Abstract
In response to hydrogen peroxide (H2O2), the transcription factor Pap1 from Schizosaccharomyces pombe regulates transcription of genes required for adaptation to oxidative stress and for tolerance to toxic drugs. H2O2 induces oxidation of Pap1, its nuclear accumulation and expression of more than fifty Pap1-dependent genes. Oxidation and nuclear accumulation of Pap1 can also be accomplished by genetic inhibition of thioredoxin reductase. Furthermore, genetic alteration of the nuclear export pathway, or mutations in Pap1 nuclear export signal trigger nuclear accumulation of reduced Pap1. We show here that a subset of Pap1-dependent genes, such as those coding for the efflux pump Caf5, the ubiquitin-like protein Obr1 or the dehydrogenase SPCC663.08c, only require nuclear Pap1 for activation, whereas another subset of genes, those coding for the antioxidants catalase, sulfiredoxin or thioredoxin reductase, do need oxidized Pap1 to form a heterodimer with the constitutively nuclear transcription factor Prr1. The ability of Pap1 to bind and activate drug tolerance promoters is independent on Prr1, whereas its affinity for the antioxidant promoters is significantly enhanced upon association with Prr1. This finding suggests that the activation of both antioxidant and drug resistance genes in response to oxidative stress share a common inducer, H2O2, but alternative effectors.
INTRODUCTION
During the last years, several oxidative stress-sensing pathways responding to fluctuations in hydrogen peroxide (H2O2) have been described. At least two independent but cross-talking pathways, that of the MAP kinase Sty1/Spc1 (with its downstream transcription factor Atf1) and the Pap1 pathways, are activated upon increased intracellular concentrations of H2O2 in the fission yeast Schizosaccharomyces pombe. The b-ZIP-containing transcription factor Pap1, homologue of mammalian c-Jun (pombe AP-1), is essential for normal tolerance to peroxides (1,2). In response to non-toxic doses of H2O2 (extracellular 70–200 μM), Pap1 triggers >2-fold the transcription of 50 genes (3). Many Pap1-dependent gene products are meant to scavenge reactive oxygen species (such as catalase, the peroxiredoxin Tpx1, the sulfiredoxin Srx1 or superoxide dismutase) (3–6), or reverse the oxidative burst (such as thioredoxin, thioredoxin reductase or some glutaredoxins) (7). However, other genes coding for efflux pumps, dehydrogenases, etc., seem to be involved in the cellular defence against multiple drugs (see below).
The molecular mechanisms underlying Pap1 activation are still focus of study. The transcription factor has a hypothetical double nuclear import signal (NLS) and a nuclear export signal (NES), which are recognized by the importin-α Imp1 (8) and the exportin Crm1 (9), respectively. The Crm1-dependent export of Pap1 prevails over the import, and therefore the transcription factor displays cytosolic localization prior to stress imposition (10). In response to H2O2, at least one intramolecular disulfide bond between two cysteine residues is formed in Pap1, which hinders its NES from the nuclear exporter Crm1, resulting in transient Pap1 nuclear accumulation and in Pap1-dependent gene expression (9,11,12). Oxidation of Pap1 by H2O2 requires the participation of the peroxide scavenger/sensor Tpx1 (6,13), and it has been postulated that the thioredoxin–thioredoxin reductase system contribute to both maintaining Pap1 in a reduced state in the absence of stress and returning it in to the inactive/reduced/cytoplasmic form once the gene response has been engaged (12,14).
Activation of Pap1-dependent antioxidant genes by H2O2 is essential to confer wild-type tolerance to the oxidant, as shown on solid plates and in liquid cultures (2,10). But, in fact, Pap1 was first isolated as conferring resistance to multiple drugs, such as brefeldin A, staurosporine or caffeine, when constitutively activated or over-expressed (1,15,16). Multidrug resistant phenotypes are often associated to constitutive activation of oxidative stress signalling pathways in several microbes. We have recently reported that the Pap1-dependent efflux pumps Hba2 and Caf5, two ABC-family transporters, are essential to trigger normal tolerance to caffeine, probably acting as efflux pumps to extrude this and other drugs from the intracellular compartment (17).
In general, three different strategies have been described to confer Pap1-dependent resistance to several drugs (Figure 1A): (i) constitutive oxidation of Pap1 by deletion of the thioredoxin reductase gene; in this particular case, Pap1 export is blocked since the protein is locked in the oxidized conformation; (ii) over-expression of the protein, with concomitant enhancement of Pap1-dependent transcription; this can be genetically accomplished by depletion of a subunit of the 26S proteasome, Pad1 (16,18), or deletion of the ubr1 gene, encoding a ubiquitin ligase which regulates nuclear Pap1 stability (19); (iii) inhibition of Pap1 nuclear export, either by chemical- (9) or temperature-(10) dependent inactivation of the essential Crm1 protein, or by depletion of the Crm1 cofactor Hba1 (20). We demonstrate here that, unexpectedly, this gain of drug resistance does not correlate with enhanced tolerance to oxidative stress. In fact, defects in nuclear export render Pap1 being insensitive to H2O2-mediated oxidation, probably due to the cytoplasmic localization of its upstream redox transmitter, Tpx1. Analysis of the transcriptome of these constitutively nuclear-expressing cell types indicate that the drug tolerance genes are being activated under basal conditions, but not the antioxidant ones. In fact, we have determined that only oxidized nuclear Pap1, but not the reduced one, interacts with the transcription regulator Prr1 and activates also antioxidant genes. The distinct regulation of these two subsets of genes may reflect an evolutionary merge of previous and independent oxidative stress and multidrug resistance responses.
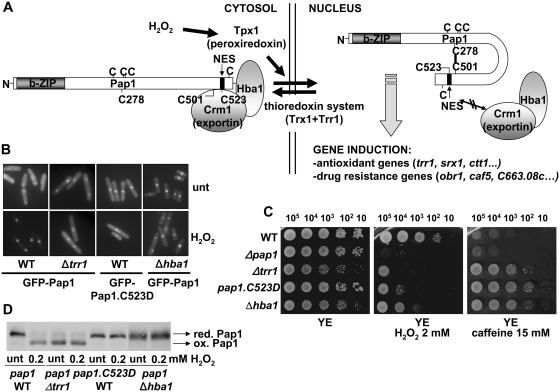
Figure 1.
Genetic inhibition of Pap1 export renders a transcription factor insensitive to H2O2 stress. (A) Schematic representation of Pap1 activation by H2O2. Upon peroxide stress, Tpx1 mediates disulfide bond formation in Pap1, which hinders the recognition by the exportin Crm1 and its cofactor Hba1 to the Pap1 NES. Nuclear accumulation of oxidized Pap1 triggers transcription both antioxidant and drug resistance genes. The relative position of the seven cysteines residues (C) in Pap1 is indicated. (B) Localization of Pap1 in wild-type and mutant strains. The cellular distribution of GFP-Pap1 was determined by fluorescence microscopy in EHH14 (WT), EHH14.C523D (Pap1.C523D), AV19 (Δtrr1) and EA33 (Δhba1) treated or not with 0.2 mM H2O2 for 5 min. (C) Constitutively nuclear Pap1 confers resistant to caffeine but not to H2O2. Strains IC2 (WT), IC1 (Δpap1), NG25 (Δtrr1), IC2.C523D (pap1.C523D) and caf1::ura4+ (Δhba1) were grown in liquid YE media, and the indicated number of cells were spotted onto plates with or without 2 mM H2O2 or 15 mM caffeine. (D) In vivo oxidation of Pap1 in wild-type and mutant strains. Strains IC2 (WT), NG25 (Δtrr1), IC2.C523D (pap1.C523D) and caf1::ura4+ (Δhba1) were treated or not with 0.2 mM H2O2 for 5 min. TCA extracts were processed by non-reducing SDS–PAGE and analysed by western blot with antibodies against Pap1. Reduced/inactive (red. Pap1) and oxidized/active (ox. Pap1) Pap1 forms are indicated with arrows.
MATERIALS AND METHODS
Yeast strains and growth conditions
The origins and genotypes of strains used in this study are outlined in Supplementary Table S1. Cells were grown in rich medium (YE) or in synthetic minimal medium as described previously (21).
Plasmids
The integrative plasmid p85.41x (11) was used to generate a Δprr1 strain expressing GFP-Pap1. The prr1 coding sequence was PCR-amplified from S. pombe cDNA using specific primers and cloned into the nmt (no message in thiamine)-driven expression vector pREP.41 (22) to yield plasmid p397.41.
RNA analysis
Total RNA from S. pombe minimal media cultures was obtained, processed and transferred to membranes as described previously (11). Membranes were hybridized with [α-32P] dCTP-labelled caf5, obr1, SPCC663.08c, trr1, srx1 or ctt1 probes, containing the complete open reading frames. We used ribosomal RNA, tfb2 or act1 as loading controls.
H2O2 sensitivity assay
For survival on solid plates, S. pombe strains were grown, diluted and spotted in YE5S media agar plates as described previously (17), containing 2 mM H2O2 or 15 mM caffeine.
Preparation of S. pombe TCA extracts and immunoblot analysis
To analyse the in vivo redox state of Pap1, trichloroacetic acid (TCA) extracts were prepared as described elsewhere (6). Immunoblotting was performed as described previously (23). Pap1 was immunodetected using polyclonal anti-Pap1 antibody (12). A similar protocol but without alkaline phosphatase treatment was followed to obtain TCA extracts to detect Prr1-HA. Immunoblotting was performed using monoclonal anti-HA antiserum (12CA5).
Fluorescence microscopy
Fluorescence microscopy and image capture was performed as described before (12).
Chromatin immunoprecipitation
To test the binding of Pap1 and Prr1 to all six promoters, the indicated strains were grown in minimal media, and chromatin isolation and immunoprecipitation was performed as described previously (24) but with 10 min instead of 20 min for cross-linking and 1 μl of polyclonal anti-Pap1 and monoclonal anti-HA antiserum (12CA5) (40 and 10 μg, respectively, of each antiserum). Also, the specific primers for Pap1 dependent genes, amplifying promoters, corresponded to the following positions with respect to the translation initiation sites: −222 to −117 of the trr1 gene; −359 to −258 of the srx1 gene; −506 to −403 of the ctt1 gene; −238 to −164 of the caf5 gene; −398 to −322 of the obr1 gene and −121 to −44 of the SPCC663.08c gene. Control primers, spanning an intergenic region of S. pombe chromosome I (position 465 226 to 465 326) were also used. Results were expressed as a percentage of the input. The error bars (SD) were calculated from biological triplicates.
Co-immunoprecipitation analysis
Cells from 100 ml of rich media cultures at an OD600 of 0.5 (1 × 107 cells) were pelleted and re-suspended in lysis buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40), and lysed with two 60-s pulses in a cryogenic grinder (6770 Freezer/Mill; SPEX SamplePrep). Lysates were centrifuged for 5 min at 6000g and supernatants transferred to fresh microtubes. Prr1-GFP was immunoprecipitated from cleared supernatants by adding 10 μl of GFP-Trap beads (Chromotek) for 1 h at 4°C. Immunoprecipitates were washed twice with dilution buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA), and samples were then treated with 0.04 U/µl of alkaline phosphatase for 30 min at 37°C, to avoid broad bands after electrophoresis. Proteins were released from immunocomplexes by boiling for 5 min in sodium dodecyl sulfate (SDS) loading buffer. Samples were separated by 8% SDS–polyacrylamide gel electrophoresis (PAGE) and detected by immunoblotting with polyclonal anti-Pap1 (12) or anti-GFP antiserum, raised against a fusion protein GST-GFP purified from E. coli, following standard rabbit immunization procedures.
RESULTS
Cells expressing constitutively nuclear Pap1, regardless of its oxidation state, are resistant to multiple drugs but not to oxidative stress
While Pap1 has been designed to remain in the cytosol in the absence of oxidative stress (Figure 1A), induction of its oxidation by deletion of the thioredoxin reductase-coding gene can render a constitutively oxidized and nuclear Pap1 (Δtrr1, Figure 1B and D). Alternatively, defects in the Crm1-dependent export machinery (i.e. cells lacking the Crm1 cofactor Hba1) or alteration of the NES in Pap1 (i.e. Cys-to-Asp mutation of residue 523 of Pap1, expressed from the pap1 chromosomal locus) can block Pap1 in the nucleus in the absence of stress (Figure 1B). In all three cases (Δtrr1, Δhba1 and pap1.C523D strains), constitutively nuclear Pap1 significantly enhances S. pombe resistance to caffeine, as previously described (Figure 1C). Unexpectedly, while cells lacking Trr1 display some sensitivity to peroxides probably due to the lack of an active thioredoxin reducing system, cells lacking Hba1 or expressing constitutively nuclear Pap1.C523D are not resistant to peroxides, and, in fact, display more sensitivity than wild-type cells (Figure 1C).
Constitutively nuclear Pap1 is not sensitive to oxidation by H2O2
We analysed the redox state of Pap1 in the four strain backgrounds. As shown before, Δtrr1 cells display oxidized Pap1 even prior to stress, what explains its constitutively nuclear localization (12) (Figure 1D). Surprisingly, inhibition of Pap1 export by mutating the NES or by Hba1 depletion renders a constitutively nuclear Pap1 which is unable to become oxidized in the presence of H2O2 stress (Figure 1D). We have determined that Tpx1, the H2O2 sensor that initiates the redox relay towards Pap1 (Figure 1A), has cytoplasmic localization (our unpublished data), and we suspect that this may cause the inability of constitutively nuclear Pap1 to become oxidized in the presence of peroxide stress; that explains why cells expressing constitutively reduced Pap1 have more sensitivity to H2O2 than wild-type cells (compare WT to pap1.C523D or Δhba1; Figure 1C).
Constitutively nuclear Pap1 can bind and activate a subset of Pap1-dependent genes
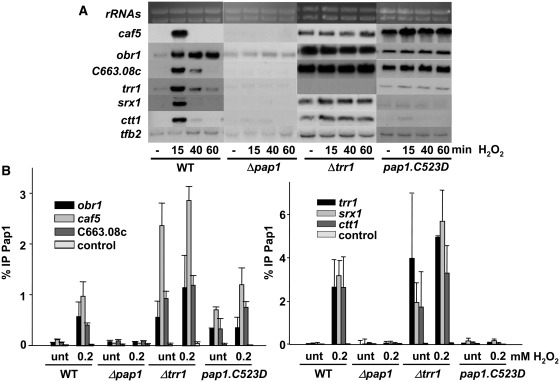
We decided to study how constitutively nuclear Pap1 altered the pattern of induction of the transcriptional response to peroxides. We analysed by northern blot the activation of several genes which expression is strongly triggered in a Pap1-dependent manner by low doses of H2O2: the ctt1, srx1 and trr1 genes, coding for important antioxidant activities (catalase, sulfiredoxin and thioredoxin reductase, respectively); and the caf5, obr1, and SPCC663.08c, coding for an ABC-containing efflux pump, an ubiquitinated histone-like protein and a putative dehydrogenase, respectively. As observed in Figure 2A, cells lacking Trr1 (and therefore expressing oxidized nuclear Pap1) display constitutive expression of all Pap1-dependent genes (except trr1), whereas cells expressing constitutively reduced and nuclear Pap1 (pap1.C523D) only display constitutive activation of caf5, obr1 and SPCC663.08c, and are unable to trigger the antioxidant genes upon stress. Similar results were observed in cells lacking Hba1 (Supplementary Figure S1A).
Figure 2.
Constitutively reduced/nuclear Pap1 can only bind and activate a subset of Pap1-dependent genes. (A) Stress-dependent transcriptional analysis of wild-type and mutant strains. Cultures of strains IC2 (WT), IC1 (Δpap1), NG25 (Δtrr1) and IC2.C523D (pap1.C523D) were treated or not with 0.2 mM H2O2 for the indicated times. Total RNA was obtained and analysed by northern blot with probes for trr1, srx1, ctt1, caf5, obr1 and SPCC663.08 c. Ribosomal RNA (rRNAs) and tfb2 are shown as loading controls. (B) Oxidized/nuclear Pap1 is recruited to all Pap1-dependent promoters. Cultures of the strains used in Figure 2A were treated with 0.2 mM H2O2 for 5 min. ChIP experiments using anti-Pap1 antibody, coupled to quantification by real-time PCR, were performed using primers covering only promoter regions of trr1, srx1, ctt1, caf5, obr1 and SPCC663.08c genes. Primers of an intergenic region were used as a negative control (control). Error bars (SD) for all ChIP experiments were calculated from biological triplicates.
Reduced Pap1, when located in the nucleus, was therefore unable to trigger RNA polymerase II-dependent transcription at some promoters. To test whether reduced/nuclear Pap1 was capable of binding DNA in vivo, we used chromatin immunoprecipitation (ChIP) assay at all six promoters, and observed that oxidized/nuclear Pap1 constitutively binds to both sets of promoters (Δtrr1, Figure 2B left and right panels). On the contrary, reduced/nuclear Pap1 constitutively binds to the obr1, caf5 and SPCC663.08c promoters (pap1.C523D, Figure 2B, left panel), but not to the antioxidant promoters (pap1.C523D, Figure 2B, right panel). Similar results were observed in cells lacking Hba1 (Supplementary Figure S1B). It is worth mentioning that the phenotypes of strain Δhba1 are less severe than that of cells expressing Pap1.C523D, probably due to the non-essential character of Hba1 regarding Crm1-mediated export.
The transcription factor Prr1 is essential for Pap1-dependent activation of the antioxidant, but not the drug tolerance, genes
We have established that there are at least two subsets of Pap1-dependent genes: the drug resistance ones only require nuclear Pap1, independent of its redox state, whereas the antioxidant genes need Pap1 to be oxidized as well. We decided to search for other transcription factors which could mediate this different pattern of gene induction. The response regulator Prr1, homologous to the Saccharomyces cerevisiae SKN7, has been shown to be important for the oxidative stress response in S. pombe, since a strain lacking this transcription factor is very sensitive to H2O2 (25) (Figure 3A). In its absence, induction of some Sty1-Atf1- and/or Pap1-dependent genes is severely compromised (25,26).
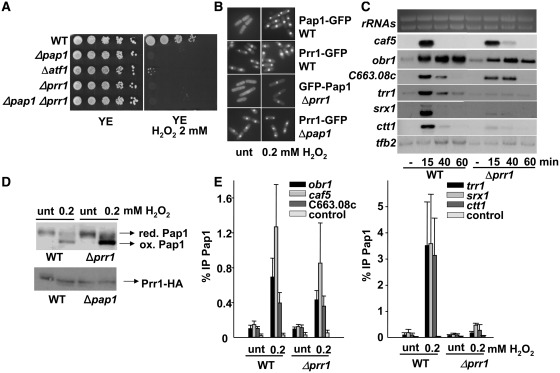
Figure 3.
The response regulator Prr1 is required for the activation of the antioxidant, but not the drug resistance Pap1-dependent genes. (A) Prr1 is sensitive to H2O2 stress. Serial dilutions from cultures of strains IC2 (WT), IC1 (Δpap1), MS46 (Δatf1), MC16 (Δprr1) and MC18 (Δpap1Δprr1) were spotted onto rich plates with or without H2O2. (B) Prr1 has constitutively nuclear localization. The cellular distribution of Pap1 or Prr1 was determined by fluorescence microscopy of strains MC41 (prr1-GFP), MC42 (pap1-GFP), IC81 (Δprr1 with integrative nmt::GFP-pap1) and IC83 (Δpap1 prr1-GFP) treated or not with 0.2 mM H2O2 for 5 min. (C) Northern blot analysis of Pap1-dependent genes. Total RNA from strains IC2 (WT) and MC16 (Δprr1) was obtained from cultures treated with 0.2 mM H2O2 for the indicated times, and analysed as described in Figure 2A. (D) In vivo oxidation of Pap1 is Prr1-independent. The redox state of Pap1 in IC2 (WT) and MC16 (Δprr1) strains, and of Prr1 in MC40 (prr1-HA) and IC64 (Δpap1 prr1-HA) strains were analysed by western blot after non-reducing electrophoresis, with or without stress. (E) ChIP analysis of Pap1 recruitment to stress promoters in strains IC2 (WT) and MC16 (Δprr1) was performed as described in Figure 2B.
To deeply characterize the function of Prr1 regarding Pap1-dependent gene induction, we first determined that Prr1, which is constitutively nuclear regardless the presence of Pap1, did not affect the nuclear accumulation of Pap1 upon H2O2 stress (Figure 3B). Importantly enough, the absence of Prr1 allowed H2O2-dependent activation of caf5, obr1 and SPCC663.08c, but not of the antioxidant genes (Figure 3C). Since those are the genes which require oxidized Pap1 for activation (Figure 2A), we tested whether Prr1 was affecting Pap1 oxidation in the presence of peroxides. As shown in Figure 3D, cells lacking Prr1 were perfectly able to oxidize Pap1. However, whereas the binding of Pap1 to the drug tolerance genes was unaffected in Δprr1 cells (Figure 3E, left panel), its recruitment to the antioxidant genes was severely impaired (Figure 3E, right panel). We conclude that Prr1 facilitates binding of oxidized Pap1 to one subset of promoters, but not to the other.
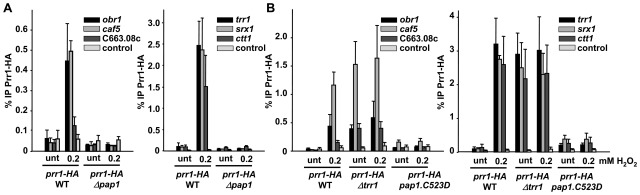
We then analysed the capacity of Prr1 to bind to both sets of promoters by ChIP. As observed in Figure 4A, Prr1 is recruited to all six promoters after mild oxidative stress in a Pap1-dependent manner. Importantly enough, Pap1 is only able to drag Prr1 to DNA in its oxidized form: cells lacking Trr1 constitutively display Prr1 bound to all promoters, whereas in cells expressing Pap1.C523D, which is nuclear and cannot become oxidized, Prr1 is not detected at DNA either before or after stress imposition (Figure 4B).
Figure 4.
Prr1 is recruited to all Pap1-dependent promoters in an oxidized Pap1-dependent manner. (A) Strains MC40 (prr1-HA) and IC64 (Δpap1 prr1-HA) were treated or not with 0.2 mM H2O2 for 5 min. ChIP of Prr1 using anti-HA antibody was performed as described in Figure 2B. (B) Same as in Figure 4A, with strains MC40 (prr1-HA), PG7 (Δtrr1 prr1-HA) and PG3 (pap1.C523D prr1-HA).
Oxidized Pap1 interacts with Prr1, and that interaction is required for the activation of antioxidant genes
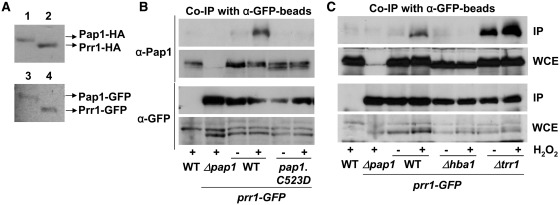
Our experiments predict that oxidized Pap1 may form a complex with Prr1 at the nucleus, and that such association may be required for binding of Pap1 to some, but not all, promoters. We first evaluated the relative concentrations of cellular Pap1 and Prr1 by tagging each gene at their chromosomal loci with either HA or GFP, and therefore maintaining their own promoters. Dismissing a putative effect of both HA and GFP tags on protein stability, the amount of cellular Prr1 is slightly higher than that of Pap1 (Figure 5A), what would suggest that all Pap1 molecules, once accumulated at the nucleus, could potentially associate to Prr1 in vivo. We then tested whether Pap1 and Prr1 interact in vivo by immunoprecipitating Prr1-GFP, and using polyclonal antibodies against Pap1. The Prr1-GFP-expressing strain was completely wild-type regarding gene induction and sensitivity to peroxides (data not shown). As shown in Figure 5B, a clear co-immunoprecipitation between Prr1-GFP and Pap1 was detected only after stress imposition. Importantly enough, only oxidized/nuclear Pap1 was able to interact with Prr1, since cells lacking Hba1 or expressing the non-exportable Pap1.C523D protein did not co-immunoprecipitate (Figure 5B and C). On the contrary, the association between Prr1-GFP and Pap1 was constitutive in Δtrr1 cells, where Pap1 is oxidized even prior to stress imposition (Figure 5C).
Figure 5.
Oxidized Pap1 and Prr1 interact in vivo. (A) Pap1 and Prr1 are expressed at similar levels in cells. Pap1 and Prr1 were tagged at their genomic loci with HA (upper panel) or GFP (lower panel), and protein concentration was compared from 10 μg of total TCA extracts of strains IC70 (1; pap1-HA), MC40 (2; prr1-HA), MC42 (3; pap1-GFP) and MC41 (4; prr1-GFP) by western blot using antibodies against HA or GFP. (B and C) Pap1 and Prr1-GFP interact in vivo after H2O2 stress. Strains 972 (WT), IC83 (Δpap1 prr1-GFP), MC41 (WT prr1-GFP), IC97 (Pap1.C523D prr1-GFP), PG20 (Δhba1 prr1-GFP) and IC102 (Δtrr1 prr1-GFP) were treated (+) or not (−) for 5 min with 0.2 mM H2O2. Native extracts were obtained, and 2 mg of total protein extracts were immunoprecipitated with GFP-trap beads. The resulting immunoprecipitates were analysed by SDS–PAGE and blotted with anti-Pap1 or anti-GFP antibodies. As a loading control, 40 µg of whole-cell extracts were loaded (WCE).
DISCUSSION
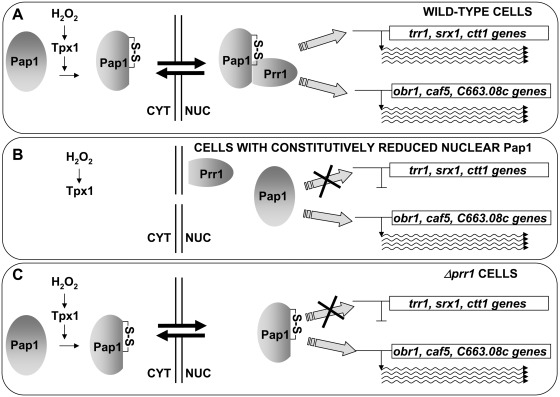
Components of oxidative stress signalling pathways have often been isolated in screens of general drug resistance, since several of these regulons include genes reported to contribute to drug export or detoxification. Here, we show that the Pap1-dependent gene expression program can be classified into two different and not overlapping subsets. The first one codes for activities responsible for multidrug resistance, and are triggered by nuclear Pap1, irrespective of its oxidation state. The second one, which includes traditional antioxidant genes, is only engaged by oxidized Pap1, which then binds to Prr1 and recognizes these promoters (Figure 6).
Figure 6.
Association of oxidized Pap1 and Prr1 is required for the activation of the antioxidant, but not the drug resistance, genes. (A) In wild-type cells, oxidation of Pap1 upon H2O2 stress induces its nuclear accumulation and its association with Prr1. The heterodimer is then able to activate both sets of promoters, the antioxidant (trr1, srx1, ctt1) and the drug resistance (obr1, caf5, c663.08c) genes. (B) In cells defective in Pap1 export (such as cells lacking Hba1 or expressing Pap1.C523D), the transcription factor cannot be oxidized by H2O2, cannot associate with Prr1 and can only trigger transcription of the drug resistance genes. (C) Similarly, in cells lacking Prr1, H2O2-oxidized Pap1 will only be able to activate drug resistance genes.
We have classified as ‘drug tolerance genes’ all those which are expressed upon nuclear Pap1 accumulation, but which do not require Pap1 oxidation or the presence of Prr1: caf5, obr1 and SPCC663.08c. In fact, over-expression of the efflux pump Caf5 alone seems to be sufficient to explain the drug resistant phenotype (17). The obr1 gene product, one of the first polypeptides which Pap1-dependent over-expression was detected in drug resistant strains (27), was soon discarded as mediator of the multidrug resistance phenotype (15), although its function is still unknown. Regarding the uncharacterized gene product SPCC663.08c, it has weak similarity to rat carbonyl reductase 1, which using a short chain dehydrogenase domain may catalyse the reduction of prostaglandins, steroids and ketone-containing xenobiotics.
Prr1 belongs to the two-component systems, which in S. pombe consists on three histidine kinases, a unique phosphotransmitter protein Mpr1 and two response regulators, Mcs4 and Prr1 [for a review, see (28)]. It has recently been shown that the upstream two-component regulation of Prr1 is required for the response of cells to high levels of H2O2, which is mediated by the Sty1-Atf1 pathway (29). On the contrary, we suspect that regulation of Prr1 by Mpr1 is not required for activation of Pap1-dependent genes, since cells lacking the phosphotransmitter Mpr1 can properly induce both caf5 and trr1 (Supplementary Figure S2A), and the H2O2-dependent recruitment of Prr1 to both sets of promoters in cells lacking Mpr1 is very similar to that of wild-type cells (Supplementary Figure S2B). Similarly, phosphorylation of the Prr1 S. cerevisiae homologue, SKN7, has been shown to be dispensable to trigger the oxidative stress response genes (30). Fission yeast cells lacking Prr1 are not only sensitive to H2O2 (Prr1 contributes to the regulation of both Sty1-Atf1-global stress genes and Pap1-dependent antioxidant genes, as characterized here), but are sterile since they are unable to induce ste11 expression, coding for a master regulator of the meiotic pathway (31).
We believe that oxidation of Pap1 not only hinders its Crm1-recognizing domain which causes its accumulation at the nucleus, but also exposes a Prr1-interacting domain, which may contribute to Prr1 participating in the activation of the Pap1-dependent genes rather than on its other reported cellular functions, similarly to what has been described for the budding yeast homologues SKN7 and YAP1 (32). It is worth pointing out that these two S. cerevisiae transcription factors, SKN7 and YAP1, orthologues of Prr1 and Pap1, respectively, have been shown to co-regulate several stress genes (33). They were both described to be important for wild-type tolerance to H2O2, but differentially required for cadmium resistance, and to co-operate in the induction of only a subset of H2O2-dependent proteins (34). Even though several reports have been focused on the identification of SNK7 and YAP1 DNA binding sites (35–37), or in the protein domains involved in their putative hetero-dimerization (32), the H2O2-mediated oxidation of YAP1 has never been demonstrated to be required for its association with SKN7.
How is Prr1 contributing to the regulation of Pap1-dependent antioxidant genes? We have evidences suggesting that Prr1 enhances the access or affinity of Pap1 for the antioxidant promoters, but may also contribute to transcriptional up-regulation of these genes as a transcription factor. On one hand, Pap1 recruitment to the trr1, srx1 and ctt1 promoters is severely impaired in the absence of Prr1 (Figure 3E). We suspect that the access/affinity of Pap1 alone for the drug resistance promoters is high enough in the absence of Prr1 as to allow Pol II transcription, whereas association with Prr1 is required to enhance the recruitment of the Prr1-Pap1 complex to the antioxidant promoters. An in vivo confirmation of this hypothesis arises from the comparison of gene induction in strains expressing different concentrations of Pap1. As observed in Supplementary Figure S3AB, increasing 10-fold the endogenous concentration of Pap1 compensates the low affinity of the reduced/nuclear GFP-Pap1.C523D mutant, increases promoter occupancy, and allows the activation of the antioxidant set of genes. On the contrary, strong over-expression of Prr1 in cells lacking Pap1 does not trigger gene induction (Supplementary Figure S3C). The search for specific cis elements between both types of promoters which would validate the different affinities for Pap1 or Pap1-Prr1 is in progress. On the other hand, Prr1 may also contribute to the transcriptional up-regulation of antioxidant genes by recruiting some chromatin modifiers required at those promoters, or by establishment of specific contacts with Pol II: a greatly diminished but reproducible binding of Pap1 to antioxidant promoters can still be detected in vivo by ChIP in cells lacking Prr1 (Figure 3E, right panel), whereas up-regulation of the mRNA antioxidant genes is completely abolished in this strain, as determined by northern blot (Figure 3C, Δprr1 strain, trr1, ctt1, srx1 probes). That seems to indicate that the presence of Prr1, and not only of Pap1, at those promoters is required for transcriptional activation of this subset of genes. It is worth mentioning that Prr1 binds to Pap1-dependent promoters only after association with oxidized Pap1, as determined by ChIP (Figure 4).
In conclusion, while oxidation of Pap1 contributes to the activation of two distinct sets of genes, induction of the drug resistance ones can be accomplished when non-oxidized Pap1 is accumulated in the nucleus. It is worth mentioning that Pap1 responds to peroxides by means of very exposed cysteine residues, which have already been reported to also react with alkylating agents such as diethylmaleate (11). This modified Pap1 protein also localizes in the nucleus, and importantly enough only triggers the drug tolerance genes (Supplementary Figure S4A), what suggests that defence against this drug may only require a partial and smaller gene expression program for adaptation. Similarly, drugs such as leptomycin B inhibit Crm1 function through covalent modification of some essential cysteine residues in this exportin (38); under those circumstances, Pap1 accumulation specifically triggers the drug resistance, but not the antioxidant, gene response (Supplementary Figure S4B).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–4 and Supplementary References [39].
FUNDING
Spanish Ministry of Science and Innovation (BFU2009-06933), PLAN E and FEDER, by the Spanish program Consolider-Ingenio 2010 Grant CSD 2007-0020, and by SGR2009-196 from Generalitat de Catalunya (Spain) to E.H; ICREA Academia Awards (Generalitat de Catalunya) to E. H. and J.A.; pre-doctoral fellowship (FPI) and a post-doctoral contract Juan de la Cierva, respectively, from the Ministerio de Ciencia e Innovación (Spain) to I.A.C. and P.G. Funding for this work and for open access charge: Spanish Ministry of Science and Innovation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Mercè Carmona for technical assistance.
REFERENCES
- 1.Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 2.Quinn J, Findlay VJ, Dawson K, Millar JB, Jones N, Morgan BA, Toone WM. Distinct Regulatory Proteins Control the Graded Transcriptional Response to Increasing H(2)O(2) Levels in Fission Yeast Schizosaccharomyces pombe. Mol. Biol. Cell. 2002;13:805–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Wilkinson CR, Watt S, Penkett CJ, Toone WM, Jones N, Bahler J. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol. Biol. Cell. 2008;19:308–317. doi: 10.1091/mbc.E07-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagawa CW, Yamada K, Mutoh N. Role of Atf1 and Pap1 in the induction of the catalase gene of fission yeast schizosaccharomyces pombe. J. Biochem. 2000;127:233–238. doi: 10.1093/oxfordjournals.jbchem.a022599. [DOI] [PubMed] [Google Scholar]
- 5.Lee YY, Jung HI, Park EH, Sa JH, Lim CJ. Regulation of Schizosaccharomyces pombe gene encoding copper/zinc superoxide dismutase. Mol. Cell. 2002;14:43–49. [PubMed] [Google Scholar]
- 6.Vivancos AP, Castillo EA, Biteau B, Nicot C, Ayté J, Toledano MB, Hidalgo E. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl Acad. Sci. USA. 2005;102:8875–8880. doi: 10.1073/pnas.0503251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song JY, Roe JH. The role and regulation of Trxl, a cytosolic thioredoxin in Schizosaccharomyces pombe. J. Microbiol. 2008;46:408–414. doi: 10.1007/s12275-008-0076-4. [DOI] [PubMed] [Google Scholar]
- 8.Umeda M, Izaddoost S, Cushman I, Moore MS, Sazer S. The fission yeast Schizosaccharomyces pombe has two importin-alpha proteins, Imp1p and Cut15p, which have common and unique functions in nucleocytoplasmic transport and cell cycle progression. Genetics. 2005;171:7–21. doi: 10.1534/genetics.105.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudo N, Taoka H, Toda T, Yoshida M, Horinouchi S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- 10.Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1 [published erratum appears in Genes Dev. 1998;12:2650] [see comments] Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo EA, Ayté J, Chiva C, Moldón A, Carrascal M, Abián J, Jones N, Hidalgo E. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol. Microbiol. 2002;45:243–254. doi: 10.1046/j.1365-2958.2002.03020.x. [DOI] [PubMed] [Google Scholar]
- 12.Vivancos AP, Castillo EA, Jones N, Ayté J, Hidalgo E. Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 2004;52:1427–1435. doi: 10.1111/j.1365-2958.2004.04065.x. [DOI] [PubMed] [Google Scholar]
- 13.Bozonet SM, Findlay VJ, Day AM, Cameron J, Veal EA, Morgan BA. Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J. Biol. Chem. 2005;280:23319–23327. doi: 10.1074/jbc.M502757200. [DOI] [PubMed] [Google Scholar]
- 14.Benko Z, Sipiczki M, Carr AM. Cloning of caf1+, caf2+ and caf4+ from Schizosaccharomyces pombe: their involvement in multidrug resistance, UV and pH sensitivity. Mol. Gen. Genet. 1998;260:434–443. doi: 10.1007/s004380050914. [DOI] [PubMed] [Google Scholar]
- 15.Turi TG, Webster P, Rose JK. Brefeldin A sensitivity and resistance in Schizosaccharomyces pombe. Isolation of multiple genes conferring resistance. J. Biol. Chem. 1994;269:24229–24236. [PubMed] [Google Scholar]
- 16.Benko Z, Fenyvesvolgyi C, Pesti M, Sipiczki M. The transcription factor Pap1/Caf3 plays a central role in the determination of caffeine resistance in Schizosaccharomyces pombe. Mol. Genet. Genomics. 2004;271:161–170. doi: 10.1007/s00438-003-0967-3. [DOI] [PubMed] [Google Scholar]
- 17.Calvo IA, Gabrielli N, Iglesias-Baena I, Garciá-Santamarina S, Hoe KL, Kim DU, Sansó M, Zuin A, Pérez P, Ayté J, et al. Genome-wide screen of genes required for caffeine tolerance in fission yeast. PLoS One. 2009;4:e6619. doi: 10.1371/journal.pone.0006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimanuki M, Saka Y, Yanagida M, Toda T. A novel essential fission yeast gene pad1(+)-positively regulates pap1(+)-dependent transcription and is implicated in the maintenance of chromosome structure. J. Cell Sci. 1995;108(Pt 2):569–579. doi: 10.1242/jcs.108.2.569. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura K, Taki M, Tanaka N, Yamashita I. Fission yeast Ubr1 ubiquitin ligase influences the oxidative stress response via degradation of active Pap1 bZIP transcription factor in the nucleus. Mol. Microbiol. 2011;80:739–755. doi: 10.1111/j.1365-2958.2011.07605.x. [DOI] [PubMed] [Google Scholar]
- 20.Castillo EA, Vivancos AP, Jones N, Ayté J, Hidalgo E. Schizosaccharomyces pombe cells lacking the Ran-binding protein Hba1 show a multidrug resistance phenotype due to constitutive nuclear accumulation of Pap1. J. Biol. Chem. 2003;278:40565–40572. doi: 10.1074/jbc.M305859200. [DOI] [PubMed] [Google Scholar]
- 21.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- 22.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 23.Zuin A, Vivancos AP, Sansó M, Takatsume Y, Ayté J, Inoue Y, Hidalgo E. The glycolytic metabolite methylglyoxal activates Pap1 and Sty1 stress responses in Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:36708–36713. doi: 10.1074/jbc.M508400200. [DOI] [PubMed] [Google Scholar]
- 24.Sansó M, Vargas-Pérez I, Quintales L, Antequera F, Ayté J, Hidalgo E. Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res. 2011;39:6369–6379. doi: 10.1093/nar/gkr255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohmiya R, Kato C, Yamada H, Aiba H, Mizuno T. A fission yeast gene (prr1(+)) that encodes a response regulator implicated in oxidative stress response. J. Biochem. 1999;125:1061–1066. doi: 10.1093/oxfordjournals.jbchem.a022387. [DOI] [PubMed] [Google Scholar]
- 26.Greenall A, Hadcroft AP, Malakasi P, Jones N, Morgan BA, Hoffman CS, Whitehall SK. Role of fission yeast Tup1-like repressors and Prr1 transcription factor in response to salt stress. Mol. Biol. Cell. 2002;13:2977–2989. doi: 10.1091/mbc.01-12-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toda T, Shimanuki M, Saka Y, Yamano H, Adachi Y, Shirakawa M, Kyogoku Y, Yanagida M. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol. Cell Biol. 1992;12:5474–5484. doi: 10.1128/mcb.12.12.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vivancos AP, Jara M, Zuin A, Sansó M, Hidalgo E. Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol. Genet. Genomics. 2006;276:495–502. doi: 10.1007/s00438-006-0175-z. [DOI] [PubMed] [Google Scholar]
- 29.Quinn J, Malakasi P, Smith DA, Cheetham J, Buck V, Millar JB, Morgan BA. Two-Component Mediated Peroxide Sensing and Signal Transduction in Fission Yeast. Antioxid. Redox. Signal. 2011;276:153–165. doi: 10.1089/ars.2010.3345. [DOI] [PubMed] [Google Scholar]
- 30.He XJ, Mulford KE, Fassler JS. Oxidative stress function of the Saccharomyces cerevisiae Skn7 receiver domain. Eukaryotic Cell. 2009;8:768–778. doi: 10.1128/EC.00021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohmiya R, Yamada H, Kato C, Aiba H, Mizuno T. The Prr1 response regulator is essential for transcription of ste11+ and for sexual development in fission yeast. Mol. Gen. Genet. 2000;264:441–451. doi: 10.1007/s004380000305. [DOI] [PubMed] [Google Scholar]
- 32.Mulford KE, Fassler JS. Association of the Skn7 and Yap1 transcription factors in the Saccharomyces cerevisiae oxidative stress response. Eukaryotic Cell. 2011;10:761–769. doi: 10.1128/EC.00328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan BA, Banks GR, Toone WM, Raitt D, Kuge S, Johnston LH. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 35.Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toone WM, Jones N. AP-1 transcription factors in yeast. Curr. Opin. Genet. Dev. 1999;9:55–61. doi: 10.1016/s0959-437x(99)80008-2. [DOI] [PubMed] [Google Scholar]
- 37.He XJ, Fassler JS. Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 2005;58:1454–1467. doi: 10.1111/j.1365-2958.2005.04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl Acad. Sci. USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leupold U. Genetical methods for Schizosaccharomyces pombe. Methods Cell. Physiol. 1970;4:169–177. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.