Abstract
Pancreatic ductal adenocarcinoma (PDAC) has the lowest survival rate of all cancers and shows remarkable resistance to cell stress. Nuclear protein 1 (Nupr1), which mediates stress response in the pancreas, is frequently upregulated in pancreatic cancer. Here, we report that Nupr1 plays an essential role in pancreatic tumorigenesis. In a mouse model of pancreatic cancer with constitutively expressed oncogenic KrasG12D, we found that loss of Nupr1 protected from the development of pancreatic intraepithelial neoplasias (PanINs). Further, in cultured pancreatic cells, nutrient deprivation activated Nupr1 expression, which we found to be required for cell survival. We found that Nupr1 protected cells from stress-induced death by inhibiting apoptosis through a pathway dependent on transcription factor RelB and immediate early response 3 (IER3). NUPR1, RELB, and IER3 proteins were coexpressed in mouse PanINs from KrasG12D-expressing pancreas. Moreover, pancreas-specific deletion of Relb in a KrasG12D background resulted in delayed in PanIN development associated with a lack of IER3 expression. Thus, efficient PanIN formation was dependent on the expression of Nupr1 and Relb, with likely involvement of IER3. Finally, in patients with PDAC, expression of NUPR1, RELB, and IER3 was significantly correlated with a poor prognosis. Cumulatively, these results reveal a NUPR1/RELB/IER3 stress-related pathway that is required for oncogenic KrasG12D-dependent transformation of the pancreas.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) has the highest mortality rate and the lowest overall survival of all cancers (less than 3%–4% at 5 years). The incidence of PDAC almost equals its mortality rate and is increasing every year, with more than 38,000 predicted new cases in the United States and 65,000 in Europe. Surgery is the most effective treatment, but the mean life expectancy of the 15%–20% of patients who present with a resectable tumor is only 15–18 months (1). Even for patients eligible for surgery, aggressive metastasis often appears after operation, and survival of patients with metastatic disease is only 3–6 months (2). Chemotherapy and radiotherapy offer limited benefit for patients undergoing surgery in metastatic disease (3). Most strategies tested so far for the treatment of PDAC have consisted of using therapies that show some efficacy in other carcinomas, but none of them improved significantly the overall survival of PDAC patients. Hence, in order to develop new, efficient therapies against PDAC, future research must take into account PDAC specificities such as its remarkable resistance to cell stress, notably to the stress induced by chemotherapy and radiotherapy (4), and the preponderant presence of a fibrotic stroma, which favors tumor progression by creating a barrier against drug delivery and immune cell infiltration (5). Besides a very abundant stroma, PDAC presents with poor vascularization, compared with other carcinomas (6), indicating that pancreatic cancer cells face a particularly adverse microenvironment with severe deprivation of nutrients and oxygen. Therefore, survival of malignant pancreatic cells is conditioned by their ability to develop a pro-survival stress response to these deprivations, whose efficiency will be pivotal in the selection of the most aggressive cell clones. The molecular mechanisms underlying the outstanding resistance of PDAC cells to stress are currently unknown. During the last decade, we have focused our studies on the functional characterization of Nupr1, a chromatin protein whose expression is induced by most cellular stress in vitro and whose inactivation leads to increased sensitivity to pancreatic and liver stress in vivo (reviewed in ref. 7). Nupr1 is a potential mediator of PDAC resistance to cell stress, not only because it is highly expressed during the progression of PDAC and other cancers (8–12) and induced in pancreatic cancer cells by many endogenous and exogenous stresses agents (13–22), but also because it is one of the best-characterized stress sensors in the pancreas. For instance, we reported Nupr1 antiapoptotic function against the anticancer drug gemcitabine (23), which is at present the first choice for PDAC treatment. Nupr1 regulates in part the cell response to stress through the modulation of target gene expression (24). In addition, Nupr1 regulates chromatin accessibility through its interaction with proteins implicated in histone acetylation/deacetylation and may thereby modulate the genetic response to stress (25).
Transgenic mice bearing a constitutively activated KrasG12D allele under the control of cre-inducible LSL-KrasG12D driven by pancreas-specific Pdx1-cre or Ptf1a+Cre(ex1) transgene develop precancerous ductal lesions known as pancreatic intraepithelial neoplasia (PanIN) that eventually develop into invasive adenocarcinoma (26–28). The histology and kinetics observed in these mice’s pancreata faithfully model the early human pathology, thus offering the possibility to perform in-depth studies of the biology of the pancreatic malignancy. Consequently, we assessed in this study the impact of Nupr1 deficiency in the development of pancreatic precancerous lesions in Pdx1-cre;LSL-KrasG12D mice. We also investigated the role of Nupr1 in the survival of pancreatic cancer cells in response to stress, which led us to discover a Nupr1-driven molecular cascade that involves the alternative RelB-dependent NF-κB pathway and its genetic target IER3 in PanIN formation. Finally, a significant correlation between expression of Nupr1, RelB, and IER3 and poor prognosis of patients with PDAC was found. These results expand our understanding of the molecular machinery that mediates the early steps of pancreatic carcinogenesis, knowledge that has both mechanistic importance and biochemical relevance.
Results
Nupr1 deletion prevents PanIN development in KrasG12D mice.
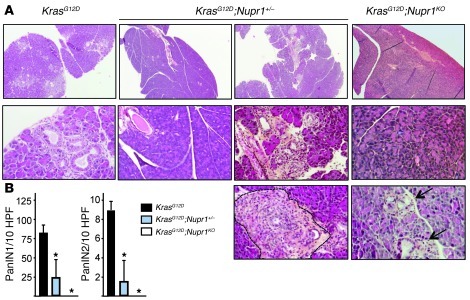
In order to elucidate the role of Nupr1 during pancreatic tumorigenesis, we inbred Pdx1-cre;LSL-KrasG12D mice with Nupr1KO mice developed in our laboratory (29). As previously reported (26–28), we observed that the pancreas of KrasG12D mice develop numerous PanIN lesions from 13 weeks of age (Figure 1A). By contrast, KrasG12D;Nupr1KO pancreas of mice of the same age did not develop PanINs at all, and only 1 sample of 5 developed acinar-ductular metaplasia (ref. 30 and Figure 1B). Of note, only 3 of 9 Nupr1-heterozygous (Nupr1+/–) KrasG12D pancreata analyzed developed PanINs. However, in this hypomorphic condition, PanINs remained scarce and appeared sprinkled among foci of acinar-ductular metaplasia. In fact, a single focus of insular-ductular lesions was observed in KrasG12D;Nupr1+/– pancreas (Figure 1A), indicating that even partial deficiency of Nupr1 may hamper PanIN formation. KrasG12D;Nupr1+/+ pancreata from mice followed up to 18 weeks showed that almost all the tissue was replaced by PanINs, whereas no lesions, or very scarce and small lesions, were observed in pancreata from KrasG12D;Nupr1–/– mice (data not shown). It was concluded that Nupr1 expression is essential for PanIN formation in a KrasG12D background.
Figure 1. Nupr1 expression is necessary for PanIN development.
(A) Pancreata from KrasG12D, KrasG12D;Nupr1+/–, and KrasG12D;Nupr1KO 13-weeks-old littermate mice were H&E stained. A total of 6 sections were collected from the top, middle, and bottom of pancreas specimens for analysis of histological features (representative microphotographs in A) and quantification of PanIN lesions (B). As expected, pancreata from KrasG12D mice displayed multiple PanIN1 and, to a lesser extent, PanIN2 lesions. In contrast, pancreata from littermate KrasG12D;Nupr1KO mice were devoid from such lesions, and only scarce acinar-ductular metaplasia was observed in 1 specimen (arrows). Most KrasG12D;Nupr1+/– pancreata were also PanIN-free, except for a few low-grade PanINs associated with metaplasia. The dotted line indicates insular-ductular metaplasia of a Nupr1-heterozygous specimen. Original magnification: upper panels, ×40; lower panel panels, ×100. (B) Frequency of low-grade PanINs in KrasG12D, KrasG12D;Nupr1+/–, and KrasG12D;Nupr1KO pancreata. PanIN numbers are given per ×200 field. Values are expressed as mean ± SEM of 5 pancreata from KrasG12D and KrasG12D;Nupr1KO mice. *P ≤ 0.05 relative to the number of PanINs in pancreas from KrasG12D mice.
Nupr1 expression is required for the survival of pancreatic cancer cells subjected to a stress.
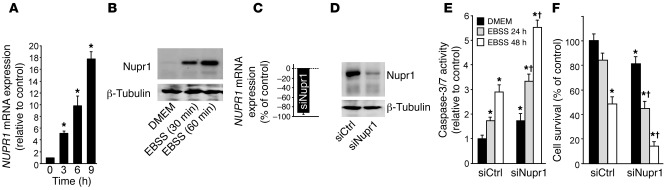
Since Nupr1 expression is strongly activated after cell insult, we hypothesized that this chromatin-binding protein favors PanIN development by converting stress signals into a program of gene expression that empowers malignant pancreatic cells with resistance to the stress induced by a change in their microenvironment. To test this hypothesis, we exposed pancreatic cancer cells to nutrient deprivation, a situation particularly stressful in these desmoplasia-rich and poorly vascularized cells. We cultured human PDAC cells in non-supplemented Earle’s balanced salt solution (EBSS) medium (free of glucose, amino acids, lipids, and growth factors). Using quantitative RT-PCR (qRT-PCR), we could demonstrate that, under this nutrient-deprived setting, Nupr1 mRNA is increased up to 19-fold after 9 hours of exposure to stress compared with levels observed in cells cultured in conventional culture medium (Figure 2A). Western blot analyses revealed a concomitant increase in the level of the Nupr1 protein (Figure 2B). To gain better insight into the role of Nupr1 in the defense mechanism, we investigated the impact of Nupr1 deficiency on the response of pancreatic cells to the stress induced by nutrient deprivation. To this end, we inactivated Nupr1 with an siRNA in pancreatic cancer cells (Figure 2, C and D) and monitored cell survival and caspase-3/7 activity. We observed a significant increase in caspase-3/7 activity in the unstressed Nupr1-depleted cells compared with controls (1.0 ± 0.1– vs. 1.8 ± 0.2–fold), indicating that Nupr1 depletion per se enhances apoptosis (Figure 2E). In agreement with this finding, Nupr1 depletion led to decreased cell survival even when cells were cultured in conventional medium (100% ± 11% vs. 80.5% ± 10%) (Figure 2F). More importantly, Nupr1-depleted cells were far more sensitive to starvation than control cells, since cell survival after 48 hours decreased from 50% ± 5% in controls to 12.5% ± 3% in Nupr1-depleted cells, respectively; Figure 2F), and caspase-3/7 activation was concomitantly enhanced (Figure 2E).
Figure 2. Nupr1 protects pancreatic cancer cells from stress-induced cell death.
(A) MiaPaCa2 cells were cultured in EBSS for 3, 6, and 9 hours, and NUPR1 mRNA expression was measured by qRT-PCR and expressed relative to cells cultured with serum-supplemented medium (DMEM). Cyclophilin was used as housekeeping invariable control for relative transcript level normalization. (B) MiaPaCa2 cells were cultured in conventional or EBSS medium for 30 or 60 minutes. Nupr1 expression was analyzed by Western blot. (C and D) MiaPaCa2 cells were transfected with siNupr1 or siCtrl. After 48 hours, NUPR1 mRNA expression was measured by qRT-PCR, and Nupr1 protein expression by Western blot. (E and F) Cells were transfected with siNupr1 or siCtrl and cultured in conventional or EBSS medium for 24 or 48 hours. Cell viability was assessed by cell counting using Countess (Invitrogen), and caspase-3/7 activity was measured using the Apo-ONE fluorescence assay. Values are expressed as mean ± SEM of triplicate experiments repeated twice. *P ≤ 0.05 relative to siCtrl-transfected cells and cultured in conventional medium; †P ≤ 0.05 relative to siNupr1-transfected cells.
Nupr1-induced RelB expression protects pancreatic cancer cells from cell death triggered by nutrient deprivation.
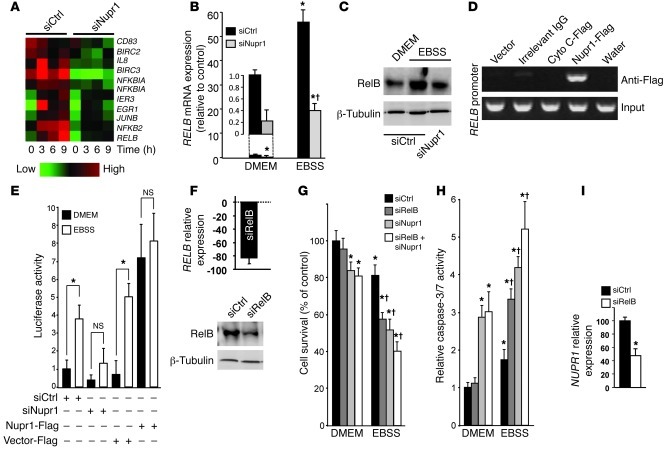
Since Nupr1 belongs to the HMG family of chromatin remodelers with transcriptional cofactor activity, Nupr1 could increase cell survival in a nutrient-deprived microenvironment by activating the expression of pro-survival genes. To test that hypothesis, we performed an Affymetrix-based large-scale transcriptome analysis of pancreatic cancer cells transfected with control siRNA (siCtrl) or siNupr1 and cultured for 3, 6, or 9 hours in EBSS. Nutrient deprivation resulted in a massive modification of the gene expression pattern of pancreatic cancer cells. In agreement with above-mentioned data, Nupr1 expression was increased by starvation-induced stress (data not shown). More importantly, we observed that siRNA-induced Nupr1 depletion affected the activation of key pro-survival genes, including several members of the NF-κB family (RELB, NFKB2, and NFKBIA; Figure 3A) and their target genes (BIRC2, BIRC3, IL8, JUNB, EGR1, and IER3). Real-time qRT-PCR showed that RELB mRNA expression was activated 56-fold after 9 hours of starvation, but this activation was dramatically inhibited when Nupr1 was depleted with a specific RNAi (Figure 3B). Western blot analysis showed that nutrient deprivation increased the level of the RELB protein about 8-fold, whereas in Nupr1-depleted cells the increase was only 1.6-fold under the same conditions (Figure 3C). ChIP assays demonstrated that Nupr1 directly binds the endogenous RELB promoter (Figure 3D), supporting a role for this chromatin-associated HMG protein in the regulation of NF-κB gene expression. Complementary luciferase-based gene reporter assays, using a construct containing the RELB promoter region, demonstrated that activation of the RELB promoter, which occurred under the experimental stress condition studied here, is dependent on Nupr1 expression. EBSS-induced stress inhibited the RELB promoter (3.8 ± 0.6– vs. 1.3 ± 0.7–fold) when Nupr1 was depleted from pancreatic cancer cells. Conversely, the activity of the promoter was strongly enhanced in Nupr1-overexpressing cells under conventional culture conditions (0.8 ± 0.7– vs. 7.2 ± 1.7–fold) (Figure 3E).
Figure 3. Nupr1 activates RelB expression to promote cell survival upon stress.
MiaPaCa2 cells were transfected with the indicated siRNAs and cultured in conventional or EBSS medium for the times indicated. (A) Heat map showing relative expression of NF-κB family genes and targets. (B and C) qRT-PCR and Western blot showing increased RelB mRNA and protein expression, respectively, in Nupr1-depleted cells compared with controls. Cyclophilin and tubulin were housekeeping controls for mRNA and protein load, respectively. (D) Cells were transfected with pCDNA3 vectors containing a Flag-tagged Nupr1, an irrelevant cytochrome c (Cyto C), or an empty vector (Empty). ChIP was performed using an anti-Flag antibody or a irrelevant IgG. Top: Occupancy of Nupr1 on the RELB promoter; bottom: DNA input (10%). (E) Cells were transfected with combinations of a RELB promoter-Luc vector, siCtrl, siNupr1, Nupr1-Flag, or Empty-Flag pCDNA3 vector, and pSV40-RL as indicated. After 9 hours, luciferase activity was determined and expressed as the ratio of specific luciferase activity to an internal standard. (F) RelB expression was measured by qRT-PCR (right) and Western blot (left). (G and H) Cell viability and caspase-3/7 activity were measured after 24 hours as described in Figure 2. (I) NUPR1 mRNA was measured after 24 hours by qRT-PCR. In E and I, values are expressed as mean ± SEM of triplicate, from 2 independent experiments. *P ≤ 0.05 relative to siCtrl-transfected cells cultured in conventional medium; †P ≤ 0.05 relative to siNupr1-transfected cells.
Next, we evaluated whether RelB plays a protective role in pancreatic cancer cells against the stress induced by nutrient deprivation. To this end, RelB expression was inhibited in pancreatic cancer cells using a specific siRNA (Figure 3F), alone or in combination with Nupr1 depletion with siNupr1, before culture in EBSS and assessment of cell survival and caspase-3/7 activity (Figure 3, G and H). RelB-depleted cells displayed a significant decrease in cell survival (96.0% ± 7.2% vs. 57.1% ± 4.3%) and enhanced caspase-3/7 activation compared with controls (1.2 ± 0.1– vs. 3.4 ± 0.3–fold) after the treatment with EBSS. Combined depletion of Nupr1 and RelB only slightly enhanced the effects on cell survival observed when they were targeted separately (P = 0.04). These results show that RelB and Nupr1 are both involved in the survival response against starvation-induced cell death. The increase in Nupr1 expression that occurs in response to nutrient deprivation is also partially dependent on RelB, as indicated by the decrease in NUPR1 mRNA levels observed in siRelB-treated cells (Figure 3I), suggesting a positive feedback regulation between Nupr1 and RelB.
RelA/p65 expression is upregulated by starvation but is not necessary to PDAC cell survival in nutrient-deprived conditions.
Studies performed during the last two decades have established the pro-survival effect of the NF-κB pathway in cancer and inflammation. These studies have primarily focused on characterizing the function of the dimeric transcription factor complex known as NF-κB1, which is composed of a single unit of two molecules, RelA/p65:p50, named the classical pathway. However, other, less numerous studies revealed the existence of a second form of NF-κB (NF-κB2), which differs from the conventional NF-κB1 complex by the replacement of RelA by the structurally related yet functionally distinct RelB protein. The first aim of our investigation was to identify which Rel isoform linked the stress-protective function of Nupr1 to an NF-κB pathway. Nupr1 depletion did not affect the 4-fold increase in RelA/p65 mRNA concentration induced by starvation (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI60144DS1), and siRNA-mediated depletion of RelA/p65 (Supplemental Figure 1B) did not significantly alter either cell survival or caspase-3/7 activity in cells grown in conventional or EBSS medium (Supplemental Figure 1, C and D). Hence, NF-κB1 biosynthesis is not linked to Nupr1 function in the context of starvation-induced stress. On the other hand, siRelA/p65-starved cells presented with reduced expression of Nupr1 (Supplemental Figure 1E), establishing that although Nupr1 does not couple to the classical NF-κB1 pathway, the latter contributes, at least in part, to regulating Nupr1 expression in nutrient-deprived conditions. Thus, these results establish a hierarchical order within this Nupr1-mediated stress resistance pathway and indicate that RelB but not NF-κB1 is a major downstream effector responsible for survival.
IER3 is an antiapoptotic target of RelB and Nupr1 involved in cell survival upon exposure to a stress.
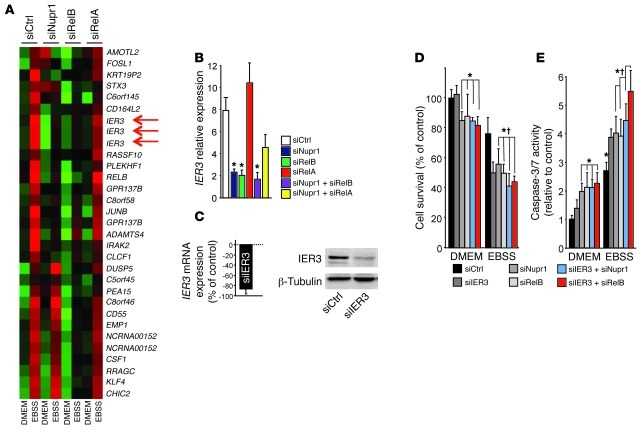
Since RelB, and not RelA/p65, is essential to the Nupr1-mediated survival mechanism that takes place upon nutrient deprivation–induced stress, we made the hypothesis that the two NF-κB transcription factors should activate different sets of genes, among which a pivotal pro-survival gene would be exclusively dependent on RelB. In order to test this hypothesis, an Affymetrix microarray analysis was performed using pancreatic cancer cells transfected with siNupr1, siRelB, siRelA/p65, or siCtrl (Figure 4A). Results were analyzed using conventional statistical methods for microarray processing, including clustering (Cluster and TreeView at www.rana.lbl.gov/EisenSoftware.htm), ontological analysis (Ingenuity Systems at www.ingenuity.com), and semantic-based pathway reconstruction algorithms (Ingenuity and Pathway Assist at www.ariadnegenomics.com) to identify key prosurvival genes of this Nupr1/RelB-mediated pathway, which ultimately supports the resistance and growth of pancreatic cancer cells exposed to environmental stress. Among RelB-dependent genes with known antiapoptotic activity, we focused our attention on the immediate early response 3 (IER3) gene, also known as IEX-1. IER3 is an important target gene for RelB in non-pancreatic tissues. How the coupling of these two important regulators of cell survival occurs remains, however, poorly understood. Results shown in Figure 3A demonstrate that IER3 expression upon starvation is also dependent on Nupr1. In addition, qRT-PCR analysis demonstrated that IER3 is induced by cell starvation in a RelB- and Nupr1-dependent manner, but independent of RelA/p65 expression (Figure 4B). Next, we investigated the impact of IER3 on PDAC cell survival in a nutrient-deprived microenvironment. We depleted MiaPaCa2 cells in IER3 using a specific siRNA (Figure 4C), and cell survival and caspase-3/7 activity were monitored after cells were grown in EBSS (Figure 4, D and E). IER3 depletion was without effect in cells cultured in conventional medium but resulted in massive cell death in EBSS, as already observed when RelB and/or Nupr1 were depleted. Combined deletion of IER3 and Nupr1 or RelB did not significantly increase cell death upon starvation, suggesting that Nupr1, RelB, and IER3 participate in a common survival pathway.
Figure 4. Expression of IER3 is dependent on Nupr1 and RelB, but not on RelA/p65, and protects from stress-induced cell death.
(A) Heat map showing RelB- and RelA/p65-dependent gene expression in response to nutrient deprivation. A P value of 0.05 or less was required for establishment of differential gene expression. Red arrows indicate 3 different probes hybridizing to the IER3 transcript. (B) Cells were transfected with siCtrl, siNupr1, siRELB, or siRELA alone or combined and cultured in conventional or EBSS medium for 24 hours. IER3 mRNA was measured by qRT-PCR. (C) Cells were transfected with IER3-specific siRNA or siCtrl, and IER3 mRNA was measured after 24 hours by qRT-PCR (right) and IER3 protein expression by Western blot (left). (D and E) MiaPaCa2 cells were transfected with siCtrl, siNupr1, siRelB, and siIER3 alone or in combination, and cultured in conventional or EBSS medium for 24 hours. Cell viability and caspase-3/7 activity were measured as described in Figure 2. Cyclophilin was used as a housekeeping control for calculation of relative transcript levels. Values are expressed as mean ± SEM of triplicate, and experiments were repeated twice. *P ≤ 0.05 relative to siCtrl-transfected cells cultured in conventional medium; †P ≤ 0.05 relative to siNupr1-transfected cells.
Because siRNA transfection can generate off-target effects depending on their nucleotide sequence, we repeated several experiments using a supplementary set of siRNAs designed against Nupr1, RelB, and IER3 with very similar results (see Supplemental Figure 2). Along the same lines, because results may be conditioned by the specificity of the cell line used as model, some experiments were repeated in Panc1 cells, whose genetic background is different from that of MiaPaca2 cells. Results in MiaPaCa2 and Panc1 were very similar, as shown in Supplemental Figure 3. When pancreatic cancer cells were subjected to a milder stress than culture in EBSS, such as glucose starvation, the Nupr1/RelB/IER3 cascade remained necessary to protect from cell death, as shown in Supplemental Figure 2. In addition, when cells were treated with gemcitabine, this pathway also played a protective role (Supplemental Figure 2). Together, these data strongly suggest that Nupr1, RelB, and IER3 activation is a cell death–protective cascade against all stress agents, independent of their strength. Finally, to confirm that Nupr1, RelB, and IER3 proteins are involved in the same cell-protective pathway, a genetic rescue was designed. Cells transduced with lentivirus designed to overexpress RelB or IER3 were treated with Nupr1 siRNA and incubated in the presence of EBSS, and cell survival was monitored. No protective effect was observed with the empty vector LtvEmpty, but an almost complete recovery was observed in LtvRelB- or LtvIER3-transduced MiaPaca2 (Supplemental Figure 2) and Panc1 (Supplemental Figure 3) cells.
Nupr1 is required for RelB and IER3 expression in KrasG12D pancreas.
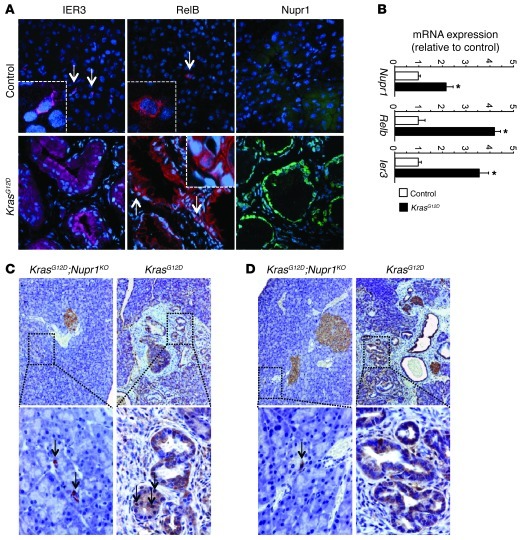
Increased resistance to stress, including the stress induced by nutrient deprivation, is a functional hallmark of pancreatic cancer. We investigated whether the Nupr1/RelB/IER3 survival pathway could account for the pathophysiological behavior of exocrine pancreatic cells in vivo. First, we looked by immunohistochemistry at the expression of the Nupr1, RelB, and IER3 proteins in precancerous PanIN lesions formed in the pancreas of 18-week-old KrasG12D mice carrying wild-type Nupr1 alleles. Our goal was to infer the location and time for the activation of the Nupr1/Relb/IER3 pathway as a surrogate marker for determining when these genes are expressed during pancreatic carcinogenesis. These experiments should reveal when and where these genes are translated to make a potentially functionally active protein. Figure 5A shows in KrasG12D mice that PanIN lesions — considered to be an initiating event associated with increased survival and resistance to cell death and metabolic stress — displayed concomitant expression of Nupr1, RelB, and IER3 proteins. Second, we measured mRNAs encoding Nupr1, RelB, and IER3 in the pancreas of 8-week-old KrasG12D mice (a time in which PanINs are almost absent) and found that expression of all 3 genes was activated before PanINs became visible (Figure 5B). These results indicate that the Nupr1-mediated pathway described here might be operational under conditions where KrasG12D-mediated growth and survival promote initiation of pancreatic carcinogenesis. Based upon these results, we performed appropriate genetic crosses to address the question whether Nupr1 deficiency affects the expression of RelB and IER3 proteins in PanINs from KrasG12D mice. Figure 5, C and D, show that, in contrast to Nupr1-expressing KrasG12D pancreas, RelB and IER3 proteins were absent in the pancreas of KrasG12D;Nupr1KO mice. Together, these results suggest that expression of RelB and IER3 participate in Nupr1-mediated promotion of PanIN formation.
Figure 5. Nupr1, RelB, and IER3 are expressed in PanIN lesions.
(A) Immunofluorescence staining of Nupr1, RelB, and IER3 on frozen pancreas sections from 13-week-old KrasG12D and control mice. Nuclei were stained with DAPI (blue). Original magnification, ×200; insets, ×600. (B) mRNA expression of Nupr1, Relb, and Ier3 in 8-week-old KrasG12D and control mice. *P < 0.05. (C and D) Pancreas sections from 13-week-old KrasG12D and KrasG12D;Nupr1KO mice were immunostained with antibodies against RelB (C) and IER3 (D) proteins. Original magnification, top panels, ×100; bottom panels, ×200.
PanIN development is delayed in the RelB-deleted KrasG12D pancreas.
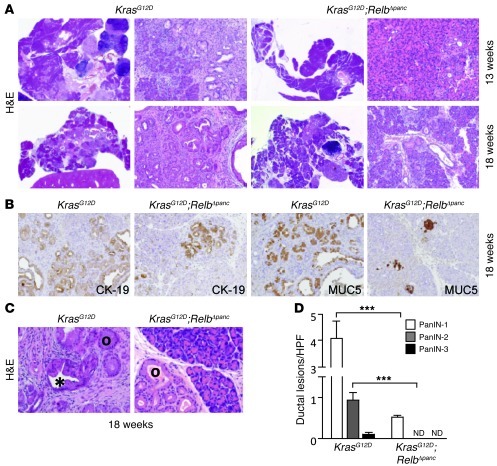
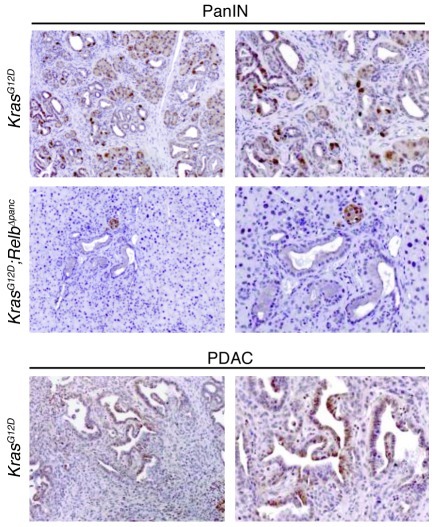
In order to test the putative role of the Nupr1/RelB/IER3 cascade in PanIN development, we introduced pancreas-specific deletion of RelB into a KrasG12D background. We inbred a Ptf1a+Cre(ex1) strain (28) with mice carrying a floxed Relb allele. Recombination between the two loxP sites mediated by the Cre recombinase led to complete inactivation of the Relb gene via deletion of exon 4. Ptf1a+Cre(ex1) mice homozygous for the floxed Relb allele displayed no apparent abnormalities in either the embryonic or adult pancreas. For simplicity, these mice are designated RelbΔpanc (pancreas-specific Relb knockout). These mice were crossed with KrasG12D mice to obtain KrasG12D;RelbΔpanc animals expressing activated KrasG12D in the pancreas that concomitantly carry deleted Relb alleles. We performed both histopathological and immunohistological analyses of the pancreata harvested from these mice at 13 and 18 weeks of age. As shown in Figure 6A, KrasG12D mice, which also carry a wild-type Relb locus, developed, as previously reported, numerous PanIN lesions at 13 weeks of age. In contrast, pancreata of age-matched KrasG12D;RelbΔpanc mice seldom developed this type of lesion. In these animals, PanIN lesions were histopathologically indistinguishable from PanINs observed in control animals, as documented by the conserved expression of epithelial markers such as cytokeratin-19 (CK-19) and MUC5 in lesions from both KrasG12D and KrasG12D;RelbΔpanc animals (Figure 6B). This effect was more evident at 18 weeks of age, when the pancreas from KrasG12D mice was massively replaced by PanIN lesions, whereas KrasG12D;RelbΔpanc mice still displayed extensive areas of normal parenchyma (Figure 6, C and D). Surrogate marker analysis by immunohistochemistry readily detected IER3 immunoreactivity within PanIN lesions derived from 18-week-old KrasG12D mice (Figure 7), with no obvious correlation between PanIN grade and level of IER3 expression. Expression of RelB and IER3 was also readily detected in a few rare spontaneous adenocarcinomas that developed in KrasG12D mice (Figure 7). In contrast, expression of IER3 was either negligible or undetected in pancreata from KrasG12D;RelbΔpanc mice, in agreement with our results in vitro. These genetic experiments demonstrate that, as for Nupr1 deletion, inactivation of the Relb locus delays the development of PanIN lesions in KrasG12D pancreas and that this delay is associated with the absence of IER3. These data lend further support to the existence of a hierarchical pathway arrangement (Nupr1/RelB/IER3) of the key pro-survival genes.
Figure 6. RelB is essential for PanIN progression.
(A) Representative H&E-stained pancreas sections from 13- and 18-week-old mice of the indicated genotypes. Original magnification, right panel images of the KrasG12D and KrasG12D;RelbΔpanc, ×20, left panel images; ×100. (B) Immunostaining of pancreas from KrasG12D and KrasG12D;RelbΔpanc mice with CK-19 and MUC5 antibodies. Original magnification, ×100. (C) Black circles indicate PanIN-1, and asterisk indicates PanIN-2. Original magnification, ×200. (D) Number of PanINs per ×200 field. Values are expressed as mean ± SEM for 6 pancreata from KrasG12D;RelbΔpanc and KrasG12D mice. ***P ≤ 0.05 relative to the number of PanINs in pancreata from KrasG12D mice.
Figure 7. IER3 expression in PanINs is dependent on RelB.
Top: PanIN lesions from KrasG12D and KrasG12D;RelbΔpanc mice were immunostained with antibodies against the IER3 protein. Bottom: Samples from pancreatic adenocarcinoma (PDAC) developed on KrasG12D mice were also immunostained with antibodies against the IER3 protein. Original magnification, ×100 (left panels) and at ×200 (right panels).
Expression of Nupr1, RelB, and IER3 in patients with PDAC.
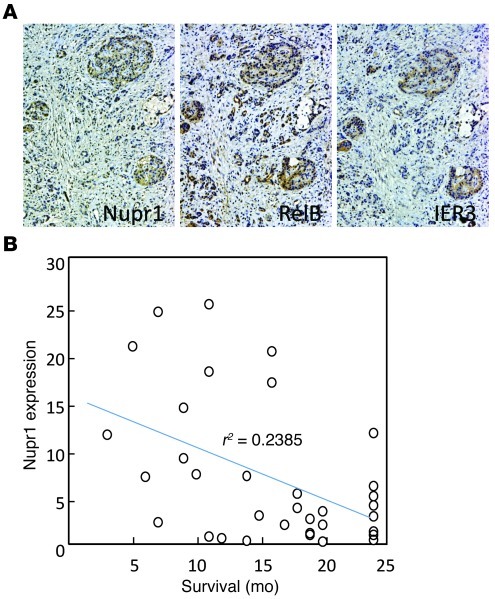
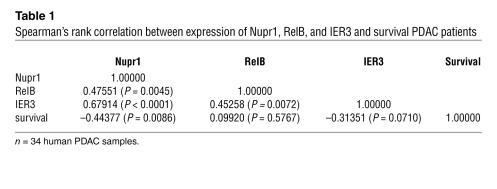
We investigated the relationship between the expression levels of the 3 proteins involved in this pathway and the prognosis of patients with PDAC. To this end, levels of the 3 proteins were assessed by immunohistochemistry with tissue microarray (TMA) containing PDAC samples from 34 patients with a follow-up of 24 months. First, we found coexpression of the 3 proteins (Figure 8A), with a significant correlation of the intensities of their expressions (Table 1). Second and more important, as shown in Figure 8B, an inverse correlation was found between Nupr1 expression and time of survival. These data support the hypothesis by which expression of Nupr1, RelB, and IER3 facilitates the progression of PDAC and serves as a marker of poor prognosis.
Figure 8. Nupr1, RelB, and IER3 expression in human PDAC.
(A) Successive PDAC samples were stained with anti-Nupr1, anti-RelB, and anti-IER3 antibodies, revealed with the avidin-peroxidase complex, and counterstained with hematoxylin. Original magnification, ×100. (B) Nupr1 expression values in TMA are plotted against survival time for 34 patients.
Table 1 .
Spearman’s rank correlation between expression of Nupr1, RelB, and IER3 and survival PDAC patients
Discussion
Nupr1 was originally identified a decade ago as a product of a gene inducible in injured pancreatic tissue (acute pancreatitis) (18). Additional studies from our laboratory have implicated this protein in the response of human epithelial cells to various stimuli known to induce stress (7). In addition, recent experimental evidence demonstrates that Nupr1 is overexpressed in a variety of human cancers (8, 9, 12, 31), suggesting its activation during pancreatic carcinogenesis (10, 11). However, whether such expression is relevant to the process of neoplastic transformation and malignant histogenesis (PanIN formation) remains to be determined. We designed the current study to test the hypothesis that expression of Nupr1 in pancreatic cells under stress conditions promotes pancreatic carcinogenesis. We designed genetic experiments based on the inactivation of Nupr1 and of its primary target genes, both in cultured pancreatic cancer cells and in genetically engineered mice. Nupr1 and its target pathways were shown to promote the survival and growth of pancreatic cancer cells as well as PanIN formation in vivo. Our studies support the existence of a pathway involved in the process of pancreatic carcinogenesis in addition to other previously reported pathways (32, 33). Nupr1 binds chromatin and regulates, through modulation of gene expression of RelB, the antiapoptotic factor IER3, which is a key component of the non-conventional NF-κB pro-survival pathway, although at this time direct activation by Nupr1 of IER3 expression cannot be formally excluded. The antiapoptotic role of IER3 in vivo was established by Zhang and colleagues using a transgenic Ier3 allele, which, when expressed in T lymphocytes, expands their lifespan and promotes their activation, resulting in the development of a lupus-like disease (34). Here, we show that IER3 inactivation leads to diminished survival of pancreatic cancer cells submitted to nutrient deprivation. In addition, blocking IER3 expression alone or together with Nupr1 or RelB led to the same results, indicating that these 3 factors act through the same pathway. As the development of pharmacological tools to manipulate cell death responses in vitro and in vivo, including those that target NF-κB pathways, is a major focus of modern experimental therapeutics, these results have significant translational potential.
Successive studies on pancreatic cancer have contributed to the description of a multistep pattern of tumor progression through accumulation of DNA mutations, changes in DNA methylation, and expression of many large and small RNA molecules. This model, adapted from Kinzler and Vogelstein’s genetic paradigm for colon cancer (35), influenced pancreatic cancer research during the last two decades. However, the study of protein-protein interaction at the chromatin-promoter level, which is not genetic in nature but interacts with genetic mutations to facilitate PanIN development, has remained an underrepresented area of study in pancreatic cancer research. This work provides evidence that fills the gap in the existing knowledge, through the demonstration that chromatin-promoter interactions underlie the promotion of PanIN lesion by a mutant Kras allele. Such a mechanism supports the recently proposed genetic-epigenetic model for PanIN formation. This model predicts that genetic mutations activate aberrant chromatin proteins required for the expression of the genes required for generation of neoplastic lesions (36). Furthermore, through genetic crosses between Nurp1- and RelB-knockout mice in the context of Kras activation (Nurp1–/–;KrasG12D and RelbΔpanc;KrasG12D), we have determined that the Nurp1/RelB/IER3 pathway is required for the initiation and early progression of pancreatic carcinogenesis. To date, very few chromatin proteins have been shown to be operational during early carcinogenesis. Hence, our data provide mechanistic insight into a previously poorly understood process. However, because Nupr1, RelB, and IER3 are not exclusively expressed in pancreatic epithelial cells (see Figure 5), we cannot exclude another, still-unknown role of these proteins in PanIN development.
The NF-κB family is a critical regulator of many cellular processes, among which cell survival is the most widely acknowledged. The most studied heterodimer is RelA/p65:p50 (the classical pathway), believed to be preferentially activated in somatic cells. However, the noncanonical RelB-dependent NF-κB pathway was found to be involved in survival processes of several cell types, often of hematopoietic origin but also of epithelial origin (37). For instance, RelB expression was found to be necessary for protecting thymocytes from differentiation-induced apoptosis (38), for protecting kidney cells from renal ischemia-reperfusion–induced injury (39), and for protecting prostate cancer–derived cells against apoptosis induced by ionizing radiation (40). Together, our data indicate that Nupr1-induced RelB upregulation is sufficient to trigger an IER3-based antiapoptotic mechanism in PDAC cells upon stress, without a primary need for RelA/p65 activation. All these observations are in agreement with the fact that, in pancreatic adenocarcinoma, Nupr1 is overexpressed (11, 21) and RelB is constitutively activated (41, 42). The antiapoptotic factor IER3 is overexpressed in pancreatic cancer (43), and its antiapoptotic effect was confirmed in several cancer-derived cell types (34, 44–48). In addition to its direct antiapoptotic effect, IER3 binds the C-terminal region of RelA/p65 and inhibits its transcriptional activity (49), suggesting that IER3 may also provide negative feedback to the NF-κB pathway in response to stress. It is interesting to note that while activation of Nupr1 leads to RelB overexpression, a positive feedback mechanism in turn enhances the expression of Nupr1 (Figures 3I). This was expected, because an NF-κB–responsive element (NRE) is present in the promoter region of the Nupr1 gene (50) and its functionality is well documented (17). Collectively, these results help us to better understand the mechanisms by which the NF-κB–mediated transcriptional pathways are involved in the regulation of cell survival, cell growth, and cancer.
We then investigated whether this pathway could be involved in human PDAC. We studied expression of Nupr1, RelB, and IER3 in a group of patients with PDAC and found a significant correlation between the expressions of these proteins, in particular Nupr1, and poor prognosis of patients. This is in agreement with our previous studies showing an inverse correlation between Nupr1 and apoptosis in PDAC samples (10). However, although the correlation coefficient is –0.44377 with a P value of 0.0086, this study was retrospective and therefore needs to be confirmed in a prospective way.
In conclusion, we show in this work that the chromatin factor Nupr1 is essential for the survival of pancreatic cancer cells exposed to a stress. Mechanistically, the Nupr1 protein activates the expression of the NF-κB family member RelB, which in turn induces the expression of the antiapoptotic factor IER3. Like that of Nupr1, expression of both RelB and IER3 protects PDAC cells from the consequences of stress. In vivo, Nupr1 and RelB expression is critical for the development of precancerous PanIN lesions in KrasG12D mice, thus supporting the implication of the Nupr1/RelB/IER3 cascade in cancer cell development within the tumor microenvironment. This study unravels a cell stress response pathway that accounts for the remarkable resistance of pancreatic cancer cells to stress. Finally, a significant correlation between expression of Nupr1, RelB, and IER3 and the poor prognosis of patients with PDAC was found. It is hoped that description of this pathway will open up new perspectives on the involvement of the alternative RelB-based NF-κB pathway during pancreatic adenocarcinoma development. We believe these findings will promote studies attempting to block the Nupr1/RelB/IER3 cascade as a novel anticancer therapy.
Methods
Cell culture and in vitro stress induction.
MiaPaCa2 cells obtained from ATCC were maintained in DMEM (Invitrogen) supplemented with 10% FBS at 37°C with 5% CO2. Cells were at about 70% confluence for all experiments. To avoid additional stress, all media were warmed to 37°C and cells were washed twice with warm PBS before addition of EBSS (Invitrogen), which is completely devoid of glucose, amino acids, and growth factors.
siRNA transfection.
MiaPaCa2 and Panc1 cells were plated at 70% confluence in 100-mm dishes. INTERFERin reagent (Polyplus-transfection) was used to perform siRNA transfections according to the manufacturer’s protocol. Nupr1, RelA/p65, RelB, and IER3 were knocked down using 140 ng of specific siRNAs. Scrambled siRNA targeting no known gene sequence was used as negative control. The sequences of Nupr1-specific siRNA [siNupr1 r(GGAGGACCCAGGACAGGAU)dTdT and siNupr1#2 r(AGGUCGCACCAAGAGAGAA)dTdT] were previously reported (51), and RelA/p65- [r(GAUCAAUGGCUACACAGG)dTdT], RelB- [siRelB r(GGAUUUGCCGAAUUAACAA)dTdT and siRelB#2 r(CUGCGGAUUUGCCGAAUUAUU)dTdT], and IER3- [siIER3 r(GGAAGGAGAGCGUCGUUAA)dTdT and siIER3#2 r(AAGGCUUCUCUUUCUGCUG)dTdT] specific siRNAs were from QIAGEN.
DNA microarray.
Total RNA (15 μg) was isolated and reverse transcribed for hybridization to the human oligonucleotide array U133 Plus 2.0 (GeneChip, Affymetrix) as described previously (52). Arrays were processed using the Affymetrix GeneChip Fluidic Station 450 (protocol EukGE-WS2v5_450) and scanned using a GeneChip Scanner 3000 G7 (Affymetrix). GeneChip Operating Software (Affymetrix GCOS v1.4) was used to obtain chip images, with quality control performed using the AffyQCReport software. Microarray data were deposited in the GEO database (accession number GSE35463).
Cell viability and caspase-3/7 assay.
To detect caspase-3/7 activity, an Apo-ONE fluorescence assay was performed according to the manufacturer’s (Promega) instructions. Data were normalized to cellular enzymatic activity CellTiter-Blue (Promega). To validate viability, we counted cells after metabolic stress induction using a cell counter (Countess, Invitrogen).
Luciferase assay.
MiaPaCa2 cells were plated at 70% confluence in 6-well plates and 24 hours later transiently transfected with 3 μg total DNA using FuGENE 6 transfection reagent (Roche Applied Science). The human 1.7-kb Relb promoter (–1,694 to +1) was cloned into the luciferase reporter pGL3 basic vector and transfected with one of the following expression vectors: pcDNA3-Flag containing the human Nupr1 cDNA sequence or pcDNA3-Flag without the cDNA insert, siNupr1, or scrambled siRNA. Co-transfection of Renilla luciferase under the control of the SV40 early enhancer/promoter region (pSV40-RL, Promega) was used to normalize for transfection efficiency. Medium was replaced 24 hours later with fresh DMEM or EBSS and incubated for an additional 9-hour period. All transfections were performed at least two times, in triplicate.
ChIP assay.
MiaPaCa2 cells were transfected with the Nupr1-Flag, irrelevant cytochrome c–Flag, or empty vector–Flag constructs with subsequent ChIP assay performed using the EZ-ChIP kit (Millipore) according to the manufacturer’s instructions. Input DNA was collected after pre-clearing lysate with protein G/agarose beads prior to immunoprecipitation with the anti-FLAG (M2) antibody (Sigma-Aldrich) or with a non-relevant IgG. PCR was performed using TaKaRa LA Taq with GC Buffer II, according to the manufacturer’s suggestions (Takara Bio Inc). A 255-bp region of the RelB promoter was amplified by PCR using specific primers (5′-ATGGATGGCAGGTGTAGAGCCC-3′, 5′-TGCTCTGGACGAGACAACTGAG-3′).
Lentivirus constructs.
Full-length RelB and IER3 cDNA were amplified by PCR using (pcDNA3-RelB) and (pcDNA3-HA-IER3-1) expression plasmids as template. Primers for PCR were designed to include EcoRI and SmaI or XhoI and ApaI restriction sites for RelB or IER3, respectively. After digestion, fragments were subcloned into the PLVX-DsRed-Monomer-N1 lentiviral vector (Clonetech). Lentivirus particles (LtvEmpty, LtvRelB, or LtvIER3) were produced by transient transfection of 293T cells according to standard protocols. MiaPaCa2 and Panc-1 cells were grown at 50%–60% confluence and infected with the supernatant containing viruses. Cells were used for experiments 72–96 hours after transduction.
RNA extracts/real-time qRT-PCR.
Cells were washed once with PBS, and cell lysis was performed using a total RNA extraction kit (Norgen Biotek) according to the manufacturer’s protocol. RNA from pancreas of 8-week-old Pdx1-cre;LSL-KrasG12D or control mice was purified following the procedure of Chirgwin et al. (53). cDNA was obtained using the ImProm-II Reverse Transcription System (Promega). Sequences of the primers used to amplify human and mouse genes are indicated in Supplemental Table 1.
Immunoblotting.
Protein extraction was performed on ice using total protein extraction buffer: 50 mM HEPES pH 7.5, 150 mM NaCl, 20% SDS, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton, 25 mM NaF, 10 μM ZnCl2, 50 mM DTT. Before lysis, protease inhibitor cocktail at 1:200 (Sigma-Aldrich, NUPR1340), 500 μM PMSF, 1 mM sodium orthovanadate, and 1 mM β glycerophosphate were added. Protein concentration was measured using a BCA Protein Assay Kit (Pierce Biotechnology). Protein samples (80 μg) were denatured at 95°C and subsequently separated by 12.5% SDS-PAGE gel electrophoresis. After transfer to nitrocellulose membrane and blocking with 1% BSA, samples were probed with one of the following antibodies: RelA/p65 and RelB (Santa Cruz Biotechnology Inc.), IER3 (rabbit anti-serum to recombinant full-length GST-IER3 was obtained from Françoise Porteu, INSERM U1016, Institut Cochin, Paris, France) (47), β-tubulin and vimentin (Sigma-Aldrich), and monoclonal Nupr1 (developed in our laboratory).
Confocal microscopy.
Ten-micrometer cryosections of pancreatic tissue were dried and fixed with acetone. After nonspecific binding site blockade with 3% bovine serum albumin, 10% fetal calf serum, and 10% goat serum for 30 minutes, tissue sections were labeled 1 hour at room temperature with primary antibodies or control antibodies, followed by incubation for 30 minutes at room temperature with secondary antibodies and SYTOX Blue when nuclei staining was required. Primary antibodies were the same as those for immunoblotting. Slides were mounted in ProLong Gold (Invitrogen) and observed with a Zeiss LSM 510 confocal microscope. Images were analyzed using Adobe Photoshop 7.0.
Histology and immunohistochemistry.
Pancreatic sections were fixed in 4% paraformaldehyde and paraffin embedded. H&E staining and immunohistochemistry were performed using standard procedures. Sections were stained with antibodies MUC5 (Chemicon) and CK-19 (Developmental Studies Hybridoma Bank).
Animals.
Mice bearing a homozygous deletion of exon 2 of the Nupr1 gene were described previously (54). The Pdx1-cre;LSL-KrasG12D mice were provided by R. Depinho (Dana-Faber Cancer Institute, Boston, Massachusetts, USA) and resulted from crossbreeding of the following strains: Pdx1-Cre (55) and LSL-KrasG12D (26). Mouse strains and generation of RelbΔpanc mice will be published elsewhere (56). Because animals are from different genetic backgrounds, we systematically used littermate control and experimental mice.
Patients and tissue microarray.
PDAC samples were formalin-fixed surgical specimens obtained from the Pathology Department of Aix-Marseille University. Two-year follow-up data were recorded from 34 patients (see Supplemental Table 2). The procedure for construction of TMAs was as previously described (57, 58). Briefly, cores were punched from the selected paraffin blocks and distributed in new blocks, with 2 cores of 0.6-mm diameter for each tumor. TMA serial tissue sections were prepared 24 hours before immunohistochemical processing and stored at 4°C. The dilution of each antibody was determined by pre-screening on the full 4-μm-thick sections before use on TMA sections. The immunoperoxidase procedures were performed using an automated Ventana BenchMark XT autostainer. Measurements of immunoprecipitate densitometry in cores were made for each marker in an individual core after digitization and “cropping” of microscopic images as previously reported (57, 58).
Statistics.
Statistical analyses were performed using the unpaired 2-tailed Student t test. A P value less than or equal to 0.05 was considered significant. All values are expressed as mean ± SEM. qRT-PCR data are representative of at least 3 independent experiments with technical duplicates completed. For microarray analysis, background subtraction and normalization of probe set intensities were performed using robust multiarray analysis (RMA). To identify differentially expressed genes, gene expression intensities were compared using a moderated t test and a Bayes smoothing approach developed for a low number of replicates. To correct for the effect of multiple testing, the false discovery rate was estimated from P values derived from the moderated t test statistics. The analysis was performed using the affylmGUI graphical user interface for the limma microarray package ( www.bioconductor.org). Statistical analysis of the relationship between TMA results and survival data was conducted using the SAS version 9.3 computer program (SAS Institute Inc.). Spearman’s rank correlation coefficient was applied for the analysis of significance of the correlation.
Study approval.
Care and manipulation of mice were performed in accordance with national and European legislation on animal experimentation and were approved by the Aix-Marseille University Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank M. Barbacid and C. Guerra for their comments; M.N. Lavaut and C. Loncle for technical assistance; and P. Berthezene for statistical analysis. This work was supported by grants from La Ligue contre le Cancer, Institut national du cancer (INCa), and INSERM to J.L. Iovanna; by German Cancer Aid (grant 107195), the German Federal Ministry of Education and Research (grant 01GS08115), the Lustgarten Foundation (RFP05-14 and 06-12), and the German Research Foundation (grant SI 1549/1-1) to R.M. Schmid; and from the NIH (DK52913) to R. Urrutia. C.E. Cano and T. Hamidi are supported by La Ligue contre le Cancer; M.J. Sandi is supported by l’Association pour la Recherche sur le Cancer; and G. Lomberk by the Fraternal Order of Eagles for Cancer Research.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(6):2092–2103. doi:10.1172/JCI60144.
References
- 1.Sultana A, et al. Systematic review, including meta-analyses, on the management of locally advanced pancreatic cancer using radiation/combined modality therapy. Br J Cancer. 2007;96(8):1183–1190. doi: 10.1038/sj.bjc.6603719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Reilly EM, Lutz MP, Neuhaus P. Accomplishments in 2008 in the management of localized pancreatic cancer. Gastrointest Cancer Res. 2009;3(5 Supplement 2):S37–S42. [PMC free article] [PubMed] [Google Scholar]
- 3.Burris H, Storniolo AM. Assessing clinical benefit in the treatment of pancreas cancer: gemcitabine compared to 5-fluorouracil. Eur J Cancer. 1997;33(suppl 1):S18–S22. doi: 10.1016/s0959-8049(96)00324-3. [DOI] [PubMed] [Google Scholar]
- 4.Wong HH, Lemoine NR. Pancreatic cancer: molecular pathogenesis and new therapeutic targets. Nat Rev Gastroenterol Hepatol. 2009;6(7):412–422. doi: 10.1038/nrgastro.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esposito I, et al. Tumor-suppressor function of SPARC-like protein 1/Hevin in pancreatic cancer. Neoplasia. 2007;9(1):8–17. doi: 10.1593/neo.06646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goruppi S, Iovanna JL. Stress-inducible protein p8 is involved in several physiological and pathological processes. J Biol Chem. 2010;285(3):1577–1581. doi: 10.1074/jbc.R109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito Y, et al. Expression of p8 protein in breast carcinoma; an inverse relationship with apoptosis. Anticancer Res. 2005;25(2A):833–837. [PubMed] [Google Scholar]
- 9.Ito Y, et al. Expression of p8 protein in medullary thyroid carcinoma. Anticancer Res. 2005;25(5):3419–3423. [PubMed] [Google Scholar]
- 10.Su SB, et al. Overexpression of p8 is inversely correlated with apoptosis in pancreatic cancer. Clin Cancer Res. 2001;7(5):1320–1324. [PubMed] [Google Scholar]
- 11.Su SB, et al. Expression of p8 in human pancreatic cancer. Clin Cancer Res. 2001;7(2):309–313. [PubMed] [Google Scholar]
- 12.Ree AH, Pacheco MM, Tvermyr M, Fodstad O, Brentani MM. Expression of a novel factor, com1, in early tumor progression of breast cancer. Clin Cancer Res. 2000;6(5):1778–1783. [PubMed] [Google Scholar]
- 13.Bratland A, et al. Expression of a novel factor, com1, is regulated by 1,25-dihydroxyvitamin D3 in breast cancer cells. Cancer Res. 2000;60(19):5578–5583. [PubMed] [Google Scholar]
- 14.Garcia-Montero A, Vasseur S, Mallo GV, Soubeyran P, Dagorn JC, Iovanna JL. Expression of the stress-induced p8 mRNA is transiently activated after culture medium change. Eur J Cell Biol. 2001;80(11):720–725. doi: 10.1078/0171-9335-00209. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Montero AC, et al. Transforming growth factor beta-1 enhances Smad transcriptional activity through activation of p8 gene expression. Biochem J. 2001;357(pt 1):249–253. doi: 10.1042/0264-6021:3570249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang YF, Vaccaro MI, Fiedler F, Calvo EL, Iovanna JL. Lipopolysaccharides induce p8 mRNA expression in vivo and in vitro. Biochem Biophys Res Commun. 1999;260(3):686–690. doi: 10.1006/bbrc.1999.0953. [DOI] [PubMed] [Google Scholar]
- 17.Kallwellis K, Grempler R, Gunther S, Path G, Walther R. Tumor necrosis factor alpha induces the expression of the nuclear protein p8 via a novel NF kappaB binding site within the promoter. Horm Metab Res. 2006;38(9):570–574. doi: 10.1055/s-2006-950503. [DOI] [PubMed] [Google Scholar]
- 18.Mallo GV, et al. Cloning and expression of the rat p8 cDNA, a new gene activated in pancreas during the acute phase of pancreatitis, pancreatic development, and regeneration, and which promotes cellular growth. J Biol Chem. 1997;272(51):32360–32369. doi: 10.1074/jbc.272.51.32360. [DOI] [PubMed] [Google Scholar]
- 19.Motoo Y, Iovanna JL, Mallo GV, Su SB, Xie MJ, Sawabu N. P8 expression is induced in acinar cells during chronic pancreatitis. Dig Dis Sci. 2001;46(8):1640–1646. doi: 10.1023/A:1010637100933. [DOI] [PubMed] [Google Scholar]
- 20.Passe CM, Cooper G, Quirk CC. The murine p8 gene promoter is activated by activating transcription factor 4 (ATF4) in the gonadotrope-derived LbetaT2 cell line. Endocrine. 2006;30(1):81–91. doi: 10.1385/ENDO:30:1:81. [DOI] [PubMed] [Google Scholar]
- 21.Su SB, Motoo Y, Iovanna JL, Xie MJ, Sawabu N. Effect of camostat mesilate on the expression of pancreatitis-associated protein (PAP), p8, and cytokines in rat spontaneous chronic pancreatitis. Pancreas. 2001;23(2):134–140. doi: 10.1097/00006676-200108000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Liu Z, Zhang S. Tissue distribution, developmental expression and up-regulation of p8 transcripts on stress in zebrafish. Fish Shellfish Immunol. 2009;28(4):549–554. doi: 10.1016/j.fsi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Giroux V, et al. p8 is a new target of gemcitabine in pancreatic cancer cells. Clin Cancer Res. 2006;12(1):235–241. doi: 10.1158/1078-0432.CCR-05-1700. [DOI] [PubMed] [Google Scholar]
- 24.Cano CE, Iovanna JL. Stress proteins and pancreatic cancer metastasis. ScientificWorldJournal. 2010;10:1958–1966. doi: 10.1100/tsw.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gironella M, et al. p8/nupr1 regulates DNA-repair activity after double-strand gamma irradiation-induced DNA damage. J Cell Physiol. 2009;221(3):594–602. doi: 10.1002/jcp.21889. [DOI] [PubMed] [Google Scholar]
- 26.Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 28.Siveke JT, Einwachter H, Sipos B, Lubeseder-Martellato C, Kloppel G, Schmid RM. Concomitant pancreatic activation of Kras(G12D) and Tgfa results in cystic papillary neoplasms reminiscent of human IPMN. Cancer Cell. 2007;12(3):266–279. doi: 10.1016/j.ccr.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Vasseur S, et al. p8 improves pancreatic response to acute pancreatitis by enhancing the expression of the anti-inflammatory protein pancreatitis-associated protein I. J Biol Chem. 2004;279(8):7199–7207. doi: 10.1074/jbc.M309152200. [DOI] [PubMed] [Google Scholar]
- 30.Hruban RH, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66(1):95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 31.Ito Y, et al. Expression and cellular localization of p8 protein in thyroid neoplasms. Cancer Lett. 2003;201(2):237–244. doi: 10.1016/j.canlet.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Activated K-ras and INK4a/Arf deficiency cooperate during the development of pancreatic cancer by activation of Notch and NF-kappaB signaling pathways. PLoS One. 2011;6(6):e20537. doi: 10.1371/journal.pone.0020537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Morton JP, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A. 2010;107(1):246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Schlossman SF, Edwards RA, Ou CN, Gu J, Wu MX. Impaired apoptosis, extended duration of immune responses, and a lupus-like autoimmune disease in IEX-1-transgenic mice. Proc Natl Acad Sci U S A. 2002;99(2):878–883. doi: 10.1073/pnas.022326699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87(2):159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 36.Lomberk G, Mathison AJ, Grzenda A, Urrutia R. The sunset of somatic genetics and the dawn of epigenetics: a new frontier in pancreatic cancer research. Curr Opin Gastroenterol. 2008;24(5):597–602. doi: 10.1097/MOG.0b013e32830b111d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dejardin E. The alternative NF-kappaB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem Pharmacol. 2006;72(9):1161–1179. doi: 10.1016/j.bcp.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Guerin S, Baron ML, Valero R, Herrant M, Auberger P, Naquet P. RelB reduces thymocyte apoptosis and regulates terminal thymocyte maturation. Eur J Immunol. 2002;32(1):1–9. doi: 10.1002/1521-4141(200201)32:1<1::AID-IMMU1>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 39.Feng B, et al. Small interfering RNA targeting RelB protects against renal ischemia-reperfusion injury. Transplantation. 2009;87(9):1283–1289. doi: 10.1097/TP.0b013e3181a1905e. [DOI] [PubMed] [Google Scholar]
- 40.Josson S, Xu Y, Fang F, Dhar SK, St Clair DK, St Clair WH. RelB regulates manganese superoxide dismutase gene and resistance to ionizing radiation of prostate cancer cells. Oncogene. 2006;25(10):1554–1559. doi: 10.1038/sj.onc.1209186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishina T, Yamaguchi N, Gohda J, Semba K, Inoue J. NIK is involved in constitutive activation of the alternative NF-kappaB pathway and proliferation of pancreatic cancer cells. Biochem Biophys Res Commun. 2009;388(1):96–101. doi: 10.1016/j.bbrc.2009.07.125. [DOI] [PubMed] [Google Scholar]
- 42.Wharry CE, Haines KM, Carroll RG, May MJ. Constitutive non-canonical NFkappaB signaling in pancreatic cancer cells. Cancer Biol Ther. 2009;8(16):1567–1576. doi: 10.4161/cbt.8.16.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasada T, et al. Prognostic significance of the immediate early response gene X-1 (IEX-1) expression in pancreatic cancer. Ann Surg Oncol. 2008;15(2):609–617. doi: 10.1245/s10434-007-9669-0. [DOI] [PubMed] [Google Scholar]
- 44.Bruheim S, Xi Y, Ju J, Fodstad O. Gene expression profiles classify human osteosarcoma xenografts according to sensitivity to doxorubicin, cisplatin, and ifosfamide. Clin Cancer Res. 2009;15(23):7161–7169. doi: 10.1158/1078-0432.CCR-08-2816. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez S, Perez-Perez MM, Hernando E, Serrano M, Cordon-Cardo C. p73beta-Mediated apoptosis requires p57kip2 induction and IEX-1 inhibition. Cancer Res. 2005;65(6):2186–2192. doi: 10.1158/0008-5472.CAN-04-3047. [DOI] [PubMed] [Google Scholar]
- 46.Ria R, et al. Gene expression profiling of bone marrow endothelial cells in patients with multiple myeloma. Clin Cancer Res. 2009;15(17):5369–5378. doi: 10.1158/1078-0432.CCR-09-0040. [DOI] [PubMed] [Google Scholar]
- 47.Garcia J, Ye Y, Arranz V, Letourneux C, Pezeron G, Porteu F. IEX-1: a new ERK substrate involved in both ERK survival activity and ERK activation. EMBO J. 2002;21(19):5151–5163. doi: 10.1093/emboj/cdf488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu MX, Ao Z, Prasad KV, Wu R, Schlossman SF. IEX-1L, an apoptosis inhibitor involved in NF-kappaB-mediated cell survival. Science. 1998;281(5379):998–1001. doi: 10.1126/science.281.5379.998. [DOI] [PubMed] [Google Scholar]
- 49.Arlt A, et al. IEX-1 directly interferes with RelA/p65 dependent transactivation and regulation of apoptosis. Biochim Biophys Acta. 2008;1783(5):941–952. doi: 10.1016/j.bbamcr.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 50.Vasseur S, et al. Structural and functional characterization of the mouse p8 gene: promotion of transcription by the CAAT-enhancer binding protein alpha (C/EBPalpha) and C/EBPbeta trans-acting factors involves a C/EBP cis-acting element and other regions of the promoter. Biochem J. 1999;343(pt 2):377–383. doi: 10.1042/0264-6021:3430377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carracedo A, et al. Cannabinoids induce apoptosis of pancreatic tumor cells via endoplasmic reticulum stress-related genes. Cancer Res. 2006;66(13):6748–6755. doi: 10.1158/0008-5472.CAN-06-0169. [DOI] [PubMed] [Google Scholar]
- 52.Archange C, et al. The WSB1 gene is involved in pancreatic cancer progression. PLoS One. 2008;3(6):e2475. doi: 10.1371/journal.pone.0002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 54.Malicet C, Lesavre N, Vasseur S, Iovanna JL. p8 inhibits the growth of human pancreatic cancer cells and its expression is induced through pathways involved in growth inhibition and repressed by factors promoting cell growth. Mol Cancer. 2003;2:37. doi: 10.1186/1476-4598-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 56.Powolny-Budnicka I, Riemann M, Tänzer S, Schmid RM, Hehlgans T, Weih F. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in γδ T cells. Immunity. 2011;34(3):364–374. doi: 10.1016/j.immuni.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 57.Charpin C, et al. Validation of an immunohistochemical signature predictive of 8-year outcome for patients with breast carcinoma. Int J Cancer. doi: 10.1002/ijc.27371. [published online ahead of print January 3, 2012]. doi:10.1002/ijc.27371. [DOI] [PubMed] [Google Scholar]
- 58.Giusiano S, et al. Immunohistochemical profiling of node negative breast carcinomas allows prediction of metastatic risk. Int J Oncol. 2010;36(4):889–898. doi: 10.3892/ijo_00000567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.