Background: Kre6 is required for fungal β-1,6-glucan synthesis.
Results: Genetic and protein-protein interactions between ER residents (Rot1, Keg1, calnexin cycle member homologues) and Kre6 are required for folding and plasma membrane localization of Kre6.
Conclusion: Action of ER chaperon-like proteins is essential for β-1,6-glucan synthesis.
Significance: Kre6 was identified as the first potential target of yeast calnexin homologue.
Keywords: Cell Wall, Chaperone Chaperonin, Endoplasmic Reticulum (ER), ER-associated Degradation, Plasma Membrane, Calnexin Cne1, Keg1, Kre5, Rot1, Skn1
Abstract
Saccharomyces cerevisiae Kre6 is a type II membrane protein essential for cell wall β-1,6-glucan synthesis. Recently we reported that the majority of Kre6 is in the endoplasmic reticulum (ER), but a significant portion of Kre6 is found in the plasma membrane of buds, and this polarized appearance of Kre6 is required for β-1,6-glucan synthesis. An essential membrane protein, Keg1, and ER chaperon Rot1 bind to Kre6. In this study we found that in mutant keg1-1 cells, accumulation of Kre6 at the buds is diminished, binding of Kre6 to Keg1 is decreased, and Kre6 becomes susceptible to ER-associated degradation (ERAD), which suggests Keg1 participates in folding and transport of Kre6. All mutants of the calnexin cycle member homologues (cwh41, rot2, kre5, and cne1) showed defects in β-1,6-glucan synthesis, although the calnexin chaperon system is considered not functional in yeast. We found synthetic defects between them and keg1-1, and Cne1 co-immunoprecipitated with Keg1 and Kre6. A stronger binding of Cne1 to Kre6 was detected when two glucosidases (Cwh41 and Rot2) that remove glucose on N-glycan were functional. Skn1, a Kre6 homologue, was not detected by immunofluorescence in the wild type yeast, but in kre6Δ cells it became detectable and behaved like Kre6. In conclusion, the action of multiple ER chaperon-like proteins is required for proper folding and localization of Kre6 and probably Skn1 to function in β-1,6-glucan synthesis.
Introduction
The cell wall is essential for fungi as the physical support of the cell, barrier against various attacks from the outside, and interface to communicate with the environment. The cell wall of Saccharomyces cerevisiae is composed of mannoproteins, β-1,3-glucan, β-1,6-glucan, and chitin. Mannoproteins are delivered from the ER2 to the cell wall via the secretion pathway. β-1,3-Glucan and chitin are synthesized from UDP-sugars by their synthases in the plasma membrane (PM). However, the polymerase of β-1,6-glucan has not yet been uncovered in the PM (1).
A number of genes whose mutants show reduction in β-1,6-glucan content have been reported (2, 3), and their gene products are localized in the intracellular secretion pathway from the ER to PM. Cne1, Cwh41, Keg1, Kre5, Rot1, and Rot2 are in the ER, Kre11 is a component of the secretion factor TRAPPII at the Golgi, Kre1 is a glycosylphosphatidylinositol-anchor protein on the PM, and Kre9 and its homologue, Knh1, are secreted (2).
Kre6 (killer toxin resistant 6) and its homologue Skn1 (suppressor of kre null 1) are type II membrane proteins and important candidates that may directly participate in β-1,6-glucan synthesis because they are homologous to family 16 glycoside hydrolase and may participate in transglycosylation that elongate nascent short glucans (4). We raised rabbit antiserum against an N-terminal fragment of Kre6 and detected the intrinsic untagged Kre6 in the wild-type cells by immunofluorescence microscopy. Clear Kre6-specific signals were found mainly in the PM of growing buds, as in the case of Kre6-3HA that was detected by anti-HA monoclonal antibody. For an unknown reason, the majority of Kre6 in the ER was not detected by indirect immunofluorescence staining. This polarized localization is apparently required for β-1,6-glucan synthesis (5).
Folding of nascent secretary proteins in the ER occurs with the help of general chaperons including Kar2 and Rot1 (6), and several quality control systems are also working there. The calnexin cycle is a system composed of 4 proteins; glucosidase I, glucosidase II, UDP-glucose:glycoprotein glucosyltransferase (UGGT), and calnexin. Two glucosidases remove glucose from the N-glycan Glc3Man9GlcNAc2 transferred to the asparagines of nascent polypeptides, but if the polypeptides are not correctly folded, UGGT adds glucose again, and then the molecular chaperon calnexin binds to the polypeptide to help its folding (7–9). The calnexin cycle works in a number of eukaryotes including mammals, but it is considered not to work in budding yeast (10). The homologues of four members of the calnexin cycle, Cwh41 (glucosidase I), Rot2 (glucosidase II), Kre5 (UGGT), and Cne1 (calnexin) exist in the ER, and mutants of their genes show reduction in cell wall β-1,6-glucan content (3). A decrease of Kre6 protein in cwh41Δ cells has been reported (11). Calcofluor White hypersensitivity of cwh41Δ cell and growth defects at the non-permissive temperature of a temperature-sensitive kre5 mutant were suppressed by introduction of multicopy KRE6 (12). These genetic interactions suggest that Kre6 may be a target of the yeast calnexin cycle member homologues.
We considered that the appearance of Kre6 in the growing buds should require its correct folding and exit from the ER and examined Keg1, ER chaperons, and calnexin cycle member homologues to find further relationships with Kre6. We also examined if the Kre6-homologue Skn1 has similar characteristics to play a compensatory role in the absence of Kre6.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Media
S. cerevisiae strains used in this study are listed in Table 1. Tagging of Skn1, Cne1, and Kre5 with three copies of the HA or six copies of myc epitope at their C termini was done by homologous recombination, as described previously (5). For a description of genotypes in this paper, when the recombinant gene SKN1-3HA was integrated at the original SKN1 locus by selection of linking HIS3 marker, it was indicated as SKN1-3HA:HIS3 to clarify that HIS3 is not at its original chromosomal locus but is linked to SKN-3HA. As Kre5 has an ER retention signal sequence (HDEL) at the C terminus, HA epitope was inserted in the adjacent upstream of HDEL. The keg1-1 plasmid was made by cloning the PCR amplification fragment from AKY17 (keg1-1) genomic DNA using appropriate primers, as described previously (13). The nucleotide sequences were confirmed.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotypea and plasmid | Origin |
|---|---|---|
| BY4741 | MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| BY4742 | MATα, his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| Y00349 | as BY4741, cne1Δ::kanMX4 | Euroscarf |
| Y00597 | as BY4741, ubc7Δ::kanMX4 | Euroscarf |
| Y01065 | as BY4741, alg5Δ::kanMX4 | Euroscarf |
| Y02098 | as BY4741, pep4Δ::kanMX4 | Euroscarf |
| Y03369 | as BY4741, rot2Δ::kanMX4 | Euroscarf |
| Y04395 | as BY4741, cwh41Δ::kanMX4 | Euroscarf |
| Y05574 | as BY4741, kre6Δ::kanMX4 | Euroscarf |
| AKY17 | as BY4741, keg1-1:LEU2 | Nakamata et al. (13) |
| KTY284 | as BY4741, KRE6-3HA:LEU2 | Kurita et al. (5) |
| KTY432 | MATa, SKN1-3HA:LEU2 his3Δ1 ura3Δ0 | This study |
| KTY209 | as Y05574, SKN1-3HA:LEU2 | This study |
| KTY236 | MATa, keg1Δ::kanMX4 his3Δ1 leu2Δ0, pCA120 (CEN, URA3 myc6-KEG1) | This study |
| RMY11 | as KTY236, kre6::LEU2 | This study |
| KTY658 | as RMY11, SKN1-3HA:HIS3 | This study |
| KTY659 | as KTY236, SKN1-3HA:HIS3 | This study |
| KNY15 | as AKY17, pAK56 (2μ, URA3 ROT1) | Nakamata et al. (13) |
| KTY203 | as AKY17, pCA69 (CEN, URA3 KEG1) | Nakamata et al. (13) |
| KTY205 | as AKY17, pRS316 (CEN, URA3) | Nakamata et al. (13) |
| KTY656 | as KTY203, kre6Δ::kanMX4 | This study |
| KTY496 | MATa, keg1Δ::kanMX4 ura3-52::GFP-KEG1:URA3 KRE6-3HA:LEU2 his3Δ1 | This study |
| KTY498 | MATα, keg1Δ::kanMX4 ura3-52::GFP-KEG1:URA3 his3Δ1 leu2Δ0 | This study |
| KTY500 | MATa, keg1Δ::kanMX4 ura3-52::GFP-keg1-1:URA3 KRE6-3HA:LEU2 his3Δ1 | This study |
| KTY502 | MATα, keg1Δ::kanMX4 ura3-52::GFP-keg1-1:URA3 his3Δ1 leu2Δ0 | This study |
| KTY449 | as BY4742, KRE6-3HA:LEU2 | This study |
| KTY678 | MATa, keg1Δ::kanMX4 leu2Δ0, pKT119 (CEN, HIS3 GFP-keg1-1) pAK56 (2μ, URA3 ROT1) | This study |
| KTY679 | MATa, keg1Δ::kanMX4 leu2Δ0, pKT119 (CEN, HIS3 GFP-keg1-1) pRS426 (2μ, URA3) | This study |
| KTY682 | as KTY678, KRE6-3HA:LEU2 | This study |
| KTY683 | as KTY679, KRE6-3HA:LEU2 | This study |
| KTY638 | MATα, keg1-1:LEU2 pep4Δ::kanMX4 his3Δ1 ura3Δ0 | This study |
| KTY640 | MATa, keg1-1:LEU2 ubc7Δ::kanMX4 his3Δ1 ura3Δ0 | This study |
| KTY294 | as BY4742, CNE1-3HA:LEU2 | This study |
| KTY331 | MATα, keg1Δ::kanMX4CNE1-3HA:LEU2 his3Δ1, pCA120 (CEN, URA3 myc6-KEG1) | This study |
| KTY379 | as KTY236, KRE5-3HA-HDEL:LEU2 | This study |
| KTY429 | as BY4742, KRE5-3HA-HDEL:LEU2 | This study |
| KTY672 | as Y04395, CNE1-3HA:LEU2 | This study |
| KTY674 | as Y03369, CNE1-3HA:LEU2 | This study |
| KTY512 | MATα, keg1-1:LEU2 SKN1-3HA:HIS3 ura3Δ0 | This study |
| KTY519 | MATa, SKN1-3HA:HIS3 leu2Δ0 ura3Δ0 | This study |
| KTY525 | MATa, ubc7Δ::kanMX4 SKN1-3HA:HIS3 leu2Δ0 ura3Δ0 | This study |
| KTY527 | MATa, keg1-1:LEU2 ubc7Δ::kanMX4 SKN1-3HA:HIS3 ura3Δ0 | This study |
a Gene A::B indicates that B replaced A or B was inserted in A at the original locus of A. Gene A:B indicates that A and B are linked at the original locus of A, because B was used as the selection marker for modification of gene A on the chromosome.
A diploid (as BY4743, keg1Δ::kanMX4/KEG1 SKN1-3HA:LEU2/SKN1 leu2Δ0/leu2Δ0, pCA120 (CEN, myc6-KEG1 URA3) was sporulated, and the progeny having SKN1-3HA:LEU2 was named KTY432. A diploid (as BY4743, keg1Δ::kanMX4/KEG1) was transformed with pCA120 and sporulated, and the progeny having keg1Δ::kanMX his3Δ1 leu2Δ0 and pCA120 was named KTY236. A diploid (as BY4743, keg1Δ::kanMX4/KEG1 ura3-52/ura3Δ0 KRE6-3HA:LEU2/KRE6) was transformed with pKT69 (GFP-KEG1:URA3) or pKT78 (GFP-keg1-1:URA3) to integrate the tagged gene at the chromosomal ura3-52 locus and sporulated. The progeny having keg1Δ::kanMX4 ura3-52::GFP-KEG1:URA3 KRE6-3HA:LEU2 his3Δ1 was named KTY496. Also, the progeny as KTY496 but without KRE6-3HA:LEU2 was named KTY498. The progeny having keg1Δ::kanMX4 ura3-52::GFP-keg1-1:URA3 KRE6-3HA:LEU2 his3Δ1 was named KTY500. Also, the progeny as KTY500 without KRE6-3HA:LEU2 was named KTY502. KTY236 was transformed with pKT119 (CEN, GFP-keg1-1 HIS3), and pCA120 was removed by propagation on a 5-fluoroorotic acid (5-FOA) plate. Then it was transformed with either pAK56 (2μ, URA3 ROT1) or pRS426 (2μ, URA3) and named KTY678 or KTY679, respectively. A diploid (as BY4743, keg1-1/KEG1 LEU2/leu2Δ0 pep4Δ::kanMX4/PEP4) was sporulated. The progeny with keg1-1 his3Δ1 LEU2 and ura3Δ0 was named KTY638. KTY640 was constructed by a similar procedure to that of KTY638. A diploid (as BY4743, keg1Δ::kanMX4/KEG1 CNE1-3HA:LEU2/CNE1, pCA120) was sporulated, and the progeny having keg1Δ::kanMX4 CNE1-3HA:LEU2 and pCA120 was named KTY331.
A diploid made by mating AKY17 with BY4742 was sporulated, the progeny having MATα keg1-1:LEU2 was obtained, and then the SKN1 allele was replaced with SKN1-3HA:HIS3 by homologous recombination (KTY512). A diploid made by mating KTY512 with Y00597 was sporulated, and the progenies having SKN1-3HA:HIS3 (KTY519), SKN1-3HA:HIS3 ubc7Δ::kanMX4 (KTY525), or SKN1-3HA:HIS3, KEG1::keg1-1:HIS3 ubc7Δ::kanMX4 (KTY527) was obtained. Yeast and Escherichia coli were grown and used, as described previously (5).
Antibodies, Immunoblotting, and Indirect Immunofluorescence
Antiserum against Scs2 was kindly provided by Dr. Satoshi Kagiwada (Nara Women's University, Nara, Japan). Anti-HA and anti-myc mouse monoclonal antibodies and anti-Gas1, anti-GFP, and anti-Kre6 rabbit polyclonal antibodies were described previously (5, 14). For immunoblotting, anti-Kre6 antiserum was used at a dilution of 1/500, and the other antibodies were used at a dilution of 1/1000. The intracellular localizations of proteins were observed by indirect immunofluorescence, as described previously (5).
Immunoprecipitation
The physical interaction between proteins were examined by immunoprecipitation, as described previously (13). 1% Triton X-100 or 1% digitonin was used as a detergent according to the experiments. SDS-PAGE samples were boiled for 1 min, except those to detect Keg1, which were incubated at 37 °C for 5 min.
Sucrose Density Gradient Fractionation
The intracellular localizations of proteins were determined by sucrose density gradient fractionation. Aliquots of each fraction were analyzed by SDS-PAGE followed by immunoblotting (5).
RESULTS
Keg1 Is Required for Both Folding and Polarized Localization of Kre6
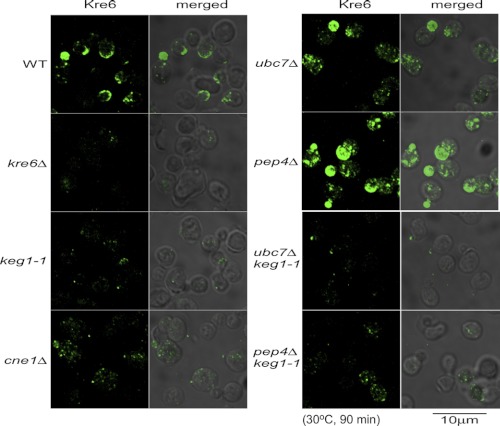
KEG1 encodes a Kre6-binding ER membrane protein, and the temperature-sensitive keg1-1 mutant (H126L substitution) shows a defect in β-1,6-glucan synthesis similar to kre6Δ mutants (13). At first, we sought to observe Kre6 in keg1-1 cells grown at the semi-permissive temperature. When cells were grown at 25 °C to a log phase and then at 30 °C for 90 min, the cell morphology was not normal, and the immunofluorescence signal was extremely weak (Fig. 1, panel keg1-1). Faint polarized dots were found in several cells, but they were much fewer than in the wild-type cells (panel WT).
FIGURE 1.
Indirect immunofluorescence staining images of Kre6 in the cells of mutants that showed alteration in the stability or localization of Kre6. The strains were grown overnight at 25 °C until A600 nm = 0.5 and then shifted to the semi-permissive temperature, 30 °C, and grown for 90 min. The immunofluorescence signals of the wild type (WT) and seven mutant cells using anti-Kre6 antiserum were captured under the same experimental condition (Kre6, left panels). The Nomarski images are merged to show the cells (merged, right panels). The bar indicates 10 μm.
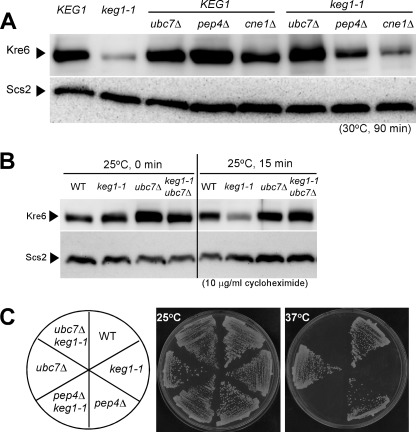
One reason for the faint signal was that the amount of Kre6 was reduced in the keg1-1 cells. The same amounts of total proteins from the wild-type and keg1-1 cells grown at 30 °C for 90 min were analyzed by SDS/PAGE and immunoblotting (Fig. 2A). The keg1-1 cells had less Kre6 than the wild-type KEG1 cells. When grown at 25 °C, the keg1-1 cells had a similar amount of Kre6 to the wild-type cells (Fig. 2B). However, the signal of Kre6 in the keg1-1 mutant decreased more rapidly than in the wild type after protein synthesis was blocked by the addition of cycloheximide (Fig. 2B). This suggests that degradation of Kre6 is accelerated when Keg1 is not fully functional.
FIGURE 2.
The amount and stability of Kre6 protein in various mutant cells and the colony-forming activity of these mutants. A, the strains were grown overnight at 25 °C until A600 nm = 0.5 and then shifted to the semi-permissive temperature, 30 °C, and grown for 90 min. Kre6 protein in the total cell lysate was detected by immunoblotting using anti-Kre6 antiserum. The immunoblotting bands of Scs2 are shown as the loading control. B, stability of Kre6 protein at the permissive 25 °C was examined by immunoblotting after protein synthesis was blocked by adding 10 μg/ml cycloheximide at A600 nm = 0.5. Degradation in the keg1-1 mutant was retarded by ubc7Δ. Scs2 was used as the loading control. C, Colony formation of keg1-1 at 37 °C was not rescued by pep4Δ or ubc7Δ mutation that reduced degradation of Kre6.
To see the degradation system of Kre6, we introduced the null mutant allele of UBC7 or PEP4, which is involved in ER-associated degradation (ERAD) or vacuolar proteolysis, respectively. The amount of Kre6 slightly increased in the pep4Δ keg1-1 double mutant compared with in keg1-1, and the ubc7Δ keg1-1 strain had nearly a similar amount of Kre6 as the wild type at 30 °C (Fig. 2A). As a slight increase in the amount of Kre6 was also observed in the pep4Δ KEG1 strain, stabilization of Kre6 polypeptide in the absence of vacuolar proteolysis is likely not specific in the keg1-1 mutant, and ERAD is responsible for the instability of Kre6 in the keg1-1 strain. The rapid degradation of Kre6 in the keg1-1 cells was also prevented by ubc7Δ mutation (Fig. 2B). These results suggest that proper folding of Kre6 does not occur efficiently in the keg1-1 mutant, and Kre6 in immature conformation is removed by ERAD.
The colony-forming activity at 37 °C was not recovered in the keg1-1 ubc7Δ double mutant (Fig. 2C), which indicates that not the stability of polypeptide, but proper folding of Kre6 with the association of Keg1 is required. The immunofluorescence signal of Kre6 in the cell clearly increased in the ubc7Δ or pep4Δ single disruptant, but the images were distinct from those of the wild type and apparently abnormal (Fig. 1, panels ubc7Δ and pep4Δ). The large spherical structure in the pep4Δ cells is likely to be the vacuole, but the nature of the others is currently unknown. The immunofluorescence images of Kre6 in the ubc7Δ keg1-1 and pep4Δ keg1-1 double mutants were indistinguishable from those in the keg1-1 single mutant (Fig. 1). Polarized signals of Kre6 were not observed. This is consistent with the observation that the colony-forming activity was not suppressed even when the degradation of Kre6 was suppressed (Fig. 2C).
keg1-1 Has Defect in Binding to Kre6
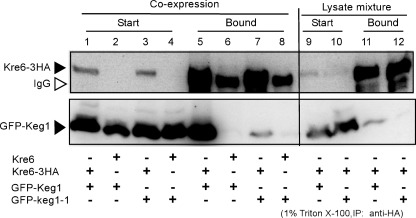
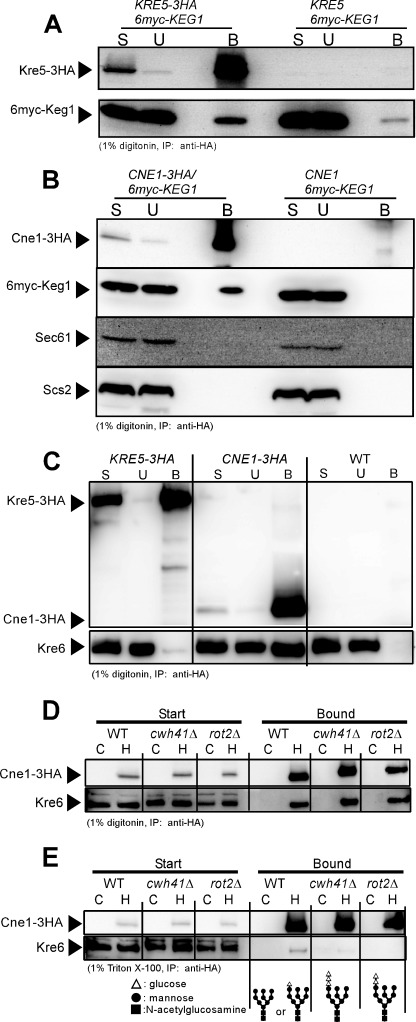
Next, we investigated the effect of keg1-1 mutation in the protein-protein interaction with Kre6. We constructed strains that had Kre6-3HA and either GFP-Keg1 or GFP-keg1-1 proteins. Kre6-3HA was immunoprecipitated from the cell lysate solubilized with 1% Triton X-100, and the presence of GFP-Keg1 and GFP-keg1-1 was examined. Although the amounts of GFP-Keg1 and GFP-keg1-1 were similar in the solubilized samples (Fig. 3, lanes 1–4), the amount of GFP-keg1-1 in the Kre6-3HA immunoprecipitate (lane 7) was much smaller than that of GFP-Keg1 (lane 5), which suggests the amino acid replacement H126L in keg1-1 reduces interaction with Kre6.
FIGURE 3.
Protein-protein interactions between Kre6-3HA and GFP-Keg1 or GFP-keg1-1. Kre6-3HA was immunoprecipitated (IP) by anti-HA monoclonal antibody from the cleared cell lysate containing 1% Triton X-100, and Kre6-3HA and GFP-Keg1 or GFP-keg1-1 in the starting materials (Start) and in the immunoprecipitate (Bound) were detected by immunoblotting. Materials derived from 22.5-fold more cells were loaded for the Bound sample than for the Start sample in SDS-PAGE. Lanes 1 and 5, GFP-KEG1 KRE6-3HA; lanes 2 and 6, GFP-KEG1 KRE6; lanes 3 and 7, GFP-keg1-1 KRE6-3HA; lanes 4 and 8, GFP-keg1-1 KRE6; Lanes 9 and 11, mixture of the lysates of GFP-KEG1 cells and KRE6-3HA cells; lanes 10 and 12, mixture of the lysates of GFP-keg1-1 cells and KRE6-3HA cells. Samples for SDS-PAGE were solubilized at 37 °C because boiling makes Keg1 insoluble. This condition resulted in the appearance of the immunoglobulin (open arrowhead) close to Kre6-3HA.
In addition, we examined if the Keg1-Kre6 interaction has a biological significance. When the Triton X-100 lysates that had either GFP-Keg1 or Kre6-3HA were prepared and then mixed on ice, little co-immunoprecipitation of GFP-Keg1 was found (Fig. 3, lanes 11 and 12) in comparison to in the lysate from the cells that had both proteins (lanes 5 and 7). This indicates that binding of Keg1 and Kre6 requires some biological process in living yeast cells and is not the result of binding by sole affinity between the proteins.
ER-membrane General Chaperon Rot1 Participates in Folding of Kre6
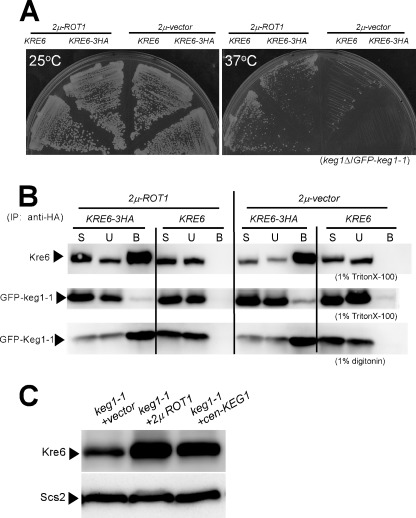
Previously, we found ROT1 as a multicopy suppressor gene of colony formation of keg1-1 mutant at the non-permissive temperature (13). As the suppression of colony formation occurred when keg1-1 was tagged with GFP (Fig. 4A), we examined if more Kre6 binds to keg1-1 in the presence of multicopy ROT1. No increase in the amount of GFP-keg1-1 co-immunoprecipitated with Kre6-3HA was detected in the 1% Triton X-100 lysate even in the presence of the multicopy ROT1 (Fig. 4B). If Triton X-100 was replaced with digitonin that had been reported to have less influence in protein-protein interactions than Triton X-100 (18), similar co-precipitation of Kre6-3HA and GFP-keg1-1 was detected with either single or multicopy ROT1. It is likely that the ability to bind to Kre6 is reduced but not completely damaged in keg1-1 (Fig. 4B).
FIGURE 4.
Role of ROT1 in the interaction of Kre6-keg1-1 and stabilization of Kre6. A, the GFP-keg1-1 cells carrying the chromosomal keg1Δ KRE6 or keg1Δ KRE6-3HA genes in the presence or absence of multicopy ROT1 gene were grown on yeast extract/peptone/dextrose plates at 25 or 37 °C. B, the cleared lysates containing either 1% Triton X-100 or 1% digitonin were prepared from the cells shown in A, and immunoprecipitates (IP) by anti-HA monoclonal antibody were analyzed by immunoblotting using anti-Kre6 or anti-GFP antibodies. Materials derived from 15-fold more cells were loaded for the bound sample (B) than for the start sample (S) and the unbound sample (U) in SDS-PAGE. C, the amount of Kre6 in the keg1-1mutant was examined after introduction of 2μ-ROT1 or CEN-KEG1 at 27.5 °C. The strains were grown overnight at 25 °C until A600 nm = 0.5 and then shifted to 27.5 °C and grown for 90 min.
On the other hand, the amount of Kre6 protein in keg1-1 mutant was restored to the wild-type level in the presence of multicopy ROT1 (Fig. 4C), which is consistent with the report that Rot1 assists the folding of Kre6 (6). As binding of Kre6 and keg1-1 in 1% Triton X-100 was not recovered by multicopy ROT1 (Fig. 4B), the effect of overproduced Rot1 to repair the mutant keg1-1 seems inefficient.
Genetic Interactions between KEG1 and Calnexin Cycle Member Homologue Genes
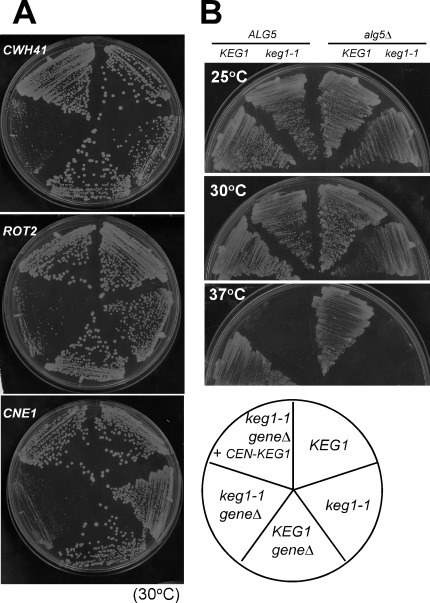
β-1,6-Glucan is decreased in mutants of calnexin cycle member homologues (2, 3, 19). The null alleles of kre6Δ and cwh41Δ that encodes glucosidase I are synthetically lethal. The amount of Kre6 is reduced in the cwh41Δ strain, and Calcofluor White hypersensitivity of cwh41Δ was suppressed by multicopy KRE6 (11). Therefore, it is suggested that the decrease of β-1,6-glucan in the cwh41Δ cells is a secondary effect of the decrease of Kre6. We sought to examine if Keg1 is also concerned in these genetic interactions. We constructed double mutants of calnexin cycle member homologues with keg1-1. All combinations of keg1-1 with cne1Δ, rot2Δ, or cwh41Δ resulted in synthetic growth defects at the semi-permissive temperature of 30 °C (Fig. 5A). This suggests Keg1 and calnexin cycle member homologues have related functions.
FIGURE 5.
Genetic interactions between keg1-1 and null alleles of genes related to β-1,6-glucan. A, shown is colony formation of the cells carrying combinations of keg1-1 and null mutations of calnexin cycle member homologue genes, cwh41Δ (glucosidase I), rot2Δ (glucosidase II), and cne1Δ (calnexin) at the semi-permissive 30 °C on a yeast extract/peptone/dextrose plate. B, shown is colony formation of strains carrying combinations of keg1-1 and alg5Δ that suppress the phenotype of cwh41Δ.
It has been reported that the calnexin cycle does not work in budding yeast (10). However, because the null mutant of KRE5 that encodes UGGT homologue is lethal (12), it should play some essential role in the cell. The synthetic lethality of cwh41Δ kre6Δ was completely suppressed by disruption of ALG5 that encodes dolichol-P-glucose synthase (11). As the decrease of Kre6 in the cwh41Δ strain was recovered by alg5Δ, it is considered that Kre6 escapes from degradation if its N-glycan chains carry no glucose residues by a yet unknown mechanism (11). We examined if the temperature-sensitive growth of keg1-1 mutant is similarly suppressed by alg5Δ. As shown in Fig. 5B, the alg5Δ keg1-1 double mutants grew more slowly, and no suppression was observed.
Physical Interactions of Calnexin Cycle Member Homologues with Kre6 and Keg1
In the calnexin cycle, calnexin recognizes α-1,3-glucose residue added to the N-glycan modification of nascent proteins and binds the immature proteins and supports their folding (7–9). We constructed strains that had a 3HA tag at the C terminus of Cwh41, Rot2, Kre5, and Cne1 and examined their interactions with GFP-Keg1 and Kre6 by immunoprecipitation. As Kre5 has the ER retention signal HDEL at the C terminus, 3HA was inserted in front of it. No co-precipitation of 6myc-Keg1 was detected if the cell lysate was solubilized with 1% Triton X-100 (data not shown). When the detergent was replaced with 1% digitonin, 6myc-Keg1 was detected in the immunoprecipitates of Kre5-3HA and Cne1-3HA (Figs. 6, A and B). As Cne1 is a molecular chaperone homologue, we tested other ER membrane proteins Sec61 and Scs2, but these were not detected in the immunoprecipitate (Fig. 6B). This suggests that Keg1 may associate with Kre5 and Cne1, and the complex may assist the maturation of Kre6. No signals of 6myc-Keg1 or Kre6 were detected in the immunoprecipitate of either Cwh41-3HA or Rot2-3HA (data not shown).
FIGURE 6.
Protein-protein interactions between calnexin cycle member homologues and Keg1 or Kre6. A, immunoblots of the start (S), unbound (U), and bound (B) materials of the immunoprecipitation experiment using anti-HA monoclonal antibody from the cleared cell lysates containing 1% digitonin are shown. Kre5-3HA and 6myc-Keg1 were detected by anti-HA and anti-myc monoclonal antibodies, respectively. Materials derived from 40-fold more cells were loaded for the B sample than for the others in SDS-PAGE. B, similar experiments to A were done to see the interaction between Cne1-3HA and 6myc-Keg1. As Cne1 may be a chaperon, ER membrane proteins Sec61 and Scs2 were also analyzed as the controls. Materials derived from 80-fold more cells were loaded for the B sample than for the others in SDS-PAGE. C, interaction between Kre6 and Kre5-3HA or Cne1-3HA was examined, as in A. Kre6 was detected by anti-Kre6 antiserum. Materials derived from 50-fold more cells were loaded for the B sample than for the others in SDS-PAGE. D, interaction between Cne1-3HA and Kre6 was examined by immunoprecipitation from the cleared cell lysate of calnexin cycle mutants in the presence of 1% digitonin. Lanes C and H indicate that the cell lysates were prepared from the wild-type CNE1 and HA-tagged CNE1-3HA strains, respectively. Materials derived from 30-fold more cells were loaded for the Bound sample than for the Start sample in SDS-PAGE. E, similar experiments to D were done by replacing digitonin with Triton X-100. Materials derived from 190-fold more cells were loaded for the Bound sample than for the Start sample in SDS-PAGE. The expected molecular structures of N-glycan in the strains used are shown at the bottom.
Fig. 6C shows that in the 1% digitonin lysate, Kre5 and Kre6 were not co-precipitated, but a significant amount of Kre6 was found in the precipitate of Cne1. This Kre6-Cne1 interaction was also observed in the absence of glucosidase I (cwh41Δ) and glucosidase II (rot2Δ), i.e. in the presence of 2 or 3 glucose residues on the N-glycan in 1% digitonin (Fig. 6D), which is not consistent with the general rule that calnexin recognizes N-glycan with single glucose. The structures of N-glycan are illustrated at the bottom of Fig. 6E. When digitonin was replaced with Triton X-100, co-precipitation of Kre6 and Cne1 was not observed in cwh41Δ or rot2Δ mutants even if 6-fold more samples than routine experiments were loaded to the gel. In contrast, a weak but significant signal of Kre6 was detected in the wild type sample (Fig. 6E). The same results were obtained in four independent experiments, which indicates that the number of glucose residues has some effect on the interaction of Kre6 and Cne1. This is consistent with the general binding rule of calnexin that recognizes single glucose on the N-glycan. Therefore, Kre6 is the first candidate of the substrate of calnexin-homologue Cne1, although the calnexin cycle is considered to not work in S. cerevisiae (10). The amount of Kre6 was not reduced in the cne1Δ strain, and no significant difference was found between the keg1-1 single and keg1-1 cne1Δ double mutants (Fig. 2A). Therefore, no folding defect of Kre6 susceptible to ERAD is likely to occur in the cne1Δ mutant.
Localization of Kre6 in Calnexin Homologue Mutant
Indirect immunofluorescence staining microscopy of Kre6 revealed that its polarized signals were not observed in the cne1Δ cells (Fig. 1, panel cne1Δ), although a similar amount of Kre6 as the wild type was found (Fig. 2A). This suggests that the defect of β-1,6-glucan in the cne1Δ mutant is caused by a defect of localization of Kre6 in the growing PM.
Skn1 Localizes at Polarized Growing Region of Cell as Kre6 Does in Absence of Kre6
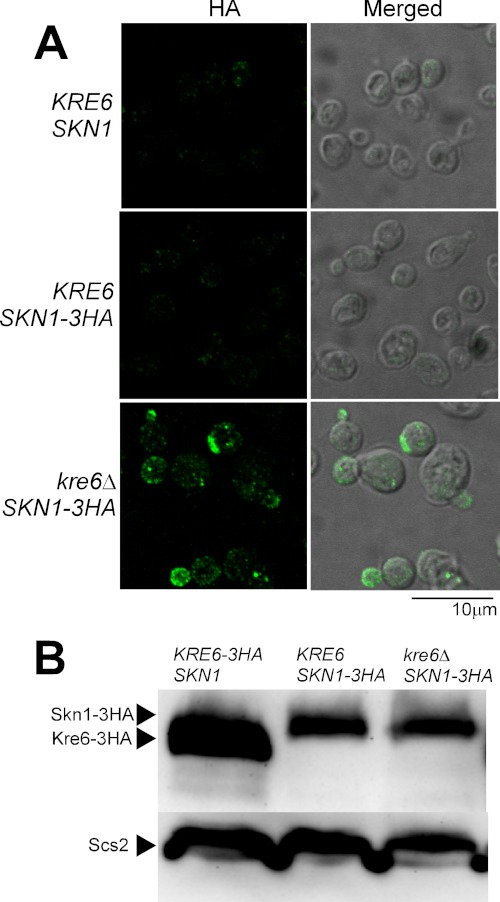
Next, we examined the localization of Kre6-homologue Skn1 to address its predicted roles in β-1,6-glucan synthesis. SKN1 was discovered as a suppressor gene of the kre6Δ phenotypes, K1 killer toxin resistance, and slow growth defect (15). Skn1 has a high amino acid sequence identity with Kre6; 63% as a whole and 86% in the C-terminal lumenal domain that has homology to family 16 glycoside hydrolase (15, 16). As the kre6Δ skn1Δ double null mutant is lethal, these proteins should have a duplicated essential function. As the skn1Δ single mutant shows no apparent phenotypes, Kre6 usually plays a major role. In our previous report we showed that the majority of Kre6 is in the ER, but a significant portion moves to the secretary vesicles and PM at the growing region of the cells, such as the bud tips. It should be emphasized that this appearance in the growing PM is essential for the function of Kre6 in β-1,6-glucan synthesis (5). Although Skn1 is expected to have a similar characteristic, no experimental data are available, except that a tagged Skn1 has been detected by immunoblotting (17). Therefore, we sought to determine its cellular localization. The position and kind of tags as well as the production rate had a large influence on the localization of Kre6. As the tagging of 3HA at the C terminus of Kre6 encoded on the chromosomal gene had similar indirect immunofluorescence images as those stained by anti-Kre6 antiserum, we constructed a strain with a sole chromosomal SKN1 gene tagged with 3HA at the C terminus.
The immunofluorescence image of the KRE6 SKN1-3HA cells was similar to the image of the wild-type cells without HA used as the background control (Fig. 7A). However, when KRE6 was replaced with the kre6Δ null allele, a number of dots of HA signal was detected in the growing regions of the cell, especially in the small buds. These images are quite similar to those of Kre6 in the wild-type cells (Fig. 1, panel WT). Therefore, Skn1 is likely to fulfill the loss of Kre6 not only in its function but also in its localization.
FIGURE 7.
Immunofluorescence images and amounts of Skn1-3HA protein in the presence or absence of its homologue Kre6. A, the epitope-tagged Skn1-3HA was expressed from the original chromosomal SKN1 locus in either KRE6 or kre6Δ background and detected by indirect immunofluorescence staining with mouse anti-HA monoclonal antibody (HA, left panels). The Nomarski images are merged to show the cells (Merged, right panels). The images of wild-type KRE6 SKN1 cells were included to show the background signals without HA epitope. The bar indicates 10 μm. B, the same amount of lysates from KRE6-3HA SKN1, KRE6 SKN1-3HA, or kre6Δ SKN1-3HA cells were subjected to SDS-PAGE and immunoblotting using anti-HA antibody. The immunoblotting bands of Scs2 are the loading control.
The amounts of Skn1-3HA estimated from the signal intensity of immunoblotting were similar in the presence and absence of KRE6 (Fig. 7B). This is consistent with the report that the amount of SKN1 mRNA does not change by the disruption of KRE6 (15). The amount of Kre6-3HA was larger than that of Skn1-3HA when detected by the same anti-HA antibody.
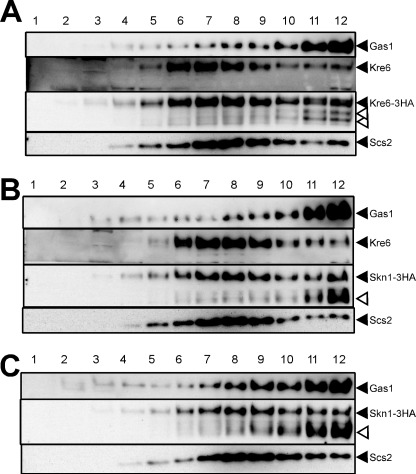
Intracellular localization was further analyzed by fractionation of the cell lysate by sucrose density gradient fractionation in the presence of 10 mm EDTA. To check the effect of 3HA tagging, fractionation of Kre6-3HA was done as a control (Fig. 8A). As we previously reported about the intrinsic untagged Kre6, a majority of Kre6-3HA was in fractions 7–10 as the ER-marker Scs2, and a significant amount was also found in fractions 11–12 in which PM-marker Gas1 had the peak. If Kre6-3HA was detected by immunoblotting using anti-HA antibody, two minor bands (open arrowheads in Fig. 8A) were detected in front of the major band (filled arrowhead). As these bands were not detected by the anti-Kre6 antiserum that was raised using the N-terminal peptide of 84 amino acids, they were likely degradation products losing their N-terminal regions. In contrast to the absence of signals in immunofluorescence microscopy, Skn1-3HA was clearly detected by cell fractionation and immunoblotting even in the presence of KRE6 (Fig. 8B). The distribution of Skn1-3HA was similar to that of Kre6-3HA, but the amount of possible degradation products was more abundant than in the case of Kre6-3HA, especially in fractions 11–12 (Fig. 8B, open arrowhead). As no bands of degraded polypeptides were detected when the cell lysate was directly subjected to SDS-PAGE and immunoblotting (Fig. 7B), degradation should have occurred during the cell fractionation despite the addition of various protease inhibitors. If the degradation products are included, similar amounts of Skn1-3HA are apparently present in the ER and PM fractions. In the kre6Δ cells, Skn1-3HA was similarly distributed (Fig. 8C). These results indicate that similar amounts of Skn1-3HA are present in the ER and PM in the presence or absence of Kre6.
FIGURE 8.
Subcellular localization of Kre6-3HA and Skn1-3HA. A, the lysate of cells carrying chromosomal KRE6-3HA at the original KRE6 locus (KTY284) was fractionated by sucrose density gradient centrifugation in the presence of 10 mm EDTA. Aliquots of fraction 1 (top) to 12 (bottom) were analyzed by SDS-PAGE, and proteins of interest were detected by immunoblotting using the antibodies, as shown on the right with filled triangles. Kre6-3HA was detected either by anti-Kre6 antibodies (Kre6) or by anti-HA monoclonal antibody (Kre6-3HA). Open arrowheads indicate bands of possible degradation products detected by anti-HA antibody. B, the lysate of cells carrying chromosomal SKN1-3HA at the original SKN1 locus (KTY432) was fractionated, and proteins of interest were detected by immunoblotting, as in A. C, the lysate of SKN1-3HA kre6Δ cells (KTY209) was fractionated and analyzed, as in B.
Skn1 Also Binds to Keg1 as Kre6 Does
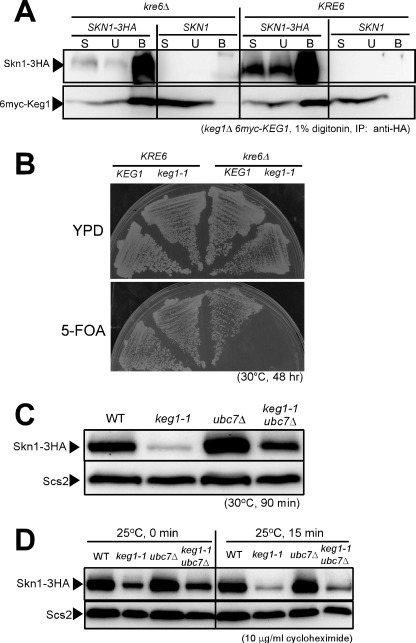
The ER-resident essential membrane protein Keg1 is co-immunoprecipitated with Kre6 when membranes are solubilized with 1% Triton X-100 (13). We did similar experiments to detect an interaction between Skn1-3HA and 6myc-Keg1, but no signal of 6myc-Keg1 was detected in the precipitate of Skn1-3HA (data not shown). We repeated the experiments after replacing Triton X-100 with digitonin. Then, a signal of 6myc-Keg1 was detected in the precipitate of Skn1-3HA. The signal intensity was the same whether the cell had either the KRE6 or kre6Δ allele (Fig. 9A). These results indicate both Kre6 and Skn1 bind to Keg1, in a non-competitive manner, although Kre6-Keg1 has stronger interaction than Skn1-Keg1 in respect to the influence of detergents.
FIGURE 9.
Protein-protein interaction between Keg1 and Skn1, synthetic lethality of keg1-1 and kre6Δ, and stability of Skn1 protein in keg1-1 mutant cells. A, detection of 6myc-Keg1 in the immunoprecipitate (IP) of Skn1-3HA from the cleared cell lysate containing 1% digitonin is shown. Materials derived from 25-fold more cells were loaded for the bound sample (B) than for the start sample (S) and the unbound sample (U) in SDS-PAGE. B, the cells of KRE6 KEG1, KRE6 keg1-1, kre6Δ KEG1, and kre6Δ keg1-1, harboring the removable plasmid that complements keg1-1 (CEN URA3 KEG1), were grown on either yeast extract/peptone/dextrose (YPD) or 5-FOA plates at the semi-permissive temperature of 30 °C for 48 h. C, the strains were grown overnight at 25 °C until A600 nm = 0.5 and then shifted to the semi-permissive temperature, 30 °C, and grown for 90 min. Skn1-3HA protein in the total cell lysate was detected by immunoblotting using anti-HA monoclonal antibody. The immunoblotting of Scs2 is shown as the loading control. D, stability of Skn1-3HA protein at the permissive 25 °C was examined by immunoblotting after protein synthesis was blocked by adding 10 μg/ml cycloheximide at A600 nm = 0.5. Scs2 was used as the loading control.
Keg1 Is Also Required for Proper Folding of Skn1
ER membrane protein Keg1 likely helps the folding of Kre6 that binds to Keg1, as described in this paper. To reveal whether Keg1 has a similar role on Skn1, we examined the effect of the replacement of KEG1 with the keg1-1 allele in kre6Δ SKN1cells. A KEG1 URA3 CEN plasmid that is removable by 5-FOA was introduced in the keg1-1mutant, and its KRE6 was replaced with kre6Δ::kanMX4 by homologous recombination. This and the control strain were grown on 5-FOA plates (Fig. 9B). At the semi-permissive temperature for keg1-1 (30 °C), the KRE6 SKN1 keg1-1 strain formed colonies, but the kre6Δ SKN1 keg1-1 strain did not when KEG1 was removed by growing on 5-FOA plates. As the kre6Δ SKN1 KEG1 strain had colony-forming activity, Skn1 can fulfill the loss of Kre6 if Keg1 is active. The synthetic lethality of keg1-1 and kre6Δ is consistent with the idea that Keg1 is also required for Skn1 as for Kre6.
Next, we examined the role of Keg1 on the amount of Skn1-3HA in the cell. As shown in Fig. 9C, the signal of Skn1-3HA was much weaker in the keg1-1 mutant than in the wild type when grown at 30 °C for 90 min, as in the case of Kre6 (Fig. 2A). Also, the signal intensity was significantly recovered by the introduction of ubc7Δ mutation. Skn1-3HA was more rapidly degraded in the keg1-1 mutant than in the wild type after the addition of a protein synthesis inhibitor, and this degradation was partly prevented by the ubc7Δ mutation (Fig. 9D). These results indicate ERAD is also responsible for degradation of Skn1-3HA in the keg1-1 mutant, although the suppression of degradation by the ubc7Δ mutation seems somewhat less effective than in the case of Kre6.
DISCUSSION
The process of cell wall β-1,6-glucan synthesis is one of the most mysterious problems in yeast cell biology. It is still uncertain where it is actually synthesized and which enzyme directly catalyzes the reaction. Kre6 is believed to be a key protein because it has the structural motif of glycoside hydrolase (4), and its loss results in a severe defect in β-1,6-glucan synthesis (3). As Skn1 and Kre6 have high amino acid sequence identity and their genes are synthetically lethal, they are considered to share an essential functional role (15). We reported that a part of Kre6 is delivered to the growing PM, and this polarized deposition of Kre6 is essential for β-1,6-glucan synthesis (5).
If Skn1 has a same function as Kre6, it is not unusual to expect that Skn1 would show the same localization as Kre6 in the cell. However, no indirect immunofluorescence staining signals of Skn1-3HA was observed in the wild type. However, in the kre6Δ disruptant, similar images to Kre6 were obtained (Fig. 7A). As similar amounts of Skn1-3HA were detected in the kre6Δ and KRE6 cells by immunoblotting (Fig. 7B), we first speculated that Skn1-3HA is localized only in the ER where Kre6 is also not detectable by indirect immunofluorescence in the presence of Kre6, but it can move to the growing PM regions in the absence of Kre6. Fractionation of cells by sucrose density gradient centrifugation indicated that similar amounts of Skn1-3HA are present in the fractions corresponding to the ER and PM in the presence and absence of Kre6. Possible degradation products of Skn1-3HA and Kre6-3HA were found in heavier fractions, especially in fractions 11–12. These bands with the 3HA epitope should have been produced by proteolysis during cell fractionation, because they were not detected if the cell lysate was directly subjected to SDS-PAGE. The PM fractions are likely to have more proteolytic activity that digests the N-terminal regions of Kre6 and Skn1 than other fractions. Although the mechanism is currently unclear, these proteins are not detected by standard methods of indirect immunofluorescence staining. We suspect some interacting molecules prevent the approach of antibodies to these undetectable proteins, as already discussed in the previous paper (5). If so, it should be helpful to develop a method to disrupt the masking structures during the process of indirect immunofluorescence staining for future analysis. Alternatively, the local concentration of antigens may influence the staining intensity of immunofluorescence. In the case of Skn1-3HA on the PM, the lack of immunofluorescence signal may be explained if Skn1-3HA is dispersed in the PM in the presence of Kre6, but it becomes concentrated in the buds in the absence of Kre6.
We have shown that the N-terminal cytoplasmic region is responsible for the localization of Kre6 at the polarized growing regions (5). Li et al. (16) reported that the N-terminal fragment of Kre6 binds to Las17 and Sla1 on the PM, and if this fragment is replaced with that of Skn1, the chimera Skn1-Kre6 does not complement the phenotype of Δkre6. These findings indicate that intracellular distribution of Kre6 and Skn1 could be different, although their lumenal domains have high amino acid sequence identity and probably catalyze similar reactions.
Kar2/BiP is an essential molecular chaperon in the ER lumen. Takeuchi et al. (20) screened for synthetically lethal genes with kar2-1 and found mutations in KRE6, SKN1, and ROT1. The essential ER membrane protein Rot1 binds to Kar2, and their in vitro anti-aggregation assay using multiple denatured proteins indicated that Rot1 is also a general chaperon as Kar2 (6). In a temperature-sensitive rot1-2 (G45E) mutant, Kre6 showed a defect in folding and glycosylation and was susceptible to ERAD (6). These findings indicate that both soluble chaperon Kar2 and membrane chaperon Rot1 are concerned in the folding of Kre6. Additionally, the essential ER membrane protein Keg1 binds to Kre6 and Skn1 in a biologically significant manner, and folding of Kre6 and Skn1 is defective in the keg1-1 mutant where these proteins are susceptible to ERAD. Deposition of Kre6 in the growing regions like bud tips, which was detected by immunofluorescence, was not observed in this mutant. We could not examine Skn1-3HA in the keg1-1 mutants by immunofluorescence because kre6Δ and keg1-1 were synthetically lethal (Fig. 9B). It is likely that Keg1 plays an essential role responsible for the folding in the ER and deposition to the PM of Kre6 and possibly Skn1.
As we described in the Introduction, the calnexin cycle is highly conserved in the eukaryote, but it is not considered to be functional as a protein folding system in budding yeast (10) and instead is required for β-1,6-glucan synthesis (2, 3, 19). The activities of Cwh41 (glucosidase I) and Rot2 (glucosidase II) were demonstrated (19, 21). Cne1 (calnexin) binds to Glc1Man9 oligosaccharide and has in vitro protein-folding activity (22). CNE1 is also a multicopy suppressor of rot1ts phenotype (20). Kre6 has five possible N-glycosylation sites (374NGT, 461NQS, 538NFT, 563NVT, and 691NLT) predicted by the amino acid sequence of the C-terminal lumenal region, and the presence of glycosylated Kre6 protein has been demonstrated previously (6, 17), although it is not known which of the predicted sites are actually glycosylated. Under “Results” we demonstrated that Cne1 binds to Kre6 in a similar manner as calnexin, which prefers single glucosylated N-glycan in the 1% Triton X-100 lysate, although the signal was very weak (Fig. 6E). This is the first case that has suggested preference of the number of glucose residues for binding, although a similar amount of Kre6 was co-precipitated with Cne1-3HA from the 1% digitonin lysates of the glucosidase-deficient mutants and wild type (Fig. 6D). As Kre6 was not localized in the growing buds in cne1Δ cells (Fig. 1), Cne1 may be concerned in the deposition of Kre6 to the growing PM regions.
Mutants of KRE5 show the most severe defect in β-1,6-glucan synthesis among the calnexin cycle member homologue mutants. Although Kre5 has a similarity to UGGTs, it is in the fungal subgroup distinct from the major UGGTs in the phylogenetic relationships (23), and its enzymatic activity has not yet been detected (10). Castro et al. (24) reported that a KRE5 homologue of Schizosaccharomyces pombe had UGGT activity, but it could not rescue the lethality of S. cerevisiae kre5Δ null mutation. From these facts, it is proposed that Kre5 may have a different activity from the classical UGGT that adds single glucose from UDP-glucose to N-glycan of malfolded polypeptides, and it may make a nascent β-1,6-glucan chain on N-glycan or glycosylphosphatidylinositol anchor of hypothetical carrier proteins in the secretary pathway (2). If Kre5 is the β-1,6-glucan synthase that adds glucose to yet uncharacterized primer molecules and Kre6 and Skn1 are the transglycosidases that make longer glucan chains, regulation of their activity and cellular localization should be very important. In this report the ER-membrane protein Keg1 was co-precipitated with Kre5, Kre6, Skn1, and Cne1 from the digitonin lysate. Cne1 was also co-precipitated with Kre6, and the polarized localization of Kre6 was not observed in the keg1-1 and cne1Δ mutants. These protein-protein interactions or a temporary complex in the ER may play roles in depositing of nascent glucan-bound primer molecules to the PM. Further analysis of these interrelated proteins will reveal the mechanism of fungal β-1,6-glucan synthesis.
Acknowledgment
We thank Dr. Satoshi Kagiwada for anti-Scs2 antiserum.
This work was supported by a grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science (to Y. N. and K. Y.) and a grant from the Noda Institute of Scientific Research (to Y. N.).
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- PM
- plasma membrane
- UGGT
- UDP-glucose:glycoprotein glucosyltransferase
- 5-FOA
- 5-fluoroorotic acid.
REFERENCES
- 1. Lesage G., Bussey H. (2006) Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shahinian S., Bussey H. (2000) β-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 35, 477–489 [DOI] [PubMed] [Google Scholar]
- 3. Pagé N., Gérard-Vincent M., Ménard P., Beaulieu M., Azuma M., Dijkgraaf G. J., Li H., Marcoux J., Nguyen T., Dowse T., Sdicu A. M., Bussey H. (2003) A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163, 875–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Montijn R. C., Vink E., Müller W. H., Verkleij A. J., Van Den Ende H., Henrissat B., Klis F. M. (1999) Localization of synthesis of β1,6-glucan in Saccharomyces cerevisiae. J. Bacteriol. 181, 7414–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kurita T., Noda Y., Takagi T., Osumi M., Yoda K. (2011) Kre6 protein essential for yeast cell wall β-1,6-glucan synthesis accumulates at sites of polarized growth. J. Biol. Chem. 286, 7429–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takeuchi M., Kimata Y., Kohno K. (2008) Saccharomyces cerevisiae Rot1 is an essential molecular chaperone in the endoplasmic reticulum. Mol. Biol. Cell 19, 3514–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) N-glycan structures. Recognition and processing in the ER. Trends. Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 8. Caramelo J. J., Parodi A. J. (2008) Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283, 10221–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lederkremer G. Z. (2009) Glycoprotein folding, quality control and ER-associated degradation. Curr. Opin. Struct. Biol. 19, 515–523 [DOI] [PubMed] [Google Scholar]
- 10. Fernández F. S., Trombetta S. E., Hellman U., Parodi A. J. (1994) Purification to homogeneity of UDP-glucose:glycoprotein glucosyltransferase from Schizosaccharomyces pombe and apparent absence of the enzyme fro Saccharomyces cerevisiae. J. Biol. Chem. 269, 30701–30706 [PubMed] [Google Scholar]
- 11. Abeijon C., Chen L. Y. (1998) The role of glucosidase I (Cwh41p) in the biosynthesis of cell wall β-1,6-glucan is indirect. Mol. Biol. Cell 9, 2729–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levinson J. N., Shahinian S., Sdicu A. M., Tessier D. C., Bussey H. (2002) Functional, comparative and cell biological analysis of Saccharomyces cerevisiae Kre5p. Yeast 19, 1243–1259 [DOI] [PubMed] [Google Scholar]
- 13. Nakamata K., Kurita T., Bhuiyan M. S., Sato K., Noda Y., Yoda K. (2007) KEG1/YFR042w encodes a novel Kre6 binding endoplasmic reticulum membrane protein responsible for β-1,6-glucan synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282, 34315–34324 [DOI] [PubMed] [Google Scholar]
- 14. Sato K., Noda Y., Yoda K. (2009) Kei1. A novel subunit of inositolphosphorylceramide synthase, essential for its enzyme activity and Golgi localization. Mol. Biol. Cell 20, 4444–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roemer T., Delaney S., Bussey H. (1993) SKN1 and KRE6 define a pair of functional homologs encoding putative membrane proteins involved in β-glucan synthesis. Mol. Cell. Biol. 13, 4039–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H., Pagé N., Bussey H. (2002) Actin patch assembly proteins Las17p and Sla1p restrict cell wall growth to daughter cells and interact with cis-Golgi protein Kre6p. Yeast 19, 1097–1112 [DOI] [PubMed] [Google Scholar]
- 17. Roemer T., Paravicini G., Payton M. A., Bussey H. (1994) Characterization of the yeast (1→6)-β-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J. Cell Biol. 127, 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vallée B., Riezman H. (2005) Lip1p. A novel subunit of acyl-CoA ceramide synthase. EMBO J. 24, 730–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shahinian S., Dijkgraaf G. J., Sdicu A. M., Thomas D. Y., Jakob C. A., Aebi M., Bussey H. (1998) Involvement of protein N-glycosyl chain glucosylation and processing in the biosynthesis of cell wall β-1,6-glucan of Saccharomyces cerevisiae. Genetics 149, 843–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeuchi M., Kimata Y., Hirata A., Oka M., Kohno K. (2006) Saccharomyces cerevisiae Rot1p is an ER-localized membrane protein that may function with BiP/Kar2p in protein folding. J. Biochem. 139, 597–605 [DOI] [PubMed] [Google Scholar]
- 21. Simons J. F., Ebersold M., Helenius A. (1998) Cell wall 1,6-β-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17, 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu X., Azakami H., Kato A., (2004) P-domain and lectin site are involved in the chaperone function of Saccharomyces cerevisiae calnexin homologue. FEBS Lett. 570, 155–160 [DOI] [PubMed] [Google Scholar]
- 23. Herrero A. B., Magnelli P., Mansour M. K., Levitz S. M., Bussey H., Abeijon C. (2004) KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryot. Cell 3, 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castro O., Chen L. Y., Parodi A. J., Abeijón C. (1999) Uridine diphosphate-glucose transport into the endoplasmic reticulum of Saccharomyces cerevisiae. In vivo and in vitro evidence. Mol. Biol. Cell 10, 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]