not yet been considered in replication studies. A listing of the 200 top statistical associations is provided in Supplemental Table S2 and the complete results from the association analyses are available (http://research.cchmc.org/ARDec11). Table S2 includes additional SNPs in the regions discussed above and in genes where associations have been reported in other autoimmune diseases but not yet investigated in JIA. For example, there is evidence of association at 2p14, where the most significant SNP in JIA (rs268132) is located within intron 1 of SPRED2. This gene encodes the sprouty-related, EVH1 domain-containing protein 2, which has been shown to regulate growth factor-induced activation of the MAP kinase cascade and was reported as a rheumatoid arthritis risk locus (20). For rheumatoid arthritis, the most significant SNP, rs934734, is also in intron 1 and in high LD with the JIA associated SNP, rs268132 (r2=0.78). Similarly, rs423847 (OR=1.42, P=2.4×10-5) in ANTXR2 has evidence for association with JIA. This gene located at 4q21 encodes capillary morphogenesis protein-2 and has been associated with ankylosing spondylitis (rs4333130) (21). Our findings also include SNPs in PTPRS (receptor protein-tyrosine phosphatase sigma), a gene which has been associated with ulcerative colitis (22) and NOS2, which encodes inducible nitric oxide synthase and is a susceptibility loci for psoriasis (23). Finally, a SNP (rs2548997, P=1.82×10-4) near the IRF1 gene is within the 5q31.1 region, which also harbors IL-4, IL-5 and IL-13 is supported for association. This region was studied early on for association in JIA and produced conflicting results (24, 25) perhaps due to population substructure or JIA phenotype heterogeneity. Thus our findings argue for further studies in this important region which includes genes related to a polarization of cytokine repertoires, a phenomenon that has been reported in JIA (26).
The results confirm and firmly establish the role of genetic influences on the susceptibility to JIA and continue to provide evidence that common genetic variation is very important in explaining JIA risk. The estimated genetic variance explained by common variation is higher than estimates for other complex genetic traits such as Crohn’s disease but similar to type 1 diabetes which also has onset in childhood (32). In adult arthritis, it is estimated that the 27 non-HLA loci confirmed to date and reported by Stahl et al in 2010 (20) account for only 10.7% of the genetic susceptibility (33). We estimated that about one third of JIA risk can be attributed to common genetic variation and that the extended MHC region accounts for approximately one quarter of the heritable risk (0.25=0.08/0.32). Thus, continued mining of JIA GWAS data in expanded cohorts and for individual JIA subtypes is warranted and will likely lead to the discovery of additional susceptibility loci.
As noted before, the disparate relative sample sizes among the Replication cohorts tended to allow the UK cohort to dominate the results. Weighting each cohort equally in the meta-analysis (not removing UK cohort but weighting equally) yielded increased statistical evidence in all of the regions in Table 2 and for all SNPs except rs10995450 whose evidence was comparable. Recognizing that the WTCCC is not a North American cohort, suggests further research is needed to determine whether the differing results were likely sampling variation, false associations or population differences.
In summary, the novel JIA associated loci identified here in an agnostic scan merit study in other JIA and autoimmune disease cohorts. Furthermore, integrative analyses of expression and association data will further accelerate the discovery of the underlying genetic architecture of JIA and ultimately lead to molecular profiles potentially relevant to diagnosis, outcomes and therapeutic response.
Supplementary Material
Acknowledgements
We gratefully acknowledge contributions from physicians at CCHMC, the Medical College of Wisconsin, Schneider Children’s Hospital and Children’s Hospital of Philadelphia for the collection of patient samples and the assistance of Sandy Kramer for patient recruitment at Cincinnati Children’s Hospital Medical Center and coordination of clinical information. In addition, we also acknowledge David R. McWilliams, Ph.D. for the Manhattan Plots. Computing support was provided by the Wake Forest School of Medicine Center for Public Health Genomics. This study makes use of data generated by the Wellcome Trust Case-Control Consortium 2 and funded under award 085475. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk. The normal control DNA collection in the Discovery cohort including all genotypes was supported and made available by Cincinnati Children’s Hospital Medical Center.
Funding
This work was supported by the National Institutes of Health [RC1-AR-058587, P30-AR-473639 to SDT, K23-AR-50177 to SP, R01-AR-057106 to CDL and SDT, P01-AR048929 and N01-AR-42272 to DNG]; the Federal Ministry of Education and Research Germany (BMBF) [grants 01GM0907 and 01 ZZ 0403 to JPH]; the Val A. Browning Foundation; the Texas Scottish Rite Hospital for Children [grant 0305756 to CW and MP]; the Arthritis Foundation and Arthritis Research UK [grant reference no:17552 to WT].
Footnotes
No financial support or other benefits from commercial sources have been received for the work reported in the manuscript. None of the authors have financial interests related to the work or in conflict with the work.
References Cited
- 1.Petty RE, Southwood TR, Baum J, Bhettay E, Glass DN, Manners P, et al. Revision of the proposed classification criteria for juvenile idiopathic arthritis: Durban, 1997. J Rheumatol. 1998;25(10):1991–4. [PubMed] [Google Scholar]
- 2.Petty RE, Cassidy JT. Chronic Arthritis in Childhood. In: Cassidy JT, Petty RE, Laxer RM, Lindsley CB, editors. Textbook of Pediatric Rheumatology. 6th ed Elsevier; Philadelphia: 2011. pp. 211–235. [Google Scholar]
- 3.Prahalad S, Shear ES, Thompson SD, Giannini EH, Glass DN. Increased prevalence of familial autoimmunity in simplex and multiplex families with juvenile rheumatoid arthritis. Arthritis Rheum. 2002;46(7):1851–6. doi: 10.1002/art.10370. [DOI] [PubMed] [Google Scholar]
- 4.Saurenmann RK, Rose JB, Tyrrell P, Feldman BM, Laxer RM, Schneider R, et al. Epidemiology of juvenile idiopathic arthritis in a multiethnic cohort: ethnicity as a risk factor. Arthritis Rheum. 2007;56(6):1974–84. doi: 10.1002/art.22709. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan DB, Cassidy JT, Petty RE. Pathogenic implications of age of onset in juvenile rheumatoid arthritis. Arthritis Rheum. 1975;18(3):251–5. doi: 10.1002/art.1780180309. [DOI] [PubMed] [Google Scholar]
- 6.Glass DN, Giannini EH. Juvenile rheumatoid arthritis as a complex genetic trait. Arthritis Rheum. 1999;42(11):2261–8. doi: 10.1002/1529-0131(199911)42:11<2261::AID-ANR1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 7.Hollenbach J, Thompson SD, Bugawan T, Ryan M, Sudman M, Marion M, et al. Juvenile idiopathic arthritis and HLA class I and class II interaction and age-at-onset effects. Arthritis Rheum. 2010;62(6):1781–1791. doi: 10.1002/art.27424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray KJ, Moroldo MB, Donnelly P, Prahalad S, Passo MH, Giannini EH, et al. Age-specific effects of juvenile rheumatoid arthritis-associated HLA alleles. Arthritis Rheum. 1999;42(9):1843–53. doi: 10.1002/1529-0131(199909)42:9<1843::AID-ANR8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 9.Prahalad S, Ryan MH, Shear ES, Thompson SD, Giannini EH, Glass DN. Juvenile rheumatoid arthritis: linkage to HLA demonstrated by allele sharing in affected sibpairs. Arthritis Rheum. 2000;43(10):2335–8. doi: 10.1002/1529-0131(200010)43:10<2335::AID-ANR22>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Behrens EM, Finkel TH, Bradfield JP, Kim CE, Linton L, Casalunovo T, et al. Association of the TRAF1-C5 locus on chromosome 9 with juvenile idiopathic arthritis. Arthritis Rheum. 2008;58(7):2206–7. doi: 10.1002/art.23603. [DOI] [PubMed] [Google Scholar]
- 11.Hinks A, Barton A, Shephard N, Eyre S, Bowes J, Cargill M, et al. Identification of a novel susceptibility locus for juvenile idiopathic arthritis by genome-wide association analysis. Arthritis Rheum. 2009;60(1):258–63. doi: 10.1002/art.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prahalad S, Hansen S, Whiting A, Guthery SL, Clifford B, McNally B, et al. Variants in TNFAIP3, STAT4, and C12orf30 loci associated with multiple autoimmune diseases are also associated with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60(7):2124–30. doi: 10.1002/art.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinks A, Eyre S, Ke X, Barton A, Martin P, Flynn E, et al. Overlap of disease susceptibility loci for rheumatoid arthritis (RA) and juvenile idiopathic arthritis (JIA) Ann Rheum Dis. 2009 doi: 10.1136/ard.2009.110650. ard.2009.110650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson SD, Sudman M, Ramos PS, Marion MC, Ryan M, Tsoras M, et al. The susceptibility loci juvenile idiopathic arthritis shares with other autoimmune diseases extend to PTPN2, COG6, and ANGPT1. Arthritis Rheum. 2010;62(11):3265–76. doi: 10.1002/art.27688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnes MG, Grom AA, Thompson SD, Griffin TA, Pavlidis P, Itert L, et al. Subtype-specific peripheral blood gene expression profiles in recent-onset juvenile idiopathic arthritis. Arthritis Rheum. 2009;60(7):2102–12. doi: 10.1002/art.24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83(1):132–5. doi: 10.1016/j.ajhg.2008.06.005. author reply 135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31(2):390–2. [PubMed] [Google Scholar]
- 18.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39(10):1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 19.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reveille JD, Sims AM, Danoy P, Evans DM, Leo P, Pointon JJ, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42(2):123–7. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muise AM, Walters T, Wine E, Griffiths AM, Turner D, Duerr RH, et al. Protein-tyrosine phosphatase sigma is associated with ulcerative colitis. Curr Biol. 2007;17(14):1212–8. doi: 10.1016/j.cub.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Stuart PE, Nair RP, Ellinghaus E, Ding J, Tejasvi T, Gudjonsson JE, et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet. 2010;42(11):1000–4. doi: 10.1038/ng.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donn RP, Barrett JH, Farhan A, Stopford A, Pepper L, Shelley E, et al. British Paediatric Rheumatology Study Group Cytokine gene polymorphisms and susceptibility to juvenile idiopathic arthritis. Arthritis Rheum. 2001;44(4):802–10. doi: 10.1002/1529-0131(200104)44:4<802::AID-ANR136>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 25.Fife MS, Gathercole L, Ogilvie EM, Stock CJ, Mack LF, Donn RP, et al. No evidence for genetic association of interferon regulatory factor 1 in juvenile idiopathic arthritis. Arthritis Rheum. 2007;56(3):972–6. doi: 10.1002/art.22425. [DOI] [PubMed] [Google Scholar]
- 26.Scola MP, Thompson SD, Brunner HI, Tsoras MK, Witte D, Van Dijk MA, et al. Interferon-gamma:interleukin 4 ratios and associated type 1 cytokine expression in juvenile rheumatoid arthritis synovial tissue. J Rheumatol. 2002;29(2):369–78. [PubMed] [Google Scholar]
- 27.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petty RE, Cassidy JT. Chronic Arthritis in Childhood. In: Cassidy JT, Petty RE, Laxer RM, Lindsley CB, editors. Textbook of Pediatric Rheumatology. 6 ed Elsevier; Philadelphia: 2011. pp. 211–235. [Google Scholar]
- 29.Ma W, Du J, Chu Q, Wang Y, Liu L, Song M, et al. hCLP46 regulates U937 cell proliferation via Notch signaling pathway. Biochem Biophys Res Commun. 2011;408(1):84–8. doi: 10.1016/j.bbrc.2011.03.124. [DOI] [PubMed] [Google Scholar]
- 30.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–40. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffin TA, Barnes MG, Ilowite NT, Olson JC, Sherry DD, Gottlieb BS, et al. Gene expression signatures in polyarticular juvenile idiopathic arthritis demonstrate disease heterogeneity and offer a molecular classification of disease subsets. Arthritis Rheum. 2009;60(7):2113–2123. doi: 10.1002/art.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88(3):294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orozco G, Eyre S, Hinks A, Bowes J, Morgan AW, Wilson AG, et al. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann Rheum Dis. 2011;70(3):463–8. doi: 10.1136/ard.2010.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Packham JC, Hall MA. Long-term follow-up of 246 adults with juvenile idiopathic arthritis: functional outcome. Rheumatology (Oxford) 2002;41:1428–1435. doi: 10.1093/rheumatology/41.12.1428. [DOI] [PubMed] [Google Scholar]
- 35.Adib N, Hyrich K, Thornton J, Lunt M, Davidson J, Gardner-Medwin J, et al. Association between duration of symptoms and severity of disease at first presentation to paediatric rheumatology: results from the Childhood Arthritis Prospective Study. Rheumatology (Oxford) 2008;47:991–995. doi: 10.1093/rheumatology/ken085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.