Abstract
In plant organelles, RNA editing alters specific cytidine residues to uridine in transcripts. All of the site-specificity factors of RNA editing identified so far are pentatricopeptide repeat (PPR) proteins. A defect in a specific PPR protein often impairs RNA editing at multiple sites, at which the cis-acting elements are not highly conserved. The molecular mechanism for sharing a single PPR protein over multiple sites is still unclear. We focused here on the PPR proteins OTP82 and CRR22, the putative target elements of which are, respectively, partially and barely conserved. Recombinant OTP82 specifically bound to the −15 to 0 regions of its target sites. Recombinant CRR22 specifically bound to the −20 to 0 regions of the ndhB-7 and ndhD-5 sites and to the −17 to 0 region of the rpoB-3 site. Taking this information together with the genetic data, we conclude that OTP82 and CRR22 act as site-specificity factors at multiple sites in plastids. In addition, the high-affinity binding of CRR22 to unrelated cis-acting elements suggests that only certain specific nucleotides in a cis-acting element are sufficient for high-affinity binding of a PPR protein. The cis-acting elements can therefore be rather divergent and still be recognized by a single PPR protein.
INTRODUCTION
Pentatricopeptide repeat (PPR) proteins form a large protein family that is particularly prevalent in terrestrial plants: 450 members are encoded in the Arabidopsis thaliana genome (1). PPR proteins are defined by tandem arrays of a degenerate 35 amino acid motif, the PPR motif (2). Most PPR proteins are predicted to localize to plastids or mitochondria (2). Genetic and biochemical evidence indicates that these proteins have a range of essential functions in posttranscriptional gene regulation (1). The most probable explanation for their divergent roles is that they are sequence-specific RNA binding proteins that recruit effector enzymes to the target RNA (3) or serve as a protective cap to stabilize upstream and downstream RNA (4). The PPR protein family is divided into the P and PLS subfamilies (2); the latter accounts for roughly half of the members in Arabidopsis and is specific to land plants, although some have been found in the protist Naegleria gruberi (5,6). On the basis of differences in the C-terminal motifs, the PLS subfamily is further classified into the PLS, E and DYW subclasses (5). The expansion of the PLS subfamily is correlated with the specific characteristics of plant organelles, particularly RNA editing (5,7,8).
In flowering plants, RNA editing is a process that alters specific cytidine (C) residues to uridine (U) in plastid and mitochondrial transcripts (9). The reverse reaction, from U to C, is observed in a few plant lineages. RNA editing is found in nearly all lineages of terrestrial plants. The exception is a subclass of early land plants, the marchantiids, namely Marchantia polymorpha (10). In Arabidopsis, 34 sites are edited in plastids (9,11) and more than 450 sites are edited in mitochondria (12–14). One puzzling question of plant RNA editing is how a C target of editing is distinguished from other C residues. Pioneer works using the plastid transformation technique and in vitro RNA editing systems have revealed that editing sites in plastids are recognized via cis-acting elements that in most cases, are located within ∼30 nt surrounding the editing site (15,16). The case is similar in mitochondria (17). The site-specificity factor that recognizes the cis-acting element was initially discovered by the work that focused on the Arabidopsis nuclear mutants defective in photosynthetic electron transport (18). Subsequently, a biochemical study concluded that a PPR protein, chlororespiratory reduction 4 (CRR4), acts as the site-specificity factor for recognizing RNA editing site 1 of the plastid ndhD transcript (ndhD-1), which encodes a subunit of the NADH dehydrogenase-like complex (19). Many other PPR proteins required for RNA editing in plastids and mitochondria have since been identified; almost all of them belong to either the E or DYW subclass (20–27). Additional experimental evidence to show that a PPR protein acts as a site-specificity factor is the observation that the PPR proteins PpPPR71, which is essential for ccmF-2 RNA editing in Physcomitrella patens mitochondria, and OTP87, which is essential for nad7 and atp1 RNA editing events in Arabidopsis mitochondria, bind to the region surrounding the target site (28,30). It is therefore generally accepted that a PPR protein recognizes each editing site in plant organelles.
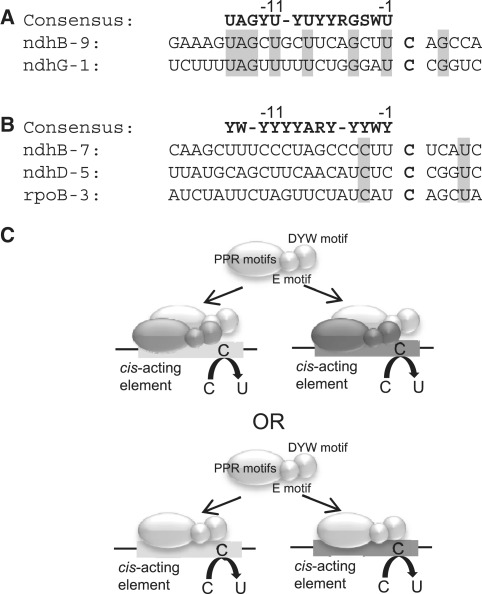
A defect in a single PPR protein often impairs RNA editing of multiple sites (20–22,27,29). The hypothesis that a site-specificity factors can be shared by multiple sites was originally suggested by the finding that high-level accumulation of RNA including a single editing site decreases editing efficiency at other sites (31,32). Subsequently, this hypothesis was confirmed by using in vitro RNA editing systems with chloroplast extracts (33). In addition, transcripts encompassing two editing sites, ZMrpoB C467 and ZMrps14 C80, competed against each other for editing activity in vitro (34). An analysis of the target sites of five PPR proteins involved in RNA editing of multiple sites revealed an unambiguous 15-nt consensus, providing sufficient specificity to define the edited sites in the plastid transcriptome (20). An in silico search of the Arabidopsis mitochondrial genome revealed that two sites recognized by OTP87 contained sufficient conserved nucleotides to be unambiguously defined as a binding site consensus; moreover, biochemical evidence for the specific binding of OTP87 to multiple editing sites was provided (30). However, the molecular mechanism by which a trans-acting factor is shared by multiple sites is still not fully clear, because the cis-acting elements recognized by a single PPR protein are often not conserved. The most prominent examples are the plastid targets of the PPR protein CRR22: the ndhB-7, ndhD-5 and rpoB-3 sites (35). In contrast to previous reports, in which multiple target sequences recognized by a single PPR protein have been almost identical (36,37) or partially conserved (20,27,38), the sequences surrounding these editing sites appear divergent (Figure 1). It was proposed that the DYW motif could catalyze the editing reaction, because it contains some residues that are conserved in the human C deaminase, APOBEC-1 (7). Since the catalytically active form of APOBEC-1 is an asymmetric homodimer (39), DYW-subclass PPR protein may likewise dimerize. Since different editing sites targeted by the same PPR protein do not share high conservation, it is plausible that the PPR protein plays different roles at these editing sites. In this model, two PPR proteins would be heterodimeric: one would act as a site-specificity factor and the other would function to provide the DYW motif for the editing reaction (Figure 1C). This may be the case for CRR22, because it is required for RNA editing events in which the putative cis-acting elements are not conserved (Figure 1B). Alternatively, CRR22 may recognize all the elements as a genuine site-specificity factor. To answer the question of whether a PPR protein involved in the editing of multiple sites with low levels of conservation binds specifically to its different targets, we selected CRR22. We also analyzed OTP82, which is required for two RNA editing events at sites with putative cis-acting elements that are only partially conserved (Figure 1A).
Figure 1.
Two models to explain the function of a single PPR protein that functions at multiple sites. Alignments of the nucleotide sequences in the region surrounding the editing sites affected in (A) otp82 and (B) crr22 are shown. The alignments include the sequences from −20 to +5 around the edited C (bold), with identical nucleotides shown in shaded boxes. Consensus sequences of the 15 nt immediately upstream of the edited C were identified by bioinformatics analysis (20) and are shown above the target sequences. In the consensus, full conservation of nucleotides (A, U, G and C), conservation of purines (A or G = R) or pyrimidines (U or C = Y), and conservation of the number of hydrogen bonding groups (A or U = W, G or C = S) are indicated. (C). In the upper model, each cis-acting element is recognized by the indicated PPR protein (medium and dark gray) and a second PPR protein (light gray) functions as a binding partner shared via the formation of a heterodimer at each site. The PPR protein may have additional functions other than RNA recognition, such as providing the DYW motif for the editing reaction. In the lower model, the PPR protein acts as a site-specificity factor at multiple sites.
MATERIALS AND METHODS
Expression and purification of rOTP82 and rCRR22
DNA fragments encoding mature OTP82 and CRR22 were amplified from genomic DNA by PCR using the primers rOTP82-FW (5′-CACAATTGCAAAACTCTTC-3′) and rOTP82-RV (5′-CCAGTAGTCATTGCAGGAAC-3′), and rCRR22-FW (5′-TGTTCTTCTCTGAAAGAAC-3′) and rCRR22-RV (5′-CCAGTAATCTCCGCACG-3′), respectively. These fragments were cloned in-frame into pBAD/Thio-TOPO vector (Invitrogen). The recombinant OTP82 and CRR22 (rOTP82 and rCRR22) were expressed in Escherichia coli LMG194 as fusion proteins with thioredoxin at the N-terminus and V5 and Hisx6 epitope tags at the C-terminus. Escherichia coli cultures were grown at 30°C in LB medium with carbenicillin to an OD650 of 0.45 and cooled on ice for 20 min. Protein expression was induced by the addition of 0.02% arabinose, and cultures were incubated for 6 h at 18°C. Harvested cells were suspended in cold lysis buffer [50 mM Tris-HCl (pH 7.5), 0.3 M NaCl, 7 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride and 0.01% CHAPS]. Cells were lysed at 4°C by sonication, cooled on ice for 5 min, and sonicated six times. The lysate was cleared by centrifugation at 15 000g for 30 min. The following steps were performed at 4°C. The rOTP82 and rCRR22 were purified by binding to Ni-NTA Agarose (Qiagen). To remove particulates, the protein was resolved on a Superdex 200 column (GE Healthcare) in buffer E [20 mM HEPES-KOH (pH 7.9), 60 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 17% glycerol and 2 mM dithiothreitol]. The fractionated rOTP82 and rCRR22 were concentrated with a centrifugal concentrator and dialyzed in buffer E. The proteins were stored in aliquots at −80°C. The protein concentration was determined by the method of Bradford. Bovine serum albumin was used as the standard.
Preparation of RNA probes
All RNAs used in this study were synthesized as synthetic oligonucleotides. Each synthetic oligonucleotide was 5′-end labeled by incubation with T4 polynucleotide kinase (Takara) and [γ-32P]-ATP at 37°C for 1 h and then extracted by ethanol precipitation (40).
Gel mobility shift assays
A binding reaction was performed by mixing various amounts of rOTP82 or rCRR22 (the protein concentrations are indicated in the respective figures) with the 32P-labeled RNA probe (0.1 nM) in a total volume of 20 µL of solution containing 5 mM HEPES-KOH (pH 7.9), 7 mM MgCl2, 2.5 mM dithiothreitol, 150 mM KCl, 4.3% glycerol, 0.25 mM EDTA and 0.3 nM yeast tRNA. Unlabeled competitor RNAs were preincubated with the protein for 5 min before the labeled RNA was added. The reaction mixture was incubated for 15 min at 25°C. The samples were then loaded onto an 8% non-denaturing polyacrylamide gel (acrylamide: N,N′-methylene bisacrylamide at 29:1) and electrophoresed in TBE buffer (89 mM Tris, 89 mM boric acid and 2 mM EDTA) at 4°C. The gel was dried and exposed to an imaging plate (Fuji Film) overnight. The image was visualized with a BAS1000 image analyzer (Fuji Film). The equilibrium Kd was determined from the concentration of protein at which 50% of the RNA probe was bound.
RESULTS
Expression and purification of recombinant OTP82 and CRR22
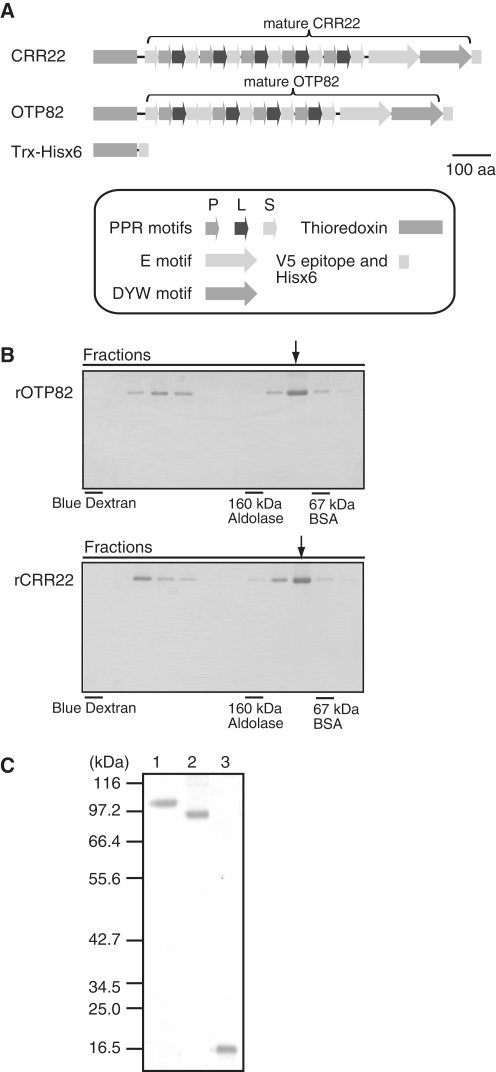
We selected OTP82 and CRR22 for in vitro RNA binding assays of the proteins involved in editing of multiple sites in plastids (35,41). OTP82 and CRR22, respectively, contain 14 and 16 PPR or PPR-like (P, L, L2 and S) motifs. Each also contains E and DYW motifs, which are characteristic of the DYW subclass (Figure 2A) (1). To produce recombinant proteins, mature OTP82 and CRR22 (i.e., lacking their transit peptides) were expressed as fusion proteins with thioredoxin (18 kDa) at the N-terminus and V5 and Hisx6- epitope tags at the C-terminus (Figure 2A). Their molecular weights were predicted to be ∼95 kDa and ∼101 kDa, respectively. As is often observed in the expression of recombinant PPR proteins (2,42), we found that expression of OTP82 and CRR22 was extremely low and that almost all the protein was in inclusion bodies when expressed in E. coli with standard growth and induction according to the manufacturer's protocol (data not shown). As described in the ‘Materials and Methods’, the induction and purification conditions were optimized to produce soluble protein that could be used for in vitro RNA-binding assays. As reported by Williams-Carrier et al. (42), a low concentration of CHAPS detergent in the lysis buffer improved protein solubility. We could not purify untagged proteins, because elimination of thioredoxin caused substantial protein aggregation (data not shown). The rOTP82 and rCRR22 were subjected to a final purification step on a gel filtration column before being used in the RNA-binding assay. rOTP82 and rCRR22 were mainly eluted from the gel filtration column at a position between the BSA (67 kDa) and aldolase (160 kDa) markers (Figure 2B). This result is consistent with the predicted molecular weights of their monomeric forms; it was therefore unlikely that rOTP82 and rCRR22 formed homomultimers. As reported in CRP1 (42), aliquots of rOTP82 and rCRR22 were often present in the void column fractions eluted ahead of the monomeric species, presumably as microaggregates (Figure 2B). Only the fraction of the monomeric form (Figure 2B, fraction indicated by arrowhead) was used for the in vitro RNA-binding assays. Purified proteins were detected on an SDS-polyacrylamide gel as bands at ∼95 kDa (for OTP82) and ∼100 kDa (for CRR22) (Figure 2C). These values were close to the predicted molecular masses. The purity of proteins was confirmed by visual inspection of the stained gel (Figure 2C). They had a low A260/A280 ratio (∼0.6), indicating negligible contamination of nucleic acids.
Figure 2.
Purification of rOTP82 and rCRR22. (A) The domain organization of rOTP82 and rCRR22 fusion proteins. (B) Elution of rOTP82 and rCRR22 from a Superdex 200 gel filtration column. rOTP82 and rCRR22 were purified with Ni-NTA Agarose and then fractionated on a Superdex 200 gel filtration column. Equal proportions of contiguous column fractions were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue. Size standards BSA (67 kDa) and aldolase (160 kDa) are indicated. (C) The fractions indicated by the arrowheads in Figure 2 (B), and purified rTrx, were analyzed by SDS-PAGE. Six hundred nanograms of rOTP82, rCRR22 and rTrx was loaded onto the gel, and the gel was stained with Coomassie Brilliant Blue. The positions of molecular mass standards are indicated to the left of the gel. Lane 1 contains rCRR22, lane 2 contains rOTP82 and lane 3 contains rTRX.
OTP82 acts as a site-specificity factor at two editing sites in plastids
On the basis of the available information (15,16,19,28,30,33,43,44), most cis-acting elements for RNA editing appear to be ∼20-nt upstream and, in some case, include ∼10-nt downstream of the editing site. It is unlikely that regions farther upstream than ∼40 nt function as cis-acting elements. OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts (the ndhB-9 and ndhG-1 sites) (41). The sequences surrounding the ndhB-9 and ndhG-1 sites show only partial sequence identity on the primary sequence, and when pyrimidines are distinguished from purines, the sequences contain sufficient conserved nucleotides to be unambiguously defined as consensuses (Figure 1A). To test whether OTP82 binds directly to the sequences surrounding its target sites, as do CRR4, PpPPR71 and OTP87, we performed gel mobility shift (GMS) assays with purified rOTP82.
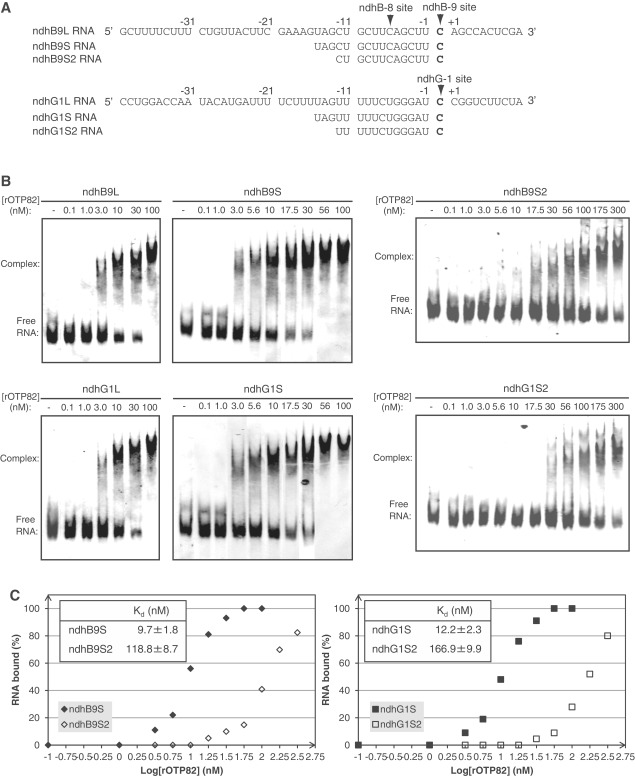
We selected the region (−40 to +10) surrounding the editing site; this was probably sufficient to contain all the cis-information (Figure 3A, ndhB9L RNA). The ndhG1L RNA covered the −40 to +10 region surrounding the editing site of ndhG-1 (Figure 3A). The probe RNAs were labeled with 32P and incubated with the purified rOTP82. Under stringent binding conditions (in the presence of 150 mM KCl, 0.3 nM yeast tRNA), the rOTP82-RNA complex was detected as a band that migrated more slowly than free RNA in the gel. The retarded band was detected upon increasing the amount of rOTP82 (Figure 3B). A 300-fold excess of rOTP82, relative to the ndhB9L and ndhG1L probes, was required for retardation (Figure 3B). A 1000-fold excess of rOTP82 was needed to shift all of the RNA (Figure 3B).
Figure 3.
GMS assays with rOTP82 and target RNAs. (A) The RNA sequences used as probes are shown. Editing sites of ndhB-9 and ndhG-1 are indicated in bold and are marked with arrowheads. (B) GMS assays were performed with the indicated concentrations of rOTP82 and labeled RNAs (ndhB9L, ndhB9S, ndhB9S2, ndhG1L, ndhG1S and ndhG1S2), as described in the ‘Materials and Methods’ section. (C) Equilibrium Kd of rOTP82 for the ndhB9S and ndhB9S2 probes (left), and for the ndhG1S and ndhG1S2 probes (right). The rOTP82 concentrations and fraction of RNA bound in each lane are plotted. The Kd calculation assumed a 1:1 interaction between the RNA and protein. The Kd value and each data point are means ± SD of three experiments performed with the same rOTP82 preparation. All of the GMS assays were performed with the same preparation of rOTP82 and within 2 weeks after purification.
Biochemical evidence suggests that most of the sequence recognition is concentrated within the 15 nt immediately upstream of the editing site (16,33,45,46). To narrow down the OTP82 binding site and examine the necessity of the region downstream of the editing site for binding, RNA-binding activity was determined by using ndhB9S and ndhG1S probes, which covered the respective −15 to 0 regions surrounding the ndhB-9 and ndhG-1 sites (0 represents the edited C) (Figure 3A). The retarded band was detected with increasing amounts of rOTP82 (Figure 3B). A 300-fold excess of rOTP82 was required for retardation of both probes, the same as when the ndhB9L and ndhG1L probes were used (Figure 3B). A 560-fold excess of rOTP82 was needed to shift all of the RNA (Figure 3B). To eliminate the possibility that thioredoxin binds to the RNA probes, the same RNA probes were incubated with recombinant thioredoxin (rTrx) lacking the OTP82 sequence. Even a 1000-fold excess of rTrx (100 nM) added to the RNA (0.1 nM) did not induce any band shift (Supplementary Figure S1), indicating that the RNA-binding activity was specific to rOTP82. To estimate the relative binding affinity of rOTP82 for the ndhB9S and ndhG1S RNAs, a constant low concentration of labeled RNA probe was incubated with a range of protein concentrations. The equilibrium Kd was estimated to be 9.7 nM for ndhB9S RNA and 12.2 nM for ndhG1S RNA, on the basis of the protein concentration at which half of each RNA was bound (Figure 3B and C).
To test whether the 16 nt was the minimum sequence efficiently recognized by OTP82, we eliminated the conserved distal UAG (Figure 3A, ndhB9S2 and ndhG1S2 RNAs). Truncation of 3 nt caused a substantial loss of binding affinity in both probes (Figure 3B). The equilibrium Kd was estimated to be 118.8 nM for ndhB9S2 and 166.9 nM for ndhG1S2; these values are much higher than those for ndhB9S and ndhG1S (Figure 3C). We also determined RNA-binding activity by using ndhB9S3 and ndhG1S3 probes that had proximal 3-nt truncations from the 3′-ends of ndhB9S and ndhG1S, respectively, resulting in the loss of target C sites (Supplementary Figure S2). Both 3-nt truncations caused a slight loss of binding affinity to rOTP82 (Supplementary Figure S2). We concluded that 16 nt (−15 to 0) was the minimum sequence length that was efficiently recognized by OTP82, although the proximal 13 nt (−15 to −3) retained the partial affinity for OTP82.
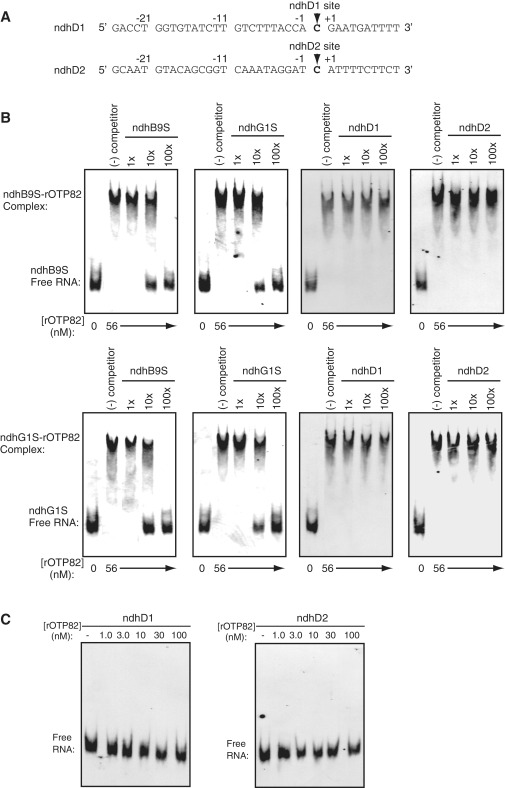
To analyze whether OTP82 bound to its targets in a sequence-specific manner, we used a set of competitor RNAs to assess interfering effects in the binding of rOTP82 to radioactive RNA probes. As competitors that do not include the OTP82-binding site, we selected two 36-nt RNAs, ndhD1 and ndhD2, which contain the CRR4 binding site and the CRR21 putative binding site, respectively (19,47). They cover the −25 to +10 regions surrounding the ndhD-1 and ndhD2 editing sites, respectively (Figure 4A). The addition of 10 times the amounts of competitor RNAs, ndhB9S and ndhG1S, to the radioactive ndhB9S probe partially blocked retardation, and a 100-fold excess of competitor RNAs completely eliminated retardation (Figure 4B). The addition of the competitor RNAs ndhB9S and ndhG1S to the radioactive ndhG1S probe also interfered with retardation (Figure 4B). In contrast, the addition of a 100-fold excess of the competitor RNAs ndhD1S and ndhD2S did not affect OTP82 binding (Figure 4B). We further confirmed that rOTP82 did not bind to the 32P-labeled ndhD1 and ndhD2 RNAs, even if a 1000-fold excess of rOTP82 was added (Figure 4C). The results of the binding experiments strongly suggested that OTP82 specifically recognized the RNA nucleotides within the −15 to 0 region surrounding the editing sites. Taken together with the genetic evidence indicating that OTP82 is required for RNA editing of ndhB-9 and ndhG-1(41), these findings indicate that OTP82 acts as a site-specificity factor at two sites in plastids.
Figure 4.
Competition assays demonstrating sequence-specific interactions between rOTP82 and the target RNAs. (A) Two RNAs used as competitors (ndhD1 and ndhD2) are shown. Editing sites of ndhD-1 and ndhD-2 are indicated in bold and are marked with arrowheads. (B) Binding reactions included radioactive ndhB9S or ndhG1S RNA with a 1-, 10- or 100-fold molar excess of the non-radioactive RNA indicated above each panel. The concentration of rOTP82 was held constant at 56 nM. (C) GMS assays were performed with the indicated concentrations of rOTP82 and radioactive RNAs (ndhD1 and ndhD2). All of the competition and GMS assays were performed with the same preparation of rOTP82 as used in Figure 3 and within 2 weeks after purification.
CRR22 acts as a site-specificity factor at three editing sites in plastids
In contrast to the cases with PPR proteins such as OTP82 and OTP87 that are required for multiple editing events (30,41), partial sequence conservation that may provide sufficient specificity to define the edited sites is not found in the sequences surrounding the target sites of CRR22 (Figure 1B) (20). Therefore, we questioned whether CRR22 truly acted as a site-specificity factor at all three sites, as do CRR4, PpPPR71, OTP87 and OTP82. To address this issue, GMS assays were performed with purified rCRR22.
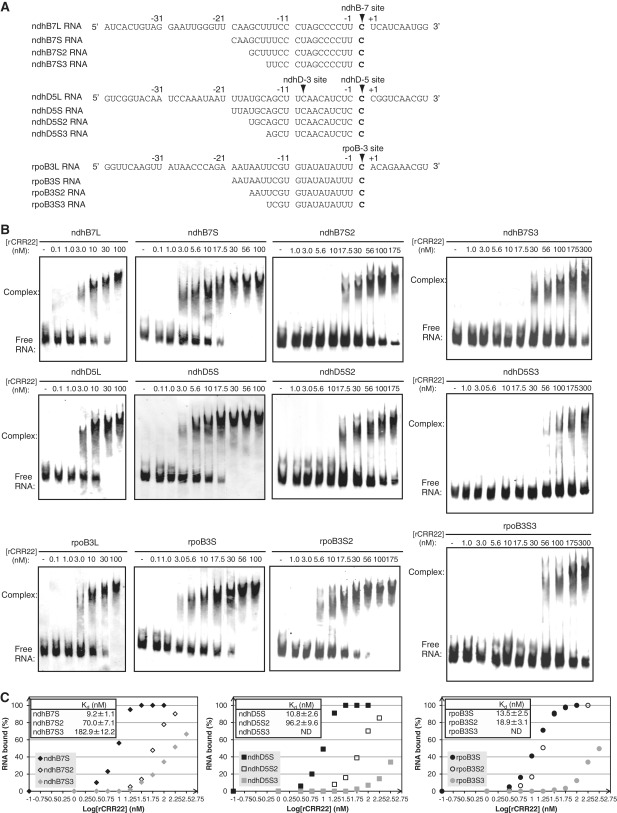
As in the case of OTP82, we first selected sufficiently long sequences surrounding the editing site as RNA probes for the GMS assay. Three 51-nt RNAs, ndhB7L, ndhD5L and rpoB3L, cover the −40 to +10 regions surrounding the ndhB-7, ndhD-5 and rpoB-3 sites, respectively (Figure 5A). A retarded band was detected upon increasing the amounts of rCRR22 (Figure 5B). A 300-fold excess of rCRR22, relative to the ndhB7L, ndhD5L and rpoB3L probes, was required for retardation (Figure 5B). To shift all of the free RNA, a 300- to 1000-fold excess of rCRR22 was needed (Figure 5B).
Figure 5.
GMS assays with rCRR22 and target RNAs. (A) RNA sequences used as probes are shown. Editing sites of ndhB-7, ndhD-5 and rpoB-3 are indicated in bold and are marked with arrowheads. (B) GMS assays were performed with the indicated concentrations of rCRR22 and labeled RNAs (ndhB7L, ndhB7S-S3, ndhD5L, ndhD5S-S3, rpoB3L and rpoB3S-S3), as described in the ‘Materials and Methods’ section. (C) Equilibrium Kd of rCRR22 for the ndhB7S-S3 (left), ndhD5S-S3 (middle) and rpoB3S-S3 (right) probes. rCRR22 concentrations and fractions of RNA bound in each lane are plotted. The Kd calculation assumes a 1:1 interaction between the RNA and the protein. The Kd values and each data point are means ± SD of three experiments performed with the same rCRR22 preparation. All of the GMS assays were performed with the same preparation of rCRR22 and within 2 weeks after purification.
rOTP82 binds specifically to the RNA nucleotides within the 16-nt sequence consisting of the target C and its upstream nucleotides (Figures 3 and 4). Similarly, the cis-acting elements for CRR22 are assumed to be located at immediately upstream region of the target Cs. Biochemical analysis of PPR10 (4) and a physically plausible consensus model of PPR protein structure built from a statistical analysis of large numbers of restorer-like PPR protein sequences (48) suggest that there is a 1-PPR motif/1-RNA nt recognition mechanism. If PPR proteins use the RNA binding mode as suggested, it appears that at least 16 nt would be required for CRR22 binding, because CRR22 harbors 16 PPR motifs. To narrow down the CRR22 binding site, RNA-binding activity was determined by using ndhB7S, ndhD5S and rpoB3S probes covering the −20 to 0 region surrounding the ndhB-7, ndhD-5 and rpoB-3 sites, respectively (Figure 5A). A retarded band was detected with increasing amounts of rCRR22 (Figure 5B). A 300-fold excess of rCRR22 was required for retardation of all probes the same as when the longer probes were used (Figure 5B). To shift all of the free RNA, a 300- to 560-fold excess of rCRR22, relative to the probe RNAs, was needed (Figure 5B). We also verified that this binding activity did not depend on rTrx (Supplementary Figure S1). The estimated equilibrium Kd was 9.2, 10.8 and 13.5 nM, for ndhB7S, ndhD5S and rpoB3S, respectively (Figure 5B and C).
To test whether 21 nt was the minimum sequence efficiently recognized by CRR22, we determined RNA-binding activity by using ndhB7S2, ndhD5S2 and rpoB3S2 probes that had proximal 3-nt truncations from ndhB7S, ndhD5S and rpoB3S2, respectively (Figure 5A). The addition of >1750 times the amount of rCRR22 to the ndhB7S2 and ndhD5S2 probes was required to detect the shift (Figure 5B), and the Kd of rCRR22 to ndhB7S2 and ndhD5S2 was 70.0 nM and 96.2 nM, respectively; these values were higher than those of rCRR22 for ndhB7S and ndhD5S (9.2 nM and 10.8 nM) (Figure 5C). In contrast, rCRR22 could also induce a shift in the rpoB3S2 probe, as efficiently as in the rpoB3S probe (Figure 5B). Kd of rCRR22 for rpoB3S2 was 18.9 ± 3.1 nM, which was similar to that of rpoBS (13.5 ± 2.5 nM) (Figure 5C). Whereas 5′ 21-nt sequences were required for efficient recognition by CRR22 at the ndhB-7 and ndhD-5 sites, 5′ 18-nt were sufficient for high affinity at the rpoB-3 site.
We analyzed the effect of a further 3-nt truncation on the affinity for CRR22 by using the ndhB7S3, ndhD5S3 and rpoB3S3 probes (Figure 5A). The further 3 nt caused a dramatic loss of binding affinity to the rpoB3S3 probe, suggesting that the proximal AAU in rpoB3S2 was required for high affinity for CRR22 (Figure 5B and C). The affinity was also drastically affected in ndhD5S3, although the effect was milder in ndhB7S3, suggesting that RNA sequences of different lengths were required for efficient recognition by CRR22 among the three targets.
We also determined RNA-binding activity by using ndhB7S4, ndhD5S4 and rpoB3S4 probes that had proximal 3-nt truncations from the 3′ ends of ndhB7S, ndhD5S and rpoB3S2, respectively (Supplementary Figure S3). All 3-nt truncations caused a slight loss of binding affinity to rCRR22 (Supplementary Figure S3).
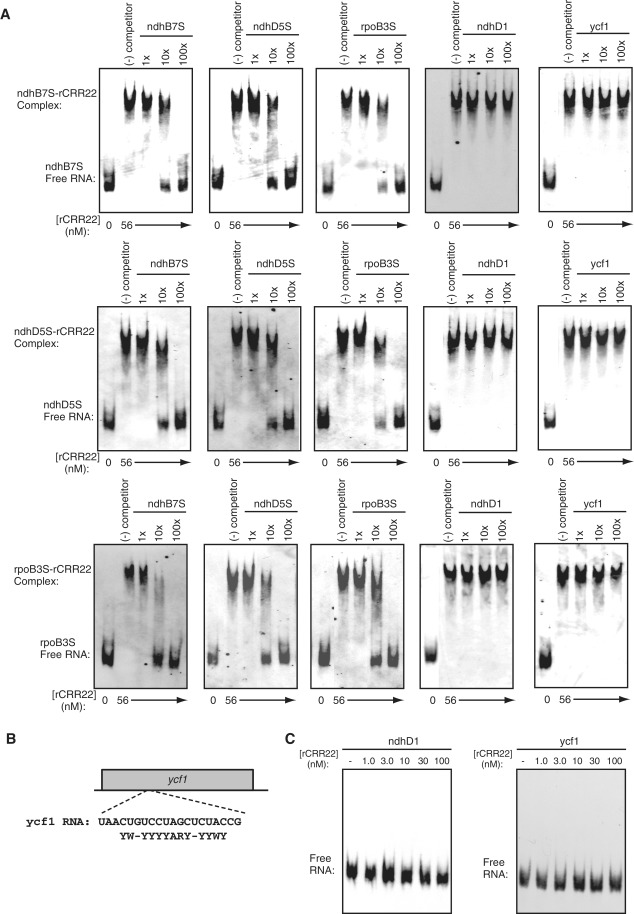
To analyze sequence-specific interactions between CRR22 and its target RNAs, competition assays were performed. The addition of 10 times the amounts of the competitor RNAs ndhB7S, ndhD5S and rpoB3S to the respective radioactive probes partially interfered with retardation, and the shifted bands were eliminated by adding a 100-fold excess of competitor RNAs (Figure 6A). As the competitor including the non-related cis-acting element that was recognized by another PPR protein, we used the 36-nt ndhD1 RNA (Figure 4A). In contrast to the ndhB7S, ndhD5S and rpoB3S competitors, the addition of a 100-fold excess of competitor RNA ndhD1 did not affect CRR22 binding (Figure 6A). Bioinformatics analysis of the target sites has revealed that sequences that show similarity to the CRR22 target RNAs are present at five positions in the plastid transcripts (psbK, ycf1, 3′-UTR atpH, ndhG and ndhA), although they are unlikely to be implicated in RNA editing (18). To test whether CRR22 bound to these RNAs, we used the 20-nt competitor RNA ycf1 (Figure 6B) in a competition assay. The ycf1 sequence was located in the coding region of the ycf1 gene and included a 15-nt sequence similar to that of the CRR22-binding region (Figures 1B and 6B). In contrast to the ndhB7S, ndhD5S and rpoB3S competitors, the addition of a 100-fold excess of competitor RNA ycf1 did not affect CRR22 binding (Figure 6A). We confirmed that rCRR22 did not bind to 32P-labeled ndhD1 and ycf1 RNAs, even if a 1000-fold excess of rCRR22 was added (Figure 6C). To further confirm the sequence-specific binding of CRR22, we tested the binding of rCRR22 to the additional negative control sequence including the putative cis-acting element, 51-nt RNA (rpoB7L), which contain the region surrounding the rpoB-7 site and confirmed that rCRR22 did not bind with rpoB7L RNA (Supplementary Figure S4). The binding experiments provided strong evidence that CRR22 specifically recognized three dissimilar sequences within the −20 to 0 regions surrounding the ndhB-7 and ndhD-5 sites and within the −17 to 0 region surrounding the rpoB-3 site. Taken together with the genetic data (35), these findings indicate that CRR22 is a genuine site-specificity factor at all three sites in plastids.
Figure 6.
Competition assays demonstrating sequence-specific interactions between rCRR22 and the target RNAs. (A) Binding reactions included radioactive ndhB7S, ndhD5S and rpoB3S RNAs, together with a 1-, 10-, or 100-fold molar excess of the non-radioactive RNAs indicated above each panel. The concentration of rCRR22 was held constant at 56 nM. (B) ycf1, the 20-nt competitor RNA used in these experiments, is shown. The 15-nt sequence, which shows similarity to the CRR22 binding region found by bioinformatics analysis (20) in the ycf1 RNA, is shown below the ycf1 sequence. In the sequence, full conservation of nucleotides (A, U, G and C), conservation of purines (A or G = R) or pyrimidines (U or C = Y), and conservation of the number of hydrogen bonding groups (A or U = W, G or C = S) are shown. (C) GMS assays were performed with the indicated concentrations of rCRR22 and radioactive RNAs (ndhD1 and ycf1). All of the competition and GMS assays were performed with the same preparation of rCRR22 as used in Figure 5 and within 2 weeks after purification.
DISCUSSION
On the basis of mutant phenotype analysis, it is clear that some PPR proteins are required for multiple RNA editing events (20–22,27,29). Since some putative cis-sequences recognized by a single PPR protein show partial sequence identity (20,22,27), multiple defects in RNA editing caused by a mutation in a PPR protein may be explained by disturbance of the function of PPR protein as a site-specificity factor. Using an in vitro binding assay, biochemical evidence has been provided for specific binding of the PPR protein OTP87 to multiple editing sites with partial sequence identity in mitochondria (30). Similarly to the mitochondrial case, here we provide evidence that OTP82 specifically binds to cis-acting elements with partial sequence identity that surround the editing sites in plastids (Figures 3 and 4). However, it is still unclear whether some PPR proteins are required as a site-specificity factor for some sites but may have additional function other than RNA recognition for the other target sites. This idea is plausible because the cis-acting elements are not always conserved sufficiently to be specifically recognized as related targets (20). As a candidate for this exception, we focused on CRR22, because the putative cis-acting elements of its targets were unrelated (Figure 1B) (20). However, our experimental results clearly indicated that CRR22 specifically recognized all three cis-acting elements (Figures 5 and 6). This result suggests that a PPR protein acts as a site-specificity factor even though its targeted cis-acting elements do not show apparent nucleotide similarity. An exception has been reported in the case of the genes encoding the DYW-subclass PPR proteins RARE1 and ECB2 (VAC1). Defects in these genes causes loss of full and partial activity, respectively, of accD-1 editing in Arabidopsis plastids (49–51). Implication of the involvement of two PPR proteins in the editing of the accD-1 site implies that one of them has a different role and/or that the two PPR proteins may form a dimer. Since CRR22 and OTP82 act as monomeric site-specificity factors in vitro (Figure 2B), heterodimer formation is unlikely to also be required for target recognition in vivo. We cannot eliminate the possibility that some PPR proteins have additional functions besides site-specific RNA recognition in RNA editing, although this case is unlikely for CRR22.
Sequence-specific binding of CRR4, PpPPR71 and OTP87 to their target RNAs has been demonstrated by in vitro RNA binding assays (19,28,30). Their binding sites reside, respectively, within the −25 to +10 region surrounding the ndhD-1 site, the −40 to +5 region surrounding the ccmF-2 site and the −30 to +10 regions surrounding the nad7-C24 and atp1-C1178 sites (19,28,30). To develop an understanding of the mechanisms of editing site recognition, more precise information of the binding site is necessary. Here, we more precisely defined the cis-acting elements required for site recognition by PPR proteins. For high-affinity binding corresponding to that of long RNA (−40 to +10) probably including all of the cis-acting element, the −15 to 0 regions surrounding the editing sites were sufficient for OTP82 (Figure 3). Such cis-acting elements recognized by CRR22 resided within the −20 to 0 regions surrounding the ndhB-7 and ndhD-5 sites and within the −17 to 0 region surrounding the rpoB-3 site (Figure 5). In all cases, the 3-nt proximal truncation cause a dramatic loss of binding affinity (Figures 3 and 5), although the 3-nt truncation from the 3′-ends affected the binding slightly (Supplementary Figures S2 and S3). Our study suggests that the cis-acting elements recognized by PPR proteins are located at immediately upstream ∼20-nt from the target site. This observation is consistent with previous biochemical results, using in vitro RNA editing systems, in which a single putative protein factor specifically binds to multiple cis-acting elements within ∼10-nt upstream of the editing sites (16,33).
Our binding experiments show that even though the target RNAs show low nucleotide identity (Figure 1B), CRR22 specifically recognizes them with almost identical affinities (Figures 5 and 6). The equilibrium Kd values for ndhB7S, ndhD5S and rpoB3S were 9.2, 10.8 and 13.5 nM, respectively (Figure 5C). These values were comparable to those of OTP82 (Figure 3C) and other PPR proteins investigated so far, including the targets of PGR3 and PpPPR38 (∼10 nM and 13.8 nM, respectively) (38,52). Recently, a physically plausible consensus model of PPR protein structure was built from a statistical analysis of large numbers of restorer-like PPR protein sequences (48). On the basis of this structure, an ∼1:1 correspondence of bases to PPR motifs was suggested (48). This structural model has not been confirmed experimentally, and we cannot eliminate the possibility that other features of a sequence, in particular, micro-structural features including a role for the RNA backbone are determinants of PPR protein affinity. However, several circumstantial evidences support this 1:1 model. The maize P-subfamily PPR protein PPR10 binds to the atpH 5′-UTR and psaJ 3′-UTR in chloroplasts, and the recognition elements are highly conserved (4). PPR10 has 18 or 19 PPR motifs and requires a 17-nt RNA segment in atpH 5′-UTR for high-affinity interaction, supporting the one-repeat-to-one-nucleotide recognition hypothesis (4). The minimum RNA length that was required for the high-affinity binding with OTP82 was 16 nt for both targets and for that with CRR22 was 21 nt for ndhB7 and ndhD5, and 18 nt for rpoB3 (Figures 3 and 5). Since any 3-nt truncation of the probes from both sides decreased the affinity (Figures 3 and 5 and Supplementary Figures S2 and S3), the minimum core sequences may be slightly shorter than this estimation. Since OTP82 and CRR22 contain 14 and 16 PPR motifs, respectively (Figure 1), our results are also consistent with the 1:1 model.
How does the 1:1 model explain the recognition of unrelated sequences by a single PPR protein? If all the nucleotides present in a cis-acting element equally contribute to and are required for high-affinity binding, a single PPR protein cannot recognize unrelated multiple sequences. Only a certain set of nucleotides in a cis-acting element may determine the affinity to a PPR protein, and only a set of PPR motifs in a PPR protein may be sufficient for binding to the target RNA. The cis-acting elements can therefore be rather divergent and still be recognized by a single PPR protein. This idea is supported by our recent observations that a single amino acid alteration in a PPR motif of PGR3 dramatically decreases the binding affinity for one target, but not another (38), and that 3 proximal nucleotides (−20 to −17) are required for high-affinity binding at the ndhB-7 and ndhD-5 sites, although this was not true for the rpoB-3 site (Figure 5). On the other hand, the equilibrium Kd values for CRR22 binding were approximately 100 times higher than that for the target of PPR10 (0.1 nM), which binds to the highly conserved elements in atpH 5′-UTR and psaJ 3′-UTR in chloroplasts (4). Small modifications or spacing in the sequence decrease the binding affinity of PPR10, implying that it interacts with the target RNA via multiple interactions between motifs and nucleotides (4). When a PPR protein recognizes a single target or highly conserved multiple targets, the majority of PPR motifs can be optimized for the single target, facilitating the high-affinity binding. In some PPR proteins like CRR22, however, it is possible that a set of PPR motifs is optimized for one target, while another set in the same protein is for another target. This idea may explain the lower affinity of CRR22 and OTP82 to their targets compared to the affinity of PPR10 to its target.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
K.O. was supported by a Grant-in-Aid from Japan Society for the Promotion of Science (22870029 and 23710220) and a grant from the Japan Science and Technology Agency (AS221Z03416). T.S. was supported by Grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (22114509 and 22247005). Funding for open access charge: a Grant-in-Aid from Japan Society for the Promotion of Science (23657032) to T.S.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Professor Ian Small (University of Western Australia) for helpful discussion in this study.
REFERENCES
- 1.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 4.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008;25:1120–1128. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- 6.Knoop V, Rüdinger M. DYW-type PPR proteins in a heterolobosean protist: plant RNA editing factors involved in an ancient horizontal gene transfer? FEBS Lett. 2010;584:4287–4291. doi: 10.1016/j.febslet.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 7.Salone V, Rüdinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small I, Knoop V, Lurin C. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581:4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- 8.Rüdinger M, Polsakiewicz M, Knoop V. Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol. Biol. Evol. 2008;25:1405–1414. doi: 10.1093/molbev/msn084. [DOI] [PubMed] [Google Scholar]
- 9.Shikanai T. RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol. Life Sci. 2006;63:698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chateigner-Boutin AL, Small I. Plant RNA editing. RNA Biol. 2010;7:213–219. doi: 10.4161/rna.7.2.11343. [DOI] [PubMed] [Google Scholar]
- 11.Chateigner-Boutin AL, Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;35:e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giegé P, Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentolila S, Elliott LE, Hanson MR. Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics. 2008;178:1693–1708. doi: 10.1534/genetics.107.073585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M. Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion. 2008;8:319–327. doi: 10.1016/j.mito.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhuri S, Maliga P. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 1996;15:5958–5964. [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamoto T, Obokata J, Sugiura M. Recognition of RNA editing sites is directed by unique proteins in chloroplasts: biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol. Cell Biol. 2002;22:6726–6734. doi: 10.1128/MCB.22.19.6726-6734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takenaka M, Neuwirt J, Brennicke A. Complex cis-elements determine an RNA editing site in pea mitchoncdria. Nucleic Acids Res. 2004;32:4137–4144. doi: 10.1093/nar/gkh763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 19.Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 2006;281:37661–37667. doi: 10.1074/jbc.M608184200. [DOI] [PubMed] [Google Scholar]
- 20.Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell. 2009;21:3686–3699. doi: 10.1105/tpc.109.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. A DYW domain-containing pentatricopeptide repeat proteins is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21:558–567. doi: 10.1105/tpc.108.064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verbitsliy D, Zehrmann A, van der Merwe JA, Brennicke A, Takenaka M. The PPR protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J. 2010;61:446–455. doi: 10.1111/j.1365-313X.2009.04076.x. [DOI] [PubMed] [Google Scholar]
- 23.Verbitskiy D, Härtel B, Zehrmann A, Brennicke A, Takenaka M. The DYW-E-PPR protein MEF14 is required for RNA editing at site matR-1895 in mitochondria of Arabidopsis thaliana. FEBS Lett. 2011;585:700–704. doi: 10.1016/j.febslet.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 24.Takenaka M. MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA editing event in the nad7 transcript in mitochondria of Arabidopsis. Plant Physiol. 2010;152:939–947. doi: 10.1104/pp.109.151175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takenaka M, Verbitskiy D, Zehrmann A, Brennicke A. Reverse genetic screening identifies five E-class PPR proteins involved in RNA editing in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 2010;285:22122–22129. doi: 10.1074/jbc.M110.128611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bentolila S, Knight W, Hanson M. Natural variation in Arabidospis leads to the identification of REME1, a pentatricopeptide repeat-DYW protein controlling the editing of mitochondrial transcripts. Plant Physiol. 2010;154:1966–1982. doi: 10.1104/pp.110.165969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohtani S, Ichinose M, Tasaki E, Aoki Y, Komura Y, Sugita M. Targeted gene disruption identifies three PPR-DYW proteins involved in RNA editing for five editing sites of the moss mitochondrial transcripts. Plant Cell Physiol. 2010;51:1942–1949. doi: 10.1093/pcp/pcq142. [DOI] [PubMed] [Google Scholar]
- 28.Tasaki E, Hattori M, Sugita M. The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. Plant J. 2010;62:560–570. doi: 10.1111/j.1365-313X.2010.04175.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim SR, Yang JI, Moon S, Ryu CH, An K, Kim KM, Yim J, An G. Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 2009;59:738–749. doi: 10.1111/j.1365-313X.2009.03909.x. [DOI] [PubMed] [Google Scholar]
- 30.Hammani K, des Francs-Small CC, Takenaka M, Tanz SK, Okuda K, Shikanai T, Brennicke A, Small I. The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J. Biol. Chem. 2011;286:21361–21371. doi: 10.1074/jbc.M111.230516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chateigner-Boutin AL, Hanson MR. Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol. Cell. Biol. 2002;22:8448–8456. doi: 10.1128/MCB.22.24.8448-8456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chateigner-Boutin AL, Hanson MR. Developmental co-variation of RNA editing extent of plastid editing sites exhibiting similar cis-elements. Nucleic Acids Res. 2003;31:2586–2594. doi: 10.1093/nar/gkg354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi Y, Matsuo M, Sakamoto K, Wakasugi T, Yamada K, Obokata J. Two RNA editing sites with cis-acting elements of moderate sequence identity are recognized by an identical site-recognition protein in tobacco chloroplasts. Nucleic Acids Res. 2007;36:311–318. doi: 10.1093/nar/gkm1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heller WP, Hayes ML, Hanson MR. Cross-competition in editing of chloroplast RNA transcripts in vitro implicates sharing of trans-factors between different C targets. J. Biol. Chem. 2008;283:7314–7319. doi: 10.1074/jbc.M709595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuda K, Chateigner-Boutin A, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009;21:146–156. doi: 10.1105/tpc.108.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai W, Okuda K, Peng L, Shikanai T. PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 2011;67:318–327. doi: 10.1111/j.1365-313X.2011.04593.x. [DOI] [PubMed] [Google Scholar]
- 39.Navaratnam N, Fujino T, Bayliss J, Jarmuz A, How A, Richardson N, Somasekaram A, Bhattacharya S, Carter C, Scott J. Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J. Mol. Biol. 1998;275:695–714. doi: 10.1006/jmbi.1997.1506. [DOI] [PubMed] [Google Scholar]
- 40.Sambrrok J, Fritsch EF, Maniatis T. Molecular Clonign: A laboratory manual. 2nd edn. Cold Spring Harbor: Cold Spring Habor Press; 1989. [Google Scholar]
- 41.Okuda K, Hammani K, Tanz SK, Peng L, Fukao Y, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J. 2010;61:339–349. doi: 10.1111/j.1365-313X.2009.04059.x. [DOI] [PubMed] [Google Scholar]
- 42.Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–1941. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bock R, Hermann M, Kössel H. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 1996;15:5052–5059. [PMC free article] [PubMed] [Google Scholar]
- 44.Reed ML, Peeters NM, Hanson MR. A single alteration 20 nt 5′ to an editing target inhibits chloroplast RNA editing in vivo. Nucleic Acids Res. 2001;29:1507–1513. doi: 10.1093/nar/29.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirose T, Sugiura M. Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J. 2001;20:1144–1152. doi: 10.1093/emboj/20.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyamoto T, Obokata J, Sugiura M. A site-specific factor interacts directly with its cognate RNA editing site in chloroplast transcripts. Proc. Natl Acad. Sci. USA. 2004;101:48–52. doi: 10.1073/pnas.0307163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl Acad. Sci. USA. 2007;104:8178–8183. doi: 10.1073/pnas.0700865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujii S, Bond CS, Small ID. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl Acad. Sci. USA. 2011;108:1723–1728. doi: 10.1073/pnas.1007667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbins JC, Heller WP, Hanson MR. A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA. 2009;15:1142–1153. doi: 10.1261/rna.1533909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu QB, Jiang Y, Chong K, Yang ZN. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 2009;59:1011–1023. doi: 10.1111/j.1365-313X.2009.03930.x. [DOI] [PubMed] [Google Scholar]
- 51.Tseng CC, Sung TY, Li YC, Hsu SJ, Lin CL, Hsieh MH. Editing of accD and ndhF chloroplast transcripts is partially affected in the Arabidopsis vanilla cream1 mutant. Plant Mol. Biol. 2010;73:309–323. doi: 10.1007/s11103-010-9616-5. [DOI] [PubMed] [Google Scholar]
- 52.Hattori M, Sugita M. A moss pentatricopeptide repeat protein binds to the 3′ end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J. 2009;276:5860–5869. doi: 10.1111/j.1742-4658.2009.07267.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.