Background: Coordinated transcription and splicing occurs at the periphery of speckles.
Results: The FF4 and FF5 domains of transcription elongation regulator 1 (TCERG1) form a structural unit that directs proteins to the periphery of speckles.
Conclusion: The FF4 and FF5 domains constitute a novel speckle periphery-targeting signal.
Significance: This speckle periphery-targeting signal might participate in the coordination of transcription and splicing.
Keywords: Alternative Splicing, Nucleus, Protein Targeting, RNA, Transcription Elongation Factors, Speckle, Targeting Signal
Abstract
Transcription elongation regulator 1 (TCERG1) is a human factor implicated in interactions with the spliceosome as a coupler of transcription and splicing. The protein is highly concentrated at the interface between speckles (the compartments enriched in splicing factors) and nearby transcription sites. Here, we identified the FF4 and FF5 domains of TCERG1 as the amino acid sequences required to direct this protein to the periphery of nuclear speckles, where coordinated transcription/RNA processing events occur. Consistent with our localization data, we observed that the FF4 and FF5 pair is required to fold in solution, thus suggesting that the pair forms a functional unit. When added to heterologous proteins, the FF4-FF5 pair is capable of targeting the resulting fusion protein to speckles. This represents, to our knowledge, the first description of a targeting signal for the localization of proteins to sites peripheral to speckled domains. Moreover, this “speckle periphery-targeting signal” contributes to the regulation of alternative splicing decisions of a reporter pre-mRNA in vivo.
Introduction
The mammalian cell nucleus is a highly dynamic organelle that contains numerous morphologically defined structures, some of which have been implicated in essential processes such as RNA biogenesis. Nuclear speckles are one of these nuclear bodies, and studies on their composition, structure, and behavior have provided useful information for understanding the functional compartmentalization of the cell nucleus (1). Speckles are enriched in pre-mRNA splicing factors and are located in the interchromatin region of the nucleoplasm of mammalian cells (2–4). They appear as 20–50 irregular regions per mammalian nucleus that are generally defined by immunofluorescence staining of RNA-processing factors, such as the serine/arginine-rich (SR)3 splicing factor SC35 (5). Speckles are dynamic structures, and they become round and increase in size upon transcriptional or splicing inhibition (6). It is believed that speckles are storage/assembly sites for splicing components and that transcription and pre-mRNA splicing do not occur within these structures. However, a significant proportion of RNA polymerase II-mediated transcription in the cell nucleus is associated with the periphery of speckles (7–9). Nascent transcript formation near the speckle compartment results in the recruitment of splicing factors from these nuclear bodies to the processing site, and this exchange rate might be regulated by continuous phosphorylation and dephosphorylation events. Thus, phosphorylation of SR proteins is necessary for their recruitment from nuclear speckles to sites of transcription in vivo (10). In summary, speckles appear to modulate the relative concentration of processing factors at active transcription sites, thus acting as an architectural integrator of the dynamic molecular associations that are involved in the coordination of transcription and RNA processing.
Although significant progress has been made on the role of speckles in gene expression, little is known about the sequence motifs responsible for the accumulation of splicing factors at the speckle region. In the case of the SR family of proteins, the RNA recognition motif (RRM) and the RS domain direct these splicing factors to the nuclear speckles (11–14). Other regions of specific splicing factors can also act as targeting signals to nuclear speckles, such as the threonine-proline repeats found in SF3b155 (15) and the arginine-, proline-, and serine-rich domains of SRm160 (16). In the case of protein kinases CrkRS and DYRK1A, the RS domain and a histidine-rich region, respectively, are required for localization to speckles (17–19). To date, no localization signal has been clearly defined to target proteins to the interface between speckles and surrounding transcription sites.
TCERG1 participates in transcriptional elongation and alternative splicing of pre-mRNAs, and a role for this protein in coordinating both processes has been proposed (20, 21). TCERG1 is composed of 1098 residues (22) and contains three WW domains at its N terminus followed by six FF domains at its C terminus. TCERG1 was first described as a transcriptional elongation regulator and was initially found in HIV-1 Tat-responsive HeLa nuclear extract fractions (22, 23). However, accumulating evidence indicates a potential role of TCERG1 in splicing and, hence, in the coupling between transcription and splicing. TCERG1 affects the alternative pre-mRNA splicing of β-globin, β-tropomyosin, CD44, and fibronectin splicing reporters (24–27) and of putative cellular targets identified by microarray analysis following TCERG1 knockdown (26). Consistent with a potential role in the coupling of transcription and splicing, TCERG1 localizes at the interface of splicing factor-rich nuclear speckles and what are presumably nearby transcription sites (21), and it associates with RNA polymerase II and with elongation and splicing components (21, 24, 28, 29).
In this study, we identified the FF4 and FF5 domains of TCERG1 as the region required to direct this protein to the periphery of nuclear speckles. We performed NMR-based analyses and observed that although the FF4 domain is folded and stable, the FF5 domain is not. However, when both domains are expressed as a pair, the folded properties of FF5 are improved. These observations suggest that both domains form a functional unit and provide insights into the nature of FF protein domains. Moreover, our data demonstrate that both of these FF domains specifically direct the localization of fused unrelated proteins to these nuclear regions. Therefore, we defined the FF4 and FF5 domains as novel targeting signals for the localization of proteins at the interface between speckles and what are presumably nearby transcription sites. Placing our data in a functional context, this “speckle periphery-targeting sequence” contributes to the regulation of alternative splicing decisions of a reporter pre-mRNA in vivo.
EXPERIMENTAL PROCEDURES
Plasmids
The intermediate vector pEF-ECFP and eukaryotic expression plasmid pEFBOS/ECFP/T7-TCERG1(1–1098) have been described previously (27). pEFBOS/ECFP/T7-TCERG1(1–662) was generated using EcoRI fragments obtained from the pEFBOST7-TCERG1 parental vector (23) by standard cloning procedures. pEFBOS/ECFP/T7-TCERG1(1–662)-FF mutants were constructed by inserting appropriate FF repeat domain sequences into the BstBI-digested pEFBOS/ECFP/T7-TCERG1(1–662) vector. The enhanced green fluorescent protein expression vectors pEGFP-FF5 and pEGFP-FF4/FF5 were obtained by inserting appropriate PCR-amplified products into the XhoI and XhoI/BamHI sites of pEGFP-C1 (Invitrogen), respectively. pEFBOS/ECFP/T7-TCERG1ΔFF4/FF5 was generated by cloning two PCR products encompassing FF1 to FF3 and FF6 sequences, respectively, into the pEFBOS/ECFP/T7-TCERG1 (1–662) plasmid by standard cloning procedures. pEFBOS/GFP/T7-TCERG1ΔFF5 was kindly provided by Mariano Garcia-Blanco (Duke University Medical Centre, Durham, NC). The phenylalanine-to-alanine mutants (F903A, F961A, F903A,F961A, F946A, and F903A,F946A) were created by mutagenesis into pEFBOS/ECFP/T7-TCERG1(1–662)-FF4/FF5 vector.
The wild-type and mutant SRSF1 expression vectors (SRSF1, RRM1, and RRM2) were described previously (14) and kindly provided by Javier F. Cáceres (MRC, Edinburgh). RRM1 and RRM2 fragments with XbaI and BamHI/XhoI sites were obtained by PCR and inserted into the same epitope-tagged expression vector as wild-type SRSF1 (pCGT7-SF2), yielding pCGT7-RRM1 and pCGT7RRM2 intermediate vectors. The RRM mutants containing FF repeat domains were created using appropriate XhoI/BglII fragments obtained by PCR from pEFBOS-ECFP-T7-TCERG1 (1–1098) and inserted into XhoI/BamHI-digested pCGT7-RRM vectors.
All constructs were verified by DNA sequencing. Plasmids were transformed into DH5α cells for selection.
Proteins
Constructs of the FF4 (878–956 fragment) and for the pair of FF4-FF5 domains (878–1022 fragment) of TCERG1 were amplified by PCR and subcloned into a pETM-11 (a gift from the European Molecular Biology Laboratory-Heidelberg Protein Expression Facility) using NcoI and HindIII sites. All constructs were confirmed by sequencing.
Unlabeled, 15N-labeled, 13C, 15N and 2H, 13C, and 15N-labeled proteins were expressed in Escherichia coli BL21 (DE3) in Luria Broth medium or minimal medium (M9) using either H2O or D2O (99.89%, CortecNet) enriched with 15NH4Cl and/or D-[13C] glucose as the sole sources of carbon and nitrogen, respectively (30). E. coli extracts were lysed using an EmulsiFlex-C5 (Avestin) cell disrupter equipped with an in-house developed Peltier temperature controller system. Soluble fusion proteins were purified by nickel affinity chromatography (HiTrap chelating High Performance column, GE Healthcare), and samples were eluted using buffer (20 mm Tris, 10 mm Imidazol, 150 mm NaCl) with EDTA. After nickel affinity purification, the proteins were cut with the Tobacco Etch Virus (TEV) protease and further purified by gel filtration on a HiLoadTM SuperdexTM 75 prepgrade (GE Healthcare). All samples were prepared in 20 mm sodium phosphate buffer, 130 mm NaCl, 0.5 mm NaN3 in 90% H2O, 10% D2O or 100% D2O (pH 5.8). To avoid aggregation of the FF4-FF5 sample, the buffer was supplemented with 5% glycerol.
Antibodies
Antibodies against the T7 tag were purchased from Bethyl and used at dilutions of 1:20,000 and 1:1000 for immunoblotting and immunofluorescence, respectively. Antibody against CDK9 (catalog no. sc-484, Santa Cruz Biotechnology) was used at a dilution of 1:500. Antibodies against U2AF65 were kindly provided by J. Valcárcel (CRG, Barcelona, Spain) and were used at a dilution of 1:500. Antibody against splicing factor SC35 (catalog no. S4045, Sigma) was used at a dilution of 1:4000. For Western blot analysis, primary antibodies were detected using HRP-conjugated secondary antibodies to rabbit and mouse (PerkinElmer Life Sciences). These were generally used at dilutions of 1:5000. For immunofluorescence, we used Alexa Fluor 488-conjugated goat anti-mouse and Alexa Fluor 647-conjugated goat anti-rabbit from Molecular Probes and were generally used at dilutions of 1:500.
Cell Culture and Transfection Assays
HeLa and HEK293T cells were grown and maintained as described previously (27). Transfection assays were carried out using protocols described previously (21, 27). Cells were transfected by using calcium phosphate and/or Lipofectamine 2000 reagent (Invitrogen) according to the protocols of the manufacturer. Empty vector was used to keep the total amount of nucleic acid constant.
Immunofluorescence, Image Processing, and Quantification
HeLa and HEK293T cells were grown on coverslips and transfected with the constructs indicated in the legends to the figures. Approximately 24 h after transfection (50–60% confluence), cells were fixed with 3.5% paraformaldehyde in PBS buffer (pH 7.4) for 45 min on ice. Cells were washed three times with PBS, permeabilized in PBS containing 0.5% Triton X-100 for 5 min at room temperature, and blocked in PBS containing 2.5% BSA overnight at 4 °C. Cells were incubated with primary antibodies at appropriate dilutions in PBS containing 0.1% BSA for 1 h at room temperature (humidity chamber) and subsequently washed extensively with 0.1% BSA in PBS and incubated with appropriate secondary antibodies under the conditions described previously. After staining, cells were rinsed five times with 0.1% BSA in PBS and three more times with PBS. Coverslips were then mounted onto glass slides using ProLong Gold antifade reagent (Molecular Probes). Images were acquired with an inverted Leica SP2 confocal microscope, using an HCX PL APO CS 40.0 × 1.25 OIL UV objective. In cases where double immunofluorescence was performed, images were all taken simultaneously. GFP, CFP, and Alexa Fluor 488 were excited with the 488-nm line of the argon laser, whereas Alexa Fluor 647 was excited with a 633-nm HeNe laser. The pinhole diameter was kept at 1 μm. Quantification analysis shown in Fig. 1B was performed by measuring and comparing the average pixel intensity of the ECFP at the nuclear speckle site and adjacent nucleoplasm of a region of interest scan for each TCERG1 mutant. Thirty-six regions of interest from three different experiments with 12 cells per experiment (72 and 108 measurements for the speckles and nucleoplasm, respectively) were quantified for each TCERG1 mutant. Statistical analysis was performed using a standard Student's t test. Acquisition software was LAS AF v2.3.6 Build 5381sps. All images were digitally processed for presentation using Adobe Photoshop CS3 extended v10.0 software.
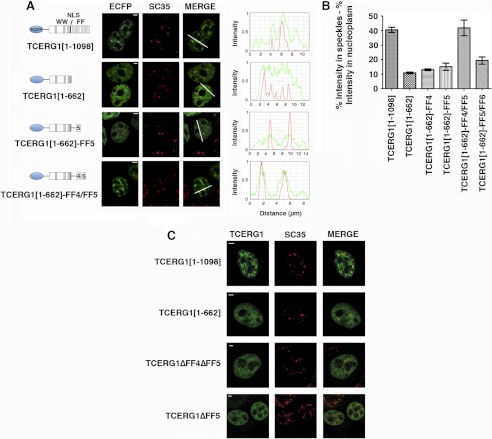
FIGURE 1.
The FF4/FF5 domains are required for efficient targeting of TCERG1 to the speckle compartment. A, dual-labeling of cells transfected with ECFP-TCERG1[1–1098] or the indicated mutant constructs (ECFP, green) with the anti-SC35 antibody (SC35, red) was performed. A diagrammatic representation of the ECFP-TCERG1 fusion proteins is shown to the left of each panel. The numbers in parentheses represent the TCERG1 amino acids contained in the construct. Shown are the three WW domains, the putative nuclear localization signal (NLS), and the six FF domains. Line scans showing local intensity distributions of TCERG1 and SC35 are shown to the right of the panels. Bars in the merged panels indicate the positions of the line scans. Scale bars, 3 μm. B, the nucleoplasmic fraction of TCERG1 proteins present in speckles was determined after measuring the fluorescence intensity in speckles relative to nucleoplasm as described under “Experimental Procedures.” The data shown are from three independent experiments. Student's t test was performed, and differences were shown to be highly significant: p(TCERG1[1–1098] versus TCERG1[1–662]) < 0.0001; p(TCERG1[1–1098] versus TCERG1[1–662]-FF4) = 0.0001; p(TCERG1[1–1098] versus TCERG1[1–662]-FF5) = 0.0011; p(TCERG1[1–1098] versus TCERG1[1–662]-FF4/FF5) = 0.8261; p(TCERG1[1–1098] versus TCERG1[1–662]-FF5/FF6) = 0.0023; p(TCERG1[1–662]-FF4/FF5 versus TCERG1[1–662]-FF4) = 0.0060; p(TCERG1[1–662]-FF4/FF5 versus TCERG1[1–662]-FF5) = 0.0106; and p(TCERG1[1–662]-FF4/FF5 versus TCERG1[1–662]-FF5/FF6) = 0.0192. The bar graph represents mean ± S.D. C, immunofluorescence analysis of cells transfected with the indicated plasmids. Images show a speckled pattern for full-length TCERG1[1–1098], but TCERG1[1–662], TCERG1[ΔFF4/FF5], and TCERG1[ΔFF5] are more diffusely dispersed throughout the nucleoplasm. Individual and merged images of the cell labeled with the indicated fluorescent proteins (FP, in green) and with the anti-SC35 antibody (SC35, red) are shown. Scale bars = 3 μm.
NMR Spectroscopy
All experiments were recorded on a Bruker Avance III 600-MHz spectrometer equipped with a z pulse field gradient unit and a triple (1H, 13C, 15N) resonance probe head. Double- and/or triple-labeled samples were prepared to obtain sequence-specific (HNCACB/HN(CO)CACB or CBCA(CO)NH/CBCANH) experiments. All spectra were processed with the NMRPipe/NMRDraw (31) software and were analyzed with Computer Aided Resonance Assignment, CARA (32). The 15N-relaxation experiments were acquired for 15N-labeled samples of FF4+FF5 (0.5 mm) and FF4 (0.5 mm) essentially as described (33). Heteronuclear {1H}-15N NOE experiment was performed using standard 2D experiments, with the reference and proton saturated spectra collected in an interleaved fashion. The values of steady-state 1H-15N NOEs were determined from the ratios of the peak intensities measured in spectra recorded either with (Is) or without (Io) presaturation during the relaxation delay as described (34). The standard deviation of the NOE was determined on the basis of measured background noise levels using the following relationship:
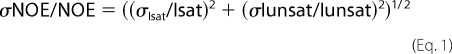 |
T1 and T2 experiments were acquired with 135 (t1) × 2048 (t2) total real points. T1 data points were obtained with 12 different relaxation periods: 20.8, 52, 104, 156, 265, 424, 520, 676, 832, 1040, 1352, and 1664 ms. Ten delay times were also sampled in the T2 experiments: 12, 24, 36, 48, 60, 72, 84, 120, 144, and 168 ms. T1 and T2 values were determined by fitting the measured peak heights to a two-parameter function of the form:
where I(t) is the intensity after a delay of time t and Io is the intensity at time t = 0. Minimization performed using the Levenberg-Marquardt optimization algorithm was used to determine the optimum value of the I0 and T1,2 parameters by minimizing the χ2 goodness of fit parameter:
where Ic(t) are the intensities calculated from the fitting parameters, Ie(t) are the experimental intensities, sI is the standard deviation of the experimental intensity measurements, and summation is performed over the number of time points recorded in each experiment.
Differential Scanning Calorimetry
Thermal denaturation was studied using differential scanning calorimetry (DSC). The experiments were performed using a VPTM DSC MicroCalorimeter (in the Polymorphism and Calorimetry Platform of the Scientific and Technical Services (SCT), Universitat de Barcelona) in 20 mm sodium phosphate buffer, 130 mm NaCl, 0.5 mm NaN3 (pH 5.8). The protein solution was heated up at a constant rate of 1 °C/min from 10 to 80° and a constant pressure. The temperature dependence of the excess heat capacity was analyzed and plotted with Origin 7.0 software (OriginLab Corp.).
RT-PCR Analysis
Approximately 2 × 106 HEK293T cells were seeded 24 h before transfection. Cells were transfected with 0.6 μg of Bcl-X2 minigene reporter (kindly provided by Benoit Chabot, University of Sherbrooke) and 1 μg of the TCERG1 derivative plasmids as indicated in the legend of Fig. 6. Total RNA was isolated from transfected cells by using the TRIzol reagent (Invitrogen). Approximately 1 μg of RNA was digested with 10 units of RNase-Free DNase (Roche). One-half of digested RNA was used for RT using the RT-Sveda primer and Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Invitrogen) according to the instructions of the manufacturer. 10% of the RT reaction was used as template together with X34 and X-Agel-R primers. Reaction products were analyzed on 2% agarose gel and quantified using Quantity One 4.5.0 software. The following primers were used: 5′-GGGAAGCTAGAGTAAGTAG-3′ (RT-Sveda1-Rev), 5′-AGGGAGGCAGGCGACGAGTTT-3′ (X34), and 5′-GTGGATCCCCCGGGCTGCAGGAATTCGAT-3′ (X-Agel-R).
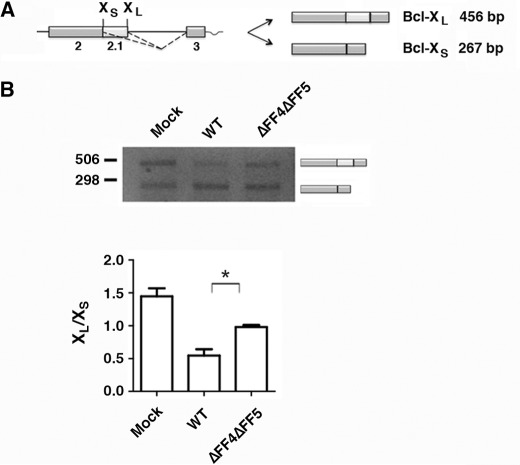
FIGURE 6.
Deletion of the FF4/FF5 domains of TCERG1 affects Bcl-x alternative splicing. A, schematic representation of the structure of the Bcl-x minigene is drawn with exons (boxes) and introns (lines). The Bcl-x pre-mRNA is alternatively spliced (dotted lines) to produce two major isoforms, Bcl-xL and Bcl-xS. B, splicing assay of the Bcl-x minigene in HEK293T cells transfected with the indicated constructs. Cells were harvested ∼44 h after transfection and processed for RT-PCR. The data are presented as the ratio of Bcl-xL to Bcl-xS from three independent experiments (mean ± S.D.). *, p < 0.05.
RESULTS
Identification of a Sequence Element Required for the Localization of TCERG1 to Nuclear Speckles
We reported previously that TCERG1 is present along the periphery of the speckles (21) but that the sequences responsible for this location have not yet been defined. In our previous study, we found that a TCERG1 protein lacking the FF5 and FF6 repeat motifs is not able to localize to speckles (21). At least three interpretations are possible: 1) FF5 is essential for the subnuclear localization of TCERG1; 2) FF6 and FF5 are both important but neither is essential; or 3) a minimum number of FF repeats are required for the localization to nuclear speckles. To identify the elements within the FF repeat motifs of TCERG1 responsible for its accumulation in nuclear speckles, we constructed a series of mutants of human TCERG1 fused to ECFP and investigated their subnuclear localization in transfected HEK293T cells by confocal laser scanning microscopy. All mutants retain the putative nuclear localization signal found in the middle of the protein (22). The localization of full-length ECFP-tagged TCERG1 was similar to that of the endogenous TCERG1 (Fig. 1A, ECFP and TCERG1[1–1098]). A TCERG1 deletion that completely eliminated the FF domains was localized in a diffuse pattern throughout the nucleoplasm without any evident accumulation on the speckles (Fig. 1A, ECFP and TCERG1[1–662]), in agreement with our data published previously (21). A mutant containing the amino terminal region and the FF5 domain of TCERG1 showed a slight enrichment in speckles, indicating that this domain might be required for targeting the protein to nuclear speckles but may not be sufficient for efficient speckle localization (Fig. 1A, ECFP, compare the signal obtained with TCERG1[1–1098] to the one obtained with TCERG1[1–662]-FF5). Addition of the FF6 domain to this mutant created a fusion protein with a staining pattern similar to the previous mutant, and it did not accumulate in nuclear speckles in significant amounts (supplemental Fig. S1A, TCERG1[1–662]-FF5/FF6). To further analyze the speckle-targeting capacity of the FF domains, we generated a chimera with the amino-terminal region fused to the FF4 and FF5 domains. Remarkably, cells expressing this chimera showed strong nuclear speckle fluorescence that was indistinguishable from that of wild-type TCERG1 (Fig. 1A, ECFP and TCERG1[1–662]-FF4/FF5). Mutant ECFP-TCERG1[1–662]-FF4, lacking FF5, was dispersed throughout the nucleoplasm (supplemental Fig. S1A). These results suggest that the combination of FF4 and FF5 is the sequence element necessary and sufficient for localization to speckles. This was not due to differential protein expression levels, as confirmed by Western blotting (supplemental Fig. S1B).
To confirm that the nuclear staining pattern of the transiently expressed proteins coincides with nuclear speckles, we carried out an immunofluorescence analysis with an antibody against the essential splicing factor SC35, which commonly serves to define nuclear speckles. Expression of wild-type and the FF4/FF5 mutant resulted in nuclear colocalization with speckles, whereas the FF5 mutant displayed a partially overlapping signal at the speckle region (Fig. 1A). The spatial relationship between wild-type and mutant TCERG1 variants relative to SC35 was identified by quantitatively scanning specific nuclear regions containing speckles and is shown on the right in Fig. 1A.
To further analyze the spatial distribution of TCERG1, we analyzed the spatial relationship between the relative spatial distributions of either wild-type or TCERG1[1–662]-FF4/FF5 and SC35 in individual nuclear speckles by confocal microscopy optical sectioning (0.3-μm sections). We found that the speckles had peripheral and internally located the TCERG1 proteins that are excluded from the core region of the speckles (supplemental Fig. S2). The analysis of those images showed that the TCERG1 peaks partially overlap, but do not coincide with, the SC35 peaks (supplemental Fig. S2), lending additional support to the observation that wild-type (21) and TCERG1[1–662]-FF4/FF5 are enriched at the speckle periphery.
We performed quantitative and statistical analyses that involved computing the proportion of signal intensity contained in speckles (see “Experimental Procedures”). Intensity measurements agreed with the above data and confirmed that FF4/FF5 is required for the efficient targeting of TCERG1 to the speckle compartment (Fig. 1B).
To further corroborate that the FF4 and FF5 domains are the targeting signal for the localization of TCERG1 to nuclear speckles, we expressed wild-type TCERG1 and FF4/FF5-deleted protein tagged with ECFP at the amino terminus and examined their nuclear localization in HEK293T cells using immunofluorescence microscopy. As shown in Fig. 1C, the TCERG1 protein with a deletion encompassing the FF4 and FF5 domains (TCERG1ΔFF4ΔFF5) exhibited diffuse localization throughout the nucleoplasm without any evident accumulation on the speckles, thus supporting the conclusion that this region of TCERG1 contains the sequence required for proper localization to speckles.
Previously, we have shown that a deletion of the FF6 domain does not affect the speckle distribution of TCERG1, which is consistent with our current data, but that a further deletion that included the FF5 domain results in perturbation of TCERG1 localization to speckles (21). To test whether the FF5 domain is essential for the proper localization of TCERG1 to speckles, we transiently transfected cells with a plasmid encoding a TCERG1 protein in which the FF5 domain had been deleted (TCERG1ΔFF5) and examined its nuclear distribution. The expressed protein was present diffusely throughout the nucleoplasm (Fig. 1C). These results indicate that a deletion of the FF5 domain disrupts the localization of TCERG1 to nuclear speckles and imply that FF5 is essential for this association.
NMR Studies of the FF4 and FF5 Domains of TCERG1
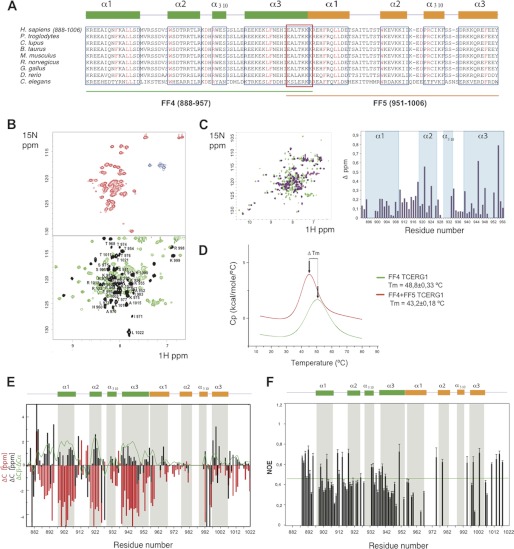
The sequence comparison of the FF4-FF5 pair of domains in different species revealed a high level of sequence conservation in this pair of domains (Fig. 2A). Remarkably, secondary structure predictions of the FF4 and FF5 domains suggested that seven residues (951–957) are shared by these two FF domains, with a fragment of the last helix of the FF4 overlapping with a part of the first helix of the FF5 domain. This prediction could imply that the FF4 and FF5 domains may not fold independently and might require the presence of the pair to acquire a stable fold. To evaluate the presence or absence of tertiary structure in the pair and to compare it with that of the independent domains, we used recombinant fragments corresponding to the FF4 and FF5 domains as well as a construct containing the FF4/FF5 pair and acquired heteronuclear single-quantum coherence (HSQC) NMR spectra at 298 K. These experiments provide information about the chemical shift dispersion of every amide proton in the sequence. Well dispersed signals are interpreted as an indication of folded samples, whereas overlapped signals usually correspond to partially folded samples. Under the experimental conditions used in this work, we observed that the independently expressed FF4 domain adopted a canonical FF fold with an α1-α2–310-α3 topology (35, 36), whereas the FF5 domain presented properties of a sample containing mixtures of partially folded and unfolded molecules. A superimposition of the HSQC spectra of the independent domains to that of the FF4/FF5 pair revealed chemical shift differences in several residues of the FF4 domain and also that the FF5 domain substantially improved its chemical shift dispersion when compared with that of the isolated FF5 domain (Fig. 2, B and C). These differences seem to indicate that in the FF4-FF5 pair, the domains may contact one another. To support this hypothesis further, we analyzed the thermodynamic behavior of the FF4/FF5 pair and compared it to that of the independent FF4 using DSC. DSC measures the enthalpy of unfolding processes because of thermal denaturation and also provides the thermal transition midpoint (Tm), which correlates with protein stability. In agreement with the NMR data, the FF4/FF5 pair presented a unique unfolding curve, suggesting that the pair unfolds in a concerted manner (Fig. 2D). The observed decrease in the midpoint thermal transition of the pair with respect to that of the FF4 domain (5.6 °C) is attributed to an aggregation process occurring in the pair that is absent in the isolated FF4 domain. The aggregation process, which affects the overall stability of the pair, probably involves the FF5 domain because of its lower stability compared with that of FF4.
FIGURE 2.
15N-HSQC NMR spectra and thermal unfolding of FF4-FF5. A, amino acid alignment of TCERG1 homologues. Secondary structure elements of FF4 and FF5 are drawn above the sequences in green and orange, respectively. Boxed sequences in blue are the residues that form the α-helices. The overlapped residues between FF4 and FF5 are shown in a red box. Strictly conserved residues are highlighted in red. B, 15N-HSQC spectra at 298 K for FF5 and FF4-FF5 constructs. The NMR data show the improvement of FF5 backbone amino acid dispersion (shown in black) observed in the pair when compared with that of the isolated construct. C, overlapped 15N-HSQC spectra at 298 K for FF4 and FF4-FF5 constructs and chemical shift differences bar representation of FF4 residues. D, thermal unfolding of the FF4 and FF4-FF5 domains as monitored by DSC. The difference in Tm is shown as an arrow connecting both maxima. E, secondary structure elements of the FF4-FF5 pair. Chemical shift distribution of Cα and Cβ. The ratio of Cα and Cβ is shown as a green line. The majorities of the values are positive, indicating the presence of a helical structure. F, heteronuclear {1H}-15N NOE. Unassigned residues, proline residues that lack a proton amide, and overlapped peaks were excluded from the analysis. Heteronuclear NOE values show that the pair of FF4-FF5 has secondary structure throughout the construct.
To identify the elements of secondary structure present in the FF4/FF5 pair, we used a (2H, 13C, and 15N) recombinant sample and acquired backbone triple-resonance experiments. The assignment was achieved up to a 70% of all possible residues. A comparison of the α and β carbon chemical shifts of the assigned residues to that of random coil values revealed positive values that correlate with the presence of α helices, characteristic of FF domains, shown in Fig. 2E as a green line. The high number of overlapped residues in the FF5 domain precluded a proper identification of all secondary structure elements (37). However, the positive 13C values obtained for the unambiguously assigned regions of the FF5 domain suggested that this domain also presents helical secondary structure. From the obtained data it seems that the first helix of FF5 domain is a continuation of the last helix of the FF4 domain, that the 310 and third helices are shorter than expected on the basis of a sequence comparison with other described FF domains, and finally, that the second helix is almost undetectable.
To further examine the structural and dynamical properties of the pair, we performed heteronuclear (34) {1H}-15N NOE relaxation experiments. Internal dynamics are directly related to folding, with less ordered proteins displaying higher internal motions. The data obtained for the assigned residues (shown in Fig. 2F) revealed that the distribution of NOE values was, on average, close for both domains in the pair. To further explore the dynamic behavior of the FF4/FF5 pair, we also performed T1 and T2 experiments and compared the obtained values to the independent FF4 domain. The correlation times obtained for the assigned residues (supplemental Fig. S3, A and B), T1, T2, and the T1/T2 ratio are predominantly in the same range, with fewer uniform data for the pair with respect to that of FF4. We attributed the observed results to the slightly higher flexibility of FF5 when compared with FF4, probably because of the presence of long loops connecting the helices in FF5, whereas the values corresponding to the secondary structured regions were more similar.
These results point out that the fourth and fifth FF domains of TCERG1 have similar properties when expressed together, suggesting that the FF5 domain is more stable in the pair. This organization is a feature specific to these two domains, as we did not observe any structural organization when FF5 was combined with FF6 (supplemental Fig. S3C).
FF4 and FF5 of TCERG1 Comprise a Novel Nuclear Localization Signal Targeting Proteins to the Periphery of Speckles
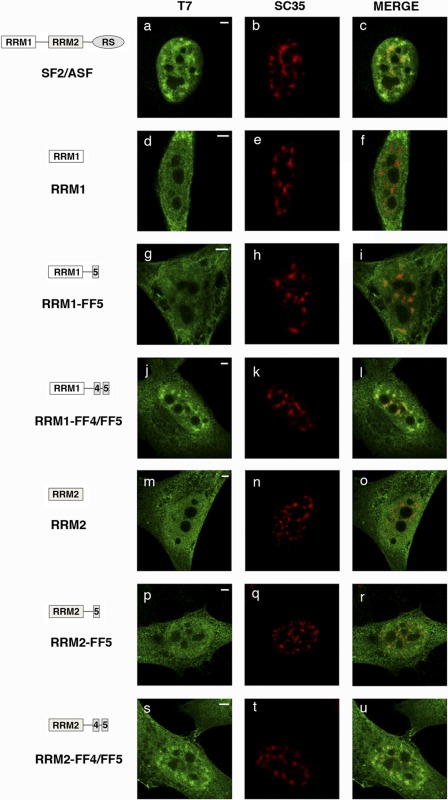
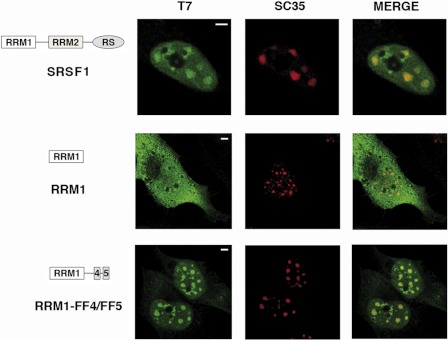
The SR protein SRSF1(formerly SF2/ASF) is an essential splicing factor that regulates alternative splicing of many pre-mRNAs and is located at the speckle region (38). SRSF1 contains two functional modules: an RS (arginine/serine-rich) domain and two RRMs. Previous work demonstrated that at least two of these domains are necessary for SRSF1 localization to nuclear speckles (14). The same work described mutant proteins carrying individual domains (RRM1 and RRM2) that localized throughout the cell without any evident accumulation on the nuclear speckles. We sought to test whether the FF4/FF5 sequence could target those SRSF1 mutants to nuclear speckles. To this end, we ligated the FF4/FF5 and FF5 sequences to the C terminus of the RRM1 and RRM2 gene coding sequences, and the fusion genes were transfected into HeLa cells. All constructs encoded proteins with a bacteriophage T7 epitope tag at their amino terminus, allowing detection of the exogenous proteins with antibodies that recognize this epitope. Immunofluorescence experiments using a T7 tag antibody showed that the transiently expressed wild-type SRSF1 protein localized exclusively in the nucleus with a typical speckled pattern (Fig. 3a). Double-labeling experiments with anti-SC35 antibodies confirmed SRSF1 localization at nuclear speckles (Fig. 3, b and c). In contrast, when individual domains were expressed (RRM1 or RRM2), the mutant protein localized throughout the cell, and colocalization with nuclear speckles was not detected (Fig. 3, d–f and m–o), which agrees with a previous report (14). Small changes in the staining pattern were observed upon addition of the FF5 region to the mutant proteins (Fig. 3, panels g–i and p–r). However, mutant proteins containing the FF4/FF5 region clearly led to speckle localization of the fusion protein (Fig. 3, j–l and s–u). Expression of the various SRSF1 constructs was nearly identical, as assessed by Western blotting (supplemental Fig. S4).
FIGURE 3.
FF4/FF5 directs SRSF1 domain-deletion mutants to nuclear speckles. Cells were transfected with the indicated plasmids and dually labeled with antibodies directed against the expressed SRSF1 protein (left column, green) and SC35 (center column, red). The merged images are also shown (right column). In all cases, colocalization of expressed proteins with the endogenous marker was assessed by confocal imaging. A diagrammatic representation of the T7-tagged SRSF1 mutants used is shown at the left of the figure. The structure of the SRSF1 domain-deletion mutants was described previously (51). Scale bars = 3 μm.
Next, we generated GFP fusion proteins containing the FF4/FF5 domains and the FF5 domain alone. The GFP-FF5 protein showed a diffuse staining pattern similar to that of the GFP alone with small punctuate areas in the nucleus (18) (supplemental Fig. S5). The GFP-FF4/FF5 protein clearly accumulated in the speckle compartment (supplemental Fig. S5), indicating that the presence of this sequence is sufficient for nuclear speckle localization of the GFP. These results, together with the analysis of the SRSF1 mutants, indicate that the FF4/FF5 region shown to be necessary for the localization of TCERG1 protein to speckles is sufficient to direct the localization of heterologous proteins to nuclear speckles.
To explore further the association of FF4/FF5-containing RRMs with speckles, we treated cells with the RNA polymerase II inhibitor α-amanitin. Upon RNA polymerase II inhibition, speckles decrease in number, enlarge, and become rounded because of the accumulation of the splicing machinery (39). We found similar changes in the immunofluorescence pattern using antibodies against SC-35 and RRM1-FF4/FF5 proteins upon transcriptional inhibition (Fig. 4). We conclude that the localization of FF4/FF5 fusion proteins to nuclear speckles is not dependent on active transcription and that the accumulation of these proteins shows behavior similar to that of splicing factors.
FIGURE 4.
Colocalization of the FF4/FF5-containing RRM1 protein with nuclear speckles is not perturbed following inhibition of transcription. HeLa cells were transfected with the indicated expression plasmids and treated with 25 μg/ml of α-amanitin for 6 h at 37 °C and then processed for immunofluorescence analysis. Dual-labeling of cells with antibodies directed against SRSF1 (T7, green) and with the SC35 antibody (red) was performed. Individual staining and merge images of the cell stained with the indicated antibodies are shown. A diagrammatic representation of the T7-tagged SRSF1 mutants used is shown at the left of the figure. Scale bars = 3 μm.
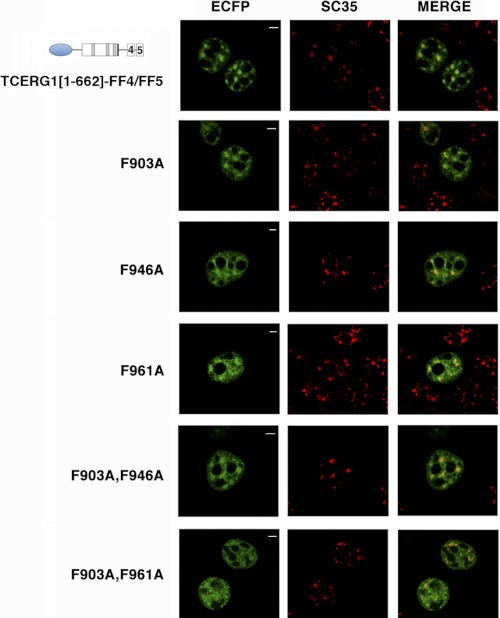
FF motifs are putative protein-protein interaction domains named for two conserved phenylalanine (F) residues (41). To study the requirement of particular residues within the FF4/FF5 domain for their nuclear targeting activity, we generated TCERG1[1–662]-FF4/FF5 constructs containing phenylalanine-to-alanine mutations at positions Phe-903, Phe-946, Phe-961, or the double mutants F903A,F946A, and F903A,F961A. We then expressed the wild-type FF4/FF5 domain and the phenylalanine-to-alanine mutants tagged with the ECFP at the amino terminus and examined their nuclear localization using immunofluorescence microscopy. As shown in Fig. 5, the TCERG1[1–662]-FF4/FF5 protein and the mutants within the FF4 domain (F903A, F946A, and the double mutant F903A,F946A) exhibited a similar nucleoplasm distribution with an increased signal in speckles. Mutation of the first phenylalanine of the FF5 domain at position 961 showed a decreased enrichment in speckles. The F903A,F961A double mutant localized in a diffuse pattern throughout the nucleoplasm without any evident accumulation on the speckles (Fig. 5). Expression of the various phenylalanine-to-alanine constructs was nearly identical, as assessed by Western blotting (supplemental Fig. S6). These data have been repeated with the same phenylalanine-to-alanine mutations in the context of a full-length TCERG1 protein (data not shown). These results demonstrate the involvement of the highly conserved phenylalanine residue within the FF5 domain in the targeting to the nuclear speckles. These results show that TCERG1 protein is able to localize to the periphery of nuclear speckles when the F-903 and F-946 residues within the FF4 domain have been mutated. This might indicate that the folding of this domain is less perturbed upon mutating these residues. This hypothesis is suggested by our NMR data (Fig. 2), which indicate a higher degree of stability for this domain.
FIGURE 5.
Strictly conserved phenylalanine residues within the FF4/FF5 domains are required for the targeting to nuclear speckles. Dual labeling of HEK293T cells transfected with TCERG1[1–662]-FF4/FF5 or the indicated phenylalanine-to-alanine mutant constructs (ECFP, green) with the anti-SC35 antibody (SC35, red) was performed. Individual and merged images of the cell are shown. Scale bars = 3 μm.
Role of the FF4/FF5 Domains in TCERG1 RNA Splicing Activity
TCERG1 is involved in the process of pre-mRNA splicing and can affect the splicing of several minigene splicing reporters (see “Introduction”). We sought to investigate the role of the FF4/FF5 domains in the regulation of alternative splicing. Recently, we have found that TCERG1 can affect alternative splicing of the apoptosis gene Bcl-x (40). Bcl-x pre-mRNA uses an alternative 5′ splice site in exon 2 to produce the antiapoptotic Bcl-xL or the proapoptotic Bcl-xS isoforms (Fig. 6A). A Bcl-x minigene was transfected into HEK293T cells in combination with vectors expressing wild-type or FF4/FF5-deleted TCERG1. The presence of different splice products was assessed by RT-PCR. As shown in Fig. 6B, cotransfection of wild-type TCERG1 with this reporter led to an increase in short-variant Bcl-xS with respect to transfection of the vector alone (Fig. 6B, WT). Furthermore, deletion of the FF4/FF5 domains reduced the ability of TCERG1 to affect 5′ splice site selection (Fig. 6B, ΔFF4ΔFF5). These results demonstrate the involvement of the FF4/FF5 domains in the determination of 5′ splice site selection in the Bcl-x gene, which supports the assumption of the FF4/FF5 unit as a functional entity. Although more work is clearly needed to show how the FF4/FF5 mutants are affecting splicing of the Bcl-x reporter, our data suggest that TCERG1 might exert its activity through its localization to the nuclear speckle region.
DISCUSSION
We have identified the sequences within the transcription and splicing-related factor TCERG1 that target it to the periphery of splicing factor-rich nuclear speckles. These consist of two contiguous FF domains, FF4 and FF5, of TCERG1: amino acids 878–1022. They are both necessary but not sufficient by themselves to achieve complete accumulation within the speckle region. We showed previously that the FF5 domain was implicated in the localization of TCERG1 to nuclear speckles (21). We show here that FF5 is necessary but not sufficient in itself to induce the accumulation of proteins to speckles. The adjacent FF4 domain is required for proper localization to the splicing speckled regions. The comparison of the TCERG1 speckle-targeting sequences with known speckle domain-targeting sequences (11–15, 17, 18) suggests that the TCERG1 speckle determinants are novel. This is consistent with the fact that the spatial distribution of TCERG1 is somewhat different from the distribution of many splicing factors that show more diffuse nucleoplasmic staining than TCERG1. The FF4 and FF5 domains of TCERG1 represent a novel nuclear speckle-targeting signal. They are necessary for TCERG1 localization and are sufficient for targeting a heterologous protein to speckles. To our knowledge, this targeting signal is the first sequence reported to direct proteins to the periphery of nuclear speckles at the interface between speckles and nearby transcription sites.
Compared with the better-characterized WW domain, the function of the FF domains is less well understood. The FF domain is an ∼60-amino acid module that contains two strictly conserved phenylalanine residues near the N terminus and C terminus, respectively. FF domains often occur in repeated arrays of four to six domains separated by linker sequences of variable length (41), and this organization is likely to be important for their biological function. Structural studies of the FF domains of TCERG1 show a flexibility that is consistent with a domain organization model that visualizes the FF domains as multifunctional units acting as a scaffold to bind to a diverse repertoire of molecules (42, 43). The regulation of FF domain interactions is an intriguing issue. The TCERG1 FF domains could have individual specificities, or they could act in concert to achieve optimal recognition of binding partners. The interactions of TCERG1 with the phosphorylated carboxy-terminal domain (CTD) of RNAPII (44) and with the splicing factor Tat-SF1 (29) via the FF domains suggest that each FF domain within a tandem repeat possesses independent binding activity, conferring an ability to mediate many different protein-protein interactions through relatively weak binding affinity contacts. The possibility of each FF domain within a repeated structure mediating distinct ligand recognition was initially supported by our own data implicating the TCERG1 FF5 domain as the critical FF domain for TCERG1 localization to the periphery of speckles (21). Here, we have presented NMR data that challenges the current view of FF domains acting as separate entities and provided evidence that the FF domains can organize in pairs to optimally achieve a relevant biological function, such as targeting to speckles. Although future investigations are necessary, our results provide a starting point for understanding the structural basis of the targeting to speckles by FF domains.
Speckles are enriched in pre-mRNA splicing factors (1, 45). However, transcription and pre-mRNA splicing take place outside of the speckle compartment (7, 8, 46, 47). We described previously that the majority of speckle-associated TCERG1 is found along the periphery of nuclear speckles (21) where newly synthesized RNA is also found (7). The distribution of TCERG1 is consistent with a potential role of this factor in linking transcription with splicing machinery. Indeed, many of the factors that interact with TCERG1, such as SF1 and U2AF65, are also present in speckles. An interesting hypothesis is that TCERG1 could recruit processing components from speckles to the nearby transcription sites. If so, interfering with the localization of TCERG1 would probably disrupt some of these interactions. This is suggested by our experiments that showed that the signals determining TCERG1 nuclear speckle localization are important for its alternative splicing function (Fig. 6). Real-time mRNA biogenesis studies will address the question of whether TCERG1 shuttles between speckles and speckle-associated transcription sites.
TCERG1 undergoes posttranslational modification by SUMOylation (27). Computer algorithms aimed at locating putative SUMO acceptor residues predict two potential SUMOylation sites within the FF4/FF5 domains. Because SUMOylation influences the targeting of proteins to different cell compartments, we investigated whether putative SUMOylation of sequences within FF4/FF5 domains affects its targeting properties. Our results show that mutation of the putative SUMO acceptor lysine residues within FF4/FF5 generates a protein that localizes to the nucleus similarly to the wild-type protein, ruling out an effect of SUMO modification for the spatial distribution of this protein in the cell.4 TCERG1 can also be modified by phosphorylation (48, 49). Computational predictions of phosphorylation sites using the NetPhos 2.0 server show that many serine and threonine residues within the FF4/FF5 domains are potential phosphorylation sites. Some of these sites are located in the loops of FF5. This is reminiscent of the phosphorylation status of SR proteins. The RS domain of SR proteins is extensively phosphorylated on serine residues, and this controls the subnuclear localization and activity of SR proteins (50). Similarly, phosphorylation events might regulate the nuclear localization and functionality of TCERG1. Moreover, TCERG1 interacts directly with phosphorylated RNAPII CTD via its FF repeats (44), and interactions between SR-related proteins and RNA polymerase II CTD have also been reported (52). Although no classical RRM has been identified, a previous study (53) described the presence of a putative RGG box in the N terminus of the Chironomus tentans homologue of TCERG1, which is also found in its human counterpart. The RGG box is a protein motif present in one class of RNA-binding proteins involved in various aspects of RNA processing (54), is characterized by the presence of an RGG triplet, and is rich in arginines and glycines. In addition, TCERG1 has highly basic FF domains, raising the possibility that some of these, such as FF1–3, FF5, and FF6, may interact with RNA. In fact, we observed nucleic acid binding activity with recombinant FF1 and FF2 proteins.5 On the basis of these observations, an exciting possibility remains that TCERG1 behaves as an SR protein in the coupling between transcription and splicing. Strikingly, the Drosophila homologue of human TCERG1 was identified as an RS domain-containing protein in a genome-wide survey of RS domain proteins (55).
Supplementary Material
Acknowledgments
We thank members of the laboratory for helpful suggestions, critical discussions, and comments. We also thank Javier Cáceres, Benoit Chabot, Mariano Garcia-Blanco, and Juan Valcárcel for providing reagents; José Luis Luque for confocal imaging assistance; Pau Martín-Malpartida for spectrum analysis; and Román Bonet for protein cloning.
This work was supported by Spanish Ministry of Science and Innovation Grants BFU2008-01599 and BFU2011-24577), by Foundation for Research and Prevention of AIDS in Spain Grant FIPSE-36768/08, and by Andalusian Government Excellence Project CVI-4626/2009) (to C. S.). This work was also supported by Spanish Ministry of Science and Innovation Grants BFU2009-08796 (to C. H. M.) and CSL2009-00080 (to M. J. M.), and by the Institute for Research in Biomedicine (to M. J. M.). This work was also supported by the European Region Development Fund, ERDF (FEDER), by a fellowship from the Spanish Ministry of Education (Formación Profesorado Universitario/FPU program) (to M. M.), and by a fellowship from the Consejo Superior de Investigaciones Científicas (Junta para la Ampliación de Estudios, CSIC/JAE program) (to N. S. H.).

This article contains supplemental Figs. S1–S6.
N. Sánchez-Hernández and C. Suñé, unpublished data.
L. Ruiz and M. J. Macias, unpublished data.
- SR
- serine/arginine-rich
- RRM
- RNA recognition motif
- TCERG1
- transcription elongation regulator 1
- ECFP
- enhanced cyan fluorescent protein
- DSC
- differential scanning calorimetry
- HSQC
- heteronuclear single-quantum correlation
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Lamond A. I., Spector D. L. (2003) Nuclear speckles. A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612 [DOI] [PubMed] [Google Scholar]
- 2. Spector D. L. (1990) Higher order nuclear organization. Three-dimensional distribution of small nuclear ribonucleoprotein particles. Proc. Natl. Acad. Sci. U.S.A. 87, 147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spector D. L., Fu X. D., Maniatis T. (1991) Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10, 3467–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang S., Spector D. L. (1992) U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc. Natl. Acad. Sci. U.S.A. 89, 305–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu X. D., Maniatis T. (1990) Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343, 437–441 [DOI] [PubMed] [Google Scholar]
- 6. Spector D. L., Schrier W. H., Busch H. (1983) Immunoelectron microscopic localization of snRNPs. Biol. Cell 49, 1–10 [DOI] [PubMed] [Google Scholar]
- 7. Wei X., Somanathan S., Samarabandu J., Berezney R. (1999) Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J. Cell Biol. 146, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cmarko D. (1999) Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell 10, 211–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moen P. T., Jr., Johnson C. V., Byron M., Shopland L. S., de la Serna I. L., Imbalzano A. N., Lawrence J. B. (2004) Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol. Biol. Cell 15, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Misteli T., Cáceres J. F., Clement J. Q., Krainer A. R., Wilkinson M. F., Spector D. L. (1998) Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H., Bingham P. M. (1991) Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell 67, 335–342 [DOI] [PubMed] [Google Scholar]
- 12. Hedley M. L., Amrein H., Maniatis T. (1995) An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. U.S.A. 92, 11524–11528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gama-Carvalho M., Krauss R. D., Chiang L., Valcárcel J., Green M. R., Carmo-Fonseca M. (1997) Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137, 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cáceres J. F., Misteli T., Screaton G. R., Spector D. L., Krainer A. R. (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138, 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eilbracht J., Schmidt-Zachmann M. S. (2001) Identification of a sequence element directing a protein to nuclear speckles. Proc. Natl. Acad. Sci. U.S.A. 98, 3849–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wagner S., Chiosea S., Nickerson J. A. (2003) The spatial targeting and nuclear matrix binding domains of SRm160. Proc. Natl. Acad. Sci. U.S.A. 100, 3269–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ko T. K., Kelly E., Pines J. (2001) CrkRS. A novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J. Cell Sci. 114, 2591–2603 [DOI] [PubMed] [Google Scholar]
- 18. Alvarez M., Estivill X., de la Luna S. (2003) DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 116, 3099–3107 [DOI] [PubMed] [Google Scholar]
- 19. Salichs E., Ledda A., Mularoni L., Alba M. M., de la Luna S. (2009) Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 5, e1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldstrohm A. C., Greenleaf A. L., Garcia-Blanco M. A. (2001) Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277, 31–47 [DOI] [PubMed] [Google Scholar]
- 21. Sánchez-Alvarez M., Goldstrohm A. C., Garcia-Blanco M. A., Suñé C. (2006) Human transcription elongation factor CA150 localizes to splicing factor-rich nuclear speckles and assembles transcription and splicing components into complexes through its amino and carboxyl regions. Mol. Cell Biol. 26, 4998–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suñé C., Hayashi T., Liu Y., Lane W. S., Young R. A., Garcia-Blanco M. A. (1997) CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol. Cell Biol. 17, 6029–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suñé C., Garcia-Blanco M. A. (1999) Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner. Mol. Cell Biol. 19, 4719–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin K. T., Lu R. M., Tarn W. Y. (2004) The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell Biol. 24, 9176–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng D., Côté J., Shaaban S., Bedford M. T. (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 [DOI] [PubMed] [Google Scholar]
- 26. Pearson J. L., Robinson T. J., Muñoz M. J., Kornblihtt A. R., Garcia-Blanco M. A. (2008) Identification of the cellular targets of the transcription factor TCERG1 reveals a prevalent role in mRNA processing. J. Biol. Chem. 283, 7949–7961 [DOI] [PubMed] [Google Scholar]
- 27. Sánchez-Alvarez M., Montes M., Sánchez-Hernández N., Hernández-Munain C., Suñé C. (2010) Differential effects of sumoylation on transcription and alternative splicing by transcription elongation regulator 1 (TCERG1). J. Biol. Chem. 285, 15220–15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goldstrohm A. C., Albrecht T. R., Suñé C., Bedford M. T., Garcia-Blanco M. A. (2001) The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell Biol. 21, 7617–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith M. J., Kulkarni S., Pawson T. (2004) FF domains of CA150 bind transcription and splicing factors through multiple weak interactions. Mol. Cell Biol. 24, 9274–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marley J., Lu M., Bracken C. (2001) A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 31. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 32. Bartels C., Xia T-H., Billeter M., Güntert P., Wüthirch K. (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6, 1–10 [DOI] [PubMed] [Google Scholar]
- 33. Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. (1992) Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy. The central helix is flexible. Biochemistry 31, 5269–5278 [DOI] [PubMed] [Google Scholar]
- 34. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 35. Gasch A., Wiesner S., Martin-Malpartida P., Ramirez-Espain X., Ruiz L., Macias M. J. (2006) The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains. J. Biol. Chem. 281, 356–364 [DOI] [PubMed] [Google Scholar]
- 36. Bonet R., Ruiz L., Morales B., Macias M. J. (2009) Solution structure of the fourth FF domain of yeast Prp40 splicing factor. Proteins 77, 1000–1003 [DOI] [PubMed] [Google Scholar]
- 37. Bonet R., Ramirez-Espain X., Macias M. J. (2008) Solution structure of the yeast URN1 splicing factor FF domain: comparative analysis of charge distributions in FF domain structures-FFs and SURPs, two domains with a similar fold. Proteins 73, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 38. Long J. C., Caceres J. F. (2009) The SR protein family of splicing factors. Master regulators of gene expression. Biochem. J. 417, 15–27 [DOI] [PubMed] [Google Scholar]
- 39. O'Keefe R. T., Mayeda A., Sadowski C. L., Krainer A. R., Spector D. L. (1994) Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124, 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Montes M., Cloutier A., Sánchez-Hernández N., Michelle L., Lemieux B., Blanchette M., Hernández-Munain C., Chabot B., Suñé C. (2012) TCERG1 regulates alternative splicing of the Bcl-x gene by modulating the rate of RNA polymerase II transcription. Mol. Cell Biol. 32, 751–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bedford M. T., Leder P. (1999) The FF domain: a novel motif that often accompanies WW domains. Trends Biochem. Sci. 24, 264–265 [DOI] [PubMed] [Google Scholar]
- 42. Murphy J. M., Hansen D. F., Wiesner S., Muhandiram D. R., Borg M., Smith M. J., Sicheri F., Kay L. E., Forman-Kay J. D., Pawson T. (2009) Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays. J. Mol. Biol. 393, 409–424 [DOI] [PubMed] [Google Scholar]
- 43. Lu M., Yang J., Ren Z., Sabui S., Espejo A., Bedford M. T., Jacobson R. H., Jeruzalmi D., McMurray J. S., Chen X. (2009) Crystal structure of the three tandem FF domains of the transcription elongation regulator CA150. J. Mol. Biol. 393, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carty S. M., Goldstrohm A. C., Suñé C., Garcia-Blanco M. A., Greenleaf A. L. (2000) Protein-interaction modules that organize nuclear function. FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 97, 9015–9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spector D. L. (2001) Nuclear domains. J. Cell Sci. 114, 2891–2893 [DOI] [PubMed] [Google Scholar]
- 46. Huang S., Spector D. L. (1996) Dynamic organization of pre-mRNA splicing factors. J. Cell Biochem. 62, 191–197 [DOI] [PubMed] [Google Scholar]
- 47. Phair R. D., Misteli T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404, 604–609 [DOI] [PubMed] [Google Scholar]
- 48. Shimada M., Saito M., Katakai T., Shimizu A., Ichimura T., Omata S., Horigome T. (1999) Molecular cloning and splicing isoforms of mouse p144, a homologue of CA150. J. Biochem. 126, 1033–1042 [DOI] [PubMed] [Google Scholar]
- 49. Yu L. R., Zhu Z., Chan K. C., Issaq H. J., Dimitrov D. S., Veenstra T. D. (2007) Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J. Proteome Res. 6, 4150–4162 [DOI] [PubMed] [Google Scholar]
- 50. Misteli T., Spector D. L. (1997) Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 7, 135–138 [DOI] [PubMed] [Google Scholar]
- 51. Cáceres J. F., Krainer A. R. (1993) Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 12, 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yuryev A., Patturajan M., Litingtung Y., Joshi R. V., Gentile C., Gebara M., Corden J. L. (1996) The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun X., Zhao J., Kylberg K., Soop T., Palka K., Sonnhammer E., Visa N., Alzhanova-Ericsson A. T., Daneholt B. (2004) Conspicuous accumulation of transcription elongation repressor hrp130/CA150 on the intron-rich Balbiani ring 3 gene. Chromosoma 113, 244–257 [DOI] [PubMed] [Google Scholar]
- 54. Burd C. G., Dreyfuss G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 55. Boucher L., Ouzounis C. A., Enright A. J., Blencowe B. J. (2001) A genome-wide survey of RS domain proteins. RNA 7, 1693–1701 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.