Abstract
Cdt1, a protein critical for replication origin licensing in G1 phase is degraded during S phase but re-accumulates in G2 phase. We now demonstrate that human Cdt1 has a separable essential mitotic function. Cdt1 localizes to kinetochores during mitosis through interaction with the Hec1 component of the Ndc80 complex. G2-specific depletion of Cdt1 arrests cells in late prometaphase due to abnormally unstable kinetochore-microtubule (kMT) attachments and Mad1-dependent spindle assembly checkpoint activity. Cdt1 binds a unique loop extending from the rod domain of Hec1 that we show is also required for kMT attachment. Mutation of the loop domain prevents Cdt1 kinetochore localization and arrests cells in prometaphase. Super-resolution fluorescence microscopy indicates that Cdt1 binding to the Hec1 loop domain promotes a microtubule-dependent conformational change in the Ndc80 complex in vivo. These results support the conclusion that Cdt1 binding to Hec1 is essential for an extended Ndc80 configuration and stable kinetochore microtubule attachment.
The cell division cycle is the process of complete and precise duplication of the entire genome in S phase followed by accurate chromosome segregation in mitosis. The formation and stability of kinetochore-microtubule (kMT) attachments during mitosis depends on the Ndc80 complex, Ndc80 (hsHec1), Nuf2, Spc24 and Spc251, 2. Hec1/Nuf2 and Spc24/25 form dimers respectively that are tethered together at the C-termini of Hec1/Nuf2 and the N-termini of Spc24/Spc25 by long alpha helical coiled-coil rod domains. In the middle of the rod domain is a “hinge” site produced by a ~40 amino acid loop in Hec1 that is thought to play a key role in MT binding dynamics and attachment3. Binding partners for the loop region of yeast Hec1/Ndc80 proteins have been identified4, 5, but the partners and function of the unique Hec1 loop domain at metazoan kinetochores remains unknown. In this study, we identify the Cdt1 protein as an essential partner for Ndc80 dynamic function through interaction with the Hec1 loop domain.
Cdt1 is required for DNA replication origin licensing, the initial step in genome duplication, which occurs in G1 phase6, 7. Proteins involved in origin licensing are not strictly DNA replication factors however; some licensing proteins have functions in transcription8, 9, the DNA damage response10, 11, centrosome duplication12, and mitosis13, 14. We report here the previously undiscovered mitotic role for the Cdt1 replication licensing protein that is distinct from its role in DNA replication.
Results
Inhibition of Cdt1 function induces mitotic arrest
Both Cdc6 and Cdt1 proteins are required for replication licensing in human cells, but we found an intriguing difference in the phenotypes of Cdc6-depleted cells compared to Cdt1-depleted cells. Cdc6-depleted cells arrest primarily in G1 as expected15, 16, but Cdt1-depleted cells arrested in both G1 and G2/M, despite the fact that both proteins cooperate in the same DNA replication step, MCM loading (Fig. 1a). The accumulation of G2 cells suggested a possible unique mitotic role for Cdt1 which we set out to test.
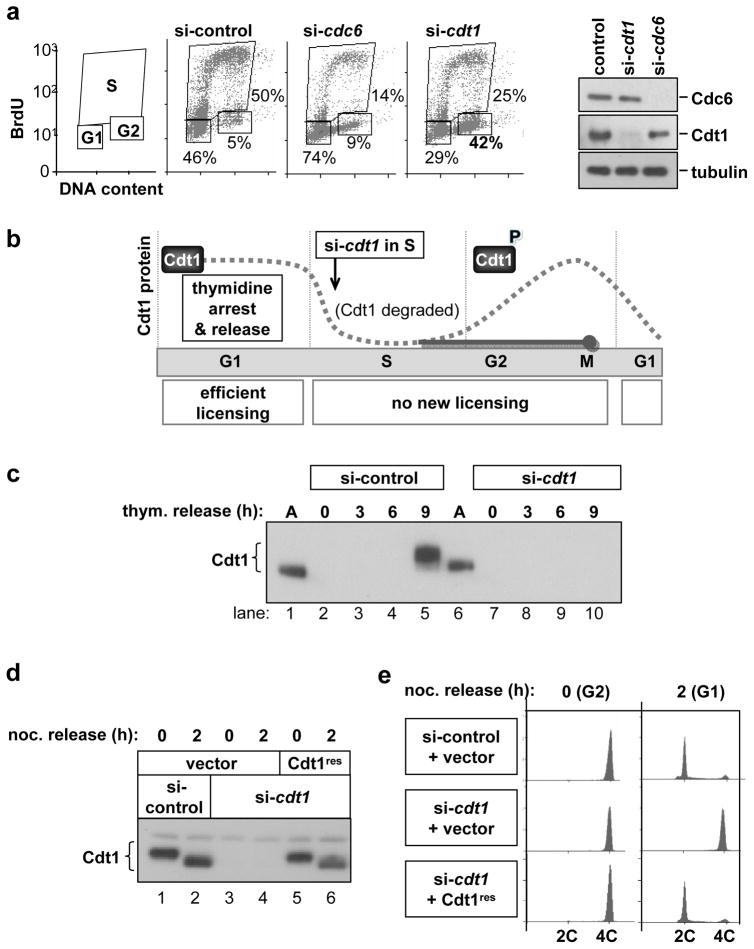
Figure 1. Cells depleted of Cdt1 after S phase do not complete cell division.
(a) Normal human fibroblasts (NHF1-htert) were transfected with siRNAs targeting cdc6, cdt1, or GFP (control) mRNAs for 72 hours. Cells were labeled with BrdU for the final hour and then analysed by flow cytometry for cell cycle position and by immunoblotting for endogenous Cdc6, Cdt1, and Tubulin. (b) Diagram of experimental design. Endogenous Cdt1 protein levels normally drop during S phase due to ubiquitin-mediated proteolysis and recover beginning in G2 (dotted gray line). Release from an early S block into cdt1 siRNA transfection medium blocks Cdt1 protein re-accumulation (solid gray line). (c) Immunoblot analysis of endogenous Cdt1 protein in cells synchronized as diagrammed in b; M phase occurs between 9 and 10 hours post-release from the second thymidine arrest. (d) Stable HeLa cell lines transduced with empty vector (lanes 1–4) or an siRNA-resistant form of Cdt1 (“Cdt1res” lanes 5 and 6) were synchronized as in b and transfected with either control siRNA targeting GFP (lanes 1 and 2) or with cdt1 siRNA (lanes 3–6). Cells were released from early S phase into nocodazole for 10 hr and then either harvested (0 hr) or released for 2 hours into G1 phase, and Cdt1 protein levels were analysed by immunoblotting. (e) Flow cytometric analysis of DNA content of cells in d.
To eliminate the possibility that a mitotic phenotype simply reflected replication errors in the preceding S phase, we took advantage of the fact that Cdt1 is actively degraded during S phase after its G1 origin licensing role is complete17 (Fig. 1b). We synchronized cells in early S phase, at which time origins are fully licensed, and released them into medium containing control siRNA or cdt1 siRNA. As expected, control cells had very low amounts of Cdt1 in S phase, and Cdt1 re-accumulated in G2 (Fig. 1c, lanes 1–5). Synchronized cells treated with cdt1 siRNA could not re-accumulate Cdt1 in G2 however (Fig. 1c, lanes 7–10). (Cdt1 is phosphorylated in G2 by stress MAP kinases17, 18) Cdt1 depletion during S phase caused no delay in S phase progression (Fig. S1a and S1b), and by 9 hours after release both control and Cdt1-depleted cells exhibited normal chromosome condensation and/or nuclear envelope breakdown (e.g. Figure 2a and data not shown). This unique experimental approach allowed us to generate cells that underwent a normal G1 and S phase but lacked Cdt1 during G2 and M phase.
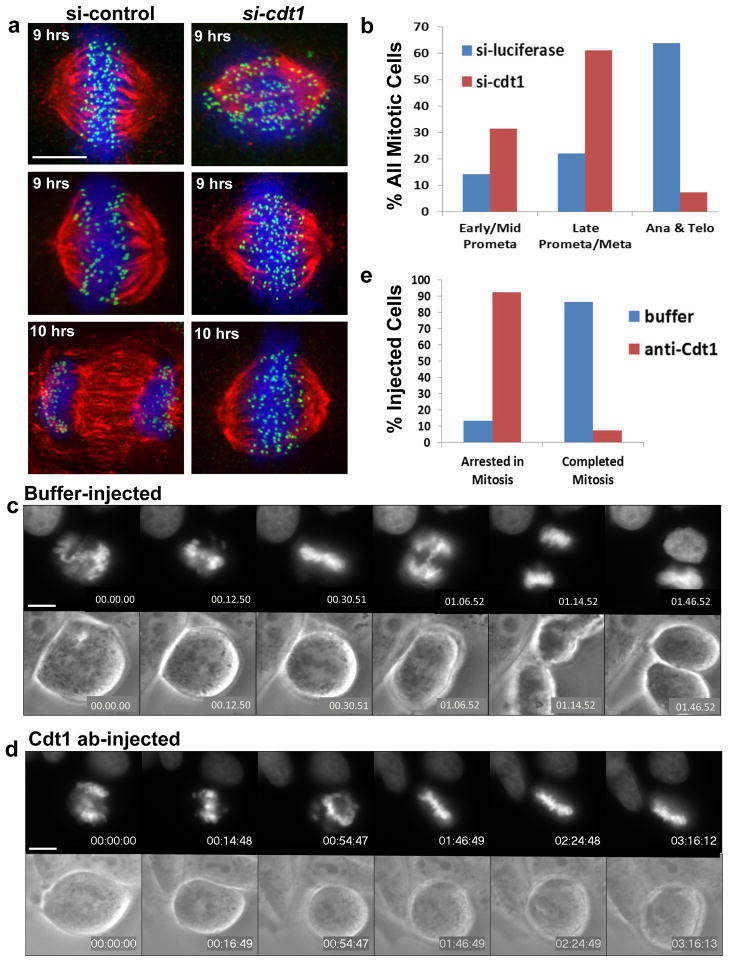
Figure 2. G2-specific Cdt1 inhibition induces mitotic arrest.
(a) HeLa cells synchronized in early S phase were released into control (luciferase) or cdt1 siRNA for either 9 hrs (top and middle panels) or 10 hrs (bottom panels) followed by fixation and staining with DAPI to label chromosomes (blue), anti-tubulin antibody to label MTs (red), and anti-Knl1 antibody to label kinetochores (green). (b) Quantification of the results at 10 hr in a by mitotic stage; n = 1500 cells. (c–e) HeLa cells stably expressing GFP-histone H2B were injected with control buffer (c, n=15) or anti-Cdt1 antibody (d, n=27). Selected frames of GFP-histone (top panels) and phase contrast images (bottom panels) are shown. (e) Quantification of the results from the microinjection experiments in c and d. Scale bars = 5 μm. See also Supplementary Movies S1–S4.
We then tested the ability of Cdt1-depleted cells to transit mitosis to G1 after a nocodazole arrest and release (Fig. 1d and 1e). Control cells completed cell division, but Cdt1-depleted cells remained arrested with G2 DNA content (Fig. 1e, si-control vs. si-cdt1). A cell line stably expressing normal Cdt1 with synonymous mutations in the siRNA target site was fully capable of mitotic progression (Fig. 1d and 1e). Unphosphorylatable Cdt118 also complemented the cell division phenotype of Cdt1-depleted cells (Fig. S1c and S1d). Cells depleted of Cdt1 during S phase did not accumulate phosphorylated H2AX by the subsequent G2, a marker of DNA damage, whereas siRNA transfection of asynchronous cultures to deplete either Cdt1 or another licensing protein, Orc6, resulted in the accumulation of phospho-H2AX-positive foci, presumably from incomplete replication and fork collapse (Figure S1e).
The majority of the synchronized Cdt1-depleted cells arrested just prior to metaphase with most chromosomes positioned near the spindle equator. A smaller fraction of cells arrested in prometaphase (Fig. 2a and 2b; Fig. S2a and S2b). In addition to Cdt1 depletion in synchronized cells, we also microinjected purified anti-Cdt1 antibody (Fig. S2c) into HeLa cells expressing GFP-tagged histone H2B during prophase or early prometaphase and conducted live imaging (n = 27). Anti-Cdt1 antibody induced both a severe delay in chromosome congression and an arrest near metaphase for the entire ~3 hr duration of imaging (Fig 2d and 2e). Control buffer-injected cells (n = 15) or control anti-mouse IgG-injected cells (n = 8) executed mitosis normally (Fig. 2c, 2e and Supplementary Movies S1 and S2). Anti-Cdt1 injection during late-prometaphase or metaphase (n = 10), resulted in 70% of the cells remaining arrested in that stage for up to 8 hours. During this period, cells progressively lost chromosome alignment but did not divide (Fig. S2d, Movie S4).
Cdt1 localizes to mitotic kinetochores through interaction with the Hec1 component of Ndc80
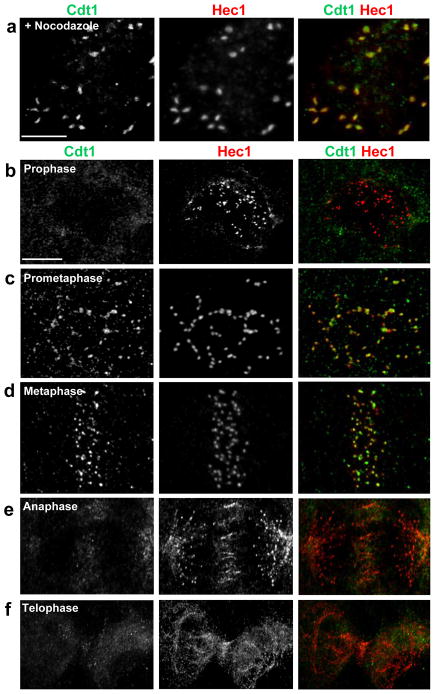
The late prometaphase arrest of Cdt1-depleted cells prompted us to examine Cdt1 localization in mitosis. Remarkably we detected endogenous Cdt1 co-localization with a known kinetochore protein, Hec1, in prometaphase in both nocodazole-treated PTK2 and HeLa cells (Fig. 3a, and data not shown). Cdt1 depletion by siRNA treatment in S phase eliminated detectable anti-Cdt1 kinetochore staining (Fig. S3a). We observed no kinetochore localization of the Cdt1 inhibitor geminin19, 20 at any stage of mitosis, (Fig. S3b). A second anti-Cdt1 antibody also detected endogenous Cdt1 at kinetochores in LLCPK1 cells (Fig. 3b–3f). Thus, Cdt1 localizes to kinetochores in control cells at a time coincident with the stage at which Cdt1-depleted cells arrest.
Figure 3. Cdt1 transiently localizes to kinetochores during prometaphase and metaphase.
(a) Nocodazole-treated PTK2 cells were immunostained with anti-Cdt1 antibody and anti-Hec1 antibody. (b–f) LLCPK1 cells at different stages of mitosis were immunostained with anti-Cdt1 antibody and anti-Hec1 antibody. Scale bars = 5 μm.
In a search for Cdt1 interacting proteins by two-hybrid screening in yeast, we identified multiple independent clones containing portions of human Hec1 (Fig. S4a); the shortest contained Hec1 amino acids 306–642 (Fig. S4a and S4b). To determine if Cdt1 and Hec1 interact biochemically, we incubated immobilized bacterially-expressed GST-Cdt1 with lysates of asynchronous HeLa cells. Endogenous Hec1 associated with GST-Cdt1 but not GST alone (Fig. 4a). Nuf2 was also retrieved from cell lysates by GST-Cdt1 (Fig. 4a) suggesting that Cdt1 can associate with the Ndc80 complex, possibly through a direct interaction with Hec1 (based on interaction of the fusions in yeast). We further confirmed the interaction between Cdt1 and Hec1 by reciprocal co-immunoprecipitation of the endogenous proteins (Fig. 4b and 4c).
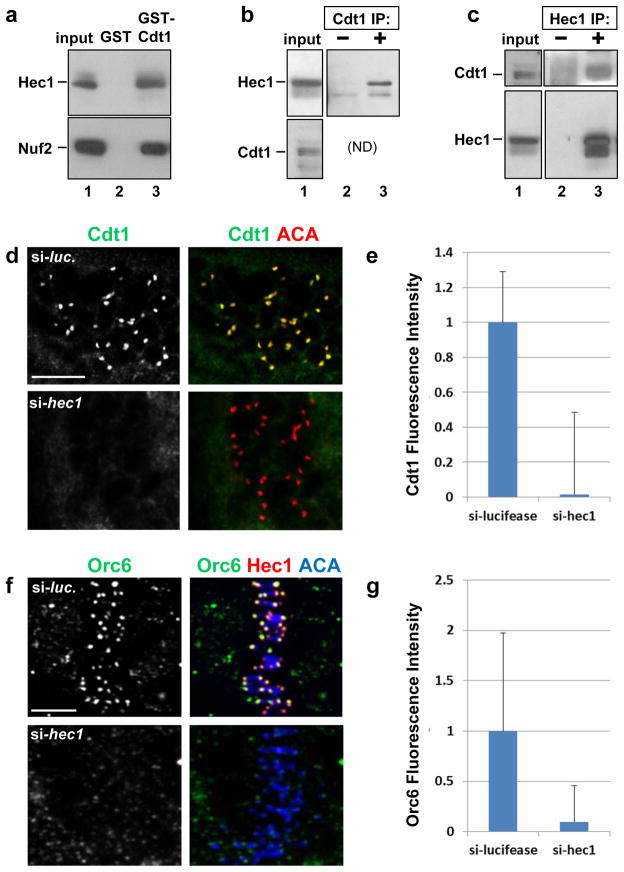
Figure 4. Hec1 is required for Cdt1 kinetochore localization.
(a) A lysate of T98G (human glioblastoma) cells was incubated with beads coated with bacterially-produced GST or GST-Cdt1, and the endogenous Hec1 and Nuf2 were detected in the input or bound fractions by immunoblotting. (b) Whole cell lysates of HeLa cells were incubated with pre-immune serum or anti-Cdt1 antibody, and endogenous Hec1 was detected in the input and immunecomplexes. (ND: Cdt1 co-migrates too closely to IgG heavy chain for detection in this IP.) (c) Whole cell lysates of HeLa cells were incubated with control serum or anti-Hec1 antibody; endogenous Cdt1 and Hec1 were detected in the input and immunecomplexes. (d) Nocodazole-treated PTK2 cells subjected to hec1 RNAi were fixed and stained with anti-Cdt1 antibody and anti-ACA antibody to mark kinetochores. (e) Quantification of Cdt1 kinetochore fluorescence intensity in d relative to control luciferase siRNA transfected cells; n = 80 kinetochores; p < 0.01. (f) HeLa cells were treated with control luciferase siRNA or hec1 siRNA, fixed and stained using anti-Orc6 antibody, anti-Hec1 antibody to monitor the Hec1 knockdown, and anti-ACA antibody to label kinetochores. (g) Quantification of Orc6 kinetochore fluorescence intensity in f relative to control luciferase siRNA transfected cells; n = 125 kinetochores; p < 0.01. Scale bars = 5 μm.
To test if Hec1 recruits Cdt1 to kinetochores in mitosis, we stained for endogenous Cdt1 in Hec1-depleted PTK2 cells. Hec1 depletion resulted in a profound loss of Cdt1 at kinetochores in nocodazole-treated prometaphase (Fig. 4d, bottom panel and Fig. 4e) and metaphase cells (Fig. S4c, bottom panel) compared to controls (Fig. 4d and S4c, top panels). We observed no Cdt1 decrease in extracts of Hec1-depleted cells (Fig. S4d), and Cdt1 depletion had no effect on Hec1 expression or kinetochore localization (Fig. S4e). Other kinetochore proteins, including components of the Mis12 complex, Knl1, and the Zwint1-RZZ complex, were also localized normally in Cdt1-depleted cells (Fig. 2a, S4e, S8a, and S8b).
Like Cdt1, ORC is essential for replication origin licensing, and the Orc2 and Orc6 subunits in particular have also been reported to localize to centromeres or kinetochores13, 14. ORC is required for Cdt1 chromatin localization in G121, and the Orc6 subunit associates with Cdt1 (Fig. S5a22). We found that, like Cdt1, Orc6 immunofluorescence at kinetochores was also substantially reduced in Hec1-depleted cells (Fig. 4f and 4g). Orc6 depletion had no effect on Hec1 or Cdt1 kinetochore localization however (Fig. S5d and S5e). Interestingly, depletion of Cdt1 resulted in a ~50% decrease in the intensity of Orc6 kinetochore staining (Fig. S5b and S5c). To compare the reported mitotic defects induced by Orc6 inhibition14 to those produced by depleting Cdt1, we first calculated the mitotic indices of asynchronous cells depleted of Orc6. Orc6 is not degraded in S phase like Cdt1, so we could not employ the same synchronization strategy to remove Orc6 only before mitosis. Orc6 depletion caused a ~50% increase in the number of mitotic cells compared to controls (Fig. S5f) by producing a four-fold increase in the number of telophase cells (Fig. S5g). In addition, cells depleted of Orc6 exhibited normal cold-stable kMTs (Fig. S5h). These observations support the conclusion14, 23 that Orc6 has a role late in mitosis during cytokinesis distinct from the role of Cdt1 during prometaphase or metaphase.
The Hec1 loop domain is required for Cdt1 localization and anaphase onset
Our two-hybrid analysis indicated that the Cdt1-binding region of Hec1 includes a short interruption in the coiled-coil pattern postulated to be a flexible “loop”3 (Fig. S4a and 4b). To test if this domain is required for Cdt1 binding, we replaced the normal loop sequence with a sequence of alternative amino acids of similar length, which we term “Hec1 LoopMUT” (Fig. 5a). A similar sequence provides a flexible linker in a well-studied GFP-PCNA fusion24. Both wild type Hec1 (wt) and Hec1 LoopMUT were tagged with GFP at their C-termini. Strikingly, Hec1 LoopMUT-GFP failed to co-immunoprecipitate with Cdt1 whereas wt Hec1-GFP bound Cdt1 readily (Fig. 5b, compare lanes 4 and 5). Both GFP-tagged wt and LoopMUT localized normally in PTK2 and HeLa cells (Fig. 5c and 5e). We then tested Cdt1 localization in PTK2 cells depleted of endogenous Hec1 but expressing siRNA-resistant versions of wt Hec1-GFP or Hec1 LoopMUT-GFP. Cdt1 localized to kinetochores normally in cells expressing wt Hec1-GFP (Fig. 5c, top panel), but was strikingly undetectable at kinetochores in cells expressing Hec1 LoopMUT-GFP (Fig. 5c, bottom panel). Orc6 kinetochore targeting was also dependent on the Hec1 loop (Fig. 5d).
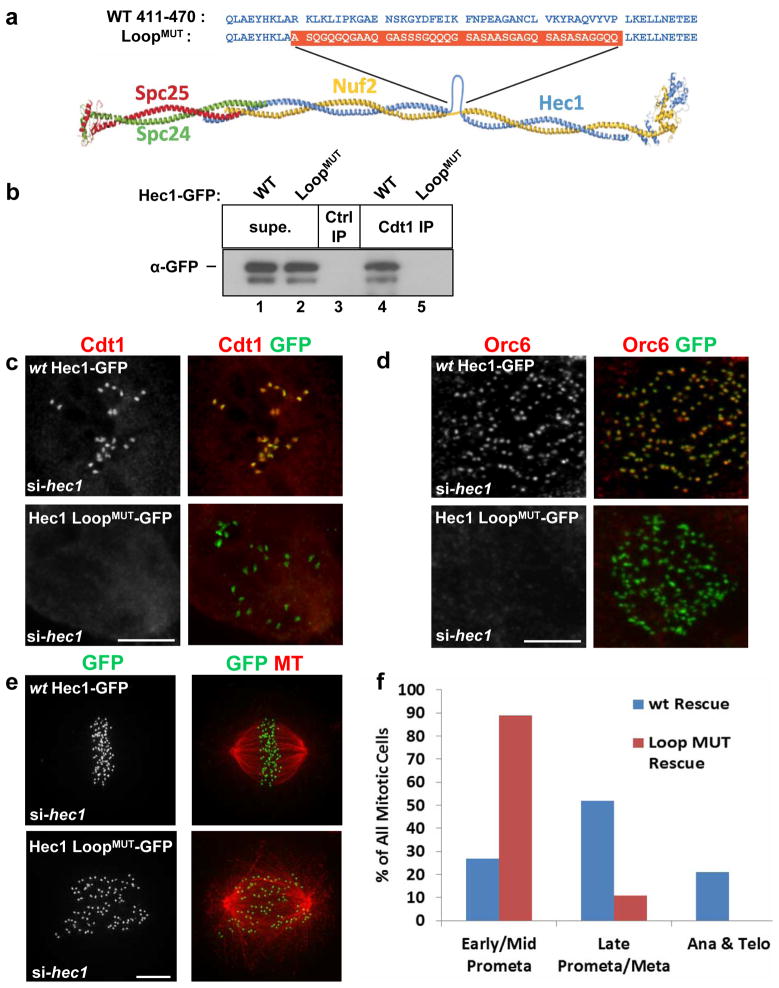
Figure 5. Cdt1 targeting to kinetochores depends on the flexible loop region of Hec1.
(a) Diagram of the Ndc80 complex showing the loop region and the construction of the Hec1 loop replacement mutant,“Hec1 LoopMUT”, (adapted from Ciferri et al., 200835). (b) Endogenous Cdt1 was immunoprecipitated from lysates of asynchronously growing HeLa cells transfected with Hec1-GFP plasmids; “Ctrl IP” indicates the use of normal mouse serum as a control. Hec1-GFP in the bound (“Cdt1 IP”) and unbound (“Supe.”) fractions was detected with anti-GFP antibody. (c) PTK2 cells were treated with hec1 siRNA followed by transfection with either wt Hec1-GFP or Hec1 LoopMUT-GFP constructs. The cells were treated with nocodazole then fixed and stained using anti-Cdt1 antibody and anti-GFP antibody. (d) As in c except that HeLa cells were stained with anti-Orc6 and anti-GFP antibodies. (e) As in c except that HeLa cells were stained with anti-tubulin antibody to mark MTs and anti-GFP antibody to mark Hec1-GFP at kinetochores. (f) Quantification of mitotic stages in Hec1-depleted cells expressing ectopic wt Hec1-GFP or Hec1 LoopMUT-GFP stained with DAPI; n = 125 GFP-expressing cells. Scale bars = 5 μm. See also Supplementary Figure S8 and Supplementary Movies S5 and S6.
HeLa cells expressing only Hec1 LoopMUT-GFP bore striking similarities to Cdt1-depleted cells. Only 14% reached late prometaphase while 86% remained in early to mid-prometaphase (Figs. 5e and 5f, Movie S6 and Fig. S6b) unlike cells expressing wt Hec1-GFP (12/13 cells, Movie S5 and Fig. S6a). Live-imaging of cells expressing only Hec1 LoopMUT-GFP also revealed a 100% mitotic arrest phenotype (n = 29). The phenotypes of Hec1 LoopMUT-GFP cells were more severe than Cdt1-depleted cells, which could be due to quantitative differences in effectiveness of the different siRNAs in synchronized vs. asynchronous cultures, defects in spindle structure from Hec1 perturbation, or to additional functions of the loop apart from Cdt1 binding. The range of phenotypes in both circumstances was similar however (Fig. S6c and S6d) suggesting that the Hec1 LoopMUT phenotype might be largely explained by failure to recruit Cdt1 to kinetochores.
The absence of Cdt1 at kinetochores causes defective kinetochore-MT attachment and Mad1-dependent arrest
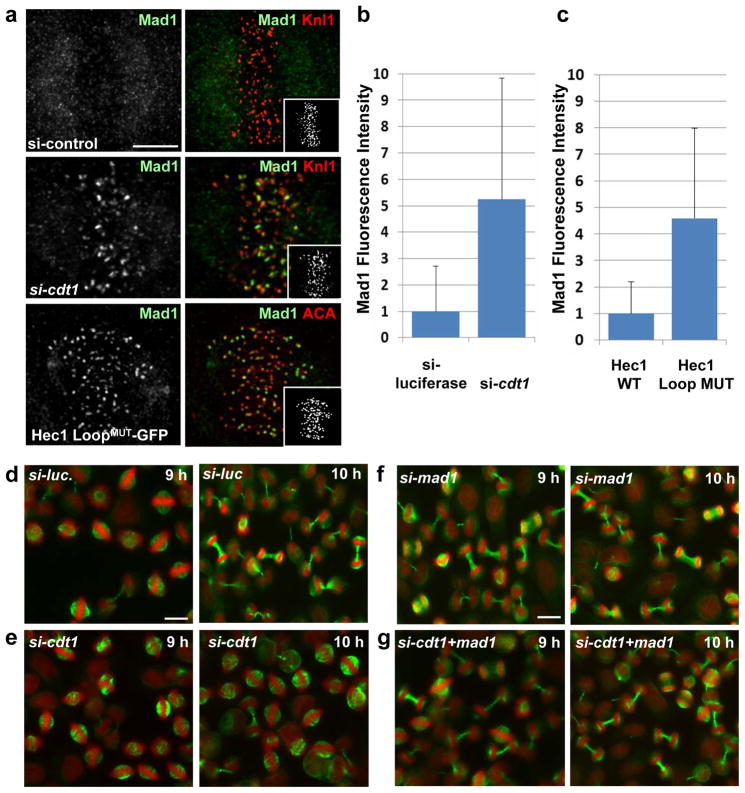
These phenotypes prompted us to probe the spindle assembly checkpoint status of arrested cells. In Cdt1-depleted cells, kinetochore localization of the checkpoint protein, Mad1, was 5.2-fold higher than in control cells (Fig. 6a, top and middle panel and Fig. 6b). We measured a similar 4.5-fold Mad1 increase at aligned kinetochores in cells expressing Hec1 LoopMUT-GFP vs. wt Hec1-GFP (Fig. 6a, bottom panel and Fig. 6c). Kinetochore localization of another checkpoint protein, BubR1, was unaltered by Cdt1 depletion (data not shown). To test if Mad1 recruitment fully accounts for the mitotic arrest produced by Cdt1 inhibition, we co-depleted Mad1 with Cdt1. As demonstrated previously25, control cells depleted of Mad1 alone underwent premature anaphase onset (Fig. 6, compare d and f). Importantly, co-depletion of Mad1 with Cdt1 bypassed the mitotic arrest of Cdt1-depleted cells (Fig. 6, compare e and g). We thus conclude that Cdt1 depletion induces persistent Mad1-dependent spindle assembly checkpoint signaling. Of further note, synchronized cells co-depleted of both Mad1 and Cdt1did not produce many anaphase bridges relative to Mad1-depleted cells. We reasoned that unreplicated DNA segments would not be segregated properly leading to the production of anaphase chromosome bridges. The absence of increased anaphase bridges reinforces our assertion that Cdt1 is not required during S phase for complete DNA replication. On the other hand, asynchronous cells depleted of either Cdt1 or Orc6 produced many anaphase bridges (Fig. S7a and S7b and data not shown).
Figure 6. Cdt1 and the Hec1 Loopdomain are required to satisfy the spindleassembly checkpoint.
(a) Top and middle panel: Cdt1-depleted or control cells as in Fig. 2b (10 hrs after thymidine release). Bottom panel: Hec1-depleted cells expressing siRNA-resistant Hec1 LoopMUT –GFP. Cells were fixed and stained with antibodies to Mad1 to assess spindle assembly checkpoint activity and Knl1 to mark kinetochores. (b) Quantification of Mad1 kinetochore levels in mitotic cells treated with control or cdt1 siRNA; n = 125 kinetochores; p < 0.01. (c) Quantification of Mad1 levels in mitotic cells expressing Hec1 wt or LoopMUT constructs instead of endogenous Hec1; n = 150 kinetochores; p < 0.01. (d–g) Double thymidine synchronized HeLa cells were transfected with control luciferase siRNA (d), cdt1 siRNA (e), mad1 siRNA (f) and a combination of both cdt1 and mad1 siRNA (g) and then stained with DAPI (pseudo-coloured red) and anti-tubulin antibody (green) at 9 hrs or 10 hrs after S phase release.
The presence of high levels of Mad1 at kinetochores suggested that loss of Cdt1 or mutation of the Hec1 loop produces kMT (kMT) attachments that are insufficiently robust. We therefore tested the cold stability of kMTs by chilling cells at late prometaphase or metaphase before fixing them for immunofluorescence. The fluorescence of kMTs after cold treatment for Cdt1-depleted cells or Hec1 LoopMUT-GFP cells was ~50% of controls (Fig. 7a, top and middle panel and Fig. S7c). Quantification of kinetochores that made contact with cold-stable spindle MTs revealed a 77% decrease in the number of K-fibers in Cdt1-depleted cells and an 84% decrease in Hec1 LoopMUT-GFP cells relative to corresponding controls (Fig. 7b). As an additional indicator of kMT attachment, we measured the inter-kinetochore stretch (K-K) distance of aligned sister kinetochore pairs; ~1.4 μm for siRNA-transfected controls and ~1.25 μm for Hec1-depleted cells expressing wt Hec1-GFP. In contrast, the K-K distance was just 0.96 μm in Cdt1-depleted cells and 1.05 μm in cells expressing Hec1 LoopMUT-GFP (Fig. 7c). These values correspond to a 50–60% loss of kinetochore stretch since the unstretched K-K centromere length is ~0.7 μm in cells treated with nocodazole26. As in Cdt1-depleted cells, there were no discernible changes in the kinetochore localization of most other proteins tested in cells expressing Hec1 LoopMUT-GFP (Fig. S8b).
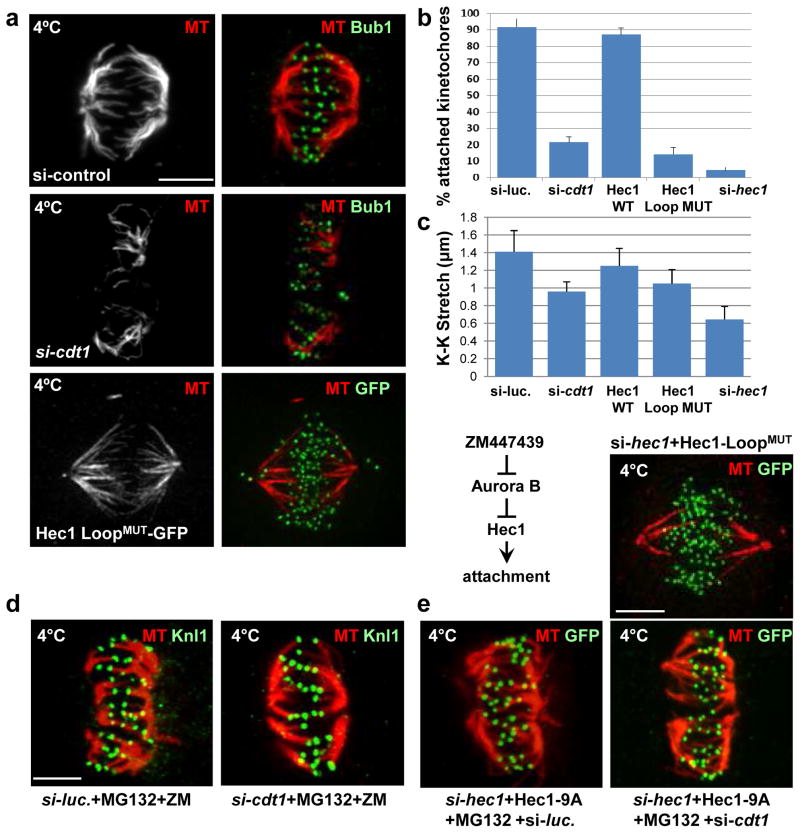
Figure 7. Cdt1 and the Hec1 Loopdomain are required for stable kMT attachments.
(a) Control cells in metaphase, Cdt1-depleted cells, and cells expressing Hec1 LoopMUT (in place of endogenous Hec1) in late prometaphase were incubated with ice-cold PBS (which allows retention of only stable kMTs) followed by fixation and staining with anti-tubulin antibody (MT) and either anti-Bub1 antibody to mark the kinetochores or anti-GFP antibody for ectopic Hec1 as indicated. (b) Quantification of the fraction of kinetochores making successful contact with K-fibers in Cdt1-depleted or Hec1 LoopMUT cells; n = 250 kinetochores (c) Inter-kinetochore (K-K) distances in the indicated cells; n = 100–125 kinetochores; p < 0.01. (d) Synchronized HeLa cells treated with either control luciferase or cdt1 siRNA as indicated were treated with both ZM447439 (Aurora B inhibitor) and MG132 (anaphase inhibitor) for hr at 8.5 hrs post-S phase release followed by cold treatment and staining with anti-tubulin antibody and anti-Knl1 antibody. (e) Synchronized HeLa cells were co-transfected with both hec1 siRNA and either a plasmid encoding unphosphorylatable 9A-Hec1- GFP (top panels) followed by MG132 treatment for hr at 8.5 hrs post S phase release or plasmid encoding Hec1 LoopMUT-GFP (bottom panel). The cells were then subjected to cold treatment at 9 hrs post-S phase release and stained with anti-tubulin antibody and anti-GFP antibody. Scale bars = 5 μm.
To determine if artificially strengthening MT attachments can compensate for Cdt1 loss or mutations in the Hec1 loop domain, we blocked Aurora B kinase-mediated Hec1 phosphorylation. Pharmacological Aurora B inhibition or mutation of the 9 phosphorylation sites in the N-terminal Hec1 tail enhances MT binding affinity at this end of the Ndc80 complex1, 2, 27–30. Treating Cdt1-depleted mitotic cells with the Aurora B inhibitor, ZM447439, caused premature anaphase onset similar to control drug-treated cells or Mad1-depleted cells, demonstrating effective Aurora B inhibition (Fig. S7f and S7g). We also coupled our synchronization and Cdt1 depletion protocol with expression of the Aurora B-resistant Hec1 (9A-Hec1-GFP) combined with endogenous Hec1 depletion. The result of this manipulation was identical to that of Aurora B inhibition (data not shown). To test if progression to anaphase in these experiments involved stabilization of kMT attachments, we treated cells with the proteasome inhibitor, MG132, to prevent anaphase onset and subjected them to the cold stability assay. ZM447439-treated or 9A-Hec1-GFP-expressing cells lacking Cdt1 retained stable kMTs (Fig 7d and 7e). These results indicated that the absence of Cdt1 at kinetochores results in failure to establish and/or maintain sufficiently strong kMT attachments.
Interaction of Cdt1 with the Hec1 loop is required for the normal conformation of the Ndc80 complex at kinetochores of bi-oriented chromosomes
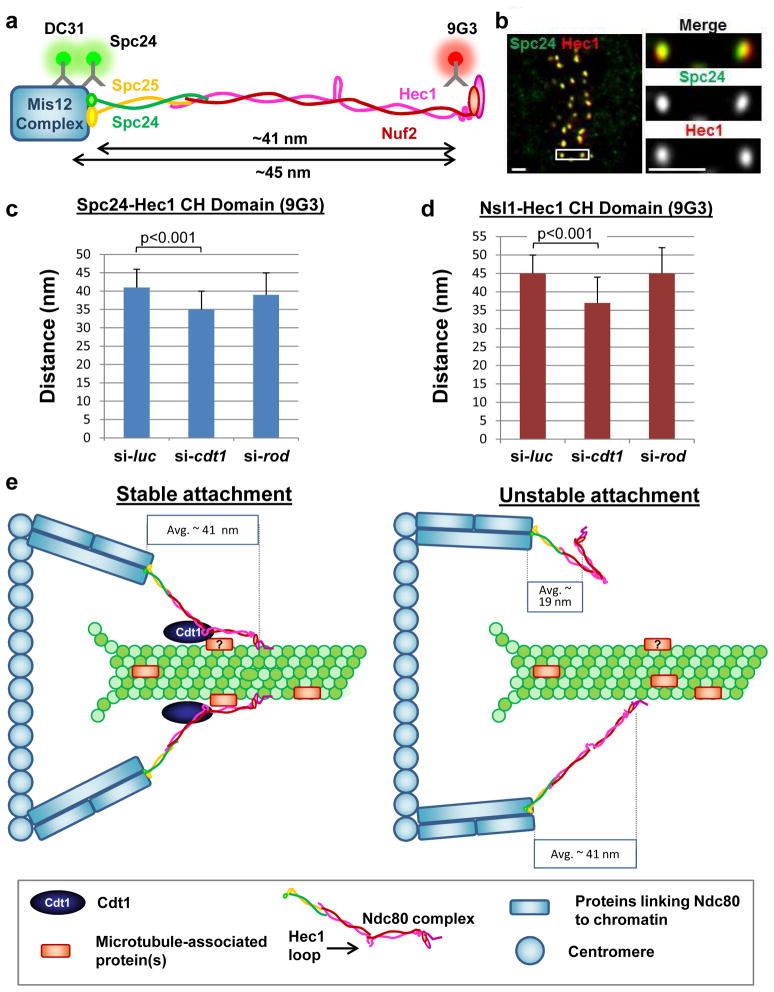
The loop region of Hec1 allows conformational changes within the Ndc80 complex3. To determine if Cdt1 influences this conformation, we employed an in vivo two-colour super resolution fluorescence microscopy technique by labeling the two ends of the Ndc80 complex (Delta analysis illustrated in Fig. 8a26). We used the 9G3 antibody whose epitope in Hec1 resides next to the N-terminal CH domain, an anti-Spc24 antibody to mark the C-terminal region of the Ndc80 complex, and a Nsl1 antibody to mark the Mis12 complex, which is about 4 nm inside Spc2426 (Fig. 8a). Measurements of the average separation, Delta, between 9G3 and the two other antibodies yielded values of ~41 and ~45 nm for kinetochores of aligned chromosomes in control cells at metaphase (Figs. 8c and 8d). After Cdt1 depletion, the corresponding Deltas were ~35 nm and ~37 nm respectively (Figs. 8c–d), values significantly different from controls (p<0.001). To determine if these reductions were a general consequence of unstable kMT attachments, we measured Delta following depletion of Rod (a component of the RZZ complex) which, like Cdt1 depletion, produces 50% reduction in kMT stability31. Unlike Cdt1 depletion, Delta values for Rod depletion (~39 nm and ~45 nm, Fig. 8c–d) were not significantly different from controls (p= 0.31 and p = 0.86 respectively); Rod-depletion reduced K-K stretch from ~1.4 μm to 1.15 μm demonstrating the effectiveness of the siRNA. In nocodazole-treated cells which lack kMTs, the distance between the ends of the Ndc80 complex along the K-K axis is short with a Delta value of ~ 17 nm28. We measured Delta values for Cdt1-depleted and nocodazole-treated cells at 19 +/− 7 nm for Spc24-9G3 and 20 nm +/−8 nm for Nsl-9G3 (n =100 sister kinetochore pairs, > 5 cells). Taken together, these results suggest that the essential mitotic function of Cdt1 is to bind to the Ndc80 complex at the Hec1 loop and maintain an extended conformation along the lattice of kMTs to stabilize end-on attachments of spindle MTs to kinetochores.
Figure 8. Cdt1 and the Hec1 loop domain are required for proper Ndc80 conformation in vivo.
(a) Diagram of the Ndc80 complex depicting the antibody epitopes used for the Delta analysis. (b) Representative image of a normal mitotic metaphase cell labeled with antibodies to the Hec1 CH domain and the Spc24 head domain. Scale bar = 1 μm. (c) Delta values (corrected for tilt28) of mean separation between Spc24 and the Hec1 CH domain (9G3 antibody) in synchronized control or Cdt1-depleted mitotic cells (9 hrs after S phase release), or in Rod-depleted cells (mitotic metaphase cells selected from an asynchronous population); n = 84, 47, and 54 kinetochore pairs respectively; (control vs. si-cdt1) p < 0.001, (control vs. si-rod) p=0.31 (not significant). (d). As in c except that Delta values of separation between Nsl1 (Mis12 complex subunit) and the Hec1 CH domain were determined; n = 74, 41, and 60 kinetochore pairs respectively; (control vs. si-cdt1) p < 0.001, (control vs. si-rod) p = 0.86 (not significant). (e) Scale model for the Cdt1-Hec1 Loop domain interaction.
Discussion
Distinct essential functions for Cdt1 in both G1 and M phases
Using a unique approach to deplete Cdt1 specifically after its replication function is completed, we unequivocally demonstrated an essential and previously unknown mitotic function for human Cdt1 in kMT attachment. This discovery raises the question of why such dual roles would have evolved. One possibility is to ensure orderly progression from S phase to M phase. Cdt1 is degraded while replication is ongoing to avoid re-licensing origins that have already fired 32, 33. We recently reported a mechanism of Cdt1 stabilisation after S phase through MAP kinase-mediated phosphorylation18. We now suggest that Cdt1 stabilisation in G2 cells evolved to facilitate its rapid re-accumulation prior to mitosis. Though yeast Cdt1 is clearly not required for anaphase34, 35, metazoan Cdt1 may have acquired its mitotic role as a means of reinforcing the dependence of a key mitotic event on the completion of genome duplication. The acquisition of functions in two different cell cycle phases has the consequence that deregulation of Cdt1 has the potential to affect both origin licensing and mitosis, and thus genome stability36, 37.
The role of the Hec1 “loop” domain
The presence of high levels of Mad1 at kinetochores of Cdt1-depleted cells or Hec1 LoopMUT–expressing cells indicates that Cdt1 and the loop are required for attachment but not for Mad1 retention, unlike cells lacking Hec1 entirely in which both attachment and Mad1 retention are defective (28, 29; data not shown). The globular CH domain and N-terminal tail of Hec1 make direct contacts with MTs, and the loop domain is not required for high affinity binding of Ndc80bonsai complexes to MTs, a construct lacking the region that includes the loop38. It was therefore somewhat surprising that mutation of the Hec1 loop domain, a site ~18 nm along the Hec1 rod domain from these direct MT contacts, had such profound negative effects on the formation of stable MT attachments. Clearly the formation of robust attachments in vivo requires not only the Hec1 CH domain and N-terminal region, but also Cdt1 interaction with the loop domain.
Recent studies in yeast have shown a requirement for the loop in yeast Hec1/Ndc80 for binding MT binding proteins (MAPs), i.e. the Dam1 complex in budding yeast or Dis1/TOG in fission yeast4, 5. The amino acid sequence of the loop is not highly-conserved3, and human Cdt1 has no detectable sequence homology to Dam1 complex members or to Dis/TOG. Nevertheless, Cdt1 could serve an analogous function by interacting with MTs or an as yet, unidentified MT-associated protein (Fig. 8e) or alternatively by silencing Aurora B-mediated k-fiber destabilisation.
A model for Cdt1-Hec1 interaction in mitosis
The Hec1 loop produces a flexible hinge in the otherwise relatively rigid Ndc80 complex. In the complete absence of MTs, the average separation, Delta, between the two ends of the Ndc80 complex is only ~19 nm26 in both controls and Cdt1-depleted cells. This average distance extends to ~41 nm for control metaphase cells when kinetochores achieve a full MT complement. In the absence of Cdt1, Ndc80 does not make this full extension producing Delta measurements ~68 – 76% between the two extremes. In contrast, Rod depletion removes the major recruitment factor for the dynein-dynactin complex and disrupts other kMT-binding sites contributed by this MT motor31, 39, 40. The fact that Ndc80 can adopt the full extended shape in Rod-depleted cells which, like Cdt1-depleted cells, have unstable kMT attachments31 argues against a general indirect effect of attachment on Ndc80 conformation. We propose that Cdt1 physically links the Hec1 loop to the MT or a MT-associated protein, and this additional attachment stabilises the extended conformation and enhances the MT affinity of the Ndc80 complex provided by the Hec1 CH domain and N-terminal tail.
Methods
Cell culture, transfection and flow cytometry
HeLa (normal and stably expressing Histone H2B) and NHF1-htert cells were cultured in DMEM (Difco or Invitrogen) supplemented with 10% fetal bovine or calf serum (Sigma), 100 U/ml penicillin and 100 mg/ml streptomycin. LLCPK1 cells were grown in DMEM with 5% FBS while PTK2 cells were cultured in MEM-alpha medium supplemented with 15% FBS. Transfections were performed with a total of 100 nM each siRNA duplex using Dharmafect 1 reagent (Dharmacon/Thermo Scientific, Lafayette, CO, U.S.A.) according to the manufacturer’s guidelines. Stable cell lines expressing Cdt1 have been described18. Flow cytometric analysis for cell cycle position was performed as in Nevis et. al., 200916. Synthetic siRNAs: si-cdt1 and si-cdc616 were synthesized by Invitrogen/Life Technologies (Carlsbad, CA, U.S.A.), si-mad1 was purchased from Dharmacon (SMART pool catalog #L-005825-00-0005), si-rod from Dharmacon (SMARTpool catalog #L006829-00-0005), si-hec1(human)28 5′-AAAAAGAACCGAAUCGUCUAGAA, si-hec1 (rat kangaroo) 5′ AAUGAGCCGAAUCGUCUAAUAdTdT, and si-orc614= AAGAUUGGACAGCAGGUCGACUU were synthesized by Dharmacon.
HeLa cells were synchronized by treatment with 2 mM thymidine for 18 hours followed by release for 9 hours and then re-treatment with 2 mM thymidine for 18 hours. Synthetic duplexed RNA oligonucleotides were s transfected into HeLa cells using the manufacturer’s instructions. Other cell manipulations include 10 μM nocodazole treatment, 3 μM ZM447439 treatment and cold treatment for 10 min with ice-cold PBS.
Antibodies
Guinea pig anti-Cdt1 is described in41 (diluted 1:3000 for immunoblots and 1:100 for immunofluorescence staining); rabbit polyclonal anti-Cdt1 was purchased from Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A. (catalog # sc28262). Other primary antibodies used in this study include anti-Tubulin monoclonal (1:20,000 for immunoblots and 1:500 for cell staining, Sigma-Aldrich, St. Louis, MO, U.S.A. #9026, clone [DM1A]), anti-Dynein IC monoclonal antibodies (1:200 for staining Sigma-Aldrich, # D5167, clone [70.1]), anti-Hec1 (1:6,000 for immunoblots and 1:500 for staining, Abcam, Cambridge, MA, U.S.A. #ab3613, clone [9G3]), Nuf2 (1:5,000 for immunoblots and 1:200 for staining, Abcam #ab17058, clone [28–37]), anti-GFP polyclonal (1:200 for staining, Invitrogen #A6455), monoclonal (1:100 for staining, Chemicon/Millipore, Billerica, MA, U.S.A., #MAB3580) or monoclonal (1:5,000 for immunoblots, Clontech, Mountain View, CA, U.S.A. #632381, clone [JL-8]), anti-Cdc6 monoclonal (1:1,000 for immunoblots Santa Cruz Biotechnology #sc-9964 clone [180.2]), anti-geminin polyclonal (1:200 for staining, Santa Cruz Biotechnology #sc-13015), anti-phospho-H2A.X(Ser139) (1:1000 for staining, Millipore #05-636, clone [JBW301]), anti-Mad1 polyclonal (GeneTex, Irvine, CA, U.S.A. #GTX109519). Alexa 488, Rhodamine Red-X, Cy5 or HRP-labeled secondary antibodies were obtained from Jackson ImmunoResearch, West Grove, PA, U.S.A. Orc6 polyclonal antibody (1:200) for staining was a generous gift of Bruce Stillman; anti-Orc6 for immunoblots (1:1000) was purchased from Santa Cruz Biotechnology (# sc-32735, clone [3A4]). Immunoblots were scanned into Adobe Photoshop and any manipulations for brightness were applied to the entire image; final figures show cropped regions and the uncropped blots are provided as Supplementary Figure S9.
Immunofluorescence microscopy, live cell imaging, and Delta analysis
Cells were typically fixed for 20 min using 4% paraformaldehyde after permeabilisation with 0.5% Triton X-100. For Spc24 staining, 2% PFA was used. For Dynein IC staining, the cells were fixed for 6 min at −20°C with ice-cold methanol. Cells were rinsed in PHEM buffer (120 mM Pipes, 50mM HEPES, 20 mM EGTA, 4 mM magnesium acetate, pH 6.9) prior to fixation. All the antibody incubations and washes were also performed in PHEM buffer plus 0.05% BSA. DAPI staining (0.1 μg/ml) was for 10 min, and cells were mounted using Prolong Antifade (Molecular Probes). All antibody incubations were conducted at 37°C for 1 h. For image acquisition, 3D stacks were obtained sequentially at 200 nm steps along the z axis through the cell using a high-resolution Nikon confocal microscopy equipped with a Yokogawa CSU10 spinning disk with image magnification yielding a 65 nm pixel size from the camera42 and an 100X/1.4NA (Planapo) DIC oil immersion objective (Nikon). Scale bars = 5 μm in all figures and supplementary figures unless otherwise defined in the figure legends. Delta analysis to measure separations at high (nm) accuracy between protein epitopes labeled with separate colours were carried out as described in Wan et al., 200926.
For microinjection, polyclonal anti-Cdt1 antiserum41 was depleted of antibodies to non-Cdt1 proteins by repeated rounds of incubation with lysates of UV-treated HeLa cells (which lack Cdt1). These antibody preparations were then concentrated by centrifugation using microconcentrators. Anti-Cdt1 antibody, control IgG, or buffer were injected into HeLa cells that stably express a GFP-fusion to histone H2B either in prometaphase or in metaphase. Injections were carried out on a Zeiss IM microscope equipped with phase optics and a 40x, 0.75 NA (Planfluar) objective at 35°C using a Sage air curtain incubator. Injections were 5% cell volume with a Narishige micromanipulator (Narishige USA, Inc., Sea Cliff, New York). Mitotic progression was monitored by live-cell imaging using both phase contrast and GFP-histone H2B fluorescence with images acquired every two minutes until the end of the duration of imaging.
For live imaging of Hec1-depleted cells, HeLa cells were plated to 50% confluency in a T25 flask and treated with siRNA to Hec1. 24 hrs post-siRNA transfection, cells were trypsinized, counted, and the Hec1 wt and mutant constructs were electroporated along with mCherry-histone H2B expression plasmid using the Lonza Nucleofector apparatus. 24 hours post-electroporation, cells were imaged by time-lapse microscopy every 4 minutes for 5 hrs using a Deltavision PersonalDV Imaging System (Applied Precision) equipped with a Photometrics CoolSnap HQ2 camera and a 60x/1.42NA (Planapo) DIC oil immersion lens (Olympus).
Protein-protein interaction assays
For two-hybrid screening, the full-length human Cdt1 cDNA was inserted into pGBT9 (Clontech). Co-transformants of yeast strain PJ69a43 with two cDNA fusion libraries (placental cDNA or thymus cDNA, Clontech) were selected on medium containing 10 mM 3-amino-1,2,4 triazole (Sigma). More than 8 million co-transformants of each library were screened; Hec1 clones were identified from both libraries. GST-pulldowns and co-immunoprecipitations were performed essentially as described in Cook et al. 200441. GST-Cdt1 was produced in BL21(DE3) purified on glutathione-Sepharose (GE Healthcare) and incubated with cell lysates prepared in Buffer 1 (50 mM HEPES pH 7.2, 33 mM potassium acetate, 0.5 mM EDTA, 0.5 mM EGTA, 0.1 % Nonidet P-40, 10% glycerol, protease and phosphatase inhibitors). Cell lysates were prepared in Buffer 1 or a 1:1 mix of Buffer 1 and CSK Buffer (10 mM PIPES, pH 7.0, 100 mM NaCl, 30 0mM sucrose, 3 mM MgCl2 plus protease and phosphatase inhibitors) digested with micrococcal nuclease prior to clarification by centrifugation, and incubated with GST protein-coated beads for 2 hours, or with antibodies for co-immunoprecipitation and protein A beads, washed three times, and then released by boiling in SDS-PAGE loading buffer.
Supplementary Material
Acknowledgments
We thank Arshad Desai for providing anit-Knl1, Nsl1, Dsn1 and Spindly antibodies, Andrea Mussacchio for anti-Mad1, Zwint1 and ZW10 antibodies, Bruce Stillman for anti-Orc6 antibody, Steven Taylor for anti-Bub1 and BubR1 antibodies, Todd Stukenberg for anti-Spc24 antibody, Tim Yen for anti-CENP-E antibody and Iain Cheeseman for anti-Ska3 antibody. We are grateful to Drs. Arshad Desai, Dhanya Cheerambathur, Todd Stukenberg, Kevin Slep and Thomas Maresca for helpful discussions and to Jeanne Mick for generating Hec1 constructs. We would also like to thank other members of the Salmon, Cook, Desai and Joseph Nevins labs for their support during this project. J.G.C. was supported by NIH K01 CA094907 and NIH GM083024, E.D.S was supported by NIHGMS GM24364 and J.G.D was supported by NIH GM088371 and a grant from the Pew Scholars Program in the Biomedical Sciences.
Footnotes
Author Contributions. D.V. and J.G.C. designed and carried out experiments, analysed data, and wrote the manuscript. S.C., L.J.R.S, and K.T.R. carried out experiments and analysed data. D.A.D.C. and J.G.C. conducted the two-hybrid screen. K.R.N. and S.C. characterized the arrest of Cdc6 and Cdt1-depleted normal fibroblasts. X.W. and D.V. conducted the Delta analyses. J.G.D., E.D.S., and J.G.C. designed experiments, analysed data, and wrote the manuscript. All authors proofread the manuscript.
References
- 1.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 2.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 3.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Current biology. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maure JF, et al. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Current biology. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machida YJ, Hamlin JL, Dutta A. Right Place, Right Time, and Only Once: Replication Initiation in Metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snyder M, He W, Zhang JJ. The DNA replication factor MCM5 is essential for Stat1-mediated transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:14539–14544. doi: 10.1073/pnas.0507479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitch MJ, Donato JJ, Tye BK. Mcm7, a subunit of the presumptive MCM helicase, modulates its own expression in conjunction with Mcm1. J Biol Chem. 2003;278:25408–25416. doi: 10.1074/jbc.M300699200. [DOI] [PubMed] [Google Scholar]
- 10.Clay-Farrace L, Pelizon C, Santamaria D, Pines J, Laskey RA. Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. Embo J. 2003;22:704–712. doi: 10.1093/emboj/cdg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida K, et al. CDC6 interaction with ATR regulates activation of a replication checkpoint in higher eukaryotic cells. J Cell Sci. 2010;123:225–235. doi: 10.1242/jcs.058693. [DOI] [PubMed] [Google Scholar]
- 12.Tachibana KE, Gonzalez MA, Guarguaglini G, Nigg EA, Laskey RA. Depletion of licensing inhibitor geminin causes centrosome overduplication and mitotic defects. EMBO Rep. 2005;6:1052–1057. doi: 10.1038/sj.embor.7400527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. Embo J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 15.Feng D, Tu Z, Wu W, Liang C. Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res. 2003;63:7356–7364. [PubMed] [Google Scholar]
- 16.Nevis KR, Cordeiro-Stone M, Cook JG. Origin licensing and p53 status regulate Cdk2 activity during G1. Cell Cycle. 2009;8 doi: 10.4161/cc.8.12.8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J Biol Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- 18.Chandrasekaran S, Tan TX, Hall JR, Cook JG. Stress-stimulated mitogen-activated protein kinases control the stability and activity of the Cdt1 DNA replication licensing factor. Molecular and cellular biology. 2011;31:4405–4416. doi: 10.1128/MCB.06163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wohlschlegel JA, et al. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 20.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 21.Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes & Development. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernal JA, Venkitaraman AR. A vertebrate N-end rule degron reveals that Orc6 is required in mitosis for daughter cell abscission. The Journal of cell biology. 2011;192:969–978. doi: 10.1083/jcb.201008125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonhardt H, et al. Dynamics of DNA replication factories in living cells. The Journal of cell biology. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Developmental cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Wan X, et al. Protein Architecture of the Human Kinetochore Microtubule Attachment Site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nature structural & molecular biology. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 28.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Current biology. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Current biology. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Current biology : CB. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Current biology : CB. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005;19:114–126. doi: 10.1101/gad.1255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishitani H, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. Embo J. 2006 doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devault A, et al. Identification of Tah11/Sid2 as the Ortholog of the Replication Licensing Factor Cdt1 in Saccharomyces cerevisiae. Curr Biol. 2002;12:689–694. doi: 10.1016/s0960-9822(02)00768-6. [DOI] [PubMed] [Google Scholar]
- 35.Hofmann JF, Beach D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. Embo J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arentson E, et al. Oncogenic potential of the DNA replication licensing protein CDT1. Oncogene. 2002;21:1150–1158. doi: 10.1038/sj.onc.1205175. [DOI] [PubMed] [Google Scholar]
- 37.Liontos M, et al. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899–10909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- 38.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. The Journal of cell biology. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varma D, Monzo P, Stehman SA, Vallee RB. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. The Journal of cell biology. 2008;182:1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cook JG, Chasse DA, Nevins JR. The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells. J Biol Chem. 2004;279:9625–9633. doi: 10.1074/jbc.M311933200. [DOI] [PubMed] [Google Scholar]
- 42.Maddox PS, Moree B, Canman JC, Salmon ED. Spinning disk confocal microscope system for rapid high-resolution, multimode, fluorescence speckle microscopy and green fluorescent protein imaging in living cells. Methods in enzymology. 2003;360:597–617. doi: 10.1016/s0076-6879(03)60130-8. [DOI] [PubMed] [Google Scholar]
- 43.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two- hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.