Abstract
Cytochrome c maturation protein E, CcmE, plays an integral role in the transfer of heme to apocytochromes c in many prokaryotes and some mitochondria. A novel sub-class featuring a heme-binding cysteine has been identified in archaea and some bacteria. Here we describe the solution NMR structure, backbone dynamics, and heme-binding properties of the soluble C-terminal domain of Desulfovibrio vulgaris CcmE, dvCcmE′. The structure adopts a conserved β-barrel OB-fold followed by an unstructured C-terminal tail encompassing the CxxxY heme-binding motif. Heme-binding analyses of wild type and mutant dvCcmE′ demonstrate the absolute requirement of residue C127 for non-covalent heme binding in vitro.
c-Type cytochromes are small, ubiquitous heme proteins that employ a conserved CxxCH motif to bind a heme prosthetic group, where both cysteines covalently bind the vinyl groups of the heme via thiols and the histidine serves as the proximal axial ligand for the metal ion. The coordination sphere of the iron is generally completed by an axial methionine or histidine. Nature has evolved at least three different systems for the post-translational addition of heme to cytochromes c (1–3). System I, relevant to this work, is generally comprised of eight cytochrome c maturation proteins (i.e., CcmABCDEFGH), and is prevalent in many prokaryotes and some protozoal and plant mitochondria. In this pathway, a small membrane-bound heme chaperone protein, CcmE, participates in the final transfer of heme to cytochrome c. CcmE is thought to form a transient covalent complex with heme through a conserved histidine (H130 in Escherichia coli) within a HxxxY motif near the C-terminus of the protein (4,5). Structural studies of the soluble periplasmic C-terminal domain (lacking the N-terminal transmembrane helix) of E. coli and Shewanella putrefaciens apoCcmE, revealed that the protein adopts a β-barrel fold immediately followed by the heme-binding motif and an unstructured C-terminal tail (6,7). Although no holoCcmE structure has been reported to date, it has been established that heme binding to E. coli CcmE occurs via a unique covalent bond between the Nδ1 of H130 and a vinyl β-carbon within the heme (8), the phenol moiety of Y134 serves as an axial ligand for the iron (9,10), and that the process occurs within a somewhat buried, hydrophobic heme-binding pocket (11).
A System I variant sub-class of CcmE proteins featuring a cysteine in place of the histidine in a CxxxY motif was predicted in archaea and some bacteria, including the sulfate-reducing Desulfovibrio species (Figure 1A) (12). Indeed, it was recently demonstrated that placing the Ccm operon from Desulfovibrio desulfuricans into an E. coli strain lacking its endogenous Ccm system still resulted in the production of holocytochrome c, and that cytochrome c maturation in this variant System I proceeds through the covalent attachment of heme to C127 in D. desulfuricans CcmE (13).
FIGURE 1.
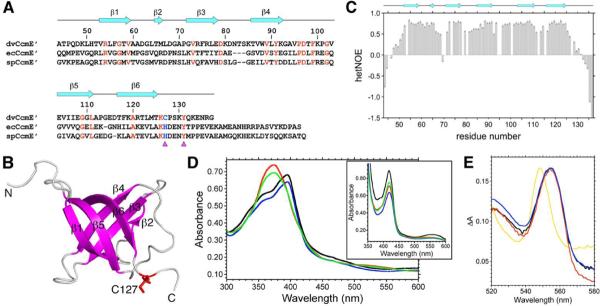
(A) Structure-based sequence alignment of the soluble C-terminal domains of D. vulgaris CcmE (dvCcmE′; residues 44 to 137), E. coli CcmE (ecCcmE′; residues 51 to 159), and S. putrefaciens CcmE (spCcmE′; residues 51 to 161). The sequence numbering for dvCcmE′ and the secondary structural elements found in the solution NMR structure of dvCcmE′Δ9 (PDB ID, 2KCT) are shown above the alignment. Identical amino acid residues are shown in red, and residues involved in heme binding are indicated below the alignment by magenta triangles. (B) Lowest energy (CNS) conformer from the final solution NMR structure ensemble of apo-dvCcmE′Δ9. The β-strands and loops are shown in magenta and grey, respectively. The heme-binding cysteine, C127, is colored red. Rendered using PyMOL (http://www.pymol.org). (C) Backbone dynamics of apo-dvCcmE′. Plot of 1H-15N heteronuclear NOE versus residue number obtained on [U-13C,15N]-apo-dvCcmE′ at a 15N Larmor frequency of 60.8 MHz. Secondary structural elements found in the solution NMR structure of dvCcmE′Δ9 are shown above the plot. (D) UV-visible absorption spectra of the ferric and ferrous (inset) forms of free heme (black), wild type dvCcmE′ (red), [C127A]-dvCcmE′ (blue), and [Y131F]-dvCcmE′ (green). (E) Pyridine hemochrome spectra (reduced – oxidized) for free hemin (black), wild type dvCcmE′ (red), [C127A]-dvCcmE′ (blue), and equine heart holocytochrome c (gold). Only cyctochrome c exhibits the expected shift of the α-band diagnostic of covalent heme binding (λmax = 549.5 nm).
Here, we present the solution NMR structure, backbone dynamics, and heme-binding properties of the soluble C-terminal domain of Desulfovibrio vulgaris CcmE, dvCcmE′. Based on solution NMR structural and 15N relaxation measurements, the apo-form of dvCcmE′ was observed to adopt a β-barrel fold characteristic of the CcmE superfamily, featuring a highly dynamic C-terminal tail that includes the CxxxY heme-binding motif. In vitro UV-visible absorption spectroscopy and heme stained gel electrophoresis studies of wild type, [C127A]-, and [Y131F]-dvCcmE′ demonstrate that non-covalent ferric heme binding requires residue C127. To our knowledge, this is the first structural representative from this variant CcmE sub-family.
The construct optimization, cloning, expression, and purification of 13C and 15N isotopically-enriched samples of Desulfovibrio vulgaris CcmE(44–128), lacking the C-terminal 9 amino-acid residues, and CcmE(44–137), dvCcmE′Δ9 and dvCcmE′, respectively (UniProtKB/TrEMBL ID, Q72D78_DESVH; NESG IDs, DvR115G and DvR115, respectively) were conducted using standard protocols of the Northeast Structural Genomics Consortium (14); see the Supporting Information for a complete description of the methods used in this work. Briefly, the solution NMR structure of apo-dvCcmE′Δ9 was determined at pH 4.5 by standard triple-resonance NMR methods and using the program CYANA 3.0 (15,16), followed by refinement in explicit water using CNS 1.1 (17,18). A summary of constraints derived from the NMR data is presented in Table S1, along with structure quality assessment statistics. The final ensemble of 20 models and constraints for dvCcmE′Δ9 were deposited into the Protein Data Bank (PDB ID, 2KCT), and NMR resonance assignments, NOESY peaks lists, 15N-1H residual dipolar couplings, and NOESY spectral FID data were deposited into the BioMagResDB (BMRB accession number, 16096). Residue-specific longitudinal and transverse relaxation rates (R1 and R2) 1H-15N heteronuclear NOE values were obtained on apo-dvCcmE′ at pH 6.5 using standard 2D gradient NMR experiments (19) (BMRB accession number, 18380). The in vitro heme-binding properties of tagless wild type, [C127A]-, and [Y131F]-dvCcmE′ were assayed by preparation of the ferric heme adduct (20), followed by UV-visible absorption spectroscopy, SDS-PAGE with heme staining (21), and pyridine hemochrome analysis (22); reduced (ferrous) heme forms were produced by addition of sodium dithionite. The apo-forms of both dvCcmE′Δ9 and dvCcmE′ are monomeric in solution under reducing conditions, based on gel filtration chromatography, static light scattering, and 15N relaxation data. The expression plasmids for dvCcmE′Δ9 and dvCcmE′ are accessible from the PSI Materials Repository (http://psimr.asu.edu/).
The solution structure of dvCcmE′Δ9 exhibits a classic β-barrel OB-fold (23) comprised of six beta strands (β1, V53-V59; β1, T65-M66; β3, G72-E78; β4, T86-K92; β5, E104-L111; β6, T117-T125) arranged in a Greek key topology (Figure 1B). The functionally important cysteine, C127, is located just beyond the final strand. The structure of dvCcmE′Δ9 is remarkably similar to structures of the CcmE′ domains from E. coli (6) and S. putrefaciens (7), in spite of the less than 30% sequence identity to the CcmE proteins from these organisms (not shown). A conserved surface, ConSurf (24), analysis of the cysteine-containing variant sub-family of the CcmE protein domain family reveals a high degree of amino-acid residue conservation in the face of the OB-fold comprising strands β1, β5, and β6, as well as the loop between strands β4 and β5 and the heme-binding C-terminus of the protein (not shown). This conserved face of the molecule most likely participates in protein:protein interactions with CcmC (25).
In order to probe the dynamics and heme-binding properties of this variant CcmE, we designed tagless constructs of dvCcmE′ containing the complete native C-terminal region of the protein. 15N NMR relaxation and 1H-15N heteronuclear NOE data (Figure 1C; Supporting Information) demonstrate that the C-terminal ca. 12 residues immediately following the last beta strand in the structure, including the functionally important heme-binding region, are intrinsically disordered in apo-dvCcmE (1H-15N hetNOE < 0.5, for residues C127 to G137; S2 order parameter < 0.5, for residues S129 to G137). In addition, several lines of experimental evidence conclusively establish that dvCcmE′ binds ferric heme in a non-covalent fashion, but is incapable of covalent heme binding in vitro. In particular, UV-visible absorption spectroscopy (Figure 1D) and SDS-PAGE followed by heme staining (Supporting Information) demonstrate that C127 is absolutely required for ferric heme binding to dvCcmE′ in vitro, whereas Y131 is not. Ferric heme adducts of wild type and [Y131F]-dvCcmE′ exhibit a dramatic Soret band shift (λmax 373 nm), whereas [C127A]-dvCcmE′ emulates free hemin (λmax 396 nm). However, reduction of heme produces identical Soret band shifts for all species (Figure 1D, inset). Moreover, pyridine hemochrome spectra (Figure 1E), gel filtration chromatography, and mass spectrometry (not shown) further demonstrate that dvCcmE′ is incapable of covalently binding heme in vitro, in contrast to the related CcmE from D. desulfuricans which was shown to covalently bind heme via the conserved cysteine when produced in vivo (13). The in vitro heme binding behavior of dvCcmE′ also contrasts that reported for wild type and [H130C]-CcmE′ from E. coli which form a covalent bond to reduced heme after initial non-covalent binding of ferric heme (26,27), although no evidence of heme binding was observed for the S. putrefaciens CcmE homolog (7).
In summary, the solution NMR structure of the C-terminal heme-binding domain of D. vulgaris CcmE heme chaperone, dvCcmE′ reported here constitutes the first three-dimensional structure from the variant CcmE sub-family featuring a CxxxY heme-binding motif. We establish that the heme-binding motif in the apo-form of dvCcmE′ is highly dynamic and intrinsically disordered in solution, and that residue C127 is required for non-covalent ferric heme binding in vitro. We postulate that covalent attachment of heme to this variant CcmE requires the intact (i.e., in vivo) Ccm machinery. Elucidating the structural and dynamic effects of heme binding to this variant sub-family of heme chaperones awaits further study.
Supplementary Material
ACKNOWLEDGMENT
We thank Julie Stevens, Alexander Eletsky, Burkard Rost, G.V.T. Swapna, Mei Jiang, Erica Foote, and Li Zhao and for valuable scientific discussions and technical support.
This work was supported by National Institute of General Medical Sciences Protein Structure Initiative Grant U54-GM094597 (to G.T.M.), and the Natural Sciences and Engineering Research Council of Canada.
Footnotes
SUPPORTING INFORMATION AVAILABLE Complete experimental methods used in this study, table of NMR and structural statistics for dvCcmE′Δ9 (Table S1), static light scattering data for dvCcmE′Δ9 (Figure S1), 15N T1 and T2 relaxation data for dvCcmE′Δ9 (Figure S2), NMR sequential connectivity map for dvCcmE′Δ9 (Figure S3), superposition of the final ensemble of 20 conformers from the solution NMR structure of apo-dvCcmE′Δ9 (Figure S4), superposition of the solution structure of dvCcmE′Δ9 with that of E. coli CcmE′ (Figure S5), ConSurf analysis of the cysteine-containing CcmE sub-family (Figure S6), assigned 1H-15N HSQC spectrum of dvCcmE′ (Figure S7), complete 15N relaxation analysis of dvCcmE′ (Figure S8), and the heme stained SDS-PAGE heme-binding analysis of wild type dvCcmE′ and mutants (Figure S9). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Allen JWA, Jackson AP, Rigden DJ, Willis AC, Ferguson SJ, Ginger ML. FEBS J. 2008;275:2385–2402. doi: 10.1111/j.1742-4658.2008.06380.x. [DOI] [PubMed] [Google Scholar]
- 2.Kranz RG, Richard-Fogal C, Taylor J-S, Frawley ER. Microbiol. Mol. Biol. Rev. 2009;73:510–528. doi: 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders C, Turkarslan S, Lee D-W, Daldal F. Trends Microbiol. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz H, Hennecke H, Thöny-Meyer L. Science. 1998;281:1197–1200. doi: 10.1126/science.281.5380.1197. [DOI] [PubMed] [Google Scholar]
- 5.Stevens JM, Uchida T, Daltrop O, Ferguson SJ. Biochem. Soc. Trans. 2005;33:792–795. doi: 10.1042/BST0330792. [DOI] [PubMed] [Google Scholar]
- 6.Enggist E, Thöny-Meyer L, Güntert P, Pervushin K. Structure. 2002;10:1551–1557. doi: 10.1016/s0969-2126(02)00885-7. [DOI] [PubMed] [Google Scholar]
- 7.Arnesano F, Banci L, Barker PD, Bertini I, Rosato A, Su XC, Viezzoli MS. Biochemistry. 2002;41:13587–13594. doi: 10.1021/bi026362w. [DOI] [PubMed] [Google Scholar]
- 8.Lee D, Pervushin K, Bischof D, Braun M, Thöny-Meyer L. J. Am. Chem. Soc. 2005;127:3716–3717. doi: 10.1021/ja044658e. [DOI] [PubMed] [Google Scholar]
- 9.Uchida T, Stevens JM, Daltrop O, Harvat EM, Hong L, Ferguson SJ, Kitagawa T. J. Biol. Chem. 2004;279:51981–51988. doi: 10.1074/jbc.M408963200. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Rubio I, Braun M, Gromov I, Thöny-Meyer L, Schweiger A. Biophys. J. 2007;92:1361–1373. doi: 10.1529/biophysj.106.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvat EM, Redfield C, Stevens JM, Ferguson SJ. Biochemistry. 2009;48:1820–1828. doi: 10.1021/bi801609a. [DOI] [PubMed] [Google Scholar]
- 12.Allen JWA, Harvat EM, Stevens JM, Ferguson SJ. FEBS Lett. 2006;580:4827–4834. doi: 10.1016/j.febslet.2006.07.073. [DOI] [PubMed] [Google Scholar]
- 13.Goddard AD, Stevens JM, Rao F, Mavridou DAI, Chan W, Richardson DJ, Allen JWA, Ferguson SJ. J. Biol. Chem. 2010;285:22882–22889. doi: 10.1074/jbc.M110.133421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acton TB, Xiao R, Anderson S, Aramini J, Buchwald WA, Ciccosanti C, Conover K, Everett J, Hamilton K, Huang YJ, Janjua H, Kornhaber G, Lau J, Lee DY, Liu G, Maglaqui M, Ma L, Mao L, Patel D, Rossi P, Sahdev S, Shastry R, Swapna GV, Tang Y, Tong S, Wang D, Wang H, Zhao L, Montelione GT. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Güntert P, Mumenthaler C, Wüthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann T, Güntert P, Wüthrich K. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 17.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Linge JP, Williams MA, Spronk CAEM, Bonvin AMJJ, Nilges M. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 19.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 20.Harvat EM, Stevens JM, Redfield C, Ferguson SJ. J. Biol. Chem. 2005;280:36747–36753. doi: 10.1074/jbc.M508355200. [DOI] [PubMed] [Google Scholar]
- 21.Thomas PE, Ryan D, Levin W. Anal. Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 22.Berry EA, Trumpower BL. Anal. Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 23.Murzin AG. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 25.Richard-Fogal C, Kranz RG. J. Mol. Biol. 2010;401:350–362. doi: 10.1016/j.jmb.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daltrop O, Stevens JM, Higham CW, Ferguson SJ. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9703–9708. doi: 10.1073/pnas.152120699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens JM, Daltrop O, Higham CW, Ferguson SJ. J. Biol. Chem. 2003;278:20500–20506. doi: 10.1074/jbc.M212925200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.