To the Editor:
Food allergy, affecting up to 8% of children in the United States,1 is most often mediated by allergen specific immunoglobulin E (IgE) antibodies in the blood. Food allergen sensitization is an intermediate phenotype measured by specific IgE blood tests or skin prick tests (SPTs) and used in the diagnosis of food allergy. Despite family history being a risk factor, no genetic variants have been conclusively identified for food sensitization or clinical food allergy.2 Using the Mexico City Childhood Asthma Study (MCCAS), we examined associations between food sensitization (based on SPTs) and single nucleotide polymorphisms (SNPs) spanning five autosomal candidate genes reviewed by Hong et al. [CD14 (cluster of differentiation 14), IL10 (interleukin 10), IL13 (interleukin 13), SPINK5 (serine peptidase inhibitor, Kazal type 5 isoform), and STAT6 (signal transducer and activator of transcription)].2
We previously conducted a genome-wide association study of asthma in MCCAS among 492 children with physician-diagnosed asthma (aged 5-17) and their parents, who were recruited from a pediatric allergy clinic in Mexico City.3 The children’s clinical evaluation included SPTs to six major food allergens that are common in the Mexican diet (milk, egg, wheat, soy, peanuts, and tree nuts). A SPT was declared positive if the largest diameter of the wheal exceeded 4mm. Testing was considered valid if the reaction to the positive control (histamine) exceeded 6 mm and also exceeded 4 mm above the negative control (glycerin).4 There were 162 trios having an asthmatic child with a positive SPT to at least one food allergen. We examined SNP associations with food sensitization in the five candidate genes in these 162 trios.2 As all probands are asthmatics, it is not possible to adjust for asthma. These associations are generalizable to the asthmatic population, but not necessarily to the general population.
Genotyping was performed on the Illumina HumanHap 550v3 BeadChip, and standard quality control filters were applied.3 For this analysis, we selected SNPs spanning five candidate gene regions (20kb upstream of the 5′ end through 20kb downstream of the 3′ end), resulting in 343 SNPs (16 CD14, 52 IL10, 39 IL13, 28 STAT6, and 208 SPINK5 SNPs). SNPs were directly genotyped or imputed using MaCH (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) with HapMap phase II release 21 reference haplotypes combined from CEU (European Americans), YRI (African Americans), and CHB+JPT (Chinese+Japanese).
PBAT v3.61 (http://www.biostat.harvard.edu/~clange/default.htm) was used to conduct family-based association tests on additive SNP genotype dosage values (estimated reference allele counts with a fractional value ranging from 0 to 2.0). To correct for multiple testing, Bonferroni correction for the total number of SNPs (P<α/M, where M is the number of independent tests) is too conservative, given our observed correlation patterns among SNPs. Therefore, we applied the widely-used Nyholt correction (P<α/Meff), which adjusts for the effective number of independent tests (Meff) based on the correlation matrix of pairwise LD among the SNPs.5,6 This method gave a significance threshold of P<0.0011 A less conservative significance threshold based on the number of candidate genes examined is P<0.01. Association results and LD patterns were plotted together using SNAP (http://www.broadinstitute.org/mpg/snap/).
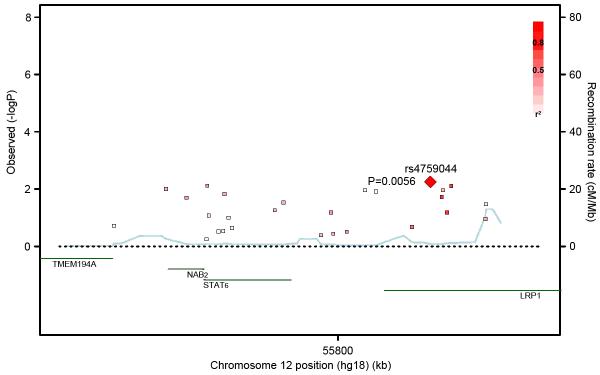
In the 162 trios with a food allergen sensitized child, no SNP associations were statistically significant at P<0.0011. However, three genotyped SNPs in or near STAT6 were associated with food sensitization at P<0.01 (Table I), two intronic SNPs in the nearby LRP1 (low density lipoprotein receptor-related protein 1) gene and a 3′ untranslated STAT6 SNP. Their minor allele frequencies were lower in the food sensitized children (39.5% for the top SNP rs4759044) compared to the children with no sensitization to the six food allergens (42.2% for rs4759044). The STAT6 and LRP1 SNPs are in moderate LD in CEU (Figure 1, r2=0.34-0.37) and weaker LD in YRI (r2=0.08-0.24) and CHB+JPT (r2=0.12-0.13). Of 28 SNPs spanning the STAT6 region, none were associated with asthma at P<0.05, despite a three-fold greater sample size and thus more statistical power (Table I). There was minimal evidence for SNP associations with food sensitization across CD14, IL10, IL13 or SPINK5 (results not shown).
Table I.
Associations of the 28 SNPs examined in or near STAT6 (±20kb) in 492 trios with an asthmatic child and the subset of 162 trios with a food sensitized child (at least one positive skin prick test to six major food allergens). The flanking regions around STAT6 included SNPs in two other genes, NAB2 and LRP1. SNPs were genotyped or imputed, and MaCH Rsq values. indicate imputation quality.
| SNPa | SNP Type | Gene | MAFb | MaCH Rsq value |
P, asthma (N=492 trios) |
P, food sensitization (N=162 trios) |
|---|---|---|---|---|---|---|
| rs17119418 | intergenic | - | 0.050 | 0.77 | 0.085 | 0.19 |
| rs2035545 | intergenic | - | 0.053 | 0.80 | 0.19 | 0.17 |
| rs324010 | intergenic | - | 0.41 | 0.92 | 0.035 | 0.010 |
| rs324020 | intronic | NAB2 | 0.48 | 0.94 | 0.034 | 0.020 |
| rs1059513c | intergenic | - | 0.074 | 0.99 | 0.35 | 0.56 |
| rs703817c | 3′ UTR | STAT6 | 0.47 | 0.99 | 0.026 | 0.0076 |
| rs324015c | 3′ UTR | STAT6 | 0.31 | 0.99 | 0.28 | 0.084 |
| rs3024975 | intronic | STAT6 | 0.060 | 0.81 | 0.25 | 0.30 |
| rs3024974 | intronic | STAT6 | 0.060 | 0.80 | 0.24 | 0.29 |
| rs841718c | intronic | STAT6 | 0.44 | 0.99 | 0.072 | 0.015 |
| rs3024971 | intronic | STAT6 | 0.029 | 0.88 | 0.077 | 0.10 |
| rs10783813 | intronic | STAT6 | 0.20 | 0.61 | 0.57 | 0.23 |
| rs324011 | intronic | STAT6 | 0.47 | 0.97 | 0.044 | 0.055 |
| rs167769c | intronic | STAT6 | 0.47 | 0.99 | 0.044 | 0.029 |
| rs324013c | intergenic | - | 0.39 | 0.99 | 0.31 | 0.41 |
| rs12368672 | intergenic | - | 0.44 | 0.90 | 0.082 | 0.066 |
| rs11172106 | intergenic | - | 0.41 | 0.91 | 0.28 | 0.36 |
| rs12298170 | intergenic | - | 0.41 | 0.88 | 0.27 | 0.31 |
| rs12307379 | intergenic | - | 0.020 | 0.69 | 0.21 | 0.011 |
| rs7306742 | intergenic | - | 0.020 | 0.70 | 0.21 | 0.012 |
| rs11172113c | intronic | LRP1 | 0.40 | 0.99 | 0.24 | 0.21 |
| rs4759044c | intronic | LRP1 | 0.34 | 0.99 | 0.25 | 0.0056 |
| rs1385526 | intronic | LRP1 | 0.49 | 0.88 | 0.24 | 0.019 |
| rs715948c | intronic | LRP1 | 0.27 | 0.99 | 0.32 | 0.011 |
| rs4759277 | intronic | LRP1 | 0.41 | 0.97 | 0.44 | 0.066 |
| rs1466535c | intronic | LRP1 | 0.43 | 0.99 | 0.17 | 0.0077 |
| rs7968719 | intronic | LRP1 | 0.41 | 0.76 | 0.18 | 0.11 |
| rs7398375 | intronic | LRP1 | 0.30 | 0.66 | 0.37 | 0.034 |
SNPs ordered by their position on chromosome 12.
MAF among the 162 food allergen sensitized children.
Genotyped SNPs.
SNP, single nucleotide polymorphism; MAF, minor allele frequency; UTR, untranslated region
Figure 1.
Association results and linkage disequilibrium patterns of SNPs in or near STAT6 (±20kb). Correlations between the SNP with the lowest P value (rs4759044) and surrounding SNPs are shown with reference to the HapMap CEU population, with darker red shading indicating higher r2 values. Gene annotations are shown in green, and estimated recombination rates are shown in light blue.
Another 3′ untranslated STAT6 SNP (rs324015) was previously associated with risk for, and severity of, nut allergy in white atopic children from the United Kingdom,7 but this association was not corroborated for severity of food allergy in Japanese children.8 In our study of Mexican children, rs324015 gave a P value of 0.072, but its adjacent SNP (rs703817) was associated with food sensitization at P=0.0076. The implicated SNPs may tag underlying functional or regulatory variants, and it is not surprising that association patterns vary across populations with dissimilar LD patterns.
LRP1 and its protein product have been largely implicated in neurologic processes. STAT6 SNPs have been associated with food allergy,7,8 asthma,9 total IgE,10 and eosinophilic esophagitis.11 STAT6 plays a central role in mediating IL4 (interleukin 4) and IL13 signals for IgE antibody production.2 Given its biological role, STAT6 polymorphisms are more likely than LRP1 polymorphisms to influence food sensitization. Nonetheless, the association patterns between STAT6 and LRP1 SNPs reflect moderate LD with low recombination across the region (Figure 1). While our findings might not be generalizable to the general population, we provide evidence that STAT6 and LRP1 polymorphisms are associated with food sensitization in asthmatics, who are at particularly high risk for developing sensitization. Follow-up studies in Hispanics and other ethnicities could help to refine these associations with food sensitization and clinical food allergy.
Acknowledgments
The authors thank Dr. Nathan Gaddis (RTI International, Research Triangle Park, NC) for his analytic assistance.
Declaration of all sources of funding: This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (Z01 ES49019). Subject enrollment was supported in part by the National Council of Science and Technology (grant 26206-M), Mexico. Dr. Romieu was supported in part by the National Center for Environmental Health at the Centers for Disease Control.
Abbreviations
- CD14
cluster of differentiation 14
- CEU
Utah residents with Northern and Western European ancestry from the Centre d’Etude du Polymorphisme Humain collection (HapMap population)
- CHB
Han Chinese (HapMap population)
- IgE
immunoglobulin E
- IL10
interleukin 10
- IL13
interleukin 13
- JPT
Japanese (HapMap population)
- LD
linkage disequilbrium
- LRP1
low density lipoprotein receptor-related protein 1
- MAF
minor allele frequency
- MCCAS
Mexico City Childhood Asthma Study
- SNP
single nucleotide polymorphism
- SPINK5
serine peptidase inhibitor, Kazal type 5 isoform
- SPT
skin prick test
- STAT6
signal transducer and activator of transcription 6
- UTR
untranslated region
- YRI
Yoruban (HapMap population)
Footnotes
Disclosure of potential conflict of interest: The authors have no conflicts of interest to disclose.
References
- 1.Gupta RS, Springston EE, Warrier MR, Smith B, Kumar R, Pongracic J, et al. The Prevalence, Severity, and Distribution of Childhood Food Allergy in the United States. Pediatrics. 2011;128(1) doi: 10.1542/peds.2011-0204. [DOI] [PubMed] [Google Scholar]
- 2.Hong X, Tsai HJ, Wang X. Genetics of food allergy. Current Opin Pediatr. 2009;21(6):770–776. doi: 10.1097/MOP.0b013e32833252dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009;5(8):e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aas K. Some variables in skin prick testing. Allergy. 1980;35(3):250–252. doi: 10.1111/j.1398-9995.1980.tb01763.x. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 6.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amoli MM, Hand S, Hajeer AH, Jones KP, Rolf S, Sting C, et al. Polymorphism in the STAT6 gene encodes risk for nut allergy. Genes Immun. 2002;3(4):220–4. doi: 10.1038/sj.gene.6363872. [DOI] [PubMed] [Google Scholar]
- 8.Negoro T, Orihara K, Irahara T, Nishiyama H, Hagiwara K, Nishida R, et al. Influence of SNPs in cytokine-related genes on the severity of food allergy and atopic eczema in children. Pediatr Allergy Immunol. 2006;17(8):583–90. doi: 10.1111/j.1399-3038.2006.00463.x. [DOI] [PubMed] [Google Scholar]
- 9.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7(2):95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 10.Granada M, Wilk JB, Tuzova M, Strachan DP, Weidinger S, Albrecht E, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol. 2011 doi: 10.1016/j.jaci.2011.09.029. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42(4):289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]