Abstract
Background
Cyanase is an enzyme found in bacteria and plants that catalyzes the reaction of cyanate with bicarbonate to produce ammonia and carbon dioxide. In Escherichia coli, cyanase is induced from the cyn operon in response to extracellular cyanate. The enzyme is functionally active as a homodecamer of 17 kDa subunits, and displays half-site binding of substrates or substrate analogs. The enzyme shows no significant amino acid sequence homology with other proteins.
Results
We have determined the crystal structure of cyanase at 1.65 Å resolution using the multiwavelength anomalous diffraction (MAD) method. Cyanase crystals are triclinic and contain one homodecamer in the asymmetric unit. Selenomethionine-labeled protein offers 40 selenium atoms for use in phasing. Structures of cyanase with bound chloride or oxalate anions, inhibitors of the enzyme, allowed identification of the active site.
Conclusions
The cyanase monomer is composed of two domains. The N-terminal domain shows structural similarity to the DNA-binding α-helix bundle motif. The C-terminal domain has an ‘open fold’ with no structural homology to other proteins. The subunits of cyanase are arranged in a novel manner both at the dimer and decamer level. The dimer structure reveals the C-terminal domains to be intertwined, and the decamer is formed by a pentamer of these dimers. The active site of the enzyme is located between dimers and is comprised of residues from four adjacent subunits of the homodecamer. The structural data allow a conceivable reaction mechanism to be proposed.
Keywords: active site, cyanase, decamer structure, MAD phasing, monoanion and dianion inhibitors, synchrotron radiation
Introduction
Cyanase (EC 4.3.99.1) in Escherichia coli is an inducible enzyme (from the cyn operon) that catalyzes the reaction between cyanate and bicarbonate [1–6]:
The gene for cyanase has been cloned, sequenced and over-expressed, and the protein purified [4,7,8].
Although cyanate at high concentrations can be fairly toxic to E. coli, cyanate can serve as the sole source of nitrogen for growth of E. coli as a result of cyanase-catalyzed decomposition to ammonia [7,9,10]. This is probably the major function of cyanase in E. coli, rather than the detoxification of cyanate [11]. It has also been suggested that cyanate originating from carbamoyl phosphate in E. coli might have a regulatory role as a ‘by-product’ inhibitor of carbamoyl phosphate synthetase and that cyanase functions as a modulator of this effect [16,12]. Cyanate can occur naturally in some tissues as the result of nonenzymatic decomposition of carbamoyl phosphate [13], and also in the environment as a result of the dissociation of urea and photo-oxidation of cyanide [14]. The toxicity of cyanate probably arises from its reactivity with nucleophilic groups in proteins [15,16], perhaps in competition with bicarbonate/CO2 [11], although in contrast with bicarbonate the reaction of cyanate with amino groups is irreversible.
An inducible cyanase activity is present in many, but not all, species of bacteria [3,6,17–21]. A constitutive cyanase activity, similar to that of the E. coli enzyme, has been also reported in the cyanobacteria Synechococcus UTEX625, where the function may be related to providing CO2 from bicarbonate and cyanate for photosynthesis [22], and Synechocystis PCC6803 [23,24]. Cyanase activity has been reported in some plant extracts [25,26] and the gene for cyanase has been identified in at least one plant species [27].
The E. coli enzyme is the only cyanase that has been characterized in detail [4,5,28–35]. Isotope labeling studies have shown that the catalytic mechanism involves nucleophilic attack of bicarbonate on cyanate to give an anhydride transition-state intermediate, which decarboxylates to CO2 and carbamate [5]; carbamate then decarboxylates nonenzymatically [4]. The reaction takes place via a rapid equilibrium random kinetic mechanism with competitive substrate inhibition by both substrates [29]. Azide and chloride are competitive inhibitors with respect to cyanate. Oxalate and malonate, putative analogs of the proposed intermediate, are potent ‘slow-binding’ inhibitors [29].
The main goals of this study were twofold. Firstly, there is no biochemical or genetic evidence available to identify the active-site residues of the enzyme; structural information could provide this data. Secondly, the amino acid sequence of cyanase shows no significant similarity to any other protein currently deposited in the Protein Data Bank (PDB), therefore, the possibility of a unique fold was conceivable. Here, we describe the structure of E. coli cyanase determined to 1.65 Å resolution. The structure of the cyanase monomer (subunit) consists of two domains: the N-terminal domain forms a five-helix bundle and the C-terminal domain shows an unusual open fold that serves as a dimerization motif. The dimerization domain presents a novel fold composed of a four-stranded antiparallel β sheet flanked by two α helices. The dimers are assembled into a decamer, with 5/2 symmetry, to form the active enzyme. Unexpectedly, the crystal structure revealed the active site of the enzyme to be located between pairs of dimers and to be composed of residues from four adjacent subunits of the decamer.
Results and discussion
MAD structure determination
Past attempts at solving the crystal structure of cyanase were hampered by the crystal symmetry and the failure to obtain good heavy-atom derivatives. However, a mercury derivative (HgCl2) confirmed the presence of a decamer in the asymmetric unit [35]. Unfortunately, the mercury sites were poorly occupied and provided no useful phase information.
Multiwavelength anomalous dispersion (MAD) phasing is now widely used to determine increasingly larger protein structures [36,37]. The advent of third-generation synchrotron sources and fast-readout charge-coupled device (CCD) detectors has allowed large amounts of data to be collected in a very short time [38,39]. Furthermore, a number of new programs [40–43] have significantly improved the success rate for finding large numbers of anomalous scatterers automatically. This made the structure determination of cyanase (170 kDa per asymmetric unit and 40 potential selenium sites) using the MAD method a feasible option. The substructure of the selenium atoms was elucidated successfully using the Patterson-based approach [40,44] and direct methods [43], as described in the Materials and methods section. These sites were refined [45,46] and produced a fully interpretable electron-density map.
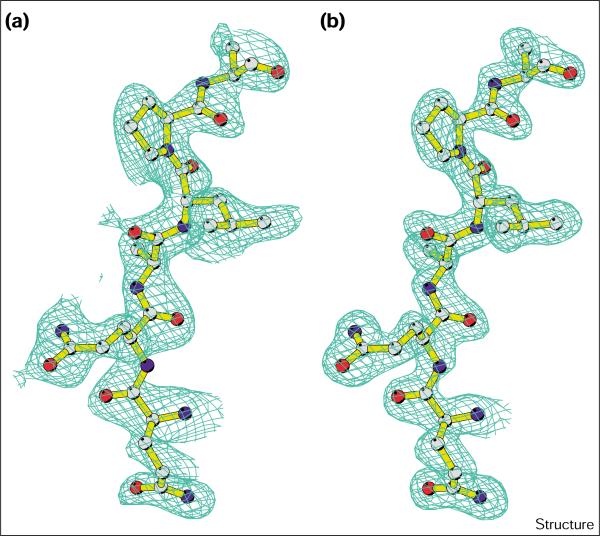
Figure 1a shows a section of the experimental map calculated from MAD phases based on 30 selenium atom sites to 2.4 Å resolution. These phases were then extended to 1.65 Å resolution together with solvent flattening [47] and used in the autochain-tracing and model-building module of ARP/wARP [48] (see the Materials and methods section). This procedure resulted in the complete automatic tracing of the polypeptide backbone of all ten subunits of the cyanase homodecamer (noncrystallographic symmetry [NCS] was not employed), as well as the positioning of over 70% of the amino acid sidechains. The high quality of the electron-density maps (Figure 1b) clearly revealed the positions of the remaining amino acids of the model. Subsequent refinement of the structure [49] was straightforward. The refined coordinates of the selenomethionine (SeMet)-labeled cyanase were used as a starting model for the analysis and refinement of the enzyme in complex with the dianion oxalate. The final crystallographic R factor for the SeMet-labeled protein is 15.0% with an Rfree of 18.9%; this structure has two chloride ions bound at the active site. The R factor for the enzyme in complex with oxalate is 13.9%, with an Rfree of 17.8%. A summary of the MAD phasing and refinement statistics is given in Tables 1 and 2.
Figure 1.
The quality of the electron density. Section of the electron-density map of cyanase calculated (a) using phases based on 30 selenium sites and (b) after density modification and phase extension to 1.65 Å resolution (see Materials and methods section for details). Atoms are shown in standard colors. (The figure was generated with Bobscript [61].)
Table 1.
Crystallographic data reduction and phasing statistics.
| Crystal* | Resolution (Å) | No. of observations | No. of unique reflections | Rmerge† | I/σI | Completeness (%) | Phasing power‡ | Anomalous Rcullis§ |
|---|---|---|---|---|---|---|---|---|
| CynSeMet (1.0332 Å) | 20–1.65 | 524,489 | 187,107 | 3.9 (19.8) | 26.4 (3.9) | 94.2 (80.6) | – | – |
| CynSeMet λ1 (1.0781 Å) | 20–2.40 | 211,456 | 55,582 | 2.7 (4.2) | 38.0 (32.0) | 87.0 (42.6) | 2.17 | 0.95 |
| CynSeMet λ2 (0.9795 Å)# | 20–2.40 | 242,516 | 63,330 | 4.5 (6.4) | 35.2 (22.9) | 97.6 (79.6) | 0.0 | 0.65 |
| CynSeMet λ3 (0.9793 Å) | 20–2.40 | 209,172 | 63,118 | 5.9 (7.9) | 36.3 (28.3) | 97.5 (80.4) | 1.29 | 0.46 |
| CynSeMet λ4 (0.9465 Å) | 20–2.40 | 242,869 | 62,595 | 2.3 (2.9) | 26.3 (24.1) | 96.7 (97.7) | 1.42 | 0.61 |
| CynSeMet-Oxl (1.0332 Å) | 20–1.65 | 436,555 | 190,231 | 4.7 (17.1) | 21.5 (4.3) | 95.4 (79.4) | – | – |
CynSeMet, selenomethionine-labeled cyanase (unit-cell parameters a = 76.3 Å, b = 81.0 Å, c = 82.3 Å, α = 70.3°, β = 72.2°, γ = 66.4°). CynSeMet-Oxl, CynSeMet soaked with sodium oxalate (unit-cell parameters a = 76.3 Å, b = 80.9 Å, c = 82.1 Å, α = 70.1°, β = 71.9°, γ = 66.4°; see text). λ1, low-energy remote data set; λ2, inflection point data set; λ3, peak data set: λ4, high-energy remote data set. The wavelengths used to collect these data are shown in parentheses.
Rmerge =Σhkl Σi = lN |〈Ihkl〉 – Iihkl| /Σhkl Σi = lN Iihkl.
Phasing power = <Fh>/<LOC>, where <Fh> and <LOC> are the root mean square heavy-atom structure factor and lack of closure, respectively.
Rcullis = Σ∥Fph(obs) ± |Fp (obs)∥ – |Fh(calc)|/Σ ∥Fph (obs)| ± |Fp (obs)∥, where Fph, Fp and Fh are the structure-factor amplitudes for the heavy-atom derivative, the native protein, and the heavy-atom contribution, respectively.
The λ2 data were taken as the reference data set for phase refinement with MLPHARE [46].
Table 2.
Refinement statistics.
| Crystal | CynSeMet | CynSeMet-Oxl |
|---|---|---|
| Resolution range (Å) | 20–1.65 | 20–1.65 |
| R factor (%) | 15.0 | 13.9 |
| Rfree (%) | 18.9 | 17.8 |
| No. of protein atoms (including double conformations) | 12,230 | 12,184 |
| No. solvent atoms | 1865 | 2464 |
| Rms deviations from ideal geometry | ||
| bond distance (1–2) (Å) | 0.018 (0.02) | 0.017 (0.02)* |
| angle distance (1–3) (Å) | 0.033 (0.04) | 0.031 (0.04) |
| planar distance (1–4) (Å) | 0.038 | 0.035 (0.05) |
| Mean B factors (Å2) | ||
| all atoms | 17.9 | 15.9 |
| protein atoms | 15.7 | 13.1 |
| water | 30.4 | 28.6 |
| chloride ions | 15.8 | – |
| sulfate ions | 44.7 | 38.4 |
| oxalate ions | – | 13.6 |
| Ramachandran plot statistics | ||
| most favored regions (%) | 95.1 | 94.6 |
| additional allowed regions (%) | 4.2 | 4.7 |
| disallowed regions (%) | 0.7 (Arg87) | 0.7 (Arg87) |
| Cruickshank diffraction-component precision index (DPI) | 0.089 | 0.078 |
CynSeMet, selenomethionine-labeled cyanase; CynSeMet-Oxl, CynSeMet soaked with oxalate.
Target values.
Structure and architecture
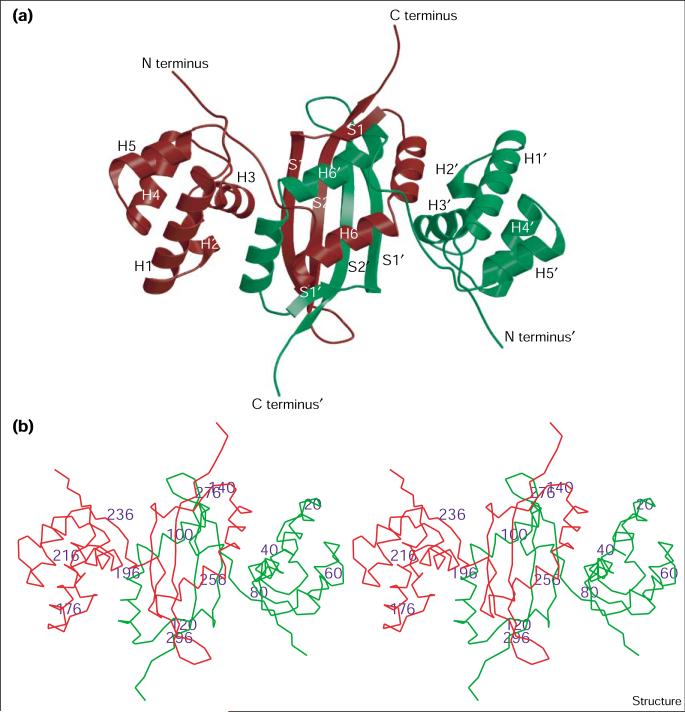
Preliminary studies of cyanase revealed a homodecamer with 5/2 symmetry [35]. The cyanase monomer (or subunit) consists of two domains (Figure 2). The N-terminal domain (residues 1–80) forms a five-helix bundle with structural homology to a common DNA-binding motif found in a number of bacterial transcription regulatory proteins [50]. The program CATH [51] revealed close structural homology between this domain and several five-helix bundle motifs present in the PDB [52]. The closest match is to the N-terminal domain of the 434 repressor (25.4% identity over 59 residues, root mean square [rms] deviation of 2.0 Å; PDB 1r69, CATH 1.20.120.90.3.1.1).
Figure 2.
The cyanase dimer. (a) Ribbon diagram of the cyanase dimer. The cyanase monomers are colored red and green. Secondary structure elements are labeled H for an α helix and S for a β strand: helix 1 (H1) consists of residues 8–24; helix 2 (H2), 29–33; helix 3 (H3), 40–47; helix 4 (H4), 55–64; helix 5 (H5), 69–76; helix 6 (H6), 92–115; strand 1 (S1), 119–134; strand 2 (S2), 140–152. A prime symbol distinguishes the two monomers. (b) Stereoview Cα trace of the cyanase dimer. The colors are as described in (a) with every 20th residue labeled. (The figure was generated with Molscript [62] and rendered with Raster 3D [63].)
The C-terminal domain (residues 91–156) consists of a two-stranded antiparallel β sheet and a single α helix of seven turns consisting of 26 amino acids. This α helix is bent in the middle by approximately 40°, giving rise to two subhelices disjoined at Val103. The monomer structure is not compact: a large cavity of approximately 12 Å diameter is formed in-between the N- and C-terminal domains. The cyanase monomer has not been observed in solution and in light of its open structure is almost certainly structurally unstable.
The cyanase dimer consists of the two C-terminal domains of the subunit monomers interlocking in a ‘hook-like’ manner by means of the two-stranded β sheet and bent α helix (Figure 2). The β-strand ‘finger’ of each monomer in the dimer extends into the approximately 12 Å cavity formed by the respective monomer subunits. The intertwining of the C-terminal domains forms a dimer interface domain, consisting of a four-stranded antiparallel β sheet and two α helices the 40° bend of which allows the tight interlocking of the two monomers. This structural motif showed no significant homology to any other structures within the PDB when analyzed using the CATH program. The formation of the dimer generates a compact dimer surface, which resembles those of globular proteins. A somewhat similar dimer-interlocking motif was observed in the structure of the trp repressor (TrpR) dimer [50]. In this case, the 68 residues of the N terminus of the monomer also formed a large opening. This hollow was filled by the twofold-related monomer unit, which gave a stable dimer structure. Such a dimer arrangement positions the α-helix bundle domains at the correct distance and in the required orientation to interact with the major groove surface of B-DNA. There is no evidence to date, however, that the cyanase dimer can interact with DNA, in fact, this structural similarity may have only evolutionary implications.
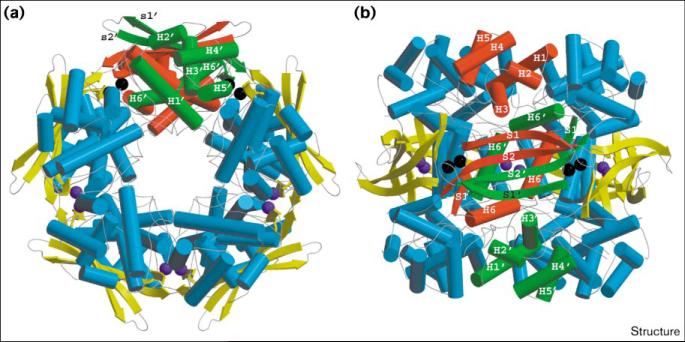
The cyanase decamer is formed by assembling five cyanase dimers into a pentamer (Figure 3). The dimers are placed in such a way that the α helices of the N- and C-terminal domains make up the cylinder walls of the decamer structure. The antiparallel β strands of the dimerization domain then form an equatorial belt around the outer surface of the decamer (Figure 3). This results in a cylindrical decamer structure of about 80 Å in length, with an outer diameter of approximately 70 Å. The central cavity of the decamer, which is open at both ends and is accessible to solvent, has a diameter of approximately 11–12 Å. The surface of the inner central channel is made up solely of α helices and is decorated predominantly by Asp86 residues, the sidechains of which extend into the channel. Other hydrophilic residues, namely Asp12, Asp85 and Arg87 as well as residues Asn7, Arg8 and Asn9, contribute to the make-up of the inner surface of the channel.
Figure 3.
Cartoon representation of the cyanase decamer. (a) View looking down the fivefold axis of the decamer. (b) View perpendicular to the fivefold axis and looking down the twofold axis. One cyanase dimer is highlighted using the same colors and labeling scheme as described in Figure 2. Ten chloride ions, which bind at the five active sites of the decamer (see text), are shown. The chloride ions shown in black are situated at the active sites to which the highlighted cyanase dimer contributes; the remaining bound chloride ions are colored purple. All other dimers are colored uniformly with α helices in cyan and β strands in yellow. (The figure was generated with Molscript [62] and rendered with Raster 3D [63].)
Earlier biochemical work aimed at locating the active site of the enzyme showed that decamer stability decreased when the single cysteine residue in each monomer was replaced with serine or glycine, thus suggesting that cysteine contributes to the stability of the decamer [32,33]. The crystal structure reveals that the single cysteine residue is located in the linker region between the N- and C-terminal domains. Moreover, the cysteine residues are not close to each other in the dimer, thus intra- and interdimer disulfide bonds are not possible (in the decamer). The sulfur atom of cysteine has been shown to participate in a variety of interactions in proteins and can provide considerable stability to the structure [53]. Here the cysteine sulfhydryl group of one subunit in the dimer hydrogen bonds to the carbonyl oxygen of Gln102 and to the amide nitrogen of Thr106 in the twofold-related subunit of the dimer. The latter interaction is known to stabilize the tertiary structure of proteins. Other studies have found that substituting buried cysteines for residues such as serine or alanine also results in the mutants being less stable than the wild type [54]. Hence, the single buried cysteine residue has a structural role, although no more so than in other proteins.
Characterization of mutations at the single histidine residue in each monomer revealed that, although not required for catalytic activity, it contributes to the stabilization of the decamer. Analysis of the structure shows His113 to be located close to the beginning of the bent α helix that makes up part of the cyanase dimerization domain. This histidine residue is tightly packed within the monomer subunit but also makes contacts with two other subunits of the decamer. Steric restraints are probably the main basis for the observed biochemical data, which showed that substitution with tyrosine or asparagine, but not with glycine, valine or leucine, had an effect on decamer stability [33].
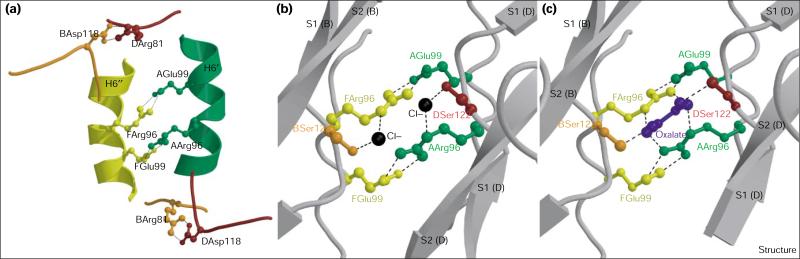
The dimers in the cyanase decamer are held together almost exclusively by salt bridges and hydrogen bonds. Twenty residues from each monomer (40 per dimer) make van der Waals and/or hydrogen-bonding interactions with residues in neighboring dimers. Of these 20 residues, 13 are highly conserved, suggesting that all cyanases may have a decameric structure. A series of salt bridges circle the equatorial β-sheet belt. As a consequence of the decamer symmetry, four salt bridges span each dimer in the decamer. These are formed by Arg81–Asp118 (located on loops) and Arg96–Glu99 (located on α helices H6 and H6′). The former salt bridge is spatially located above and below the equatorial β-sheet belt of the decamer, whereas the latter are spatially located along it (Figures 3 and 4). This results in two central, close-paired Arg96–Glu99 salt bridges, the cumulative interactions of which again resemble a belt-like structure. The Arg81–Asp118 salt bridges are arranged above and below these in a propeller-like fashion (Figure 4a).
Figure 4.
Decamer stabilization and catalytic site of cyanase. (a) Location of the four pairs of salt bridges (indicated by dashed lines) that stabilize the cyanase decamer. The dimer pairs are color coded: green (subunit A)/red (subunit D) and orange (subunit B)/yellow (subunit F). The labeling scheme is as described in Figure 2. The catalytic site of cyanase showing (b) bound chloride and (c) bound oxalate ions. The proposed catalytic residues are drawn in ball-and-stick representation using the same color scheme as in (a). Chloride and oxalate ions are in ball-and-stick representation colored in black and blue, respectively. Hydrogen-bond interactions are represented by dashed lines. The cyanase monomers are labeled by their chain (A, B, D, F), and secondary structure elements are labeled as described in Figure 2. (The figure was generated with Molscript [62] and rendered with Raster 3D [63].)
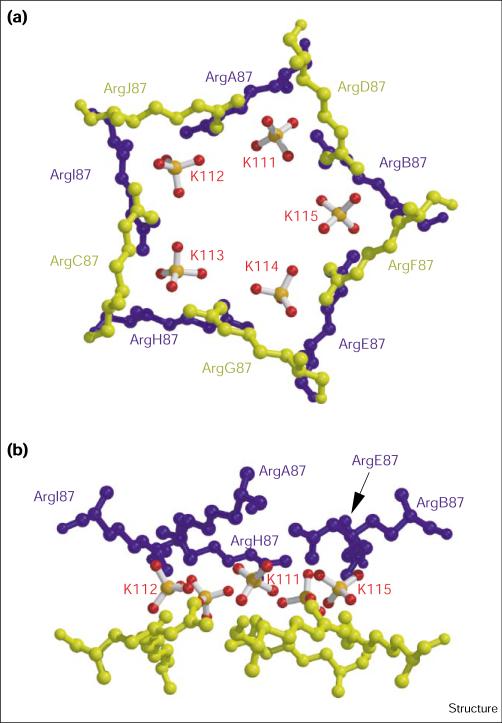
As well as being lined with water molecules, the central cavity of the cyanase decamer binds five sulfate ions through hydrogen-bonding interactions solely to Arg87 of each cyanase monomer. Each sulfate ion is bound to a cyanase dimer and two other monomers on either side of this dimer, above and below the equatorial β-sheet belt of the decamer. Thus, the five sulfates are sandwiched between two pentameric arginine ‘rings,’ which interact with each other via the NH2 groups of their guanidinium sidechains. This arrangement places the two five-membered sets of arginines at an average distance of 2.7 Å (Figure 5). The interaction of the charged oxygens (O–) of the sulfates is in the plane of the guanidinium group of the arginine, which follows the expected trend [55]. These negatively charged protein-bound sulfates provide a stabilizing counter-charge for the closely interacting positively charged Arg87 guanidinium sidechains. In addition, each cyanase subunit also binds two sulfate ions on the solvent-exposed face of the decamer through hydrogen bonds to the amide nitrogen atoms of Gly35 and Glu40. However, not all monomers show clear density for the sulfate bound at Gly35, which is likely to result from crystal packing. Interestingly, sulfate acts as a weak inhibitor of cyanase activity [30].
Figure 5.
Ball-and-stick diagram showing orthogonal views of the five Arg87 cavity-bound sulfate ions in the cyanase decamer. (a) Looking down the fivefold axis. (b) Perpendicular to the fivefold axis. Residues are labeled by their sequence number and respective monomer chain, as described in Figure 4. The bound sulfate ions are labeled as K111–K115, which follows the assignment in the coordinate file 1DW9 deposited in the PDB. The arginine residues are colored blue and yellow to highlight the delineation of the monomers as above and below the equatorial β-sheet belt formed by the dimer interface domain (see Figure 3). The sulfate ions are colored by atom type (sulfur, orange; oxygen, red). (The figure was generated with Molscript [62] and rendered with Raster 3D [63].)
The sequences of the five homologous cyanase sequences (bacterial and plant) are aligned in Figure 6, showing that the N-terminal half of the enzyme has much lower sequence conservation than the C-terminal region. Three regions, corresponding to the E. coli cyanase sequence at residues 92–99 (PxxYRxxE; in single-letter amino acid code), 114–124 (ExFGDGIxSAI) and 145–151 (TxxGKxL), show especially high conservation, suggesting potential functional relevance. These regions include the Arg96, Glu99, Asp118 and Arg/Lys81 residues that are involved in the formation of salt bridges, which probably aid decamer stability. The cyanase dimer contains extensive intermolecular contacts in the C-terminal region of the monomer subunit. Other charged residues that are conserved or partially conserved participate in intermolecular hydrogen bonds between cyanase dimers in the homodecamer structure. Arg87, which binds the sulfate ions in the central cavity of the decamer, is not conserved; three of the sequences contain valine at this position and the other contains arginine. This observation suggests that cyanases from bacteria and plants may be affected differently by sulfate ions.
Figure 6.
Alignment of the amino acid sequences for the five known cyanases. Accession numbers are given in parentheses. Residues conserved in all cyanases are in boldface and labeled with an asterisk, less conserved residues are labeled with colons and full stops. Arg96, Glu99, Ser122 (proposed active-site residues) are shown in red. Secondary structure assignments were derived using the Kabsch and Sander DSSP algorithm [64]: h/H, α helix; t/T, turn; eE, β strand; and S, bend. The aligned sequences are Synechocystis PCC6803 (GenBank accession number Q55367) [29]; Synechococcus PCC7942 (BAA19515) [28]; Aquifex aeolicus (AACO6548) [25]; E. coli (AAA23629) [8]; and Arabidopsis thaliana (BAA21660) [32].
Location of the catalytic site
Previous studies have shown that cyanase binds and is inhibited by a number of monoanion analogs of cyanate or bicarbonate and by dianion analogs of the putative transition-state intermediate [30]. Azide and chloride competitively inhibit the binding of cyanate, whereas acetate inhibits the binding of bicarbonate. Divalent anions like oxalate and malonate competitively inhibit the binding of both cyanate and bicarbonate. These observations indicate that the catalytic site must be able to accommodate two potentially negative charges during the reaction (one from cyanate and the other from bicarbonate).
We have located the catalytic site of cyanase by solving the crystal structure of cyanase in complex with two different types of cyanase inhibitor: the dianion oxalate and the monoanion chloride. X-ray diffraction data were collected from crystals soaked with either oxalate (C2Na2O4) or chloride (Tris/HCl). The protein-bound oxalate was located in electron-density maps using a difference Fourier map calculated using coefficients Foxl – Fc, αc (where Foxl are structure amplitudes from the C2Na2O4-soaked cyanase crystals and Fc, αc are the structure factors and calculated phases from the SeMet-labeled cyanase). A total of five oxalate ions were observed bound to the cyanase decamer. The chloride ions were discovered serendipitously in the catalytic site of the enzyme. Analysis of the electron-density maps calculated from the SeMet-labeled cyanase model showed ten water molecules with very low atomic displacement parameters, and some unaccounted positive electron density in the same location as the bound oxalates. These ten water molecules, which are located at the positions of the ten cis carboxylate oxygens of the five oxalate ions, were substituted for chloride ions as these ions were present in the crystallization buffer. The chloride ions refined to an average displacement parameter value of 15.8 Å2, which is comparable to the average value for the protein atoms at the ion-binding site. Therefore, the structures reveal that five catalytic sites are present in the decamer, which correlates well with the earlier observation of half-site binding [30].
Description of the catalytic site
The location of the chloride and oxalate bound to cyanase reveals the catalytic site of the enzyme. Each of the five sites is composed of amino acid residues from four subunits of two adjacent dimers. Hydrogen-bond interactions between the chloride and oxalate ions and the protein in both inhibitor complexes single out Arg96, Glu99 and Ser122 as the catalytic residues of the enzyme. These residues are conserved in the five cyanase sequences shown in Figure 6.
The five catalytic sites are located at the interface of adjacent dimers along the equator of the decamer (i.e., along the belt-like structure formed by the interlocking β strands of the C-terminal domain). The catalytic cavities are symmetrical, being bisected by five twofold NCS axes. Thus, the residues Arg96 and Glu99, from four subunits, form two interdimer salt bridges that delineate the inner surface of each catalytic site cavity (Figures 4b,c). The solvent-facing side of each cavity is flanked by a pair of serine residues (Ser122) from adjacent dimers. The same pair of dimers provides two hydrophobic leucine residues (Leu152) that shield each cavity from solvent. The electron density for these leucine residues suggest that an alternative conformation may be present and might provide a gating mechanism for the substrates and products of the enzymatic reaction.
The local twofold symmetry of the catalytic site in the complexes formed with either chloride or oxalate is not disrupted in these structures. In the chloride complex, each pair of chloride ions bound at the catalytic pocket is related by the local twofold NCS axis. In the oxalate complex, the twofold NCS axis bisects the C1–C2 bond in the plane of the oxalate ion. Thus, in this case, the two cis oxalate oxygen atoms are positioned in the same sites as the bound chloride ions and interact with the Oγ atom of Ser122 from two subunits of the same dimer. The other pair of cis-related oxygen atoms of the oxalate ion form hydrogen bonds to Arg96. When either cyanate or bicarbonate binds to the catalytic site the binding of the other substrate is predetermined. Once cyanate and bicarbonate are bound by cyanase, the local NCS symmetry of the catalytic site is broken. Whether the breakdown of this local symmetry has a role in the catalytic activity is not clear. The structure itself provides no mechanism for binding preference of cyanate or bicarbonate, which is consistent with the kinetic mechanism of cyanase: rapid equilibrium random with competitive inhibition by both substrates [29].
Implications for catalytic activity
The structures of the oxalate and chloride complexes provide the foundation for establishing a detailed understanding of the enzyme's catalytic mechanism. It is likely that bicarbonate and cyanate take up similar positions to those of the bound oxalate carboxylates. Thus, it is clear that Ser122 is important in substrate binding. The negatively charged cyanate and bicarbonate are probably stabilized by the guanidinium group of Arg96. This residue forms a salt bridge with Glu99, which in turn interacts directly via a carboxylate oxygen to a tightly bound water molecule. Protonation of the bound cyanate nitrogen by an activated water molecule bound to Glu99 is probable. Thus, the cyanate carbon atom becomes more electrophilic, and a nucleophilic attack by the bicarbonate carboxylate oxygen results in the formation of the proposed dianion intermediate [5]. This intermediate then decarboxylates to CO2 and carbamate. The structure of the catalytic site suggests that ‘on-the-enzyme’ decarboxylation of the dianion is likely, as opposed to dissociation and subsequent nonenzymatic decarboxylation [5]. Site-directed mutagenesis of these residues is required to unequivocally unravel the roles of the catalytic site residues. These studies are now in progress and will be published elsewhere.
The structure of cyanase shows the active site to be made up of two side-by-side dimers. All four subunits of these two dimers contribute to the active site of the enzyme, which is located on the twofold NCS axis that relates the two sets of dimers. The arrangement of the dimers into a pentamer of dimers to form a decameric structure is necessary to provide a solid scaffold, via a series of salt bridges, to hold the dimers forming the catalytic sites together. A consequence of the active site of the enzyme being located at a dimer–dimer interface is the observed apparent half-site binding of bicarbonate and substrate analogs and the requirement of a decamer for activity; the decamer gives rise to five catalytic sites.
Biological implications
Living organisms have developed effective mechanisms to convert some toxic chemicals into a source of nutrients. Cyanate occurs in the environment at concentrations as high as 1 mM, for example, as the result of the photo-oxidation of cyanide. Cyanate can be fairly toxic in the cell. The toxicity arises from its reactivity with nucleophilic groups in proteins. Some bacteria and plants can metabolize cyanate. An inducible enzyme, cyanase, is responsible for detoxifying cyanate and/or utilizing cyanate as a sole source of nitrogen. Cyanase catalyzes the reaction of cyanate with bicarbonate to produce ammonia and carbon dioxide. Cyanate metabolism is closely related to bicarbonate/CO2 metabolism with one major distinction: in contrast to carbon dioxide metabolism the reaction of cyanate with amino groups is irreversible.
We report here the first crystal structure of an enzyme with cyanase activity. Cyanase is active only as a decamer having 5/2 symmetry and is structurally complex. To date, there are limited examples of proteins with such symmetry. The decamer is composed of five inactive dimers, each of which shows an architecture similar to bacterial transcriptional regulators. The two monomers of each dimer are intertwined, presenting a unique fold at the dimer interface. The N-terminal domain of cyanase shows high structural homology to the DNA-binding α-helix bundle motif, and is not involved in catalysis. The interface between dimers forms a set of five symmetrically disposed active sites. These sites embody a unique set of amino acid residues that can bind and carry out the reaction between cyanate and bicarbonate. In addition to binding substrate and cofactor, cyanase can specifically accommodate a number of small monoions and dianions in its active site; these anions inhibit cyanase activity. Specific binding of anions to the protein does not require metal ions or other cofactors.
The structure of cyanase suggests that protein oligomerization might represent an alternative mechanism (in addition to others, such as gene duplication) to create new functionalities on protein interfaces from a pool of existing proteins. Adaptation of the apparent DNA-binding motif into a decamer that allows the formation of an anion-binding site provides an example of the versatility of protein surface functionality.
Materials and methods
Preparation and crystallization of SeMet-labeled protein
SeMet-labeled protein was prepared by suppressing the biosynthesis of methionine in E. coli as described elsewhere [38]. Purification was carried out as previously described, but using de-aerated buffers containing 2 mM dithiothreitol (DTT) [8]. Crystals were grown by the sitting-drop method of vapor diffusion from 2.1 M ammonium sulfate solutions buffered with 50 mM NaKPO4 (pH 7.3), and in the presence of 50 mM Tris/HCl (pH 7.3) [35]. Large sitting drops (50–100 μl) were microseeded at 18°C with wild-type crystals that produced SeMet crystals that grew to dimensions of 0.1 × 0.2 × 0.7 mm3 over five to seven days. The crystals are triclinic with unit-cell dimensions of a = 76.3 Å, b = 81.0 Å, c = 82.3 Å, α = 70.3°, β = 72.2°, γ = 66.4° (Table 1), and contain one cyanase homodecamer in the asymmetric unit, giving a Vm of 2.53 Å3/Da and a solvent content of 51%. Prior to data collection, the crystals were flash frozen in liquid nitrogen using stabilizing solution containing 25% galactose as cryoprotectant.
Data collection and reduction
All X-ray diffraction data were collected at 100K at the Structural Biology Center's undulator beamline 19ID at the Advanced Photon Source using a 3 × 3 mosaic CCD detector developed at Argonne National Laboratory [56]. A four-wavelength MAD experiment was performed at or near the K-absorption edge of selenium using data collected from a single SeMet-labeled cyanase crystal. The four wavelengths were selected on the basis of an X-ray fluorescence spectrum collected directly from the crystal, which was analyzed by the program CHOOCH [57]. This gave the absorption peak at 0.9793 Å and inflection point at 0.9795 Å; two remote data sets at 1.0781 Å and 0.9465 Å were chosen to obtain dispersive differences. Each wavelength data set was collected in a single pass using a 1° oscillation with a 3 s exposure. The four data sets were collected in the order peak, inflection, low wavelength and high wavelength remotes. A total of 360° of data were collected at each wavelength to a resolution of 2.4 Å (Table 1). After the MAD experiment, an unexposed region of the crystal was translated into the X-ray beam and a high resolution data set (1.65 Å) collected using a wavelength of 1.0332 Å. X-ray diffraction data to 1.65 Å resolution were collected in a similar manner from separate crystals (SeMet) soaked for 4 h in 5 mM sodium oxalate. All data were integrated and scaled using the HKL2000 suite [58].
MAD phasing
An initial search found 30 of the 40 selenium sites of the cyanase homodecamer using the Patterson heavy-atom search method implemented in the program suite Crystallography and NMR System [40]. The anomalous and dispersive diffraction ratios are shown in Table 1. These sites were refined with the program MLPHARE [46], giving a figure of merit of 0.8 and a clearly interpretable map. After solvent flattening and histogram matching with the program DM [47], along with phase extension against the data collected to 1.65 Å, the resulting map was of excellent quality (Figure 1). A summary of the phasing statistics is listed in Table 1.
Automatic model building and refinement
Automatic model building was performed with the program ARP/wARP 5.0 [48] using the phases output from DM at 1.65 Å. The warpNtrace procedure within ARP/wARP 5.0 produced a complete trace of the polypeptide chains of all ten subunits. In addition, it modeled over 70% of the amino acid sidechains into the electron density. A full description of the warpNtrace procedure is described elsewhere [48]. The remaining parts of the structure were built manually in the program O [59]. The resultant model was then refined with the program REFMAC [49] using data in the resolution range 20 Å–1.65 Å. The progress of the refinement was monitored using cross-validation with a 3% randomly selected set of reflections (9436). Water molecules were added with the program ARP [60], which was run in conjunction with REFMAC. The final electron-density maps were of excellent quality, with all atoms of the model placed in interpretable density. In addition, a number of sidechains with alternative conformations were seen in the model. The final refinement statistics are shown in Table 2.
Accession numbers
Atomic coordinates and structure factors have been deposited in the PDB with ID codes 1DW9 and 1DWK for cyanase with chloride and oxalate bound, respectively.
Acknowledgements
We thank Jennifer Wronski for help preparing SeMet-labeled protein and crystallization. We thank W Minor (University of Virginia) for making HKL2000 available to us before its official release and AT Brünger (Yale University) for providing a pre-release version of CNS. We would also like to thank the staff of the Structural Biology Center for helping to set up the beamline, in particular Frank Rotella, Randy Alkire and Gerd Rosenbaum, and finally Rong-guang Zhang, for help in data collection in the early stages of this project. This research was supported by the US Department of Energy, Office of Biological and Environmental Research, under contract W-31-109-Eng-38.
Footnotes
Because Structure with Folding & Design operates a ‘Continuous Publication System’ for Research Papers, this paper has been published on the internet before being printed (accessed from http://biomednet.com/cbiology/str). For further information, see the explanation on the contents page.
References
- 1.Taussig A. The effect of carbamyl phosphate and cyanate on Escherichia coli. Can. J. Microbiol. 1960;6:619–628. doi: 10.1139/m60-074. [DOI] [PubMed] [Google Scholar]
- 2.Taussig A. The synthesis of the induced enzyme, ‘cyanase,’ in E. coli. Biochim. Biophys. Acta. 1960;44:510–519. doi: 10.1016/0006-3002(60)91605-x. [DOI] [PubMed] [Google Scholar]
- 3.Taussig A, Ronen E. The effect of chloramphenicol on the induciton of cyanase in Escherichia coli. Can. J. Biochem. 1970;48:790–798. doi: 10.1139/o70-123. [DOI] [PubMed] [Google Scholar]
- 4.Anderson PM. Purification and properties of the inducible enzyme cyanase. Biochemistry. 1980;19:2882–2888. doi: 10.1021/bi00554a010. [DOI] [PubMed] [Google Scholar]
- 5.Johnson WV, Anderson PM. Bicarbonate is a recycling substrate for cyanase. J. Biol. Chem. 1987;262:9021–9025. [PubMed] [Google Scholar]
- 6.Guilloton M, Karst F. Isolation and characterization of Escherichia coli mutants lacking inducible cyanase. J. Gen. Microbiol. 1987;133:645–653. doi: 10.1099/00221287-133-3-645. [DOI] [PubMed] [Google Scholar]
- 7.Sung Y, Parsell D, Anderson PM, Fuchs JA. Identification, mapping, and cloning of the gene encoding cyanase in Escherichia coli K12. J. Bacteriol. 1987;169:2639–2642. doi: 10.1128/jb.169.6.2639-2642.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung Y, Anderson PM, Fuchs JA. Characterization of high-level expression and sequencing of the Escherichia coli K-12 cynS gene encoding cyanase. J. Bacteriol. 1987;169:5224–5230. doi: 10.1128/jb.169.11.5224-5230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guilloton MB, et al. Fuchs JA. A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J. Bacteriol. 1993;175:1443–1451. doi: 10.1128/jb.175.5.1443-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guilloton M, Karst F. Cyanate specifically inhibits arginine biosynthesis in Escherichia coli K12: a case of by-product inhibition. J. Gen. Microbiol. 1987;133:655–665. doi: 10.1099/00221287-133-3-655. [DOI] [PubMed] [Google Scholar]
- 11.Kozliak EI, Fuchs JA, Guilloton MB, Anderson PM. Role of bicarbonate/CO2 in the inhibition of Escherichia coli growth by cyanate. J. Bacteriol. 1995;177:3213–3219. doi: 10.1128/jb.177.11.3213-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson PM, Carlson JD. Reversible reaction of cyanate with a reactive sulfhydryl group at the glutamine-binding site of carbamyl phosphate synthetase. Biochemistry. 1975;14:3688–3694. doi: 10.1021/bi00687a027. [DOI] [PubMed] [Google Scholar]
- 13.Allen CM, Jones ME. Decomposition of carbamyl phosphate in aqueous solutions. Biochemistry. 1984;3:1238–1247. doi: 10.1021/bi00897a010. [DOI] [PubMed] [Google Scholar]
- 14.Hagel P, Gerding JJT, Fieggen W, Bloemendal H. Cyanate formation in solutions of urea. Biochem. Biophys. Acta. 1971;243:366–373. doi: 10.1016/0005-2795(71)90003-1. [DOI] [PubMed] [Google Scholar]
- 15.Harding JJ, Rixan KC. Carbamylation of lens proteins: a possible factor in cataractogenesis in some tropical countries. Exp. Eye Res. 1980;31:567–571. doi: 10.1016/s0014-4835(80)80015-7. [DOI] [PubMed] [Google Scholar]
- 16.Stark GR. Modification of proteins with cyanate. Methods Enzymol. 1967;11:590–594. doi: 10.1016/S0076-6879(72)25057-1. [DOI] [PubMed] [Google Scholar]
- 17.Guilloton M, Hargreaves AB. Hydrolyse du cyanate par une bacterie du genre Flavobacterium. [Cyanate hydrolysis by a bacterium of the genus Flavobacterium]. C.R. Acad. Sci. Paris. 1972:1827–1830. [PubMed] [Google Scholar]
- 18.Kunz DA, Nagappan O. Cyanase-mediated utilization of cyanate in Pseudomonas fluorescens NCIB11764. Appl. Environ. Microbiol. 1989;55:256–258. doi: 10.1128/aem.55.1.256-258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorr PK, Knowles CJ. Cyanide oxygenase and cyanase activities of Pseudomonas fluorescens NCIMB11764. FEMS Microbiol. Lett. 1989;60:289–294. [Google Scholar]
- 20.Deckert G, et al. Swanson RV. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 21.Wood AP, et al. Borodina E. A novel pink-pigmented facultative methyltroph, Methylobacterium thiocyanatum sp. nov., capable of growth on thiocyanate or cyanate as sole nitrogen sources. Arch. Microbiol. 1998;169:148–158. doi: 10.1007/s002030050554. [DOI] [PubMed] [Google Scholar]
- 22.Miller AG, Espie GS. The photosynthetic metabolism of cyanate by the cyanobacteriums Synechococcus UTEX625. Arch. Microbiol. 1994;162:151–159. [Google Scholar]
- 23.Harano Y, Suzuki I, Maeda S, Kaneko T, Tabata S, Omata T. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC6803 and Synechococcus sp. strain PCC7942. J. Bacteriol. 1997;179:5744–5750. doi: 10.1128/jb.179.18.5744-5750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko T, et al. Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 1995;2:153–166. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- 25.LoHi G. Ricerche sulla cianasi del frumenta. [Research on cyanase in wheat]. Chim. Ind. 1965;45:561–563. [Google Scholar]
- 26.Lisanti LE. Distribuzione della cianasi nei vegetali. I. Attivit cianatolitica nel Cicer arietinum. [Cyanase distribution in plants. I. Cyanatolytic activity in Cicer arietinum]. Ann. Speriment. Agaria. 1963;17:45–51. [Google Scholar]
- 27.Wambutt R, et al. Bevan M. Progress in Arabidopsis genome sequencing and functional genomics. Biotechnol. 2000;78:281–292. doi: 10.1016/s0168-1656(00)00195-4. [DOI] [PubMed] [Google Scholar]
- 28.Chin CCQ, Anderson PM, Wold F. The amino acid sequence of Escherichia coli cyanase. J. Biol. Chem. 1983;258:276–282. [PubMed] [Google Scholar]
- 29.Anderson PM, Little RM. Kinetic properties of cyanase. Biochemistry. 1986;25:1621–1626. doi: 10.1021/bi00355a026. [DOI] [PubMed] [Google Scholar]
- 30.Anderson PM, Johnson WV, Endrizzi JA, Little RM, Korte JJ. Interaction of mono- and dianions with cyanase: evidence for apparent half-site-binding. Biochemistry. 1987;26:3938–3942. doi: 10.1021/bi00387a029. [DOI] [PubMed] [Google Scholar]
- 31.Little RM, Anderson PM. Structural properties of cyanase: denaturation, renaturation, and role of sulfhydryls and oligomeric structure in catalytic activity. J. Biol. Chem. 1987;262:10120–10126. [PubMed] [Google Scholar]
- 32.Anderson PM, Johnson WV, Korte JJ, Xiong X, Sung Y, Fuchs JA. Reversible dissociation of active octamer of cyanase to inactive dimer promoted by alteration of the sulfhydryl group. J. Biol. Chem. 1988;263:5674–5680. [PubMed] [Google Scholar]
- 33.Anderson PM, Korte JJ, Holcomb TA, Cho Y, Son C, Sung Y. Formation of intersubunit disulfide bonds and properties of the single histidine and cysteine residues in each subunit relative to the decameric structure of cyanase. J. Biol. Chem. 1994;269:15036–15045. [PubMed] [Google Scholar]
- 34.Anderson PM, Korte JJ, Holcomb TA. Reaction of the N-terminal residues in cyanase with diethylpyrocarbonate. Biochemistry. 1995;33:14121–14125. doi: 10.1021/bi00251a022. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Anderson PM, Fuchs JA, Sigler PB, Joachimiak A. E. coli cyanase is a decamer with 5/2 symmetry.. Proceedings of the IV International Meeting on Crystallization of Biological Macromolecules; Freiburg, Germany. August 18-24.1991. p. 5. [Google Scholar]
- 36.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 37.Deacon AM, Ealick SE. Selenium-based MAD phasing: setting the sites on larger structures. Structure. 1999;7:R161–R166. doi: 10.1016/s0969-2126(99)80096-3. [DOI] [PubMed] [Google Scholar]
- 38.Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: ultra fast protein crystal structure determination. Acta Crystallogr. D. 1999;55:1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- 39.Walsh MA, Evans G, Sanishvili R, Dementieva I, Joachimiak A. MAD data collection — current trends. Acta Crystallogr. D. 1999;55:1726–1732. doi: 10.1107/s0907444999008392. [DOI] [PubMed] [Google Scholar]
- 40.Brünger AT, et al. Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Sheldrick GM. SHELX: applications to macromolecules. In: Fortier S, editor. Direct Methods for Solving Structures. Kluwer Academic Publishers; Dordecht, Germany: 1998. pp. 401–411. [Google Scholar]
- 42.Terwillinger TC, Berendzen J. Automated structure solution for MAD and MIR. Acta Crystallogr. D. 1999;55:905–921. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao JX. On the application of phase relationships to complex structures. XVIII. RANTAN – random MULTAN. Acta Crystallogr. A. 1983;37:35–37. [Google Scholar]
- 44.Grosse-Kuntsleve RW, Brünger AT. A highly automated heavy-atom search procedure for macromolecular structures. Acta Crystallogr. D. 1999;55:1568–1577. doi: 10.1107/s0907444999007763. [DOI] [PubMed] [Google Scholar]
- 45.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z. Maximum-likelihood refinement of heavy-atom parameters. In: Wolf W, Evans PR, Leslie AGW, editors. Isomorphous Replacement and Anomalous Scattering. SERC, Daresbury; Warrington, UK: 1991. pp. 80–86. [Google Scholar]
- 47.Cowtan K. DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 48.Perrakis A, Morris R, Lamzin A. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Schevitz RW, Otwinowski Z, Joachimiak A, Lawson CL, Sigler PB. The three-dimensional structure of trp repressor. Nature. 1985;317:782–786. doi: 10.1038/317782a0. [DOI] [PubMed] [Google Scholar]
- 51.Orengo CA, Michie AD, Jones DT, Swindel MB, Thornton JM. CATH – a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 52.Berman HM, et al. Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pal D, Chakrabarti P. Different types of interactions involving cysteine sulfhydryl group in proteins. J. Biomol. Struct. Dyn. 1998;15:1059–1072. doi: 10.1080/07391102.1998.10509001. [DOI] [PubMed] [Google Scholar]
- 54.Hiraga K, Yutani K. Study of cysteine residues in the alpha subunit of Escherichia coli tryptophan synthase. 2. Role in enzymatic function. Protein Eng. 1996;9:433–438. doi: 10.1093/protein/9.5.433. [DOI] [PubMed] [Google Scholar]
- 55.Nandi CL, Singh J, Thornton JM. Atomic environments of arginine side chains in proteins. Protein Eng. 1993;6:247–259. doi: 10.1093/protein/6.3.247. [DOI] [PubMed] [Google Scholar]
- 56.Westbrook EM, Naday I. Charge-coupled device-based area detectors. Methods Enzymol. 1997;276:233–243. [PubMed] [Google Scholar]
- 57.Evans G. 1994 CHOOCH download gage: http://lagrange.mrclmb.cam.ac.uk/doc/gwyndaf/Chooch.html.
- 58.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 60.Lamzin VS, Wilson KS. Automatic refinement for protein crystallography. Methods Enzymol. 1997;277:269–305. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
- 61.Esnouf RM. An extensively modified version of MolScript that includes generally enhanced coloring capabilities. J. Mol. Graph. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 62.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 63.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 64.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]