SUMMARY
A major cause of cell death caused by genotoxic stress is thought to be due to the depletion of NAD+ from the nucleus and the cytoplasm. Here we show that NAD+ levels in mitochondria remain at physiological levels following genotoxic stress and can maintain cell viability even when nuclear and cytoplasmic pools of NAD+ are depleted. Rodents fasted for 48 hr show increased levels of the NAD+ biosynthetic enzyme Nampt and a concomitant increase in mitochondrial NAD+. Increased Nampt provides protection against cell death and requires an intact mitochondrial NAD+ salvage pathway as well as the mitochondrial NAD+-dependent deacetylases SIRT3 and SIRT4. We discuss the relevance of these findings to understanding how nutrition modulates physiology and to the evolution of apoptosis.
INTRODUCTION
Approximately 2 billion years ago, eukaryotes evolved by subsuming a bacterial antecedent of modern mitochondria (Barile et al., 1996; Gray et al., 1999). Mitochondria still retain a variety of molecules that dictate cell survival, which at one time may have been important for the survival of the bacterial proto-mitochondrion (James et al., 1998). Elucidation of these cell survival pathways is considered key to the development of new approaches to treating a variety of human diseases including cancer and neurodegeneration (Porcu and Chiarugi, 2005).
One of the major causes of cell death due to genotoxic stress is hyperactivation of the NAD+-dependent enzyme poly(ADP-ribose) polymerase-1 (PARP-1), which depletes nuclear and cytoplasmic NAD+ causing the translocation of apoptosis inducing factor (AIF) from the mitochondrial membrane to the nucleus (Burkle, 2005; Cipriani et al., 2005; van Wijk and Hageman, 2005; Yu et al., 2002). One recent study reported that a fraction of PARP-1 is localized in mitochondria, which has led to speculation about the potential for mitochondrial NAD+ to determine cell fate (Du et al., 2003). Yet little is known about mitochondrial NAD+ biosynthesis (Barile et al., 1996; Berger et al., 2005; Kun et al., 1975), what the actual concentration of NAD+ is in mitochondria (Di Lisa and Bernardi, 2006), whether mitochondrial NAD+ levels change in response to biological stress or diet, and what impact this has on cell survival and metabolism (Porcu and Chiarugi, 2005; Viswanathan et al., 2005).
The sirtuins are a conserved family of deacetylases and mono-ADP-ribosyltransferases that use NAD+ as a co-substrate (Guarente and Picard, 2005). These unusual enzymes, which bear virtually no sequence homology to class I and II HDACs (Denu, 2005; Frye, 2000), have emerged as key regulators of cell survival and organismal longevity (Guarente and Picard, 2005). The founding member of the sirtuin family, Saccharomyces cerevisiae Sir2, is an NAD+-dependent histone deacetylase that mediates life span extension by mild stress and calorie restriction (CR) (Imai et al., 2000; Lin et al., 2000; Rogina and Helfand, 2004; Smith et al., 2000; Tanny et al., 1999). Mammals have seven sirtuins, SIRT1–7. SIRT1, a nuclear deacetylase, regulates a variety of functions including cell survival, glucose homeostasis, and fat metabolism (Guarente, 2005). There are three mitochondrial sirtuins, SIRT3–5. SIRT3 and SIRT4 were recently shown to regulate acetyl-CoA synthetase 2 (AceCS2) and glutamate dehydrogenase, respectively (Haigis et al., 2006; Hallows et al., 2006; Schwer et al., 2002), but little else is known about their biological functions.
Increased gene dosage or enhanced activity of Sir2 extends life span in S. cerevisiae, C. elegans, and D. melanogaster (Anderson et al., 2003; Kaeberlein et al., 1999; Lin et al., 2000; Rogina and Helfand, 2004; Tissenbaum and Guarente, 2001; Wood et al., 2004). Yeast Sir2 is positively regulated by PNC1, a stress- and calorie-responsive longevity gene that catalyzes the first and rate-limiting step in NAD+ biosynthesis from nicotinamide (NAM) (Anderson et al., 2003; Gallo et al., 2004). Whether or not mammals possess a functional equivalent of the PNC1 gene is not known. Clearly, finding a mammalian equivalent of a gene that governs sirtuin activity and promotes longevity has many potential implications, including our understanding of how CR extends life span in mammals.
The search for a mammalian equivalent of PNC1 has been complicated by the fact that the synthesis of NAD+ from NAM is different in mammals than in simple eukaryotes (Brenner, 2005). While yeast, worms, and flies require four steps to synthesize NAD+ from NAM, mammals require only two (Rongvaux et al., 2002). In yeast, the first step is catalyzed by Pnc1 and in mammals by the NAM phosphoribosyltransferase Nampt, also known as PBEF or visfatin (Fukuhara et al., 2004; Rongvaux et al., 2002; Samal et al., 1994). Recent studies have shown that overexpression of Nampt increases SIRT1 activity (Revollo et al., 2004) and can protect cells from death due to PARP overexpression (Pillai et al., 2005), which is consistent with the hypothesis that Nampt is a functional equivalent of Pnc1 in mammals.
In this paper, we identify NAMPT as a stress- and nutrient-responsive gene that boosts mitochondrial NAD+ levels. Mass spectrometric methods are used to accurately measure NAD+ concentrations within mammalian mitochondria and to show that NAMPT expression and mitochondrial NAD+ levels increase in vivo after fasting. Evidence is presented that increased mitochondrial NAD+ promotes cell survival during genotoxic stress and that protection is provided by the mitochondrial sirtuins SIRT3 and SIRT4. These data show that mitochondrial NAD+ is a major determinant of apoptosis and shed new light on the influence of diet on organ physiology and disease.
RESULTS
NAMPT Expression Is Induced by Cell Stress and Nutrient Restriction
Others and we have speculated that mammalian Nampt is the functional equivalent of yeast Pnc1 (Anderson et al., 2002, 2003; Bitterman et al., 2003; Revollo et al., 2004; Yang et al., 2006). This idea was based on the fact that both Pnc1 and Nampt catalyze the first and rate-limiting step in NAD+ biosynthesis from NAM (Pillai et al., 2005; Revollo et al., 2004; Rongvaux et al., 2002).
We reasoned that if NAMPT is analogous to the yeast PNC1 gene, then its expression might also be induced by cell stress and nutrient restriction. We found that human fibrosarcoma HT1080 cells cultured in serum-free conditions had ~1.5- to 2-fold the levels of Nampt relative to controls (Figure 1A). A similar increase in Nampt at both the mRNA and protein level was observed in the livers of rats that were subjected to a 48 hr fast (Figures 1B–1D). Primary cardiomyocytes exposed to hypoxia or serum-free media also had ~2-fold higher levels of Nampt (Figure 1E). Similar results were observed in primary mouse embryonic fibroblasts (MEFs) grown in serum-free medium (data not shown). Thus, NAMPT is a stress- and nutrient-responsive gene, similar in this regard to the yeast PNC1 gene.
Figure 1. Nampt Is a Stress- and Nutrient-Responsive Gene that Protects Cells against the Genotoxic Agent MMS.
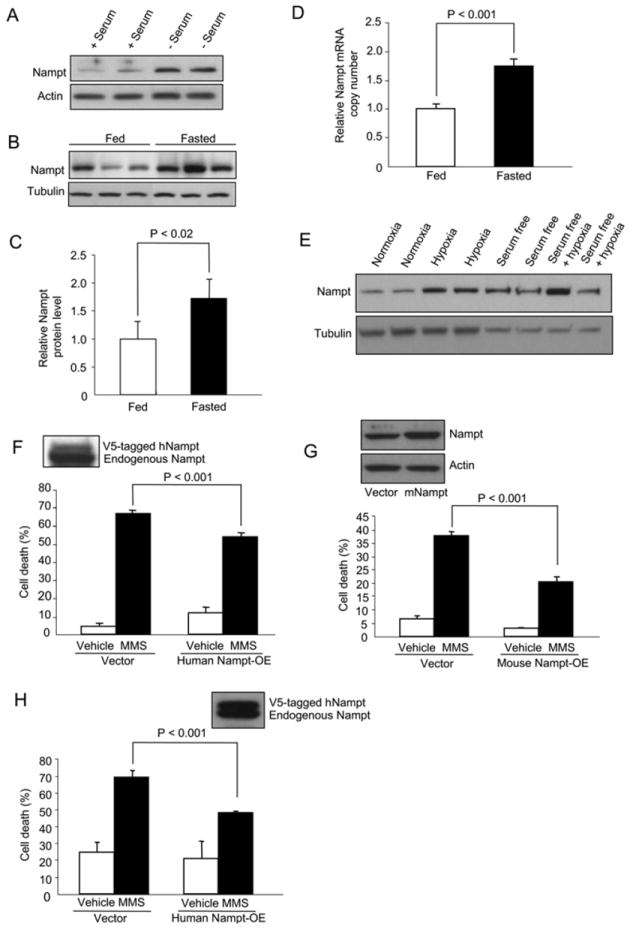
(A and B) Nampt levels in human fibrosarcoma HT1080 cells in the presence or absence of 10% FBS (A) or of liver tissue extracts from fed or 2 day-fasted Sprague-Dawley rats (B). Actin and tubulin were used as loading controls.
(C and D) Nampt protein (C) and mRNA levels of Nampt (D) in livers of fasted rats (n = 4; bars represent the mean of three experiments ± standard deviations [SD] using Student’s t test). (E) Western blot of Nampt in primary rat cardiomyocytes under hypoxia and/or serum starvation.
(F and G) Survival of HT1080 cells stably expressing human (F) or transiently expressing mouse Nampt (G) following treatment with 1.2 mM methylmethanesulfonate (MMS).
(H) Survival of human kidney HEK293 cells stably expressing human Nampt treated with MMS as in (F).
Always, bars represent the mean of three experiments ± SD.
Nampt Protects against Cell Death due to Genotoxic Stress
To mimic the upregulation of Nampt and to test what effect this has on stress resistance, we generated human Nampt-overexpressing stable cell lines from HT1080 fibrosarcoma and HEK293 embryonic kidney cells and selected lines that expressed 1.5 to 2 times the level of Nampt relative to vector controls (Figures 1F and 1H). A similar increase in Nampt expression was obtained by transiently transfecting HT1080 cells with a mouse Nampt expression construct (Figure 1G). Stable Nampt knockdown cell lines were also generated using siRNA (Figure 2A).
Figure 2. Nampt Protects against Apoptotic Cell Death Induced by Topoisomerase Inhibitors.
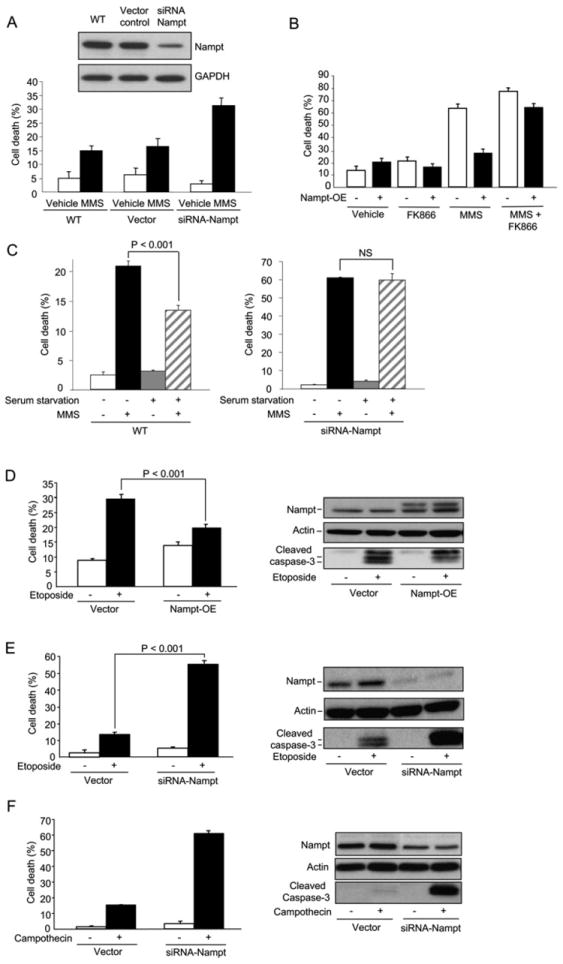
(A) Sensitivity of HT1080 with siRNA-mediated knockdown of NAMPT after MMS exposure.
(B) Stable overexpression of Nampt enhances survival of HEK293 cells following MMS treatment and the effect is blocked by the Nampt-inhibitor FK866.
(C) Survival of WT or Nampt knockdown HT1080 cells after serum deprivation for 22 hr and then exposure to MMS for 17 hr. Serum deprivation upregulates Nampt and enhances survival of WT but not Nampt knockdown HT1080 cells.
(D and E) Survival of HEK293 stably overexpressing Nampt (D) or HT1080 with siRNA knockdown of Nampt (E) following etoposide treatment.
(F) Survival of HT1080 Nampt knockdown cells after camptothecin treatment. Apoptosis was assessed by western blot analysis of cleaved Caspase-3.
Bars represent the mean of three experiments ± SD.
Cell lines were then assayed for their sensitivity to methylmethane sulfonate (MMS), a DNA alkylating agent that is known to hyperactivate PARP-1 (Horton et al., 2005). MMS treatment resulted in the death of about half the vector control cells, and the extent of cell death was greatly reduced by the PARP inhibitor, DPQ (Figure S1 available with this article online), indicating that PARP hyperactivation and depletion of NAD+ were the primary modes of death under these conditions. Although Nampt-overexpressing cells had only slightly higher levels of Nampt protein, they were substantially more resistant to MMS than controls (Figures 1F–1H). Conversely, cells with lower levels of Nampt were more sensitive to MMS (Figures 2A and Figure S2). A potent Nampt catalytic inhibitor, FK866, which binds in the active site (Drevs et al., 2003), prevented cell protection by Nampt overexpression (Figure 2B), indicating that Nampt activity is required for cell protection.
Given that Nampt levels increase in cells grown in serum-free media, we wondered whether cells in serum-free media are more resistant to MMS and, if so, whether their resistance is mediated by Nampt. As shown in Figure 2C, cells that were serum starved were more resistant to MMS, and this resistance was entirely Nampt dependent.
We also tested whether Nampt provides resistance to cell death from other types of DNA damage. Etoposide is a cancer chemotherapeutic agent that inhibits topoisomerase II, resulting in numerous double-stranded DNA breaks that trigger apoptosis. Nampt-overexpressing cells were more resistant to etoposide and had reduced levels of cleaved caspase 3, a marker of apoptosis (Figure 2D). Conversely, cells with reduced levels of Nampt were more sensitive to etoposide and had increased levels of cleaved caspase 3 (Figure 2E). The Nampt knockdown cells were also more sensitive to camptothecin, a topoisomerase I inhibitor (Figure 2F), demonstrating that the ability of Nampt to protect from cell death is not specific to MMS.
Nampt-Mediated Cell Protection Requires Mitochondrial SIRT3 and SIRT4
In light of a recent study showing that the ability of Nampt to protect against PARP-1 overexpression was SIRT1 mediated (Pillai et al., 2005), we expected that protection from MMS would be SIRT1 dependent. But neither EX-527, a SIRT1-specific inhibitor (Solomon et al., 2006), nor siRNA-mediated knockdown of SIRT1 had a significant effect on Nampt-mediated survival (Figures 3A, 3B, and S3A). Curiously though, treatment of cells with the pan-sirtuin inhibitor, sirtinol, did block the ability of Nampt overexpression to protect cells (Figure 3C), which raised the possibility that survival might be mediated by another sirtuin.
Figure 3. Nampt-Mediated Protection against Genotoxicity Requires SIRT3 and SIRT4.
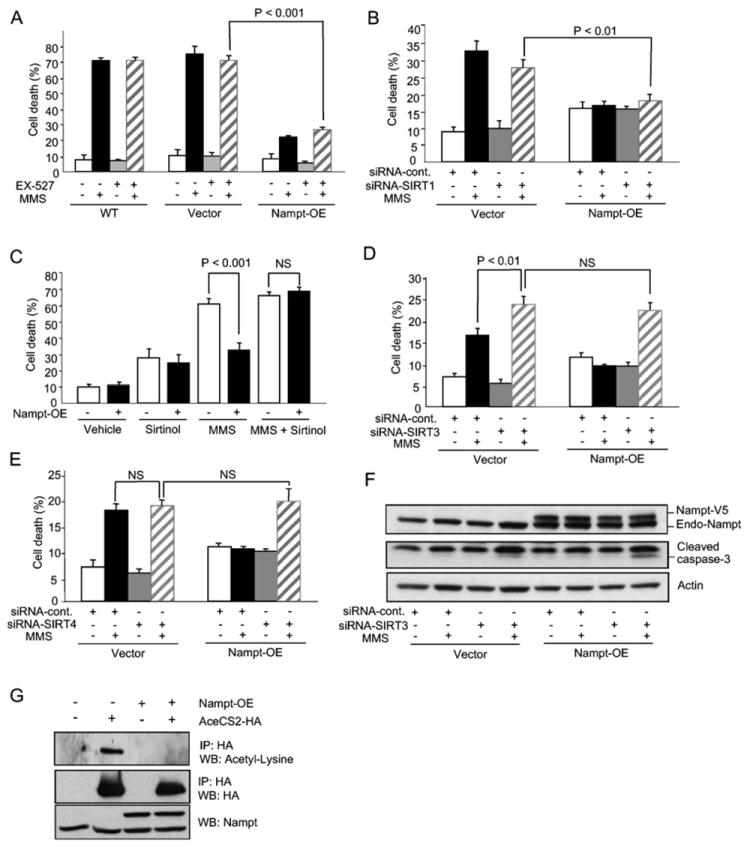
(A) Survival of HEK293 cells stably expressing Nampt following exposure to MMS in the presence or absence of the SIRT1-specific inhibitor EX-527.
(B) siRNA knockdown of SIRT1 using a pool of four siRNA oligos, compared to nontargeting siRNA controls. Cells were cotransfected with FAM-tagged fluorescent oligos and percentage cell death was determined by FACS as a ratio of PI/FAM-positive cells versus total FAM-positive cells.
(C) HEK293 cells were treated with 100 μM sirtinol, a pan sirtuin inhibitor. All experiments were carried out three times in triplicate.
(D and E) SIRT3 or SIRT4 was knocked down in HEK293 cells stably overexpressing Nampt using pools of specific siRNA oligos, and cells were then treated with MMS and scored for survival.
(F) Cells from (D) were probed by western blotting for cleaved caspase-3, an indicator of apoptosis.
(G) Immunoprecipitation (IP) of AceCS2 from cell lysates of control and Nampt-overexpressing HEK293 cells transfected with control vector or AceCS2-HA for 48 hr. The levels of acetylated AceCS2 in IPs were analyzed by western blotting.
Bars represent the mean of three experiments ± SD.
To test this idea, we knocked down each of the remaining sirtuins, SIRT2–7, using siRNA (Figures S3A–S3B) and scored survival after MMS treatment. Interestingly, the mitochondrial sirtuins, SIRT3 and SIRT4, but not the other sirtuins, were required for the ability of Nampt to protect against MMS-induced cell death (Figures 3D, 3E, and S3C–S3F). In addition, knockdown of SIRT3 sensitized wild-type (WT) cells to MMS and increased the relative abundance of cleaved caspase 3 (Figures 3D and 3F and data not shown). This effect appears to be relatively specific to MMS-induced cell death because there was no appreciable effect on sensitivity to etoposide (Figure S4).
These data indicated that overexpression of Nampt might protect cells by increasing SIRT3 activity. We tested this by monitoring the acetylation status of AceCS2, a substrate of SIRT3. As shown in Figure 3G, Nampt overexpression markedly reduced the acetylation level of AceCS2, indicating that Nampt increases SIRT3 activity. Knockdown of SIRT4, on the other hand, had no significant effect on the survival of WT cells (Figure 3E and data not shown). There was no significant increase in the survival of cells overexpressing SIRT3 or SIRT4, or in combination (data not shown), suggesting that in the absence of additional Nampt, MMS causes NAD+ levels to drop below the concentration at which these NAD+-dependent enzymes can function, a possibility we tested below (see Figure 5).
Figure 5. Mammalian Mitochondria Maintain Mitochondrial NAD+ Levels during Genotoxic Stress.
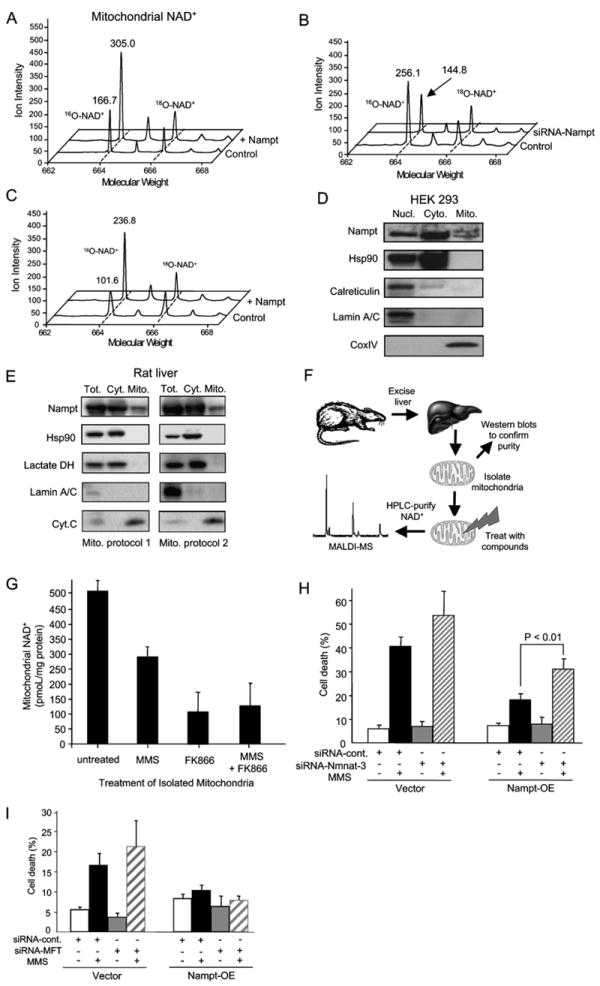
(A and B) Nampt regulates mitochondrial NAD+ levels. Mitochondrial NAD+ was isolated and analyzed as described in Figures 4B and 4C. Spectra from HEK293 are shown for vector controls and cells stably overexpressing Nampt (A), as well as spectra from HT1080 vector controls and siRNA-Nampt stable cells (B).
(C) Additional Nampt greatly attenuates mitochondrial NAD+ depletion by MMS treatment, as determined by MALDI-MS after 2 hr MMS treatment of HEK293 WT and Nampt-overexpressing cells.
(D and E) Western blotting analysis of Nampt in highly purified cytosolic and mitochondrial fractions. Mitochondiral fractions were isolated from HEK293 cells or from rat livers using two different protocols, and their purity was assessed by probing for Hsp90, calreticulin, and/or lactate dehydrogenase (exclusively cytoplasmic proteins), and CoxIV or cytochrome C (mitochondrial matrix markers). The same blot was probed for lamin A/C to test for contamination of the mitochondrial fractions with nuclei. The experiment was performed three times on HEK293 cells and on liver tissue. The same pattern was observed each time and representative blots are shown.
(F) Mitochondria from rat livers were prepared and exposed to methylmethane sulfonate (MMS), a genotoxic DNA alkylating agent, or the Nampt inhibitor FK866, or both. NAD+ levels in isolated mitochondria were determined using MALDI-MS, as above.
(G) NAD+ levels in isolated mitochondria are reduced by exposure to MMS and FK866. Similar data were obtained using a different mitochondrial isolation protocol (see Figure S5).
(H) Knocking down expression of Nmnat-3 reduces the ability of Nampt to provide resistance to MMS.
(I) Knocking down expression of a putative human mitochondrial NAD+ transporter, hMFT, does not affect survival of Nampt-overexpressing cells treated with MMS.
Bars represent the mean of three experiments ± SD.
Nampt Does Not Prevent Depletion of Total NAD+
It is generally recognized that genotoxic stress kills cells by depleting nuclear and cytosolic NAD+ pools (Burkle, 2005; Porcu and Chiarugi, 2005). To test whether Nampt protected against NAD+ depletion, we measured NAD+ concentrations in HEK293 cells after exposure to MMS using a quantitative HPLC/mass spectrometry method (HPLC/MALDI/MS) (Sauve et al., 2005). We did not attempt to measure nicotinamide levels in mitochondria because, unlike NAD+, it diffuses through membranes (van Roermund et al., 1995).
An internal NAD+ reference, 18O-NAM, was synthesized from 3-cyanopyridine, then enzymatically converted to 18O-NAD+ using a recombinant NAD+ glycohydrolase, CD38 (Figure 4A). The key advantage of the HPLC/MALDI/MS technique is that extracts are spiked with an isotopically labeled NAD+ internal reference standard so that losses of NAD+ during purification do not affect the final result. After addition of the labeled standard, the NAD+ in the cell extracts was HPLC purified and analyzed by mass spectrometry, which gave two peaks that corresponded to the two isotopomer molecular ions (Figure 4B). The higher molecular weight species, 18O-NAD+, was used to determine the quantity of the lower weight endogenous species, 16O-NAD+.
Figure 4. Nampt Regulates Total NAD+.
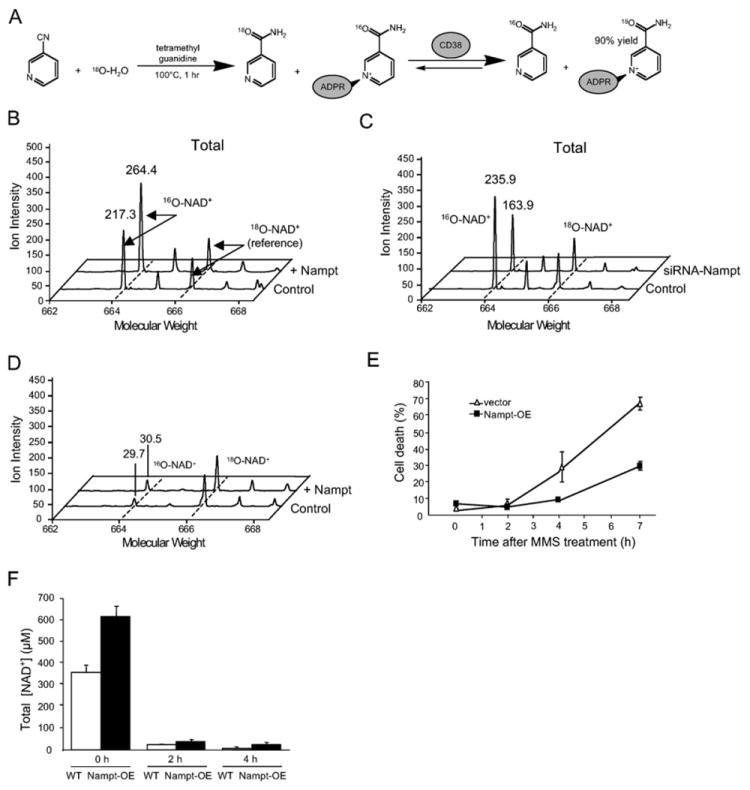
(A) Synthesis of isotope-labeled 18O-NAD+, a reference compound used in NAD+ measurement. 18O-NAM was synthesized by hydrolyzing 3-cyanopyridine in 18O-H2O and was then used as a substrate in the enzymatic reaction catalyzed by CD38, a NAD+ glycohydrolase, to generate 18O-NAD+.
(B and C) Total endogenous 16O-NAD+ and spiked-in NAD reference 18O-NAD+ were isolated by HPLC then subjected to MALDI-MS. The ion intensity of the reference peaks of 18O-NAD+ were normalized to 100 in all cases. The ratio of 16O-NAD+ peaks reflects the relative amount of NAD+ in the two samples. Experiments were performed at least three times. Total NAD+ spectra from HEK293 are shown for vector controls and cells stably overexpressing Nampt (B) as well as total NAD+ spectra from HT1080 vector controls and siRNA-Nampt stable cells (C).
(D) Overexpression of Nampt cannot prevent total cellular NAD+ depletion by MMS as determined by MALDI-MS spectra of endogenous 16O-NAD and reference 18O-NAD after 2 hr MMS treatment of HEK293 WT and Nampt-overexpressing cells.
(E) Time course of cell death induced by 1.2 mM MMS treatment. Percent cell death was determined by FACS analysis.
(F) Total cellular NAD+ as measured by MALDI-MS during the time course in (E).
Bars represent the mean of three experiments ± SD.
We estimate that the total NAD+ concentration in HEK293 cells is 365 ± 30.2 μM, which is very close to a recent estimate for mouse erythrocytes using HPLC/MS/electrospray ionization (368 μM) (Yamada et al., 2006). Interestingly, Nampt-overexpressing cells had approximately twice the total NAD+ concentration of vector control cells and, conversely, Nampt knockdown cells had approximately half (Figures 4B and 4C). These data are consistent with other studies showing that Nampt catalyzes a rate-limiting step in nucleo-cytoplasmic NAD+ biosynthesis (Pillai et al., 2005; Revollo et al., 2004).
Surprisingly, Nampt overexpression did not appreciably affect MMS-mediated depletion of NAD+ in total cell extracts (Figure 4D). To ensure that we had not simply chosen an inappropriate time point, total NAD+ levels were measured at time points just prior to, and during which, cell protection by Nampt was observed (2 and 4 hr) (Figure 4E). Again, concentrations of total NAD+ became critically low (total [NAD+] < 100 μM), irrespective of Nampt levels (Figure 4F).
Nampt-Regulated Mitochondrial NAD+ Levels Dictate Cell Survival
These data raised an intriguing question: How does Nampt protect cells from genotoxic stress if not by maintaining total NAD+ levels? An important clue came from our observation that protection by Nampt required sirtuins that reside in mitochondria. The cell lines we were using, HT1080 and HEK293, possess relatively few mitochondria compared to highly metabolically active cells such as hepatocytes and myocytes (Di Lisa and Bernardi, 2006), and hence mitochondria contribute only a minor component of the total NAD+ pool of these cells. It was plausible that the NAD+ remaining in the MMS-treated cells was primarily mitochondrial and that the increased survival in Nampt overexpressors was a consequence of increased NAD+ levels in mitochondria.
Surprisingly little is known about the precise concentration of NAD+ in mammalian mitochondria or whether changes in mitochondrial NAD+ levels impact cell survival (Porcu and Chiarugi, 2005; Viswanathan et al., 2005). This lack of knowledge appears to stem, in part, from the lack of a robust and accurate method to measure mitochondrial NAD+ concentrations. Indeed, only a few studies have attempted to measure mitochondrial NAD+ levels by any means. Using enzymatic assays, it has not been possible to accurately determine actual concentrations of NAD+ within the organelle. Levels are typically expressed not in molarity but as a nmol NAD+/mg mitochondrial protein (Di Lisa and Bernardi, 2006; Noack et al., 1992; Tobin et al., 1980).
We began by measuring actual mitochondrial NAD+ concentrations using an adaptation of the MALDI-MS technique described above (see Experimental Procedures). We estimate that the concentration of NAD+ in mitochondria of HEK293 cells is 245.6 μM, corresponding to 2053 pmol NAD+/mg mitochondrial protein. Consistent with our hypothesis, cells with additional Nampt had approximately double the concentration of NAD+ in mitochondria (Figure 5A), and there was a corresponding decrease in mitochondrial NAD+ in cells in which Nampt was knocked down (Figure 5B). Importantly, Nampt did not alter mitochondrial size or number, and losses of NAD+ during purification post-addition of the NAD+ reference would not have affected the result.
Next, we determined the effect of Nampt on mitochondrial NAD+ after MMS treatment. Strikingly, Nampt-overexpressing cells had more than double the concentration of mitochondrial NAD+ relative to MMS-treated WT controls (Figure 5C). In fact, the concentration of mitochondrial NAD+ in Nampt-overexpressing cells was higher during MMS exposure than that in WT cells that were untreated. Thus, mitochondria of Nampt-overexpressing cells retain physiological levels of NAD+ after MMS treatment, even if the rest of the cell is depleted of NAD+.
How Do Mitochondria Maintain NAD+ and Protect Cells from Apoptosis?
Next, we wondered how mitochondria maintain such high NAD+ levels during genotoxic stress. There were two plausible explanations. Mitochondria might possess an endogenous NAD+ biosynthetic pathway and/or they might import NAD+ from the cytosol. We first tested the hypothesis that Nampt is localized to mitochondria and participates in mitochondrial NAD+ synthesis. There is already good evidence for an NAD+ salvage pathway in mitochondria. Two studies showed that an NAD+ precursor can be converted to NAD+ when added to mitochondrial preparations (Barile et al., 1996; Kun et al., 1975), and, more recently, an enzyme immediately downstream of Nampt in the NAD+ salvage pathway, Nmnat3 (for NAM mononucleotide adenylyltransferase), was shown to be exclusively mitochondrial (Berger et al., 2005).
To explore whether mitochondria contain Nampt, we isolated highly pure mitochondrial fractions from either HEK293 cells (Figure 5D) or fresh rat livers, in the latter case using two different mitochondrial isolation methods (Figure 5E). The purity of the fractions was assessed by probing for cytoplasmic markers (Hsp90, calreticulin, lactate dehydrogenase), mitochondrial matrix markers (CoxIV or cytochrome c), and a nuclear membrane protein (lamin A/C). The enrichment of mitochondrial markers in the mitochondrial fractions and the absence of cytoplasmic and nuclear proteins in these fractions demonstrated that the mitochondrial preparations were highly pure. Nampt was observed in cytoplasmic and nuclear fractions, consistent with other reports (Kitani et al., 2003; Revollo et al., 2004; Rongvaux et al., 2002). In all of the mitochondrial preparations, from HEK293 cells and from liver tissue, Nampt was clearly detected. Nampt was also present in highly pure mitochondrial preparations from human lymphoblasts, HepG2 hepatocarcinoma cells, and HeLa cells (data not shown).
To add weight to these findings, we performed functional assays for Nampt activity in mitochondria. Specifically, we tested whether interfering with Nampt activity in mitochondria affects mitochondrial NAD+ levels and whether blocking mitochondrial NAD+ biosynthesis reduced the ability of Nampt to protect from genotoxic stress. First we reasoned that if Nampt activity is required to maintain mitochondrial NAD+, then inhibiting Nampt in isolated mitochondria should reduce mitochondrial NAD+ levels. On the other hand, if mitochondrial NAD+ is derived from the cytoplasm, then inhibiting Nampt activity should have no effect on the levels of NAD+ in purified mitochondria.
Mitochondria from fresh rat livers were purified and treated in vitro with MMS or FK866 or in combination (Figure 5F). NAD+ content in the mitochondria was then determined using the HPLC/mass spectrometry method utilized above for whole cells. Treatment of isolated mitochondria with MMS reduced mitochondrial NAD+ levels by ~2-fold (Figure 5G), similar to the effect of treating whole cells with MMS and consistent with hyperactivation of intra-mitochondrial PARP-1 (Du et al., 2003). Treatment of isolated mitochondria with FK866, with or without MMS, resulted in even larger decreases in mitochondrial NAD+ levels. Similar decreases in NAD+ were observed after treatment of mitochondria prepared by an alternative method (Figure S5). These data indicated that mitochondria possess an NAD+ salvage pathway and that inhibition of Nampt in the organelle results in decreased NAD+.
As a further test, we determined whether the protection provided by Nampt required an intact mitochondrial NAD+ salvage pathway. During the course of these experiments, we were fortunate that an exclusively mitochondrial NAD+ biosynthetic enzyme, Nmnat-3, was discovered (Berger et al., 2005). We reasoned that if Nampt boosts cell survival by increasing the synthesis of NAD+ in mitochondria rather than in the cytoplasm or the nucleus, then knocking down Nmnat-3 should diminish Nampt-mediated protection against MMS. If not, then knocking down Nmnat-3 should have no effect on cell protection.
Nmnat-3 was knocked down ~40% using an siRNA construct in HEK293 cells expressing a stably integrated Nampt construct (Figure S6). Cells were then tested for resistance to MMS. As shown in Figure 5H, knockdown of Nmnat-3 significantly reduced Nampt-mediated protection against MMS, demonstrating that a complete mitochondrial NAD+ salvage pathway is necessary for Nampt to provide resistance to MMS. No protection from MMS was seen by overexpressing Nmnat-3 (data not shown), which is in accordance with reports that overexpressing the cytoplasmic form of Nmnat (Nmnat-1) has no effect on NAD+ levels (Araki et al., 2004; Revollo et al., 2004).
The alternative hypothesis, that Nampt increases mitochondrial NAD+ by promoting NAD+ import from the cytoplasm, seemed less likely given our observation that mitochondria retain higher NAD+ levels than the cytoplasm during MMS treatment. Nevertheless, we searched for evidence of a mammalian mitochondrial NAD+ transporter. Although mitochondrial NAD+ transport has not been described for mammals, two mitochondrial NAD+ transporters were recently discovered in S. cerevisiae (Todisco et al., 2006). We identified a putative human homolog called hMFT (identity = 35% to yeast Ndt1; accession NM_030780; Figure S7A). Knocking down of hMFT did not, however, affect the ability of Nampt to protect from MMS (Figures S7B and 5I). We cannot rule out the possibility that another NAD+ transporter exists and that mitochondria can import NAD+ from the cytoplasm, but, taken together, we believe that the simplest explanation of the data is that Nampt is active in mitochondria.
It is generally accepted that depletion of NAD+ stimulates a number of proapoptotic pathways, including the relocalization of AIF from the outer mitochondrial membrane to the nucleus (Di Lisa and Bernardi, 2006; Porcu and Chiarugi, 2005; Yu et al., 2002). As shown in Figure 6A, Nampt overexpression suppressed translocation of AIF to the nucleus in response to MMS, demonstrating that Nampt lies upstream of this major apoptotic pathway.
Figure 6. Fasting Increases Hepatic Mitochondrial NAD+ and Nampt Levels.
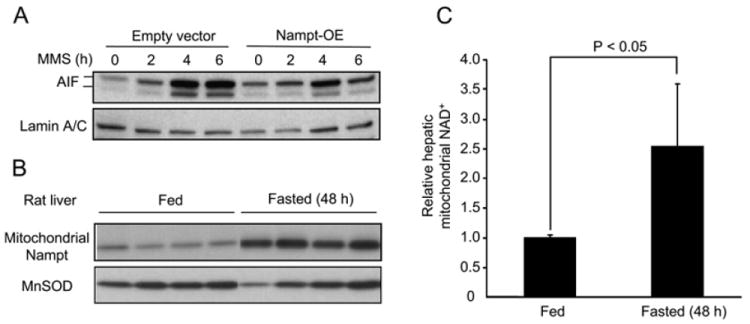
(A) Overexpression of Nampt in HEK293 cells inhibits the localization of AIF to the nucleus after MMS treatment for the times indicated, as assessed by western blotting.
(B) Western blotting analysis of Nampt in mitochondria from rats fed AL or fasted for 48 hr.
(C) Relative mitochondrial NAD+ levels in liver tissues from rats fed AL or fasted for 48 hr. Mitochondrial NAD+ levels were measured by MALDI-MS. Bars represent the mean of three experiments ± SD.
Fasting Increases Mitochondrial Nampt and NAD+ In Vivo
Finally, we tested whether these data were relevant to the in vivo situation. If our hypothesis that Nampt is a nutrient-responsive regulator of mitochondrial NAD+ is correct, we should observe increased levels of Nampt in the mitochondria of fasted rats, concomitant with increased levels of mitochondrial NAD+. To our knowledge, in vivo changes in mitochondrial NAD+ have not been examined previously. Rats were fasted 48 hr, their livers excised, and mitochondria were purified as described above. Mitochondrial fractions were divided for western blotting and for NAD+ determinations using HPLC/MS. After the 48 hr fast there was a dramatic rise in Nampt levels in the mitochondrial fractions (Figure 6B), and there was a concomitant increase in mitochondrial NAD+ levels in the mitochondrial extracts from the fasted animals (Figure 6C). Thus, our results in cell culture extrapolate meaningfully to the in vivo situation.
DISCUSSION
The importance of mitochondrial function to the rate of progression of age-related diseases such as cancer, diabetes, and neurodegeneration has become increasingly apparent in recent years (Lin et al., 2005; St-Pierre et al., 2006). Yet little is currently known about the intracellular concentration of NAD+ in mitochondria, whether it fluctuates in response to diet, or whether these changes influence key cellular functions such as apoptosis. In this study, we have accurately determined NAD+ concentrations in mammalian mitochondria, identified mitochondrial NAD+ as a determinant of cell survival, and shown that mitochondrial NAD+ levels are dramatically upregulated by nutrient restriction in vitro and in vivo. One of the more surprising findings of the study was the observation that mitochondria can maintain physiological levels of NAD+ during genotoxic stress and promote cell survival, even if NAD+ in the cytoplasm and nucleus has fallen well below normal physiological levels. We refer to the ability of mitochondria to dictate cell survival as the “Mitochondrial Oasis Effect.”
This study also shows that Nampt is a stress- and diet-responsive regulator of mitochondrial NAD+ in mammalian cells. The data strongly suggest that Nampt is both present and functional within mitochondria, directly upstream of the exclusively mitochondrial NAD+ biosynthetic enzyme Nmnat-3. Although we cannot and do not rule out other mechanisms by which mitochondria obtain NAD+, such as NAD+ import or via alternative NAD+ biosynthetic routes (Bieganowski and Brenner, 2004), the fact that Nampt activity is required to maintain NAD+ levels in isolated mitochondria is strong evidence that Nampt plays a functional role within these organelles. Given the central role of Nampt in NAD+ biosynthesis, it is likely that Nampt activity is not simply regulated at the gene expression level but at multiple levels, including by substrate availability and potentially by posttranslational modification.
Considering that numerous enzymes in mitochondria are limited by NAD+ availability, including the sirtuins SIRT3 and SIRT4, which are known to regulate GDH and AceCS2, respectively, it will be interesting to explore the potential impact of mitochondrial NAD+ levels on the metabolism and health of various organs. Perhaps diet-induced increases in mitochondrial NAD+ contribute to not only the increased resistance of calorie-restricted rodents to toxins but also the changes in fatty-acid metabolism and respiration that occur with reduced caloric intake (Ando et al., 2002; Campisi, 2003; Higami and Shimokawa, 2000; Migliaccio et al., 1999; Zhang and Herman, 2002).
The events that lead to PARP-induced apoptosis remain poorly understood, but it is known that AIF relocalization is a key event (Di Lisa and Bernardi, 2006; Yu et al., 2002). In this study, we find that overexpression of Nampt leads to an attenuation of AIF relocalization. Given that NAMPT lies upstream of AIF, it will be interesting to test whether SIRT3 or SIRT4 associate with and/or modify AIF or other determinants of apoptosis such as the permeability transition pore (PTP).
Our observation that Nampt is a stress- and nutrient-responsive gene that promotes cell survival via SIRT3 and SIRT4 lends further support to the hypothesis that NAMPT is a functional homolog of the yeast PNC1 longevity gene (Anderson et al., 2003; Bitterman et al., 2003). Transgenic mouse experiments are in progress to determine the effect of overexpressing NAMPT. We hypothesize that these animals might have increased resistance to cell stress, altered metabolism, and disease resistance (North and Sinclair, 2007).
Because NAD+ is such an ancient molecule, insights into the biology of NAD+ can provide clues about the early evolution of life on earth (Brenner, 2005). There is evidence that cells have used NAD+ as a nutrient sensor that dictates survival for a very long time, possibly predating the evolution of eukaryotes. Homologs of Nampt and Sir2 are found in bacterial relatives of mitochondria (Smith et al., 2000), and increased NAD+ levels provide bacterial resistance to heat, salt stress, and glucose restriction, for reasons that are not yet clear (Foster et al., 1990). A phylogenetic comparison of NAM-metabolizing enzymes from various species shows that vertebrates utilize a pathway more closely related to the organisms that gave rise to the first mitochondria (Andersson et al., 2003) than to S. cerevisiae, C. elegans, and D. melanogaster (Figure S8). This indicates that NAD+ levels may have controlled cell survival in the bacteria that gave rise to mitochondria, and these survival pathways have been conserved up to the present day in mammals.
In summary, we have shown that mitochondrial NAD+ levels influence cell survival following genotoxic stress and that these levels are considerably higher after nutrient deprivation. We hope that these insights into the importance of mitochondrial NAD+ will facilitate a new understanding of and the development of novel approaches to treating diseases such as cancer and neurodegeneration.
EXPERIMENTAL PROCEDURES
Cell Culture
Human embryonic kidney (HEK293) and human fibrosarcoma HT1080 cell lines were obtained from ATCC and grown in complete DMEM medium with 10% FBS and 100 mg/ml penicillin/streptomycin. To generate Nampt overexpression or empty vector stable clones, hNAMPT/pcDNA or pcDNA empty vector were transfected into HEK293 or HT1080 cells and selected with 0.5–1.0 mg/ml geneticin. To generate Nampt knockdown cells, siRNA/NAMPT/pMSCV or pMSCV empty vector were transfected into HT1080 cells, and stable clones were selected with 500 ng/ml puromycin. Primary neonatal rat cardiomyocytes were prepared as described previously (Matsui et al., 1999). Isolated cardiomyocytes were grown in complete medium (RPMI 1640, 5% fetal calf serum, 10% horse serum) to 80% confluence before cells were subjected to hypoxia and/or serum starvation. For hypoxia, medium was changed to RPMI 1640 with or without serum saturated with 95% N2/5% CO2, and then cells were placed in a 37°C airtight box saturated with 95% N2/5% CO2 for 18 hr. O2 concentrations were <0.1% (Ohmeda oxygen monitor, type 5120). To serum starve HT1080, cells were grown in DMEM medium containing no serum. Cells were harvested after 26 hr of serum starvation and Nampt expression was analyzed by western blotting.
Drug Treatments and Cell Death Assays
HEK293 and HT1080 cells at less than 85% confluency were treated with MMS (1.2 mM) for 6–8.5 hr and 8–17 hr, respectively. Transiently transfected cells were washed three times with PBS to remove residual transfection reagents before treating with MMS. HEK293 cells and HT1080 cells were exposed to etoposide for 72 hr or 46 hr at 120 or 40 μM, respectively. HT1080 cells were exposed to camptothecin at 14 μM for 23 hr. After drug treatments, cells were harvested by trypsinization, pelleted by centrifugation, and resuspended in PBS containing 3% FBS. Cell death was analyzed by FACS using propidium iodide (PI) staining. In cells transiently transfected with mNAMPT/MSCV, SIRT1–7 siRNA, Nmnat-3, or hMFT siRNA oligos and cotransfected with FAM-labeled scrambled siRNA oligos, percent cell death was determined as the proportion of PI/GFP- or PI/FAM-positive cells versus the total number of cells with green fluorescence.
Assay for Aceylated AceCS2
Control and Nampt overexpressing HEK293 cells were transfected with control vector or AceCS2-HA. Forty-eight hours post-transfections, cells were washed and lysed in IP lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-50, 400 nM TSA, 5 mM Nicotinamide, and protease inhibitors [Compelete, Roche]) for 30 min at 4°C. Lysates were cleared and subjected to immunoprecipitation with anti-HA affinity gel (Roche) for 2 hr at 4°C. Immunoprecipitations were washed three times for 15 min each in lysis buffer and resuspended in 1× SDS-PAGE buffer and analyzed by western blotting.
Cell Fractionation and Drug Treatment of Mitochondria
Unless otherwise stated, fractionation of cultured cells was performed using a differential centrifugation and sucrose gradient procedure with slight modifications (Schwer et al., 2002). For drug treatment of mitochondria, fresh mitochondria were obtained from livers excised from fed young male rats. Half of the liver homogenate was used for mitochondrial isolation using a commercially available kit as described by manufacturer’s instructions, designated Protocol 1 (Pierce Mitochondrial Isolation kit, Rockford, IL). The other half of the original homogenate was subjected to a differential centrifugation protocol to isolate mitochondria (Protocol 2). Mitochondria (500 μl) were added to a 48-well plate and treated with methylmethane sulfonate (MMS) at 1:1000 dilution and/or FK866 (10 nM) for 30 min at 37°C. Suspensions were spun down for 1 min at 14,000 rpm at 4°C and pellets were stored at −80°C prior to NAD+ analysis by HPLC/MALDI/MS. Detailed protocols are described in the Supplemental Data.
HPLC/MALDI/MS Determination of NAD+
NAD+ was determined as described previously (Sauve and Schramm, 2003), with the following modifications. HEK293 or HT1080 cells were harvested by trypsinization and counted by haemocytometer. NAD+ was extracted from whole cells or mitochondrial pellets by adding 10% perchloric acid and sonicating for 5 min on ice. The reference standard 18O-NAD+ (typically 613.4 pmol) was then added to the sample. After mixing well, samples were centrifuged for 3 min then neutralized with NaOH. NAD+ in the supernatant (100 μl sample) was separated from other cellular components by HPLC. NAD+ peaks were collected according to the standards’ retention time and dried with a lyophilizor. MALDI-MS was used to determine the peak ratio between the positively charged isotopically distinct ions, and the intensities of the 16O- and 18O-peaks were ratioed (664/666) to obtain 16O NAD+/18O NAD+ in the sample. Standards containing only 16O and 18O NAD+ (600 pmol each) were also run to determine corrections for isotopic purity and to provide calibration of the procedure. The volume of mitochondrial pellets was calculated by considering the density of the mitochondria pellets to be 1.0 μl/mg. Total cellular NAD+ concentrations were calculated by dividing the quantity of NAD+ per cell by the mean cell volume (MCV) as measured by a Coulter counter, which for HT1080 was 2183.95 fl and for HEK293 was 1691.04 fl.
Animal Experiments
To assess Nampt expression in vivo, Sprague-Dawley rats (120–150 g) were obtained from Charles River Laboratories and randomly assigned to control and experimental groups of four animals. The control group was fed ad libitum (AL) with a 78% sucrose diet prepared by Research Diets Inc. and the experimental group was fasted for 48 hr before being sacrificed. Liver tissue for RNA extraction was stored in RNAlater reagent (QIAGEN). All other samples were frozen in liquid nitrogen and stored at −80°C until use. To assess mitochondrial Nampt and NAD+ levels, Male Fischer-344 (F344) rats were bred and reared in a vivarium at the Gerontology Research Center (GRC, Baltimore, MD). From weaning (2 weeks), the rats were housed individually in standard plastic cages with beta chip wood bedding. Control animals were fed a NIH-31 standard diet AL. The procedures for preparation of mitochondria from liver are described in the Supplemental Data.
Supplementary Material
Acknowledgments
Oberdan Leo and Anthony Rongvaux kindly provided the anti-Nampt monoclonal antibody and mNAMPT/MSCV plasmids. Thanks to Carolina Smith for assisting with animals and surgery, Jill Milne for providing EX-527, and Sean Armour for technical advice. H.Y. was supported by a Harvard/Hartford Advanced Research Award, J.A.B. by an American Heart Association Award, D.L. by a National Eye Institute training grant, and J.J.C. by a Howard Hughes Medical Institute Predoctoral Fellowship. This study was supported in part by the intramural program of the National Institutes on Aging. D.A.S. was an Ellison Medical Foundation New Scholar. A.R. and T.M. are supported by grants from the NIH and the Leducq Foundation. The Sauve lab is supported by NIH grant R01 DK 073466, the Sinclair lab by NIH grants RO1GM068072, RO1AG028730, PO1 AG027916 and the Paul F. Glenn Laboratories for the Biological Mechanisms of Aging. D.A.S. and A.A.S. are consultants to Sirtris Pharmaceuticals, a company aiming to treat diseases by modulating sirtuins. D.A.S. is a cofounder of Sirtris Pharmaceuticals and sits on their advisory board and board of directors. After completing this study, H.Y. became an employee and shareholder of Sirtris Pharmaceuticals.
Footnotes
Supplemental Data
Supplemental Data include Supplemental Experimental Procedures, eight figures, and Supplemental References and can be found with this article online at http://www.cell.com/cgi/content/full/130/6/1095/DC1/.
References
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and Pnc1 govern lifespan extension by calorie restriction in S. cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Karlberg O, Canback B, Kurland CG. On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond B Biol Sci. 2003;358:165–177. doi: 10.1098/rstb.2002.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Higami Y, Tsuchiya T, Kanematsu T, Shimokawa I. Impact of aging and life-long calorie restriction on expression of apoptosis-related genes in male F344 rat liver. Microsc Res Tech. 2002;59:293–300. doi: 10.1002/jemt.10207. [DOI] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Barile M, Cozzani E, Anonide A, Usiglio D, Burroni A, Guarrera M. Is contact allergy rare in psoriatics? Contact Dermatitis. 1996;35:113–114. doi: 10.1111/j.1600-0536.1996.tb02309.x. [DOI] [PubMed] [Google Scholar]
- Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C. Evolution of NAD biosynthetic enzymes. Structure. 2005;13:1239–1240. doi: 10.1016/j.str.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Burkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cellular senescence and apoptosis: how cellular responses might influence aging phenotypes. Exp Gerontol. 2003;38:5–11. doi: 10.1016/s0531-5565(02)00152-3. [DOI] [PubMed] [Google Scholar]
- Cipriani G, Rapizzi E, Vannacci A, Rizzuto R, Moroni F, Chiarugi A. Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J Biol Chem. 2005;280:17227–17234. doi: 10.1074/jbc.M414526200. [DOI] [PubMed] [Google Scholar]
- Denu JM. The Sir2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Di Lisa F, Bernardi P. Mitochondria and ischemia-reperfusion injury of the heart: fixing a hole. Cardiovasc Res. 2006;70:191–199. doi: 10.1016/j.cardiores.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Drevs J, Loser R, Rattel B, Esser N. Antiangiogenic potency of FK866/K22.175, a new inhibitor of intracellular NAD biosynthesis, in murine renal cell carcinoma. Anticancer Res. 2003;23:4853–4858. [PubMed] [Google Scholar]
- Du L, Zhang X, Han YY, Burke NA, Kochanek PM, Watkins SC, Graham SH, Carcillo JA, Szabo C, Clark RS. Intra-mitochondrial poly(ADP-ribosylation) contributes to NAD+ depletion and cell death induced by oxidative stress. J Biol Chem. 2003;278:18426–18433. doi: 10.1074/jbc.M301295200. [DOI] [PubMed] [Google Scholar]
- Foster JW, Park YK, Penfound T, Fenger T, Spector MP. Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J Bacteriol. 1990;172:4187–4196. doi: 10.1128/jb.172.8.4187-4196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, et al. Visfatin: A protein secreted by visceral fat that mimics the effects of insulin. Science. 2004;307:426–430. doi: 10.1126/science.1097243. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and SIR2 genes–towards a mechanism. Mech Ageing Dev. 2005;126:923–928. doi: 10.1016/j.mad.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Guarente L, Picard F. Calorie restriction–the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higami Y, Shimokawa I. Apoptosis in the aging process. Cell Tissue Res. 2000;301:125–132. doi: 10.1007/s004419900156. [DOI] [PubMed] [Google Scholar]
- Horton JK, Stefanick DF, Wilson SH. Involvement of poly(ADP-ribose) polymerase activity in regulating Chk1-dependent apoptotic cell death. DNA Repair (Amst) 2005;4:1111–1120. doi: 10.1016/j.dnarep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- James SJ, Muskhelishvili L, Gaylor DW, Turturro A, Hart R. Upregulation of apoptosis with dietary restriction: implications for carcinogenesis and aging. Environ Health Perspect. 1998;106(Suppl 1):307–312. doi: 10.1289/ehp.98106s1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani T, Okuno S, Fujisawa H. Growth phase-dependent changes in the subcellular localization of pre-B-cell colony-enhancing factor. FEBS Lett. 2003;544:74–78. doi: 10.1016/s0014-5793(03)00476-9. [DOI] [PubMed] [Google Scholar]
- Kun E, Zimber PH, Chang AC, Puschendorf B, Grunicke H. Macromolecular enzymatic product of NAD+ in liver mitochondria. Proc Natl Acad Sci USA. 1975;72:1436–1440. doi: 10.1073/pnas.72.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Matsui T, Li L, del Monte F, Fukui Y, Franke TF, Hajjar RJ, Rosenzweig A. Adenoviral gene transfer of activated phosphatidylinositol 3′-kinase and Akt inhibits apoptosis of hypoxic cardiomyocytes in vitro. Circulation. 1999;100:2373–2379. doi: 10.1161/01.cir.100.23.2373. [DOI] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Noack H, Kunz WS, Augustin W. Evaluation of a procedure for the simultaneous determination of oxidized and reduced pyridine nucleotides and adenylates in organic phenol extracts from mitochondria. Anal Biochem. 1992;202:162–165. doi: 10.1016/0003-2697(92)90222-s. [DOI] [PubMed] [Google Scholar]
- North BJ, Sinclair DA. Sirtuins: a conserved key unlocking AceCS activity. Trends Biochem Sci. 2007;32:1–4. doi: 10.1016/j.tibs.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Porcu M, Chiarugi A. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol Sci. 2005;26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol. 1994;14:1431–1437. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tobin A, Djerdjour B, Journet E, Neuburger M, Douce R. Effect of NAD on malate oxidation in intact plant mitochondria. Plant Physiol. 1980;66:225–229. doi: 10.1104/pp.66.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todisco S, Agrimi G, Castegna A, Palmieri F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J Biol Chem. 2006;281:1524–1531. doi: 10.1074/jbc.M510425200. [DOI] [PubMed] [Google Scholar]
- van Roermund CW, Elgersma Y, Singh N, Wanders RJ, Tabak HF. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 1995;14:3480–3486. doi: 10.1002/j.1460-2075.1995.tb07354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJ, Hageman GJ. Poly(ADP-ribose) polymerase-1 mediated caspase-independent cell death after ischemia/reperfusion. Free Radic Biol Med. 2005;39:81–90. doi: 10.1016/j.freeradbiomed.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal Biochem. 2006;352:282–285. doi: 10.1016/j.ab.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Yang H, Lavu S, Sinclair DA. Nampt/PBEF/Visfatin: a regulator of mammalian health and longevity? Exp Gerontol. 2006;41:718–726. doi: 10.1016/j.exger.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Herman B. Ageing and apoptosis. Mech Ageing Dev. 2002;123:245–260. doi: 10.1016/s0047-6374(01)00349-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


