Abstract
The homodimeric bc1 complexes are membrane proteins essential in respiration and photosynthesis. The ~11 Å distance between the two bL-hemes of the dimer opens the possibility of electron transfer between them, but contradictory reports make such inter-monomer electron transfer controversial. We have constructed in Rhodobacter sphaeroides a heterodimeric expression system similar to those used before, in which the bc1 complex can be mutated differentially in the two copies of cyt b to test for inter-monomer electron transfer, but found that genetic recombination by cross-over then occurs to produce wild-type homodimer. Selection pressure under photosynthetic growth always favored the homodimer over heterodimeric variants enforcing inter-monomer electron transfer, showing that the latter are not competitive. These results, together with kinetic analysis of myxothiazol titrations, demonstrate that inter-monomer electron transfer does not occur at rates competitive with monomeric turnover. We examine results from others groups interpreted as demonstrating rapid inter-monomer electron transfer, conclude that similar mechanisms are likely to be in play, and suggest that such claims might need to be re-examined.
1. Introduction
The cytochrome (cyt) bc1 complexes are central components (as Complex III) of mitochondrial respiratory chains, in bacterial photosynthetic and respiratory chains, and in oxygenic photosynthesis (as cyt b6f) [1–4]. The bc1 complex operates through a Q-cycle mechanism that couples electron transfer from ubihydroquinone (QH2) to cyt c to the generation of the proton gradient that drives ATP synthesis. The Q-cycle mechanism is well characterized, providing a parsimonious explanation for the observed behavior [5–9], but a role for the dimeric nature of the complex has been controversial [10–15]. The dimer interface between cyt b subunits brings the heme bL centers within ~11 Å (for the electron transfer distance between conjugate systems) (Fig. 1), which might be expected to give rate constants much faster than the ms range of turnover (Table 1), leading to rapid electron transfer across the dimer interface. However, our previous work through titrations of QH2 oxidation at the Qo-site with myxothiazol had shown only linear titration curves diagnostic of monomeric function [12]. Recently, three groups have developed heterodimeric systems [16–18] allowing expression of two copies of the cyt b gene, and construction of strains differentially mutated so as to allow unambiguous tests of the hypothesis that electron transfer across the dimer interface occurs. All three groups have claimed to demonstrate an inter-monomer electron transfer between bL hemes by measuring the rapid kinetics expected. One alternative electron transfer scheme, the half-of-sites mechanism [10, 19–21], requires such a transfer, but any scheme depending on a central role for such a process would require major revision of the Q-cycle, and such a paradigm change demands careful scrutiny. We have investigated the inter-monomer electron transfer using a heterodimeric system constructed for R. sphaeroides and were also able to demonstrate in strains mutated to enforce inter-monomer electron transfer the rapid kinetics that they observed. However, the rapid kinetics that they attributed to the inter-monomer reaction are, in our system, accounted for by cross-over recombination to generate a functional wild-type homodimer, and selection of the native sequence to allow survival under photosynthetic growth. From these results we conclude that, if inter-monomer electron transfer occurs, it is not rapid enough to allow effective competition with a monomeric function.
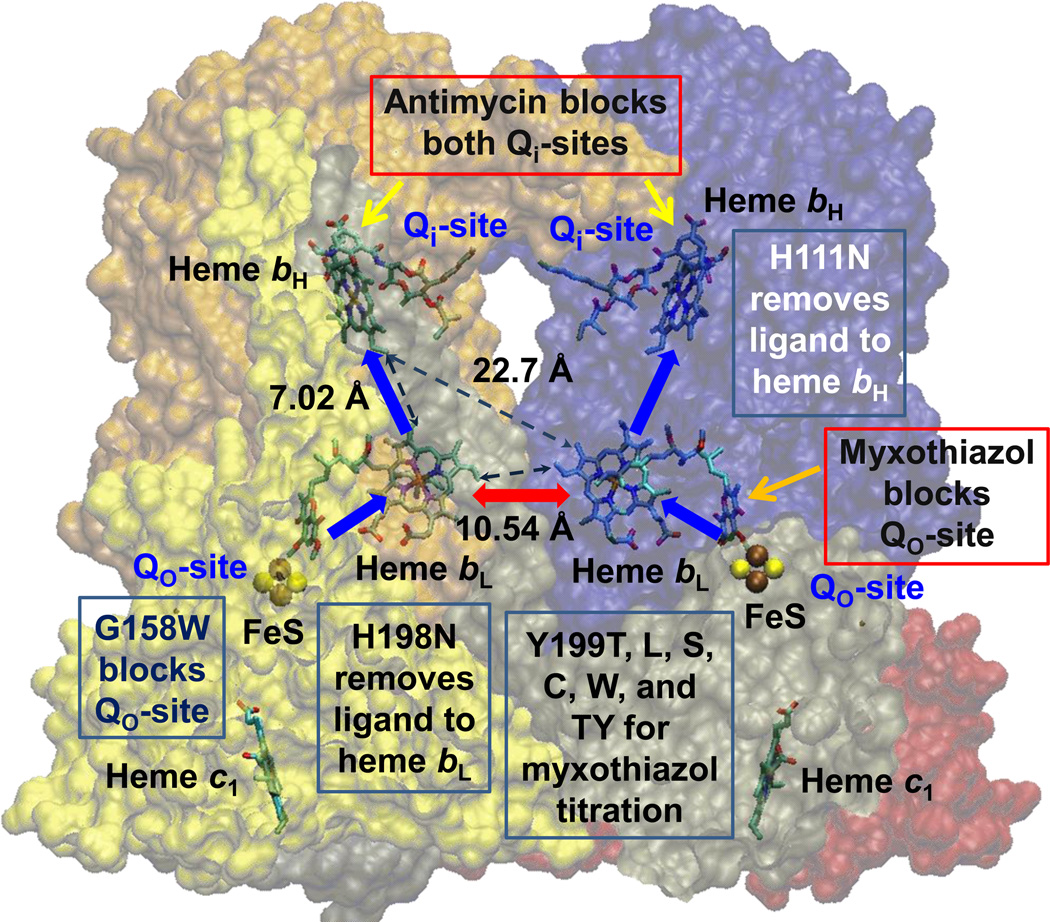
Fig. 1. Scheme to show rationale for the study of inter-monomer electron transfer.
With both Qi-sites blocked by antimycin, electrons cannot exit from heme bH. If one monomer is blocked by myxothiazol at the Qo-site, the unblocked Qo-site would deliver electrons to both monomers if inter-monomer electron transfer could occur rapidly between the bL hemes (red arrow). Blue arrows show monomeric electron transfer. Mutations used to block different partial process were G158W (Qo-site), H111N (heme bH), and H198N (heme bL). See also Table 2 and Fig S1. Structure is from PDB 2QJY [66], in which occupancy by stigmatellin defines the Qo-site, and occupancy by ubiquinone-10 defines the Qi-site.
Table 1.
Rate constants expected from structures and a Moser-Dutton treatment of distance dependence.
| heme b pairs involved and parameters assumed | Marcus-Moser-Dutton (kcat/s−1) |
|---|---|
| bL-bH through 2-vinyl (ΔG° = −60 mV, λ = 0.75 V, R = 7.02 Å) | 1.73 × 108 |
| bL-bH through 2-vinyl (ΔG° = −130 mV, λ = 0.75 V, R = 7.02 Å) | 5.68 × 108 |
| bL-bL through 4-vinyl (ΔG° = −0 mV, λ = 0.75 V, R = 10.54 Å) | 4.08 × 105 |
| bL-bL through 4-vinyl (ΔG° = −60 mV, λ = 0.75 V, R = 10.54 Å) | 1.25 × 106 |
2. Materials and Methods
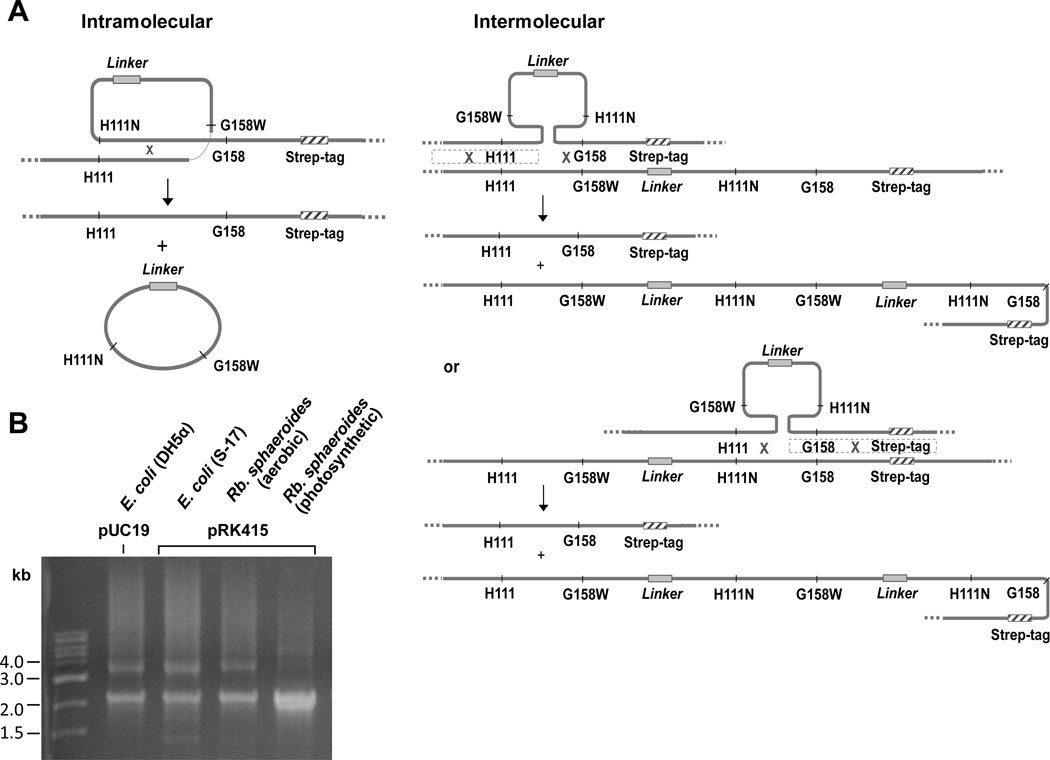
Plasmid pUC19 derivative containing fbc operon (pGB11BH6) was used as a template [22]. Mutant strains in cyt b at position 199 (Y199T, L, S, C, W, and insert Y199TY) were constructed by PCR-mediated site-directed mutagenesis [22–24]. For construction of heterodimeric expression system, linker and strep-tag sequences were inserted using PCR to generate pBST, pNT and pCT (Fig. S1). Site-directed mutagenesis at positions H111, G158 and H198 in cyt b was carried out using Transformer site-directed mutagenesis protocol. Two cyt b genes were fused using the NotI site introduced in the linker region, as described in [18]. All the mutations were verified by DNA sequencing. Growth in strain DH5α was used to amplify pUC19 derivatives. The 2.266 kb NsiI/EcoRI restriction fragment of pNT was replaced with 2.272 kb NsiI/EcoRI restriction fragment containing a factor Xa protease site and a strep-tag from pBST, generating pNTST. The 2.490 kb NotI/EcoRI restriction fragment from the pNTST was then ligated into 5.286 kb NotI/EcoRI restriction fragment of pCT to produce pBBST. Site-directed mutagenesis to generate heterodimeric constructions was performed in pNTST and pCT separately, followed by NotI/EcoRI digestion and ligation (Fig. S1). The oligonucleotide primers and R. sphaeroides strains used are listed in Tables S1 and 2.
The 5.133 kb HindIII/EcoRI restriction fragment containing the fbcFB1B2C operon was subcloned from pUC19 into pRK415, an expression vector, followed by transformation into E. coli S-17. The pRK415 derivative was then mobilized by conjugation from the S-17 into R. sphaeroides BC17, a strain in which the fbc operon had been deleted [24]. The site-directed mutations and the presence of strep-tag sequence at the C-terminus of fused cyt b in R. sphaeroides were confirmed by DNA sequencing after isolation of pRK415 and PCR amplification.
Chromatophores were prepared using the methods described earlier [25]. The computer-linked kinetic spectrophotometer was used for flash-induced kinetic spectrometric analysis as described previously [25]. The chromatophore concentration was adjusted to allow > 90 % saturation of the reaction centers by a single flash. Kinetics of redox changes of reaction center, cyt ctotal, cyt c2, cyt c1, cyt bH and cyt bL were obtained by taking the differences in the kinetics of absorbance changes measured, respectively, at 542 nm, 551-542 nm, 550–554 nm, 552-548 nm, 561–569 nm, and 566-575-0.5(561–569) nm (with small additional corrections for contributions from RC and cyt c), respectively. The measuring beam was turned off, and the reaction mixture in the cuvette was stirred between measurements.
3. Results
3.1 Myxothiazol titration of homodimeric complex to investigate inter-monomer electron transfer
We have previously demonstrated that titration curves for inhibition by myxothiazol of reduction of heme bH via Qo-site turnover in the presence of antimycin were linear, both in wild type and in strains mutated in the putative pathway for electron transfer across the dimer interface [12]. Tyr-199 (Y199) is the residue at the dimer interface in the most plausible pathway for electron transfer between the bL hemes. All strains with single mutations at position 199 in cyt b (Y199T, L, S, C, W), and insert Y199TY, grew at normal rates under anaerobic photosynthetic conditions, and all showed turnover and redox properties for the bc1 complex similar to wild-type. Properties for the strain with a threonine insert (Y199TY) were described earlier [23]. Similar linear titration curves are common in the literature for other Qo-site inhibitors when the Qo-site reaction is limiting [26–28]. Inter-monomer electron transfer would allow reduction of both bH hemes through the Qo-site of either monomer, so that strongly bowed titration curves would be expected. We suggested that the linear titrations observed were diagnostic of a monomeric mechanism [12].
We have extended these earlier studies to the titration behavior under controlled redox conditions after two flashes, and to the kinetic behavior of heme bL. These experiments test the equilibria established for the Qo-site reaction in the presence of antimycin. As shown in Fig. 2, although some small additional reduction of heme bH occurred on the second flash, it was proportionally the same throughout the titration, and there was no indication that any additional complement of heme bH became accessible to reduction through the uninhibited monomer; the titration after two flashes was still strictly linear (Fig. 2A). Instead, reduction of heme bL occurred, and the relative amplitude showed the well-characterized equilibria expected from the monomeric Q-cycle [29] (Fig. 2B). Furthermore, the same behavior was seen in all strains mutated at Y199 in the most direct electron transfer path between the bL hemes. If function of this pathway was critical to the ms kinetics of normal turnover, mutation might be expected to disrupt function, but strains Y199T, L, S, C, and W all showed close to wild-type kinetics. The kinetic behavior was similar even when the interface was modified by insertion of an extra threonine (Y199TY) to mimic the interface in the cyt b6f complex, in which the configuration is substantially different. The behavior observed in all these experiments is consistent with monomeric function in the ms range.
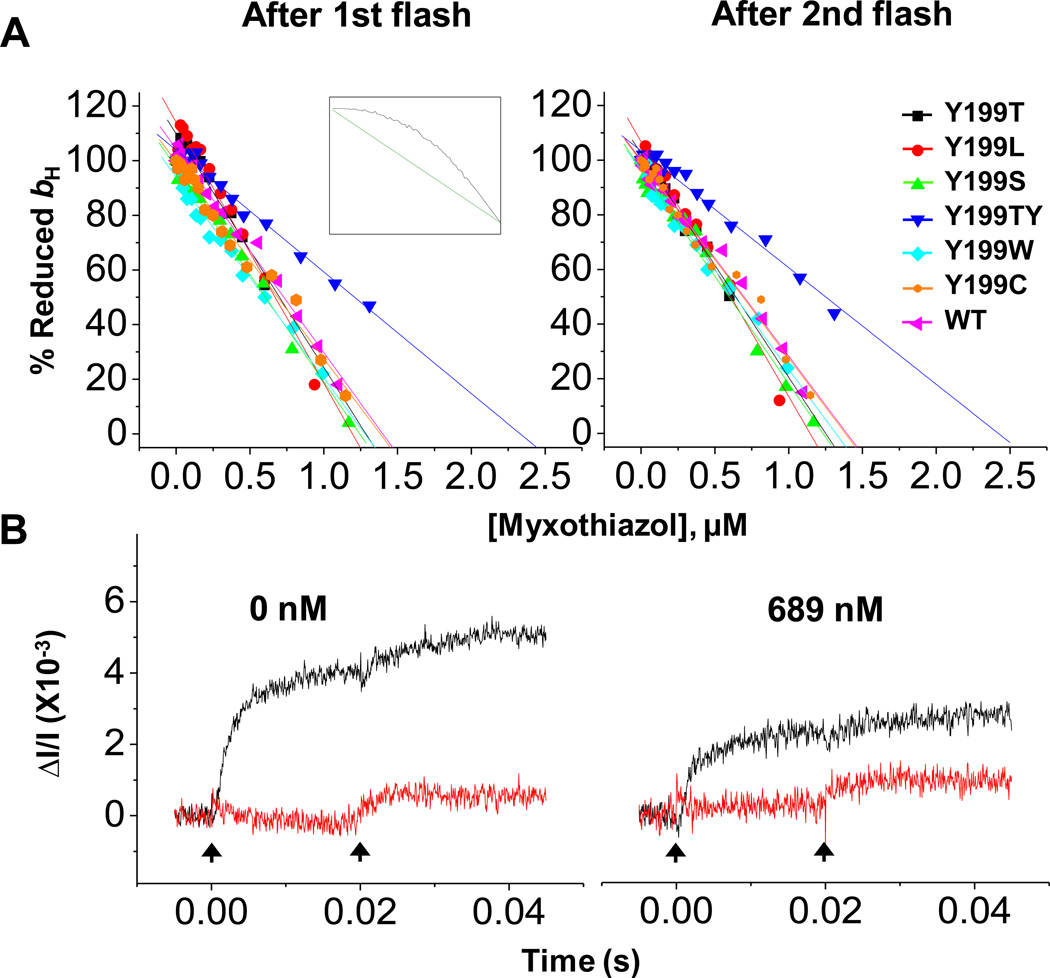
Fig. 2. Titrations of Qo-site with myxothiazol.
(A) Fraction (% of change with no inhibitor) of reduced bH at 20 ms (after 1st flash) and 50 ms (after 2nd flash) versus inhibitor concentration. Chromatophores from native and mutant strains of R. sphaeroides were poised at Eh 100 ±10 mV at pH 7.0 and 20 °C. Rates were measured in the presence of antimycin to block oxidation of heme bH through the Qi-site, as Qo-sites were titrated with myxothiazol. Insert shows titration curves expected without (straight line, green) or with (convex curve, black) inter-monomer electron transfer, from simulation (see [26]). (B) Kinetics of reduction of heme bH (black) and heme bL (red) in wild-type following two flashes. Myxothiazol was added at the concentrations shown. The vertical arrows indicate flash activation. The pattern is diagnostic of monomeric function.
3.2 Mutations in separate cyt b subunits of a heterodimeric complex to explore inter-monomer electron transfer
We have set up a heterodimeric expression system in R. sphaeroides. The general approach was similar to that used in Rhodobacter capsulatus [18], but the procedural details were adjusted to accommodate protocol differences in R. sphaeroides [22, 30] (Fig. S1 and Table S1). The fbc operon was modified so as to contain two copies of the gene for cyt b (fbcB1 and B2), separated by in-frame sequence encoding a linker span joining the two copies of cyt b in C- to N-terminal linkage, to give operon fbcFB1B2C. The linker span had the same sequence as in [18], and the downstream copy (fbcB2) was modified so as to express a factor Xa cleavage site and a strep-tag at the C-terminal end. Site-directed mutagenesis was carried out on fbcB1 and B2 separately before joining with the linker. Using this strategy, we constructed heterodimeric variants in which different partial processes in the two copies of the protein were blocked by suitable mutagenesis. Mutation bG158W at Gly-158 of cyt b introduced a tryptophan side chain into the binding volume of the Qo-site that blocked access of substrate QH2; mutation bH111N at His-111 of cyt b changed a ligand for heme bH so as to prevent incorporation of the heme [30]; and mutation bH198N eliminated a ligand to heme bL [30]. Following the nomenclature used in R. capsulatus [18], the heterodimeric construct with the wild-type sequence in both copies is indicated by B-B. Likewise, WB-BN indicates a construct with the G158W mutation in copy 1 and the H111N mutation in copy 2 of cyt b; , a construct with copy 1 inactivated by both mutations and copy 2 with wild-type sequence; and NB-BN, a construct with a heme bL knocked out through mutation H198N in copy 1 and heme bH knocked out in copy 2. We constructed a full set of combinations of these mutations (Table 2), including knockout mutations for heme bL which have not been previously reported.
Table 2.
Heterodimeric strains and their growth properties.
| Strain | Genotype | Growth properties | |||
|---|---|---|---|---|---|
| fbcB1 | fbcB2 | aerobic | photosynthetic | ||
| B-B | Wild type | Wild type | + | + | |
| NB-B | H111N | Wild type | + | + | |
| WB-B | G158W | Wild type | + | + | |
| H111N/G158W | Wild type | + | + | ||
| Wild type | H111N/G158W | + | + | ||
| WB-BN | G158W | H111N | + | + | |
| NB-BW | H111N | G158W | + | + | |
| H111N/H198N | Wild type | + | + | ||
| Wild type | H111N/H198N | + | + | ||
| NB-BN | H198N | H111N | + | + | |
| NB-BN | H111N | H198N | + | + | |
| NB-BN | H111N | H111N | + | − | |
| WB-BW | G158W | G158W | + | − | |
Table 2 shows strains that could grow photosynthetically at rates similar to wild-type, and those that failed to grow. Strains B-B, WB-BN, and other constructs previously reported [18], all showed similar behavior, all growing photosynthetically under anaerobic conditions. Strains NB-BN, and WB-BW, both failed to grow photosynthetically, but grew under aerobic conditions. These results were in line with those previously reported [18]. The turn-over of the bc1 complex in all the strains designed to express a heterodimeric complex were then assayed by comparing the kinetics of turnover of the photosynthetic chain through absorbance changes of the reaction center and the bc1 complex [6, 25, 29, 31]. We followed reduction and oxidation of the c-hemes and b-hemes in the absence and presence of antimycin, and could demonstrate in strains B-B, WB-BN, and , the same general behavior as reported by [18], as seen in example traces for strain B-B and WB-BN in Fig. 3A. The traces of WB-BN showed rapid electron transfer kinetics similar to wild-type B-B. The spectra generated from such traces also showed involvement of all the Q-cycle redox centers with behaviors similar to the wild-type (Fig. 3B).
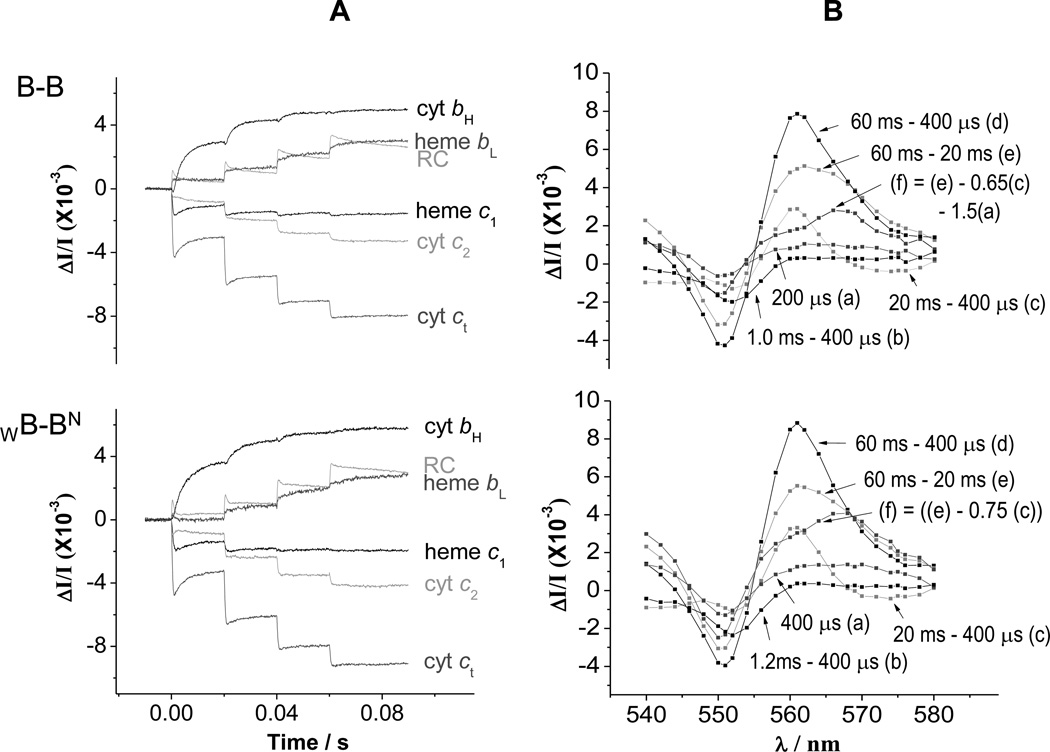
Fig. 3. Kinetics in strains containing heterodimeric constructs.
(A) Kinetic traces for the components of the photosynthetic chain, measured from difference kinetics at the following wavelengths. Reaction center (RC); 542 nm; cyt ct, 551-542 nm; cyt c2, 550–554 nm; heme c1, 552-548 nm; heme bH, 561–569 nm; heme bL, (566–575 nm) – 0.5(heme bH) (with additional small corrections for c-type hemes and RC. (B) Spectra at selected times showing involvement of hemes bH, bL, c1, c2, and RC. (a) Change 400 µs after first flash shows mainly cyt c2 oxidation; (b) over the period 0.4–1.2 ms, heme c1 oxidation dominates the change; (c) from 0.4 to 20 ms, heme bH reduction dominates the kinetics, with a derivative spectrum in the range 548–554 nm showing electron transfer from heme c1 to cyt c2; (d) the changes after the second and subsequent flashes contain contributions from all centers; (e) after the second flash, heme bL and most of the remaining heme bH go reduced; (f) subtraction of a fraction of the change (c) reveals the spectrum of heme bL reduction. RC changes contribute through a rather flat spectrum across this wavelength span, but dominate at 542 nm, where the heme changes are approximately isosbestic. Times given are after flash 1 at 0 s. Flashes are spaced 20 ms apart. Chromatophores from B-B and WB-BN cells were poised at Eh ~120 mV by addition of 2 mM ascorbate, with 2 mM KCN added to inhibit cyt oxidase activity.
Surprisingly, strains NB-BN and NB-BN (not previously reported), in which a ligand for heme bL (H198N) was mutated in one copy of cyt b and one for heme bH (H111N) in the other copy, also grew photosynthetically. The electron transfer distance for the closest available path (the diagonal from heme bL to bH across the dimer interface) is ~23 Å (Fig. 1), giving a limiting value for the rate constant for QH2 oxidation, k~10−1 s−1, so growth would be expected only if some extraordinary reconfiguration of the complex had occurred, in which case some modification of behavior would have been expected. Nevertheless, the kinetics observed were similar to wild-type, excluding this unlikely event. We therefore checked the genetic complement encoding the bc1 complex, using PCR amplification and DNA sequencing that would reveal the genotype encoding the wild-type behavior.
3.3 Homologous cross-over recombination and monomeric vs. inter-monomeric electron transfer
For all cultures used in kinetic experiments, the heterodimeric strains were checked for the presence of the expected mutations by sequence analysis after PCR amplification of the DNA encoding the heterodimeric fbc operon. The amplification was required after plasmid miniprep since the pRK415 plasmid harboring the fbc operon in R. sphaeroides is present in the cell in low copy-number. We used two sets of primers for sequencing of heterodimeric cyt b genes; one set that anneals specifically to the upstream sequence of the gene for cyt b and the linker sequence, and the other to the linker and strep-tag sequences. In all cases, sequence analysis showed in all the mutant strains the presence of the mutated sequence expected.
In order to check for the presence of intact heterodimeric fbcFB1B2C constructs in the mutant strains, we used a different set of primers that anneal to sequences upstream (in fbcF) and downstream (in fbcC) of the gene for cyt bwhich would allow us to distinguish by size of PCR products, the heterodimeric fbcFB1B2C construct from any other forms of fbc operon (Fig. 4B). Interestingly, the pattern of amplified PCR products of the heterodimeric constructs from the cells used for preparation of chromatophores tested in kinetic experiments fell into two groups, one of which showed two major bands that correspond to the sizes of heterodimeric fbcFB1B2C and homodimeric fbcFBCand the other with only one major band at the same position on the gel as homodimeric fbcFBC (Fig. 4A) Sequence analysis showed that the bands with higher molecular weight (MW) retained the heterodimeric constructs with expected mutations whereas the bands with lower MW contained a homodimeric fbc operon with wild-type sequence. The sequencing of the bands also identified in both bands the strep-tag that was used for heterodimeric construction. Since the strep-tag was introduced into the heterodimeric construction specifically for this work, the presence of strep-tag in the bands with lower MW clearly indicated that the DNA encoding the homodimeric bc1 did not come from an extraneous source, but was derived from the heterodimeric fbcFB1B2C construct.
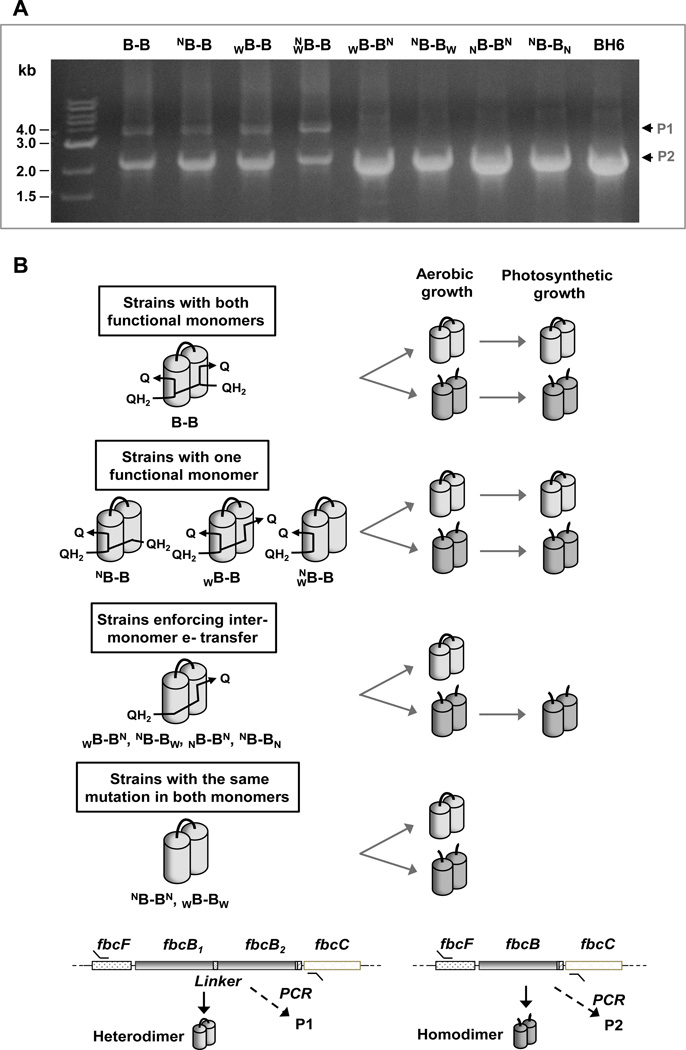
Fig. 4. Selection of functional bc1 complex variants for survival during growth.
(A) Constructs of fbc operon maintained at high abundance during photosynthetic growth. The bands represent PCR products amplified from photosynthetic cultures used in kinetic analysis. Both the heterodimeric construct (P1) and the recombinant homodimeric re-construct (P2) were maintained at high abundance in all strains with at least one functional monomer. On the other hand, in the strains which contained a mutation crippling monomeric function to enforce inter-monomer electron transfer, only the recombinant construct for the wild-type homodimer (P2) was maintained in the culture. The PCR bands from BH6 strain which contains a 6xHis-tagged homodimeric construct, was used as a control size marker. (B) Schematic representation of the selection of constructs coding for functional bc1 complex. The cultures of R. sphaeroides retain only the constructs expressing functional bc1 complex when bc1 complex is required for survival under anaerobic photosynthetic condition.
4. Discussion
4.1 Kinetic analysis
The kinetics of the partial processes measured on turnover of the Qo-site when the Qi-site is blocked by antimycin, reflect the equilibrium constant of the bifurcated reaction, and equilibria in the high and low potential chains. The equilibrium constants are given by the redox potentials of the centers involved. For oxidation of QH2 from the pool in the monomeric case, the overall equilibrium constant is given by , where subscript L and H indicate acceptors in the low and high potential chains. The value of Keq depends on the redox potentials of the acceptors available [5–7]. For the first QH2 oxidized, the acceptors are heme bH (Eo' ~40 mV) and (predominantly) ISPox (Eo' ~300 mV), and, with Q/QH2 at Eo' ~90 mV, Keq1 has a value of ~500. For the second QH2, since the two more favorable acceptors have been consumed, the acceptors are heme bL (Eo' −90 mV) and (predominantly) heme c1 (Eo' 270 mV), and Keq2 has a value ~1. If inter-monomer electron transfer occurred, the equilibria would reflect the ability of one Qo-site to deliver electrons to two equivalents of heme bH, and this would be particularly obvious from kinetics measured near the midpoint of an inhibitor titration. These conditions are important because the second flash pumps up the redox potential in the high-potential chain, increasing the driving force for heme b reduction. The linear titration shows unambiguously that the fractional reduction of heme bH is proportional to the fraction of active Qo-sites remaining. Similarly, the pattern of behavior on two flashes is that expected from a monomeric mechanism. Both are inconsistent with inter-monomer electron transfer without introduction of additional hypotheses. It might be argued that the half-of-sites hypothesis [10, 16, 32] can account for the titration data; if only one monomer is active, all heme bH would still undergo reduction. However, that would require two turnovers of the active Qo-site, and fractional inhibition would then eliminate heme bH reduction proportionately. To account for the monotonic curves observed [10, 16], the mechanism then requires that all steps, including inter-monomer electron transfer, must be at intrinsic rates much faster than the rate determining QH2 oxidation. The observation that such rapid reduction of heme bH could be observed in the presence of antimycin in heterodimeric strains mutated to enforce inter-monomer transfer [16] seemed to provide strong support for such a mechanism. The only missing component was then a plausible explanation for why the Qo-site could function in only one of the monomers. In this context, the classical Q-cycle would have to be amplified by ad hoc accretions, and the natural interpretation of the kinetic behavior that seemed to support it so parsimoniously, would have to be extensively re-interpreted. Clearly, the evidence for inter-monomer electron transfer then needs careful examination before such a major change in paradigm becomes accepted.
It might be argued that the discrepancy between the ~106 s−1 rate constant calculated from distance (Table 1) and the minimal rate (~1 s−1) suggested by the observed kinetics is so large as to discount any explanation. As noted in [12], there are several effects that might account for the difference. As argued by Shinkarev and Wraight [15], competition between alternative pathways to hemes bH and bL can explain part of the discrepancy (about 2-orders of magnitude). A more significant fraction would have to come from quantum mechanical effects [33–39]. The simple approximation from distance-dependence seems to work well in most cases [40–45], but use of the Hopfield approximation [46] in the Moser-Dutton approach [47] leads to loss of detailed balance, suggesting that the quantum mechanical adjustments are made in the wrong part of the equation [12, 48]. Winkler and Gray [49] have shown experimentally that electron transfer to the heme of cyt b562 through a direct path terminating at a liganding histidine can lead to rates lower than those expected from distance by factors ~103, and Prytkova et al. [39] explained this result using a quantum mechanical approach involving path-integrals which showed different contributions from interference effects in such direct pathways compared to edge-linked pathways. Hoffman et al. [50] have shown that electron transfer across a protein interface may be much slower than expected from distance. Other effects to be considered are local electrostatic fields, and distribution of electron or hole occupancies around the conjugate system of the heme acting as donor or acceptor; Walker [51–52] has examined the singly occupied molecular orbital of b-type hemes, and noted an asymmetrical distribution that would favor the intra-monomer case if it applied in the bc1 complex. There are therefore realistic physical mechanisms that might be invoked to explain the slow rate observed.
4.2 A genetic mechanism accounts for the rapid kinetics observed
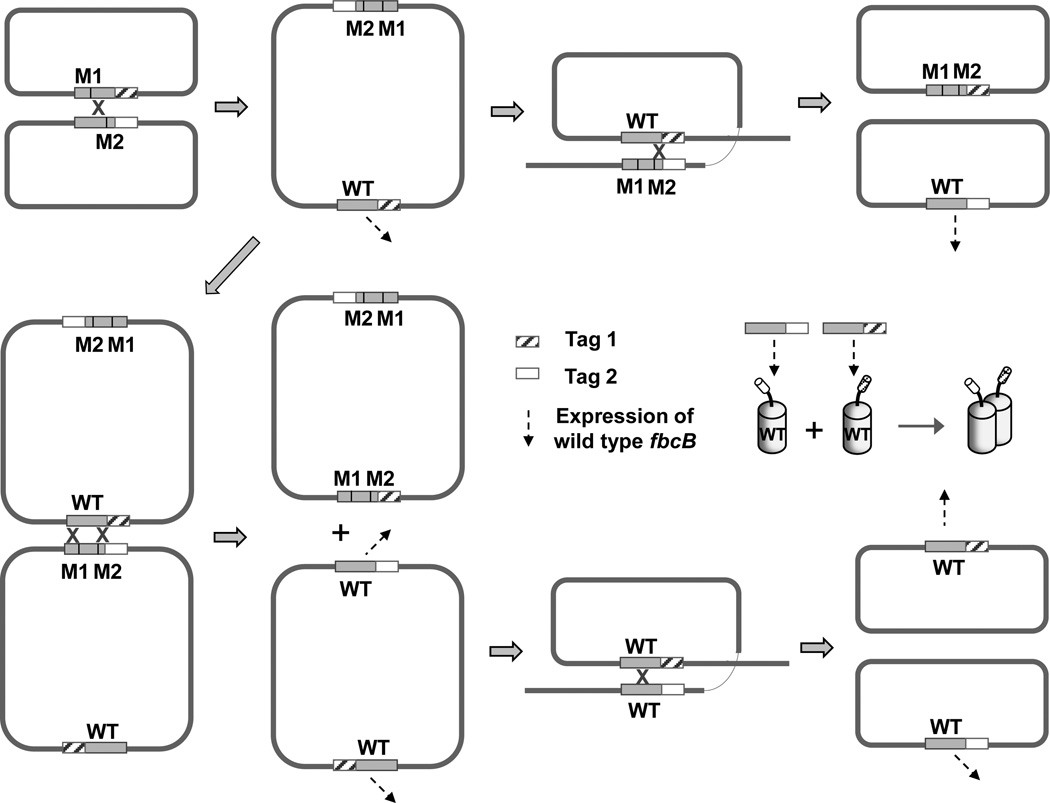
Cross-over by homologous recombination (Fig. 5A) is a well-characterized feature of genetics, ubiquitous in all realms of life [53–54], but was not addressed in [16, 18] and only minimally in [17]. Since this behavior provides an obvious mechanism for the results observed, we checked the sequence of the genetic complement encoding the bc1 complex from R. sphaeroides cells at an early stage of photosynthetic growth in order to find any recombination intermediates. In the strain, we found two homodimeric constructs containing NB and B (wild-type) both with a strep-tag in the lower MW recombination product, which are the intermediates expected by a single cross-over recombination as suggested in the mechanism in Fig. 5A. We also checked for the complement encoding the bc1 complex at different stages in the protocol used for construction of the mutant strains. As seen in Fig. 5B, both the heterodimeric construct, and homodimeric operon generated by recombination, were present even in the initial stages of the protocol. The heterodimeric fbcFB1B2C constructed in plasmids pUC19 and pRK415 in Escherichia coli strains DH5α and S-17 respectively, had been re-constructed to a homodimeric fbcFBCand both types of operon were maintained in the cells, although only the heterodimeric construct had been initially introduced into the strains. The E. coli DH5α and S-17 [55–56] are RecA-deficient strains. However, the substantial homology (~1.37 kb) of the two fbcB genes in tandem in the heterodimeric construct allowed recA-independent cross-over recombination [57–59], producing the recombinant homodimeric operon. Plasmid pRK415 is the vector used in conjugation from E. coli S-17 to R. sphaeroides so it seems possible that both types of operon were transferred to R. sphaeroides on conjugation. As noted in Materials and Methods, all plasmids encoding the bc1 complex were expressed in the BC17 strain of R. sphaeroides with chromosomal deletion of the fbc operon, so that recombination reflects only plasmid events. Although we cannot know whether a particular cell contained one or both operons, the outcome on growth of the cultures was unambiguous. In all cultures, the fbcFBC operon for expression of a wild-type homodimeric bc1 complex was the dominant DNA (Fig. 4A). In strains expressing at least one copy of a functional monomer in the heterodimer (B-B, NB-B, WB-B and ), the heterodimeric construct was also maintained on photosynthetic growth, as shown by the less prominent band. This shows that a monomeric function is sufficient to allow competitive expression. In contrast, in strains in which the heterodimeric construct encoded a complex with different crippling mutations in the two copies of cyt b (WB-BN, NB-BW, NB-BN and NB-BN), only the recombinant operon expressing a homodimeric complex with wild-type cyt b was maintained at significant levels. Although the recombinant homodimeric operon contributed the only significant PCR product in these strains and the heterodimeric construct was not seen at a level we could detect on gels (Fig. 4A), the latter was still present at a level that could provide a template for PCR amplification for sequence analysis when the primers annealing only to the heterodimeric constructs were used (see above). In support of the hypothesis of cross-over recombination, in strains in which both copies of cyt b had the same crippling mutation (NB-BN and WB-BW), no photosynthetic growth was observed, and no re-construction of the wild-type homodimeric expression system was detected.
Fig. 5. Generation of wild-type homodimer by homologous recombination in 1-plasmid approach.
(A) Schematic showing the generation of recombinant construct for homodimer from heterodimeric construct. When two genes with substantially the same sequence are present in the cell, cross-over (x marks) occurs by homologous recombination within a plasmid or between plasmids, shown here by the example of WB-BN. (B) Constructs retained at different stages of WB-BN heterodimeric construct. Only recombinant homodimeric construct with wild-type sequence remained at a significant level when grown photosynthetically. All the PCR products in the major bands contained coding for the strep-tag.
Perhaps the most interesting secondary observation was the difference between levels of the two types of operon in the WB-BN strain when grown under aerobic or anaerobic conditions in R. sphaeroides (Fig. 5B). Under aerobic conditions, R. sphaeroides can grow without a functional bc1 complex by using ubiquinol oxidase, whereas under anaerobic conditions, photosynthetic growth requires a functional bc1 complex. When grown under aerobic conditions, cultures of the WB-BN strain maintained both the heterodimeric fbcFB1B2C and the homodimeric fbcFBC constructs. However, when grown photosynthetically, the cultures maintained only the homodimeric construct expressing the wild-type bc1 complex at levels detectable on gels after unbiased PCR amplification.
From the above, it seems clear that we have observed a pretty example of micro-evolution, driven by the need for a functional bc1 complex. Cells with heterodimeric complexes enforcing inter-monomer electron transfer do not compete effectively with those containing wild-type homodimeric complexes resulting from cross-over recombination. On the other hand, cells expressing heterodimeric complexes with at least one functional monomer are sufficiently competitive as to allow the culture to maintain both expression systems. The obvious conclusion is that inter-monomer electron transfer does not occur at rates competitive with monomeric function.
Although our results pertain only to the particular expression system constructed in R. sphaeroides and our conclusions have a definitive status only in that context, the mechanism of cross-over by homologous recombination is ubiquitous. Whenever two genes with homologous sequence spans are present in the same cell, recombination might be expected [54, 57–60]. Apart from a brief note on reversion frequencies in [17], neither the question of recombination nor steps taken to ameliorate it were mentioned in the earlier papers [16–18]. However, three papers [61–63] containing further details have now become available, which make it abundantly clear that the two labs working with R. capsulatus experienced difficulties similar to those we reported. The critical areas now to be addressed are the paradigm-changing claims made in the three earlier papers, the problems exposed in this paper, the new information from the three more recent papers, and any reinterpretation of the earlier claims needed to accommodate the new data.
In electronic circuits, the bus bar feeds the active components without significant impediment through a high-conductivity path which carries the full current. Our results are clearly in contrast with the claim that in R. capsulatus strains designed to enforce inter-monomer electron transfer"…electrons moved freely within and between monomers…” through a bus bar that “…distributes electrons…within the millisecond time scale of enzymatic turnover…” [18]. The similar claim in [16] to have demonstrated in Paracoccus denitrificans “…the previously proposed half-of-sites reactivity and inter-monomeric electron transfer…” might seem on safer ground, because the 2-plasmid approach used could be less prone to recombination; the same might be said of the more modest claim that inter-monomer electron transfer occurs between bL hemes with high rate and efficiency, sufficient to sustain photosynthetic growth in R. capsulatus [17]. However, all these claims need to be re-evaluated.
For the 1-plasmid work with which our own results can be most directly compared, Czapla et al. [61–62] revealed problems from recombination similar to those demonstrated in our work. They claimed that none of their heterodimeric strains was able to grow under anaerobic photosynthesis, but it is not clear on what experimental basis. However, all strains that could grow under these conditions had reconstructed a homodimeric wild-type complex [62], which is as we had reported. The critical experimental findings from the both groups using R. capsulatus [17–18] were kinetic traces showing reduction of heme bH in the low ms range following flash excitation of the cyclic photosynthetic chain in situ in membrane vesicle preparations (chromatophores). Czapla et al. [61] reported a re-examination of rates, using steady-state measurements in proteins purified by affinity or ion-exchange chromatography from the different strains harboring 1-plasmid constructs. They showed that the complex from WB-BN was at least 10-fold slower than that from B-B. Their Fig. 4 showed the dependence of rate on [cyt c] as substrate, and complexes from all strains showed Michaelis-Menten behavior except WB-BN, where the rate was essentially independent of [cyt c]. A substantial fraction of the activity must therefore have come from some process, either non-enzymatic or of different activity, and the bc1 complex activity was much less than the turnover of 70 s−1 claimed (compared to 408 s−1 in strain B-B). Complexes with one wild-type sequence showed about half the activity of B-B complexes, indicating both that the monomer was fully functional, and that in the homodimer, both monomers function concurrently. These activities are quite compatible with our own results, but incompatible with the claims for the bus-bar model [18]. Even if the turnover of 70 s−1 was applicable, the 17% of wild-type turnover would hardly justify the claim that “…Free and unregulated distribution of electrons acts like a molecular-scale bus bar…”. From the data shown, the true turnover was likely <15 s−1. The extensive efforts to ameliorate the recombination problem led to preparations which were relatively homogeneous, so by taking appropriate measures, the approach could offer a pathway for further characterization of heterodimeric complexes. Nevertheless, all SDS-PAGE gels of preparations isolated by ionexchange still contained significant bands at the cyt b monomer size, and in strain NB-BW from a construct with only 3 residues in the linker [62], this was the dominant band, and the only band labeled by anti-Strep-tag.
The brief review by Khalfaoui-Hassan et al. [63] provides a discussion of the merits of 1-plasmid and 2-plasmid approaches, and a critique of the Osyczka group’s work based on the early experience of the Daldal group with a 1-plasmid system. This followed lines similar to our own. However, they also included an estimate for frequency of recombination in the range 10−2 when using the 1-plasmid approach, at least an order of magnitude greater than that quoted in [62].
Knowledge of the actual rates of recombination in the two systems is critical to an assessment of all other results. The pattern of band density shown by the gels in Figs. 4 and 5 suggests a high probability of recombination in the 1-plasmid system, with the reconstructed fbcFBC operon as the favored end-product even under non-selective conditions. However, these relative densities might be misleading. To compensate for the low copy-number of plasmid pRK415, all bands were derived by PCR amplification; by its nature, the time for synthesis of DNA dimers depends on the length of the sequence amplified, with the consequence that the shorter band could have been preferentially enhanced; other factors also come into play, making quantification difficult. Two possible consequences should be mentioned: i) the relative intensity of bands is not necessarily in contradiction either with the reversion frequencies quoted, or with the relative homogeneity of the protein bands reported for cultures grown under non-selective conditions in [61–62]; ii) the population of heterodimeric complexes in our cultures maintaining the operon was likely substantially higher than is apparent from the band intensities after amplification of the operon DNA in Fig. 4. Consequently, we can be quite confident of our claim for expression of heterodimeric complexes that are effective if at least one monomer is active. This is in contrast to the claim in [62] that such complexes could not support photosynthetic growth.
Rates of recombination by inter- and intra-plasmid mechanisms have been studied in some detail as a function of sequence length, distance, homology, etc. [58–60, 64], from which reversion rates in the range 10−4 seem appropriate for the 2-plasmid approach. These should be compared to reversion rates in the 10−7 range seen for spontaneous point mutations. In Fig. 6 we show examples of pathways through which wild-type sequence could have been reconstructed in either plasmid, while retaining the tags. Even with frequencies in the 10−4 range, a simple calculation shows that cultures would be well-populated by homodimeric variants with wild-type sequence. On growth under selective conditions this population would be exponentially enhanced. The question of interest in interpretation of experimental results is therefore not whether recombination occurred, but in what fraction of the population the wild-type was reconstructed. Three points from the earlier paper [17] deserve critical attention:
The growth pattern in Fig. 3A of [17] is open to interpretation. It could reflect a complete failure of the heterodimeric construct to function and two different levels of reversion, or very weak growth under control of the heterodimeric construct and a more active revertant. Neither interpretation could be used to justify a significant functionality for the heterodimeric constructs.
In either case, it is then necessary to reconciling these data with the kinetic data provided in Figs. 5 and 6 of [17]. These show in chromatophores containing heterodimeric complexes a rapid reduction of heme bH on flash activation. Unbiased expression in the same cell from the two plasmids of the expression system would yield ~25 % of each of the non-functional homodimeric complexes, and maximally ~50 % heterodimeric bc1 complex, with only one heme bH per dimer. Since the content of cyt (c1 + c2), and of RC was adjusted so as to be similar, the maximal heme bH reduction in the presence of antimycin expected would therefore be 25 % of that in wild-type. Of the six kinetic traces showing results from chromatophores prepared from such mixed samples, three assayed in the presence of antimycin showed an amplitude substantially greater than this (44 %, 67% and 61%). All showed halftimes similar (within the accuracy of the noise) to wildtype, and the one trace in which inhibitors were absent showed complete turnover. From other studies reported in many labs, strains with single-site mutations showing comparable bc1 complex activity normally grow rapidly under photosynthetic conditions. These results are therefore clearly in contradiction both with the growth characteristics, and with the amplitudes expected if the kinetics reflect the activity of heterodimeric complexes. However, we note that cross-over could occur in both plasmids, so wild-type reconstructs with a full complement of heme bH (in both monomers) might easily exceed the expected maximal amplitude, give wildtype kinetics, and account for the results.
Since recombination would generate sequence changes without loss of tag (Fig. 6), neither the isolation by sequential use of tags with different affinities, nor the labeling by tag-specific antibodies, could provide any guarantee that the sequences were those anticipated. This would also apply to the construct used in [16].
Fig. 6. Representative cross-over events generating wild type fbcB in 2-plasmid approach.
Single cross-over between two different plasmids containing mutant fbc operons with different mutations in fbcB, generates a dimeric integrant plasmid, and another round of single cross-over recombination allows production of wild-type sequences of fbcB with two different tags. Double cross-over recombination between the two different plasmids and between dimeric integrant plasmids also generates wild type fbcB sequences containing two different tags. M1 and M2 denote different mutation sites in fbcB.
In light of the later papers [61–63], one point is unambiguously clear, that despite the steps taken to mitigate reversion, none of the above results can be used to justify the claim implicit in the earlier papers [16–18] that inter-monomer electron transfer occurs at rates compatible with an essential role in normal forward electron transfer.
Since the half-of-sites mechanism [10, 16, 32] requires a rate for inter-monomer electron transfer faster than the limiting reaction at the Qo-site (otherwise, the two pathways for reduction of heme bH would resolve kinetically), the evidence in support of that hypothesis now rests on Castellani et al. [16], whose results are contradicted by all others. The monotonic reduction of heme bH in heterodimeric bc1 complex with the same kinetics as in wild-type [16] certainly appeared to support the half-of-sites hypothesis, but that conclusion would depend absolutely on an implicit assumption that the complex expressed was that designed. This would be expected only if recombination did not occur in this system, an assumption for which we can see no justification. The reversion rates would likely be similar to those indicated in the literature [17, 58–60, 63–64], and the problems above would have to be dealt with. Since the authors did not discuss the issue, it is not possible to address the question adequately, and since details about growth conditions were not provided, it is not possible to assess the selective pressure pertaining. It is quite unlikely that reversion by recombination could have been avoided; if, as suggested by the papers cited, growth was by respiration with succinate as substrate [65], that would have provided strong selective pressure for a functional bc1 complex.
More rigorous characterization of the samples used experimentally will be needed before the claims made on the basis of kinetic or spectroscopic analysis can be accepted. It would be premature to abandon the simple monomeric Q-cycle mechanism on the basis of the data reported. Our results do not exclude a functional role for inter-monomer electron transfer under some circumstances, but they do establish constraints on its scope, and raise the important question of why the reaction is so slow. If the reaction occurs, its properties in situ need to be established under circumstances in which no wild-type complex is expressed. We have shown in this study that the rapid kinetics seen when heterodimeric strains designed to enforce electron transfer across the dimer interface are grown photosynthetically, are due to cross-over recombination and selection for the wild-type homodimeric complex over the heterodimeric strains. Taken together with the failure to observe any evidence for inter-monomer electron transfer in the ms range in kinetic experiments using myxothiazol titrations, we conclude that inter-monomer electron transfer is not rapid enough to allow effective competition with monomeric function under conditions requiring normal turnover of the bc1 complex, and that monomeric function remains the simplest framework for understanding the normal function.
Highlights.
-
►
Linear titration of myxothiazol inhibition is diagnostic of a monomeric mechanism
-
►
Heterodimeric expression systems generate wild-type homodimer by recombination
-
►
Selection for survival of functional bc1 complex variants occurs during growth
-
►
Intermonomer electron transfer is not competitive with monomeric turnover
Supplementary Material
Acknowledgments
This work was supported by a grant from NIH, NIGMS PHS 5 RO1 GM35438. We acknowledge useful discussions with Artur Osyczka, Pascal Lanciano and Bernd Ludwig at the Bioenergetics GRC, 2011, at which our results were first reported.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baniulis D, Yamashita E, Zhang H, Hasan SS, Cramer WA. Structure-function of the cytochrome b6f complex. Photochem. Photobiol. 2008;84:1349–1358. doi: 10.1111/j.1751-1097.2008.00444.x. [DOI] [PubMed] [Google Scholar]
- 2.Berry EA, Guergova-Kuras M, Huang LS, Crofts AR. Structure and function of cytochrome bc complexes. Annu. Rev. Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- 3.Crofts AR. The cytochrome bc1 complex: function in the context of structure. Annu. Rev. Physiol. 2004;66:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Berry EA, Huang LS, Kim SH. Mitochondrial cytochrome bc1 complex. Subcell. Biochem. 2000;35:541–580. [PubMed] [Google Scholar]
- 5.Crofts AR. The mechanism of ubiquinol: cytochrome c oxidoreductases of mitochondria and of Rhodopseudomonas sphaeroides. In: Martonosi A, editor. The Enzymes of Biological Membranes. New York: Plenum; 1985. pp. 347–382. [Google Scholar]
- 6.Crofts AR, Meinhardt SW, Jones KR, Snozzi M. The role of the quinone pool in the cyclic electron-transfer chain of Rhodopseudomonas sphaeroides. A modified Q-cycle mechanism. Biochim. Biophys. Acta. 1983;723:202–218. doi: 10.1016/0005-2728(83)90120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crofts AR, Shinkarev VP, Kolling DR, Hong S. The modified Q-cycle explains the apparent mismatch between the kinetics of reduction of cytochromes c1 and bH in the bc1 complex. J. Biol. Chem. 2003;278:36191–36201. doi: 10.1074/jbc.M305461200. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell P. Protonmotive redox mechanism of the cytochrome b-c1 complex in the respiratory chain: protonmotive ubiquinone cycle. FEBS Lett. 1975;56:1–6. doi: 10.1016/0014-5793(75)80098-6. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell P. Possible molecular mechanisms of the protonmotive function of cytochrome systems. J. Theor. Biol. 1976;62:327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- 10.Covián R, Trumpower BL. Rapid electron transfer between monomers when the cytochrome bc1 complex dimer is reduced through center N. J. Biol. Chem. 2005;280:22732–22740. doi: 10.1074/jbc.M413592200. [DOI] [PubMed] [Google Scholar]
- 11.Crofts AR. The Q-cycle - A Personal Perspective. Photosynth. Res. 2004;80:223–243. doi: 10.1023/B:PRES.0000030444.52579.10. [DOI] [PubMed] [Google Scholar]
- 12.Crofts AR, Holland JT, Victoria D, Kolling DR, Dikanov SA, Gilbreth R, Lhee S, Kuras R, Kuras MG. The Q-cycle reviewed: How well does a monomeric mechanism of the bc1 complex account for the function of a dimeric complex?, Biochim. Biophys. Acta. 2008;1777:1001–1019. doi: 10.1016/j.bbabio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong X, Yu L, Xia D, Yu CA. Evidence for electron equilibrium between the two hemes bL in the dimeric cytochrome bc1 complex. J. Biol. Chem. 2005;280:9251–9257. doi: 10.1074/jbc.M409994200. [DOI] [PubMed] [Google Scholar]
- 14.Osyczka A, Moser CC, Daldal F, Dutton PL. Reversible redox energy coupling in electron transfer chains. Nature. 2004;427:607–612. doi: 10.1038/nature02242. [DOI] [PubMed] [Google Scholar]
- 15.Shinkarev VP, Wraight CA. Intermonomer electron transfer in the bc1 complex dimer is controlled by the energized state and by impaired electron transfer between low and high potential hemes. FEBS Lett. 2007;581:1535–1541. doi: 10.1016/j.febslet.2007.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellani M, Covian R, Kleinschroth T, Anderka O, Ludwig B, Trumpower BL. Direct demonstration of half-of-the-sites reactivity in the dimeric cytochrome bc1 complex: enzyme with one inactive monomer is fully active but unable to activate the second ubiquinol oxidation site in response to ligand binding at the ubiquinone reduction site. J. Biol. Chem. 2010;285:502–510. doi: 10.1074/jbc.M109.072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanciano P, Lee DW, Yang H, Darrouzet E, Daldal F. Intermonomer electron transfer between the low-potential b hemes of cytochrome bc. Biochemistry. 2011;50:1651–1663. doi: 10.1021/bi101736v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swierczek M, Cieluch E, Sarewicz M, Borek A, Moser CC, Dutton PL, Osyczka A. An electronic bus bar lies in the core of cytochrome bc1. Science. 2010;329:451–454. doi: 10.1126/science.1190899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Covián R, Gutierrez-Cirlos EB, Trumpower BL. Anti-cooperative oxidation of ubiquinol by the yeast cytochrome bc1 complex. J. Biol. Chem. 2004;279:15040–15049. doi: 10.1074/jbc.M400193200. [DOI] [PubMed] [Google Scholar]
- 20.Covián R, Trumpower BL. Regulatory interactions in the dimeric cytochrome bc1 complex: The advantages of being a twin. Biochim. Biophys. Acta. 2008;1777:1079–1091. doi: 10.1016/j.bbabio.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covián R, Trumpower BL. The rate-limiting step in the cytochrome bc1 complex is not changed by inhibition of cytochrome b-dependent deprotonation: implications for the mechanism of ubiquinol oxidation at center P of the bc1 complex. J. Biol. Chem. 2009;284:14359–14367. doi: 10.1074/jbc.M109.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guergova-Kuras M, Salcedo-Hernandez R, Bechmann G, Kuras R, Gennis RB, Crofts AR. Expression and one-step purification of a fully active polyhistidine-tagged cytochrome bc1 complex from Rhodobacter sphaeroides. Protein Expr. Purif. 1999;15:370–380. doi: 10.1006/prep.1998.1018. [DOI] [PubMed] [Google Scholar]
- 23.Kuras R, Guergova-Kuras M, Crofts AR. Steps toward constructing a cytochrome b6f complex in the purple bacterium Rhodobacter sphaeroides: an example of the structural plasticity of a membrane cytochrome. Biochemistry. 1998;37:16280–16288. doi: 10.1021/bi9813476. [DOI] [PubMed] [Google Scholar]
- 24.Yun C-H, Beci R, Crofts AR, Kaplan S, Gennis RB. Cloning and DNA sequencing of the fbc operon encoding the cytochrome bc1 complex from Rhodobacter sphaeroides: Characterization of fbc deletion mutants, and complementation by a site-specific mutational variant. Eur. J. Biochem. 1990;194:399–411. doi: 10.1111/j.1432-1033.1990.tb15633.x. [DOI] [PubMed] [Google Scholar]
- 25.Bowyer JR, Tierney GV, Crofts AR. Secondary electron transfer in chromatophores of Rhodopseudomonas capsulata A1a pho+. Binary out-of-phase oscillations in ubisemiquinone formation and cytochrome b50 reduction with consecutive light flashes. FEBS Lett. 1979;101:201–206. doi: 10.1016/0014-5793(79)81326-5. [DOI] [PubMed] [Google Scholar]
- 26.Bechmann G, Weiss H, Rich P. Nonlinear inhibition curves for tight-binding inhibitors of dimeric ubiquinol-cytochrome c oxidoreductase - evidence for rapid inhibitor mobility. Eur. J. Biochem. 1992;208:315–325. doi: 10.1111/j.1432-1033.1992.tb17189.x. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez-Velasco J, Crofts AR. Complexes or super complexes: Inhibitor titrations show that electron transfer in chromatophores from Rhodobacter sphaeroides involves a dimeric ubiquinol: cytochrome c2 oxidoreductase, and is delocalized. Biochem. Soc. Trans. 1991;19:588–593. doi: 10.1042/bst0190588. [DOI] [PubMed] [Google Scholar]
- 28.Tsai A-L, Kauten R, Palmer G. The interaction of yeast complex III with some respiratory inhibitors. Biochim. Biophys. Acta. 1985;806:418–426. doi: 10.1016/0005-2728(85)90249-x. [DOI] [PubMed] [Google Scholar]
- 29.Meinhardt SW, Crofts AR. The role of cytochrome b-566 in the electron-transfer chain of Rhodopseudomonas sphaeroides. Biochim. Biophys. Acta. 1983;723:219–230. doi: 10.1016/0005-2728(83)90120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yun CH, Crofts AR, Gennis RB. Assignment of the histidine axial ligands to the cytochrome bH and cytochrome bL components of the bc1 complex from Rhodobacter sphaeroides by site-directed mutagenesis. Biochemistry. 1991;30:6747–6754. doi: 10.1021/bi00241a017. [DOI] [PubMed] [Google Scholar]
- 31.Meinhardt SW, Crofts AR. Kinetic and thermodynamic resolution of cytochrome c1 and cytochrome c2 from Rhodopseudomonas sphaeroides. FEBS Lett. 1982;149:223–227. [Google Scholar]
- 32.Trumpower BL. A concerted, alternating sites mechanism of ubiquinol oxidation by the dimeric cytochrome bc1 complex. Biochim. Biophys. Acta. 2002;1555:166–173. doi: 10.1016/s0005-2728(02)00273-6. [DOI] [PubMed] [Google Scholar]
- 33.Balabin IA, Onuchic JN. Dynamically controlled protein tunneling paths in photosynthetic reaction centers. Science. 2000;290:114–117. doi: 10.1126/science.290.5489.114. [DOI] [PubMed] [Google Scholar]
- 34.DeVault D. Quantum mechanical tunnelling in biological systems. Q. Rev. Biophys. 1980;13:387–564. doi: 10.1017/s003358350000175x. [DOI] [PubMed] [Google Scholar]
- 35.Kuki A, Wolynes PG. Electron tunneling paths in proteins. Science. 1987;236:1647–1652. doi: 10.1126/science.3603005. [DOI] [PubMed] [Google Scholar]
- 36.Marcus RA, Sutin N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta. 1985;811:265–322. [Google Scholar]
- 37.Nishioka H, Kimura A, Yamato T, Kawatsu T, Kakitani T. Interference, fluctuation, and alternation of electron tunneling in protein media. 1. Two tunneling routes in photosynthetic reaction center alternate due to thermal fluctuation of protein conformation. J. Phys. Chem. B. 2005;109:1978–1987. doi: 10.1021/jp046282x. [DOI] [PubMed] [Google Scholar]
- 38.Onuchic JN, Beratan DN. A predictive theoretical model for electron tunneling pathways in proteins. J. Chem. Phys. 1990;92:722–733. [Google Scholar]
- 39.Prytkova TR, Kurnikov IV, Beratan DN. Coupling coherence distinguishes structure sensitivity in protein electron transfer. Science. 2007;315:622–625. doi: 10.1126/science.1134862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray HB, Winkler JR. Long-range electron transfer. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3534–3539. doi: 10.1073/pnas.0408029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moser CC, Dutton PL. Engineering protein structure for electron transfer function in photosynthetic reaction centers. Biochim. Biophys. Acta. 1992;1101:171–176. [PubMed] [Google Scholar]
- 42.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunnelling chains of mitochondria. Biochim. Biophys. Acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 43.Moser CC, Page CC, Chen XX, Dutton PL. Biological electron tunnelling through protein media. J. Biol. Inorg. Chem. 1997;2:393–398. [Google Scholar]
- 44.Noy D, Moser CC, Dutton PL. Darwin at the molecular scale: selection and variance in electron tunnelling proteins including cytochrome c oxidase. Biochim. Biophys. Acta. 2006;1757:90–106. doi: 10.1098/rstb.2006.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 46.Hopfield JJ. Electron transfer between biological molecules by thermally activated tunneling. Proc. Natl. Acad. Sci. U. S. A. 1974;71:3640–3644. doi: 10.1073/pnas.71.9.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moser CC, Anderson JL, Dutton PL. Guidelines for tunneling in enzymes. Biochim. Biophys. Acta. 2010;1797:1573–1586. doi: 10.1016/j.bbabio.2010.04.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crofts AR, Rose S. Marcus treatment of endergonic reactions: a commentary. Biochim. Biophys. Acta. 2007;1767:1228–1232. doi: 10.1016/j.bbabio.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler JR, Di Bilio AJ, Farrow NA, Richards JH, Gray HB. Electron tunneling in biological molecules. Pure Appl. Chem. 1999;71:1753–1764. [Google Scholar]
- 50.Hoffman BM, Celis LM, Cull DA, Patel AD, Seifert JL, Wheeler KE, Wang J, Yao J, Kurnikov I, Nocek JM. Differential influence of dynamic processes on forward and reverse electron transfer across a protein-protein interface. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3564–3569. doi: 10.1073/pnas.0408767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker FA. Magnetic spectroscopic (EPR, ESEEM, Mössbauer, MCD and NMR) studies of low-spin ferriheme centers and their corresponding heme proteins. Coord. Chem. Rev. 1999;185–186:471–534. [Google Scholar]
- 52.Walker FA. Models of the bis-histidine-ligated electron-transferring cytochromes. Comparative geometric and electronic structure of low-spin ferro- and ferrihemes. Chem. Rev. 2004;104:589–615. doi: 10.1021/cr020634j. [DOI] [PubMed] [Google Scholar]
- 53.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 54.Smith GR. Homologous recombination in procaryotes. Microbiol. Rev. 1988;52:1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/technology. 1983;1:784–791. [Google Scholar]
- 56.Taylor RG, Walker DC, McInnes RR. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 1993;21:1677–1678. doi: 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bi X, Liu LF. recA-independent and recA-dependent intramolecular plasmid recombination. Differential homology requirement and distance effect. J. Mol. Biol. 1994;235:414–423. doi: 10.1006/jmbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- 58.Bi X, Liu LF. recA-independent DNA recombination between repetitive sequences: mechanisms and implications. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:253–292. doi: 10.1016/s0079-6603(08)60365-7. [DOI] [PubMed] [Google Scholar]
- 59.Lovett ST, Drapkin PT, Sutera VA, Jr, Gluckman-Peskind TJ. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics. 1993;135:631–642. doi: 10.1093/genetics/135.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lovett ST, Hurley RL, Sutera VA, Jr, Aubuchon RH, Lebedeva MA. Crossing over between regions of limited homology in Escherichia coli. RecA-dependent and RecA-independent pathways. Genetics. 2002;160:851–859. doi: 10.1093/genetics/160.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Czapla M, Borek A, Sarewicz M, Osyczka A. Enzymatic activities of isolated cytochrome bc1-like complexes containing fused cytochrome b subunits with asymmetrically inactivated segments of electron transfer chains. Biochemistry. 2012;51:829–835. doi: 10.1021/bi2016316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Czapla M, Borek A, Sarewicz M, Osyczka A. Fusing two cytochromes b of Rhodobacter capsulatus cytochrome bc1 using various linkers defines a set of protein templates for asymmetric mutagenesis. Protein Eng. Des. Sel. 2012;25:15–25. doi: 10.1093/protein/gzr055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khalfaoui-Hassani B, Lanciano P, Lee DW, Darrouzet E, Daldal F. Recent advances in cytochrome bc1: Inter monomer electronic communication? FEBS Lett. 2011;586:617–621. doi: 10.1016/j.febslet.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chedin F, Dervyn E, Dervyn R, Ehrlich SD, Noirot P. Frequency of deletion formation decreases exponentially with distance between short direct repeats. Mol. Microbiol. 1994;12:561–569. doi: 10.1111/j.1365-2958.1994.tb01042.x. [DOI] [PubMed] [Google Scholar]
- 65.Ludwig B. Cytochrome coxidase from Paracoccus denitrificans. Methods Enzymol. 1986;126:153–159. doi: 10.1016/s0076-6879(86)26017-6. [DOI] [PubMed] [Google Scholar]
- 66.Esser L, Elberry M, Zhou F, Yu CA, Yu L, Xia D. Inhibitor-complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides. J. Biol. Chem. 2008;283:2846–2857. doi: 10.1074/jbc.M708608200. [DOI] [PubMed] [Google Scholar]
- 67.Moser CC, Page CC, Farid R, Dutton PL. Biological electron transfer. J. Bioenerg. Biomembr. 1995;27:263–274. doi: 10.1007/BF02110096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.