Abstract
Coronaviruses RNA synthesis occurs in the cytoplasm and is regulated by host cell proteins. In a screen based on a yeast three-hybrid system using the 5′-untranslated region (5′-UTR) of SARS coronavirus (SARS-CoV) RNA as bait against a human cDNA library derived from HeLa cells, we found a positive candidate cellular protein, zinc finger CCHC-type and RNA-binding motif 1 (MADP1), to be able to interact with this region of the SARS-CoV genome. This interaction was subsequently confirmed in coronavirus infectious bronchitis virus (IBV). The specificity of the interaction between MADP1 and the 5′-UTR of IBV was investigated and confirmed by using an RNA pull-down assay. The RNA-binding domain was mapped to the N-terminal region of MADP1 and the protein binding sequence to stem–loop I of IBV 5′-UTR. MADP1 was found to be translocated to the cytoplasm and partially co-localized with the viral replicase/transcriptase complexes (RTCs) in IBV-infected cells, deviating from its usual nuclear localization in a normal cell using indirect immunofluorescence. Using small interfering RNA (siRNA) against MADP1, defective viral RNA synthesis was observed in the knockdown cells, therefore indicating the importance of the protein in coronaviral RNA synthesis.
INTRODUCTION
During the replication of mammalian viruses, it is inevitable for host proteins to be involved in the viral life cycles. In fact, coronaviruses require host proteins to aid in the stages from virus entry to progeny release. Entry of the virus particle into a host cell requires the recognition of specific cell surface proteins, which act as receptors for the virus spike (S) protein (1–6). Upon entry into host cells, the ribonucleocapsid uncoats and releases the 5′-capped viral genome, a single-stranded positive-sense RNA. The genomic RNA ranges from 27 to 32 kb in length, is the largest known of its kind and is structurally similar to host mRNA (7). The replicase gene, which spans the 5′ two-thirds of the genome, is translated by host ribosomes into two large polyproteins, pp1a and pp1ab via a frameshift event (8–10). The polyproteins are autoproteolytically processed into a maximum of 16 nonstructural proteins (8,11–16), which are assembled into replication–transcription complexes, including the main enzyme RNA-dependent RNA polymerase (nsp12) (17,18). This complex is required for generating new full-length virus RNA in replication as well as subgenome-length RNAs to be used for translation of virus structural and accessory proteins. In addition to their role in RNA synthesis, these nonstructural proteins may have multiple functions, such as the suppression of host mRNA translation as well as mRNA degradation by nsp1 of SARS coronavirus (SARS-CoV; 19–21), which may play a role in the suppression of immune response mounted by the host upon infection.
The replication–transcription complex (RTC), which is located on membrane bound vesicles in the cytoplasm (22), is required for genome replication through continuous transcription and subgenomic RNA synthesis via discontinuous transcription (18,23,24). Apart from the replicase gene products, a viral structural protein, the nucleocapsid (N), is also required for efficient viral RNA synthesis (25,26). The resulting genome-size transcripts are destined to be packaged into progeny virions while the subgenomic, positive-sense transcripts are being translated into four structural proteins, spike (S), nucleocapsid (N), membrane (M) and envelope (E) proteins, as well as other accessory proteins.
In virus RNA synthesis, the replicase complex is indispensable but not an exclusive participant. Several host proteins have been identified to be able to interact with regulatory signals within the untranslated regions in the viral genome of betacoronavirus MHV. These include the polypyrimidine tract-binding protein (PTB) (27,28) with the leader sequence, hnRNP A1 (27,29,30) and hnRNP Q (31) with the 3′-UTR. More recently, poly(A)-binding protein (PABP), hnRNP Q and glutamyl-prolyl-tRNA synthetase (EPRS) were found to play a role in coronavirus RNA synthesis through their interaction with the 3′-UTR of alphacoronavirus TGEV (32). In addition, interaction of viral proteins with host proteins, such as the recently identified interaction between coronavirus nsp14 and DDX1 (33), may also play important enhancement functions in coronavirus replication and infection cycles.
In this study, we describe the interaction of a cellular protein, MADP1 (zinc finger CCHC-type and RNA binding motif 1) with the 5′-UTR of IBV and SARS-CoV, using yeast-based three hybrid screen (34) and RNA-binding assays. Subsequently, the RNA-binding domain of MADP1 and the RNA secondary structure responsible for the interaction were mapped and defined. Using indirect immunofluorescence, we confirmed that MADP1, despite being reported as a nuclear protein (35), was detected in the cytoplasm of virus-infected cells and partially co-localized with the RTCs. Upon silencing of MADP1 using siRNA, viral RNA synthesis on general has been affected, resulting in a lower replication efficiency and infectivity.
MATERIALS AND METHODS
Over-expression of Flag-tagged proteins
All wild-type and mutant MADP1 expressing constructs were based on the vector pXJ40Flag which contains both the CMV and T7 promoter and all expressed proteins were N-terminally tagged with the Flag epitope. For the over-expression of the wild-type and mutant MADP1 proteins, H1299 cells grown to 100% confluency were infected with recombinant Vaccinia-T7 virus for 2 h (h), and the constructs were transfected into the infected cells using Effectene Transfection Reagent (Qiagen). Cells were lysed with lysis buffer [140 mM NaCl, 10 mM Tris (pH 8.0), 1% NP-40] 22 h post-transfection.
Biotin-RNA pull-down assay
Template DNA was amplified from plasmid DNA encoding the 5′ end of IBV genome with various sets of primers targeting different regions of the 5′-UTR (Tables 1 and 2), with the sense primers containing the T7 promoter sequence (36). Biotinylated RNAs were in vitro transcribed with T7 RNA polymerase (Roche Applied Science) in the presence of Biotin RNA Labeling Mix (Roche Applied Science) at 37°C for 2 h. Template DNAs were removed by digestion with RNase-free DNase I (Roche Applied Science) and the labeled RNAs purified with UltraPure Phenol:Chloroform:Isoamyl Alcohol (Invitrogen) then solubilized in nuclease-free water.
Table 1.
Nucleotide sequences of primers used to amplify DNA templates for in vitro transcription
| Primer name | Sequence |
|---|---|
| T7_i1-27 | 5′-TGTAATACGACTCACTATAGGACTTAAGATAGATATTAATATATATCT-3′ |
| pT_i507-528R | 5′-TTTTTTTTTTTTTTTTTTTTTGTTGTCACTGTCTATTGTATGT-3′ |
| T7_i528-506 | 5′-TGTAATACGACTCACTATAGGGTTGTCACTGTCTATTGTATGTC-3′ |
| pT_i1-29 | 5′-TTTTTTTTTTTTTTTTTTTTTACTTAAGATAGATATTAATATATATGTAT-3′ |
| T7_i30-51 | 5′-TGTAATACGACTCACTATAGGTACACTAGCCTTGCGCTAGATT-3′ |
| pT_i99-80 | 5′-TTTTTTTTTTTTTTTTTTTTTCCTATGAGGACCAGCTGTAG-3′ |
| T7_i141-162 | 5′-TGTAATACGACTCACTATAGGGCCACCTGTCAGGTTTTTGTTA-3′ |
| pT_i140-121 | 5’-TTTTTTTTTTTTTTTTTTTTTCAGGTGCCATCCAGGGCACT-3′ |
| T7_i1-27_SL1dsmut | 5′-TGTAATACGACTGAGTATAGGACTTAAGATACTTATTAATATATATGT-3′ |
| T7_i1-26_SL1rsmut | 5′-TGTAATACGACTCACTATAGGACTTAAGATACTTATTAATATATAAG-3’ |
| pT_EGFP_F | 5′-TTTTTTTTTTTTTTTTTTTTTATGGTGAGCAAGGGCGAGG-3′ |
| T7_EGFP_510-528R | 5′-TGTAATACGACTCACTATAGGGCTGCCGTCCTCGATGTTG-3′ |
| T7_i27106-27125 | 5′-TGTAATACGACTCACTATAGGGTAACATAATGGACCTGTTG-3′ |
| LDX30 | 5′-TGATGCCGGCCACGATGCGTC-3′ |
Table 2.
Primer pairs used in amplification of DNA templates for in vitro transcription
| Fragment | Sense primer | Anti-sense primer |
|---|---|---|
| IBV 5′-UTR (+) | T7_i1-27 | pT_i507-528R |
| 5′-UTRΔ1 | T7_i1-27 | pT_i140-121 |
| 5′-UTRΔ2 | T7_i1-27 | pT_i99-80 |
| 5′-UTRΔ3 | T7_i30-51 | pT_140-121 |
| 5′-UTRΔ4 | T7_141-162 | pT_i507-528R |
| 5′-UTRΔ2M1 | T7_i1-27_SL1dsmut | pT_i99-80 |
| 5′-UTRΔ2M2 | T7_i1-26_SL1rsmut | pT_i99-80 |
| IBV 3′-UTR | T7_i27106-27125 | LDX30 |
| EGFP | T7_EGFP_510-528R | pT_EGFP_F |
Biotinylated RNA at 0.1 µM was incubated with cell lysates over-expressing EGFP, Flag-tagged MADP1 or its mutant proteins, respectively, in the presence of 10 mM dithiothreitol (DTT), 100 µg/ml yeast tRNA (Ambion) and 1 U/µl Protector RNase Inhibitor (Roche Applied Science) in a final volume of 200 µl at room temperature for 30 min. The mixtures were incubated with 40 µl (50% slurry) of streptavidin agarose beads (Sigma Aldrich) at room temperature for 30 min. The beads were collected by centrifugation and washed three times with RNase P (RP) buffer (50 mM KCl, 1 mM MgCl2, 10 mM HEPES, pH 8.0), suspended in 25 µl of sodium dodecyl sulfate (SDS) sample buffer with 100 mM DTT. Bound proteins were resolved by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and detected with appropriate antibodies.
Indirect immunofluorescence microscopy and bromo-UTP labeling
African green monkey kidney cells (Vero) grown to 50% confluency in four-chamber glass slides were transfected to over-express Flag-tagged MADP1 or vector control using Effectene for 16 h. Transfected cells were infected with wild-type IBV or mock-infected with Vero cell lysate (Vero cells with serum-free medium subjected to three freeze–thaw cycles at minus 80°C and room temperature, respectively) for 1 h. Infection was allowed to progress for 2 h after virus removal and the cells were treated with actinomycin D at 15 µg/ml (Sigma Aldrich) for 4 h; 1 mM of BrUTP (Sigma Aldrich) was transfected into the cells with SuperFECT (Qiagen) for 3 h.
Cells were fixed at 10 h post-infection with 4% paraformaldehyde for 15 min and permeabilized with 0.2% Triton-X 100 for 10 min. Treated cells were blocked in 10% goat serum, stained with primary antibodies mouse anti-BrdU and rabbit anti-Flag (Sigma Aldrich) and subsequently probed with AlexaFluor 488 anti-rabbit and 594 anti-mouse (Invitrogen) antibodies. Images were captured with Olympus Fluoview Upright Confocal microscope using a sequential laser scanning protocol.
Viral growth assays
H1299 cells grown to 30% confluency were transfected with 100 nM of either siEGFP (5′-GCAACGUGACCCUGAAGUUCdTdT-3′) or siMadp1 (5′-CAAUGACUUGUACCGGAUAdTdT-3′) using DharmaFECT 2 siRNA Transfection Reagent (Dharmacon) for 72 h. Cells were infected with recombinant IBV-Luc at a multiplicity of infectivity of ∼1 (MOI ≈ 1) and incubated for 2 h at 37°C, 5% CO2. The virus-containing medium was replaced with fresh serum-free medium and the cells were either harvested immediately (0 h) or continued to be incubated at 37°C until specific time points post infection (4, 8, 12, 16, 20 or 24 h). Infected cells were subjected to lysis, either through three freeze–thaw cycles (at −80°C and room temperature, respectively) without removal of media, or using lysis buffer after removal of media. Firefly luciferase activity which was used as an indication of viral activity for the recombinant virus was measured using Luciferase Assay System (Promega) according to manufacturer’s instructions using the cell lysates. An end-point dilution assay, the 50% tissue culture infectious dose (TCID50), the amount of virus that will produce pathological change in 50% of inoculated cell culture, of the infected cells was used as a measurement of virus titer. The TCID50 of the infected cells at each time point was determined by using the freeze–thawed infected cells. For each sample, a 10-fold serial dilution was performed and five wells of Vero cells on 96-well plates were infected with each dilution. The numbers of infected wells were collated and TCID50 of each sample was calculated using the Reed–Muench method (5).
Reverse transcription-polymerase chain reaction determination of the replication and sub-genomic transcription efficiency of IBV
Total RNAs were prepared from the infected cells at their specified time points using Trizol Reagent (Invitrogen) Reverse transcription (RT) was performed with Expand reverse transcriptase (Roche) according to the manufacturer’s instructions using the sense primer IBV leader (5′-26CTATTACACTAGCCTTGCGCT46-3′) for the detection of negative-stranded subgenomic RNA (sgRNA) and the antisense primer IBV24803-R (5′-24803CTCTGGATCCAATAACCTAC24784-3′) for the detection of positive-stranded sgRNA. Both primers were then used for PCR. If transcription of subgenomic mRNAs did occur, a 415-bp PCR product corresponding to the 5′-terminal region of subgenomic mRNA 5 and a 648-bp fragment corresponding to the 5′-terminal region of subgenomic mRNA 4 would be expected. Similarly, RT was carried out with the sense primer IBV14931-F (5′-14931GCTTATCCACTAGTACATC14949-3′) for the detection of negative-stranded genomic RNA. Sense primer IBV14931-F and the antisense primer IBV15600-R (5′-15600CTTCTCGCACTTCTGCACTAGCA15578-3′) were used for PCR. If replication of viral RNA occurred, a 670-bp PCR fragment would be expected.
Construction and selection of stable knockdown cell lines
Oligonucleotides were designed based on siMadp1 sequence and cloned into pSilencer 2.1 Neo (Ambion) according to manufacturer’s instructions. Negative control silencer construct was supplied with the cloning kit. Constructs pSilencer-NC (negative control) and pSilencer-Madp1 were transfected into H1299 cells with Effectene Transfection Reagent. Transfected cells were selected with 500 µg/ml G418 (Sigma) and the selected clones were subjected to screening for MADP1 knockdown efficiency. Selected H1299-shNC and H1299-shMadp1 stable cell lines were maintained in media containing 500 µg/ml of G418.
RESULTS
Yeast three-hybrid screening for human proteins that could interact with SARS-CoV UTRs
In order to find candidate host proteins that may be involved in the replication and transcription of coronavirus RNA, a yeast-based three-hybrid (34) screen against a human cDNA library using the 5′-UTR of SARS-CoV RNA as bait was performed. Screens were also performed using the negative sense 5′-UTR and 3′-UTR as bait. Each screen yielded about six to eight colonies which were sequenced and non-sense sequences of the candidates were eliminated. In total, the screen identified three candidates, MADP1, HAX1 and ribosomal protein L27a as binding partners to SARS-CoV positive sense 5′-UTR, negative sense 5′-UTR and negative sense 3′-UTR, respectively. Although it was interesting to find ribosomal protein L27a interacting with the anti-sense 3′-UTR, which was not required for viral protein translation, subsequent functional studies of the protein would prove to be complicated as the virus itself relies heavily on the host ribosome to translate viral proteins, necessary for the infection to proceed. Therefore, it was not chosen for further studies. HAX1 was reported to function as an anti-apoptotic protein, which was not the focus of our screen and was therefore not chosen for further studies as well. MADP1 was reported as a member of the alternative splicing pathway, which implied a possible role in facilitating distal RNA sequences to be brought into close proximity, corresponded well with current evidence on the mechanism of discontinuous transcription. Therefore, it was chosen as the sole target for this study.
The 5′-UTR of coronavirus genomic RNA interacts specifically with MADP1
The interaction between MADP1 and the coronavirus 5′-UTR was confirmed by using over-expressed Flag-tagged MADP1 in a biotin-RNA pull-down assay. Based on the efficiency of Flag-tagged MADP1 co-purification with the biotinylated RNA, the full-length, mammalian-expressed MADP1 was found to be able to interact with the 5′-UTR of IBV and SARS-CoV RNA (Figure 1A). Over-expressed Flag-tagged protein was used to facilitate detection, as there was no commercially available antibody to the protein at that time. It was noted that IBV 5′-UTR showed higher binding affinity to the Flag-tagged MADP1 than did SARS-CoV 5′-UTR (Figure 1A). The specific interaction between IBV 5′-UTR and MADP1 and its functional implication in coronavirus replication were therefore chosen for subsequent characterization.
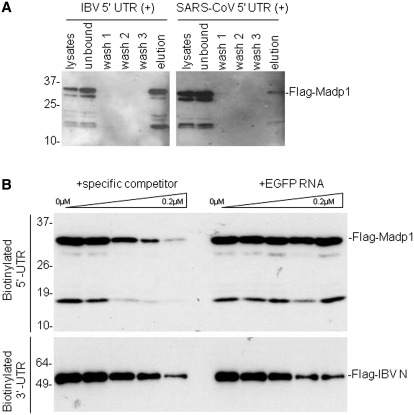
Figure 1.
MADP1 interacts specifically with IBV 5′-UTR.(A) Interaction of MADP1 with SARS-CoV and IBV 5′-UTR in a biotin-RNA pull-down assay. Total cell lysates prepared from H1299 cells over-expressing Flag-tagged MADP1 were mixed with 0.1 µM of biotinylated IBV and SARS-CoV 5′-UTR, respectively, followed by addition of streptavidin agarose beads. Unbound complexes to the beads were subsequently removed by washing and complexes that remained bound to the beads were eluted with gel loading buffer. All fractions and elute were resolved by SDS–PAGE and probed with antibody to Flag tag. (B) Competition assay for the specificity of interaction between MADP1 and IBV 5′-UTR. Total cell lysates prepared from H1299 cells over-expressing Flag-tagged MADP1 was added to mixtures of 0.1 µM biotinylated IBV 5′-UTR RNA and varying concentrations of unlabeled IBV 5′-UTR or EGFP RNA. Streptavidin agarose beads were added and treated under conditions identical to (A). Total cell lysates prepared from cells over-expressing Flag-tagged IBV N protein were added to mixtures of a fixed concentration of biotinylated IBV 3′-UTR RNA and unlabeled IBV 3′-UTR or EGFP RNA and subjected to the same treatment.
To check the specificity of the interaction, a competition assay based on the biotin-RNA pull-down assay was performed. Total cell lysates containing Flag-tagged MADP1 were incubated with 0.1 µM biotinylated IBV 5′-UTR in the presence of increasing concentrations of either unlabeled specific competitor RNA probe (IBV 5′-UTR) or unlabeled non-specific probe (EGFP RNA) composed of nucleotides 1–528 of the EGFP coding sequence, from 0 to 0.2 µM. Western blot analysis of the co-purified Flag-tagged protein showed that increasing concentrations of unlabeled specific competitor RNA led to the decreasing co-purification of MADP1 with the biotinylated RNA probe (Figure 1B). However, increasing concentrations of unlabeled non-specific competitor RNA did not result in detectable change to the efficiency of MADP1 co-purification (Figure 1B).
Simultaneously, a protein exhibiting a non-specific RNA-binding activity, the Flag-tagged IBV-N, was used as a control. Total cell lysates containing the Flag-tagged IBV N protein was incubated with 0.1 µM of the biotinylated IBV 3′-UTR, in the presence of increasing concentrations of either the unlabeled specific probe or an unlabeled non-specific probe, EGFP RNA, of an equal length. Western blot detection of the co-purified Flag-tagged N protein revealed that increasing concentrations of both unlabeled RNA probes increasingly reduced the efficiency of N protein co-purification with the biotinylated RNA probes (Figure 1B). These results confirmed that MADP1 could interact specifically with the 5′-UTR of IBV RNA.
Over-expressed Flag-tagged MADP1 translocates from the nucleus to the cytoplasm
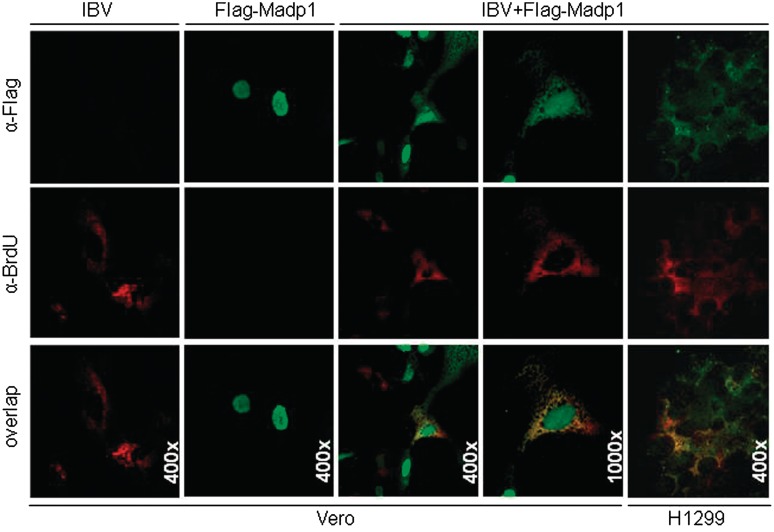
MADP1 was identified as a component of the 18 S U11/12 snRNP (37) and its subcellular localization was determined to be in the nucleoplasm (35). IBV replication and transcription, on the other hand, take place in the cytoplasm of the infected cells. Therefore, to validate the likelihood of MADP1 interacting with the viral 5′-UTR, immunofluorescence was used to track the subcellular localization of both Flag-tagged MADP1 and de novo synthesized viral RNA in both mock-infected and IBV-infected cells. Flag-tagged MADP1 was over-expressed in cultured Vero cells, which were then infected with IBV and treated with actinomycin D to inhibit host transcription. The newly synthesized viral RNA, a marker for the RTCs, was labeled with BrUTP. The cells were fixed at 10 h post-infection to allow sufficient labeling of the newly synthesized viral RNA and to minimize the formation of large syncytial cells. In uninfected cells, Flag-tagged MADP1 was localized in the nucleus exclusively (Figure 2). Upon infection by IBV, Flag-tagged MADP1 appeared to be present in the cytoplasm as well (Figure 2). Interestingly, the cytoplasmic localization pattern of Flag-tagged MADP1 appears to be partially overlapped with that for the RTCs, although further studies would be required to ascertain if MADP1 would be a part of the RTCs (Figure 2). As a negative control for the over-expressed protein, vector transfected cells probed with Flag antibody showed negative staining for the over-expressed protein (Figure 2). Similar colocalization patterns were also observed in IBV-infected H1299 cells (Figure 2).
Figure 2.
Over-expressed Flag-tagged MADP1 partially colocalized with viral RTCs. Vero cells over-expressing either Flag-tagged MADP1 or empty vector were infected with IBV, treated with actinomycin D at 3 h post-infection, and transfected with bromo-UTP at 7 h post-infection. Cells were fixed at 10 h post-infection and permeabilized with Triton-X 100. Immunofluorescence was performed with antibodies to Flag and BrdU followed by secondary antibodies conjugated with Alexa Fluor 488 and 594, respectively. Vector-transfected Vero cells infected with IBV were used as negative controls. H122 cells transfected with Flag-tagged MADP1 and infected with IBV, as described for Vero cells.
Stem–loop I of IBV 5′-UTR is required for interaction with MADP1
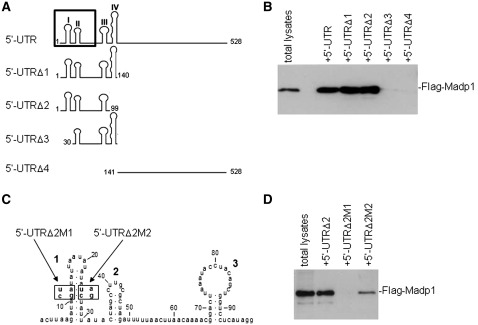
To define the segment and structural elements of IBV 5′-UTR required for its interaction with MADP1, four truncated mutant RNA fragments were synthesized, as shown in Figure 3A, by in vitro transcription. 5′-UTRΔ1 contains stem–loops I–IV (38), 5′-UTRΔ2 and 5′-UTRΔ3 spans stem–loops I to III and II to IV, respectively, whereas 5′-UTRΔ4 spans the rest of the 388 nucleotides. The biotin-labeled RNA transcripts were used in the biotin-RNA pull-down assay (Figure 3B) to check the efficiency of Flag-tagged MADP1 co-purification with RNA. Results showed that MADP1 was co-purified only with transcripts which contain stem–loops I–III of the 5′-UTR (5′-UTRΔ1 and 5′-UTRΔ2). In addition, stem–loop I appeared to be essential for interacting with MADP1 as its absence in 5′-UTRΔ3 abolished the interaction with MADP1 (Figure 3B). The rest region of the 5′-UTR (5′-UTRΔ4) did not appear to interact with MADP1 (Figure 3B).
Figure 3.
Defining the protein interaction sequence. (A) A schematic diagram of the RNA probes used to define the interaction region. Numbers denote nucleic acid residue position and roman numerals denote stem–loop number. The boundary of the leader sequence of IBV (nt. 1–64) is marked by the box on the full-length 5′-UTR. (B) Interaction of various deletion constructs of IBV 5′-UTR with MADP1. Total cell lysates prepared from H1299 cells over-expressing Flag-tagged MADP1 were mixed with RNA probes spanning different regions of IBV 5′-UTR. The RNA–protein complexes were purified with streptavidin beads, resolved by SDS–PAGE and probed with antibody to Flag tag for the presence of Flag-tagged MADP1 protein. (C) Diagram showing the two mutants of 5′-UTRΔ2 containing either two point mutations which disrupt stem–loop I (5′-UTRΔ2M1) or a mutant restoring stem–loop I in 5′-UTRΔ2M1 (5′-UTRΔ2M2). (D) The essential role of a stem–loop I in the interaction between MADP1 and IBV 5′-UTR. Total cell lysates prepared from H1299 cells over-expressing Flag-tagged MADP1 were mixed with 5′-UTRΔ2M1 and 5′-UTRΔ2M2, respectively. The RNA–protein complexes were purified with streptavidin beads, resolved by SDS–PAGE and probed with antibody to Flag tag for the presence of Flag-tagged MADP1 protein.
To confirm further the role of stem–loop I in the interaction between MADP1 and IBV 5′-UTR, two mutants were constructed, based on 5′-UTRΔ2. 5′-UTRΔ2M1 carried two-point mutations at nucleotide residues 11 and 12 from GA to CU, which would disrupt the structure of stem–loop I (Figure 3C), and 5′-UTRΔ2M2 carried additional mutations at residues 25 and 26 from UC to AG (Figure 3C), which would restore the secondary structure of stem–loop I. The mutant RNAs spanning stem–loops I–III were assessed for its ability to bind MADP1. The result indicated that the integrity of stem–loop I may be essential for the interaction between the 5′-UTR with MADP1 (Figure 3D), as the stem–loop disrupting mutation (5′-UTRΔ2M1) failed to interact with MADP1. The stem–loop restoring mutation at nucleotide residues 25 and 26 from UC to AG was able to restore partially the interaction (5′-UTRΔ2M2) (Figure 3D). This result affirmed the conclusion that the secondary structure of stem–loop I of IBV 5′-UTR is indispensable for its interaction with MADP1.
The RNA recognition motif (RRM) of MADP1 is responsible for its interaction with IBV 5′-UTR
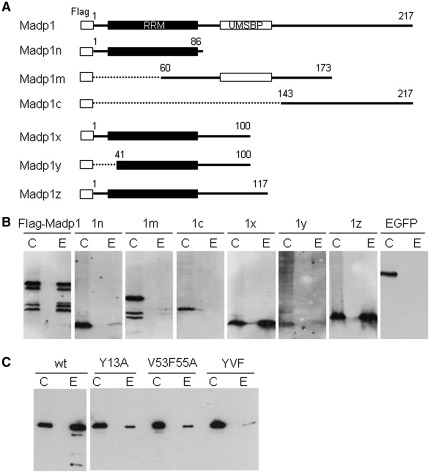
MADP1 contains two nucleic acid binding domains, the RNA recognition motif (RRM) in the N-terminal region and the universal minicircle sequence binding protein (UMSBP) in the central region. In order to identify the domain involved in the interaction between MADP1 and IBV 5′-UTR, a series of truncation mutants of the protein were created (Figure 4A). The first three mutants, Madp1n which contains the RRM domain, Madp1m spans the zinc finger domain and Madp1c contains mostly phosphorylation sites, were assessed for their ability to interact with IBV 5′-UTR. Only Madp1n retained a low level of the RNA-binding activity (Figure 4B, 1n) and negligible activity was detected for the other two truncated proteins (Figure 4B, 1m, 1c).
Figure 4.
Defining the RNA-binding domain. (A) A schematic diagram of constructs of MADP1 and its truncation mutants. Numbers denote amino acid residue positions. Conserved domains RNA Recognition Motif (RRM) and Universal Minicircle Sequence Binding Protein (UMSBP) were indicated as black and white blocks, respectively. (B) Interaction of deletion mutants of MADP1 with IBV 5′-UTR. Cell lysates prepared from H1299 cells over-expressing Flag-tagged wild-type MADP1 or its truncation mutants were used for biotin-RNA pull-down assay using the full-length IBV 5′-UTR. Both the crude lysates (labeled C) and protein bound on the streptavidin beads (labeled E) were resolved by SDS–PAGE and detected by Western blot with anti-Flag antibody. EGFP over-expressed cell lysate was included as a negative control. (C) Interaction of three MADP1 mutant constructs, Y13A, V53F55A and YVF, with IBV 5′-UTR. The three full-length MADP1 constructs with amino acid mutations at the predicted RNA-binding sites were transfected into H1299 cells and used in a biotin-RNA pull-down assay with the full-length IBV 5′-UTR.
As the RNA-binding activity for Madp1n fragment was much lower compared to the full-length protein, three more mutants were created to extend the Madp1n fragment (Figure 4A). An extension of 14 or 31 amino acid residues was made for mutants Madp1x and Madp1z, respectively. A truncation at the N-terminus by 40 residues as well as an extension by 14 amino acid residues was made for Madp1y. It was observed that both Madp1x and Madp1z bound to IBV 5′-UTR more strongly than did the full-length protein as well as Madp1n mutant protein (Figure 4B, 1x, 1z). Madp1y, on the other hand, bound weakly to the RNA fragment (Figure 4B, 1y). Hence, the 14 amino acid extension beyond the RRM (Madp1x) may have been required to preserve the integrity of the protein structure and that the 40 amino acid residues at the N-terminus of MADP1 are required for efficient RNA binding.
As the RRM domain was determined to be responsible for the interaction, information available on this domain indicated three amino acids at its active site, which interact with nucleic acid residues via their aromatic and hydrophobic side chains. For MADP1, the identified active site was composed of phenylalanine 55 and valine 53, respectively, while tyrosine 13 may have acted as an anchor for the phosphate backbone via electrostatic interactions. Hence, three mutants with either a single alanine substitution for tyrosine 13 (Y13A), a double alanine substitution for valine 53 and phenylalanine 55 (V53F55A) or triple alanine substitutions for all three residues (YVF), were constructed (Figure 4A). These three mutants were over-expressed in H1299 cells as Flag-tagged proteins, and the lysates were assessed for their respective RNA-binding affinities for full-length IBV 5′-UTR (Figure 4C). All mutants resulted in a reduction in RNA-binding affinity for the biotinylated RNA molecule and the reduction was most dramatic for triple residue mutant YVF (Figure 4C), implying cooperative binding demonstrated by the three residues. This finding confirms that the MADP1 RRM is involved in the interaction with IBV 5′-UTR.
MADP1 is required for efficient virus transcription
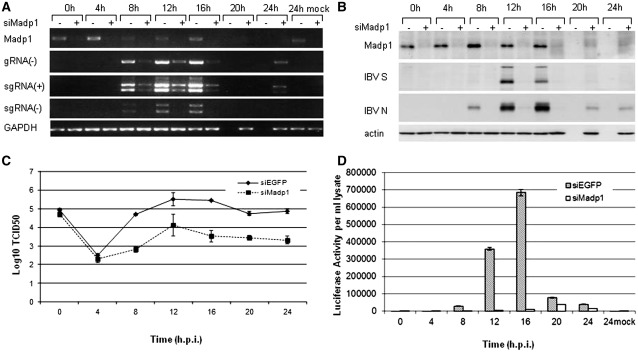
To demonstrate the significance of the interaction between MADP1 and IBV 5′-UTR, an siRNA duplex designed to silence MADP1 expression (siMadp1) and a negative control siRNA targeting EGFP protein (siEGFP) were used in time course experiments. H1299 cells were transfected twice with the siRNA duplexes in 24-h interval and infected with IBV-Luc recombinant virus (39) 3 days after the first transfection. IBV-infected samples were harvested at 0, 4, 8, 12, 16, 20 and 24 h post-infection, respectively, and the mock-infected cells were harvested at 24 h post-infection as a negative control. The levels of negative stranded genomic RNA and both the positive and negative stranded subgenomic RNA were assessed by RT-PCR (Figure 5A). Densitometric analyses identified a reduction between 40% and 80% of MADP1 mRNA was achieved by this siRNA which resulted in a reduction between 70% and 90% of negative stranded genomic viral RNA, 40–80% of negative stranded subgenomic viral RNA and 50–90% of positive stranded subgenomic viral RNA.
Figure 5.
Knockdown of MADP1 by siRNA suppresses IBV infection. (A) RT-PCR analysis of the effect of MADP1 knockdown on IBV RNA replication. H1299 cells were transfected twice with siRNA targeting either Madp1 (+siMadp1) or EGFP (–siMadp1) and infected with IBV-Luc recombinant virus 3 days after the first transfection. Samples were harvested at 4-h intervals, and mock-infected cells were used as negative control. RT-PCR analyses of the mRNA levels of MADP1, the negative strand IBV RNA (gRNA(-)), (+) and (–) mRNAs 3 and 4 (sgRNA) and control GAPDH were carried out. (B) Western blot analyses of the protein levels of Madp-1, viral proteins spike (S), nucleocapsid (N) and cellular protein actin for loading control. (C) TCID50 of total virus produced by the virus-infected cells showed that the silencing of Madp1 reduced virus titers by several folds. (D) Luciferase gene activity measured for the cell lysate indicated a dramatic drop in viral activity in the Madp1 silenced H1299 cells.
Western blot analysis also noted a reduction in the expression of viral structural genes, between 50% and 90% reduction for S and N proteins, with a reduction between 40% and 80% of MADP1 protein (Figure 5B). Virus titers as represented by the tissue culture infectious dose (Log10 TCID50) at each infection time point was reduced by a minimum of 3-fold and up to 10-fold compared to siEGFP-transfected cells beyond 4 h of infection (Figure 5C). Firefly luciferase activity of cell lysates harvested at different time points showed a minimum of 50% reduction upon the silencing of MADP1, which supports further the observation that the total viral protein production was much reduced (Figure 5D).
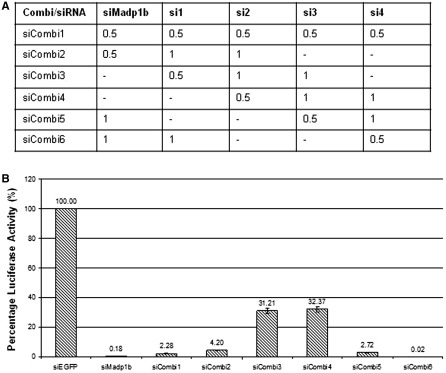
To eliminate the possibility that the phenotype observed in MADP1-silenced cells during IBV infection was due to an off-target effect of the siRNA duplex used, four additional siRNA duplexes targeting different regions of MADP1 were used in various combinations with siMadp1 (Figure 6) to check their effect on IBV infection, as illustrated by the expression of the luciferase gene (Figure 6B). All six combinations of five different siRNA duplexes resulted in a reduction in the luciferase activity of the infected cells by either 70% (siCombi 3 and 4), without siMadp1 or more than 90% (siCombi 1, 2, 5 and 6) with siMadp1, compared to negative control, siEGFP-transfected cells (Figure 6B). This implies that, in general, knocking down MADP1 with any siRNA results in a reduction of virus infection.
Figure 6.
Two hundred and fifty pico moles (250 pmol) of either siRNA to EGFP or siRNA pools against Madp1 were transfected into H1299 cells twice and infected with recombinant luciferase-IBV at 72 h after the first transfection. (A) Volumes (in microliters) of each 50 -μM siRNA used in the siRNA pools. (B) Luciferase activity of the infected cells measured at 20 h post-infection showed a decrease in viral activity after silencing Madp1 with the different siRNA pools.
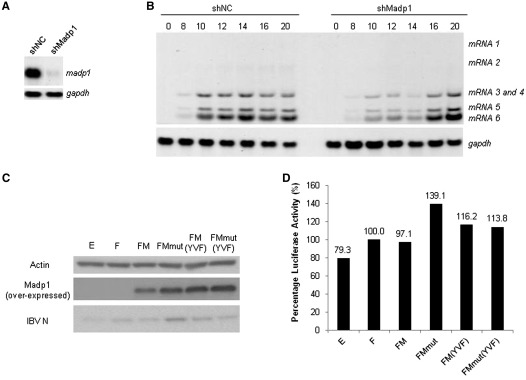
Expression of a silencing-resistant mutant MADP1 in a stable MADP1-knockdown cell clone enhances IBV replication
A stable cell clone expressing short hairpin RNA to MADP1 (shMadp1) was selected from H1299 cells and the madp1 mRNA level was confirmed using northern blot (Figure 7A). The expression of MADP1 and the effect of MADP1-knockdown on IBV infection were tested by comparing with a G418-selected cell line without expression of shMadp1 (non-targeting control, shNC). The results showed that, in general, silencing of MADP1 with shRNA reduced the amount of viral mRNA production before 16 h post-infection (Figure 7B). The amount of virus mRNA is higher in shMadp1 cells compared to shNC cells beyond 16 h of infection as infection in shNC cells progressed much faster and most cells died and detached (Figure 7B).
Figure 7.
Over-expression of siRNA-resistant MADP1 enhances IBV replication in stable MADP1-knockdown cells. (A) Northern blot analysis of H1299 cells stably transfected with either negative control shRNA (shNC) or short hairpin RNA to MADP1 (shMadp1). Total RNA was prepared from the two G418-selected stable clones and separated on 1% agarose gel and probed by a Dig-labeled specific MADP1probe. (B) Northern blot analysis of IBV RNAs in IBV-infected shNC and shMadp1 stable clones. Cells were infected with IBV-Luc at a multiplicity of ∼1, harvested at 0, 8, 10, 12, 14, 16 and 20 h post-infection, respectively. Total RNA was prepared, separated on 1% agarose gel and probed by a Dig-labeled specific probe. (C) Western blot analysis of IBV N protein expression in IBV-infected siMadp1 cells transfected with wild-type or mutant MADP1. shMadp1 stable cell clone was transfected with constructs expressing Flag-tagged wild-type MADP1 (FM) and its triple mutant [FM(YVF)], siRNA-resistant MADP1 (FMmut) and its triple mutant [FMmut(YVF)], negative controls EGFP (E), and empty vector control (F), respectively, using Lipofectamine 2000 (Invitrogen). Cells were then infected with IBV-Luc virus at a multiplicity of ∼1, and harvested at 19 h post-infection for western blot. Over-expressed Madp1 and IBV N were detected with specific antibodies, and actin was detected with a commercial antibody as a loading (Santa Cruz). (D) Firefly luciferase activities of cell lysates from above were normalized against vector-transfected control (F, treated as 100%), and expressed as a percentage to that in the control cells.
The shMadp1 cell line was then transfected with constructs expressing Flag-tagged wild type MADP1 (FM), triple residue mutant (FM(YVF)), two mRNA mutants resistant to silencing by siMadp1 based on wild-type MADP1 (FMmut) and the triple residue mutant (FMmut(YVF)), negative vector control (F) and EGFP (E), respectively. The two siRNA-resistant mutants were constructed by mutating the siRNA-targeting sequence with degenerate codons, so that the protein sequence of MADP1 was maintained. These transfected cells were subsequently infected with IBV-Luc and harvested at 19 h post-infection. Western blotting results showed an obvious increase in the amount of IBV N expression in cells over-expressing silencing-resistant wild-type MADP1 (FMmut) as well as a slight increase in cells over-expressing both normal triple residue mutant (FM(YVF)) and silencing-resistant triple residue mutant (FMmut(YVF)) (Figure 7C).
An assessment of the luciferase activity of total cell lysates showed that over-expression of triple residue mutants FM(YVF) and FMmut(YVF) resulted in a slight increase of the luciferase activity in shMadp1 cells, whereas over-expression of silencing-resistant wild-type MADP1 (FMmut) resulted in a more drastic increase of the luciferase activity in shMadp1 cells (Figure 7D).
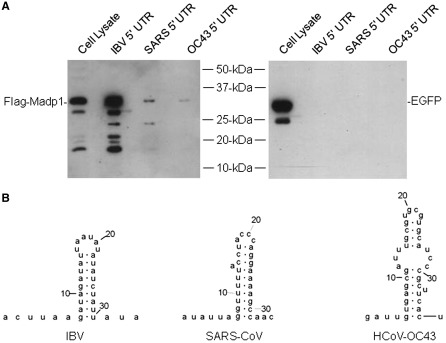
MADP1 interacts weakly with human coronavirus OC43 (HCoV-OC43) 5′-UTR
It was noted that although MADP1 interacted with both SARS-CoV and IBV 5′-UTR, the interaction was rather weak for the former. A comparison of the predicted stem–loop I structures from both coronaviruses indicated a marked difference in their primary sequence as well as the secondary structures. Hence, a third coronavirus, HCoV-OC43, whose stem–loop I which deviated further from IBV than SARS-CoV, was assessed for its binding to MADP1 (Figure 8A). It was found that the binding of MADP1 to the 5′ UTR of HCoV-OC43 was as weak, if not weaker than SARS-CoV. It was also noted that the predicted stem–loop I structure of HCoV-OC43 contained a bulge which encompassed a larger area of the stem compared to SARS-CoV (Figure 8B). Bulges were conspicuously absent from the IBV stem–loop I (Figure 8B). In addition to the differences in the secondary structures between the coronaviruses, there was a lack of sequence similarity as well (Figure 8B).
Figure 8.
Comparison of the 5′ UTR of IBV, SARS-CoV and HCoV-OC43. (A) Interaction of the 5′ UTR from IBV, SARS-CoV and HCoV-OC43 with MADP1. Total cell lysates prepared from H1299 cells over-expressing Flag-tagged MADP1 were mixed with biotinylated RNA probes of full-length 5′ UTRs of the three coronaviruses. The RNA–protein complexes were purified with streptavidin beads, resolved by SDS–PAGE and probed with antibody to Flag tag for the presence of Flag-tagged MADP1 protein. (B) The predicted secondary structures of stem–loop I from IBV, SARS-CoV and HCoV-OC43.
DISCUSSION
Previous studies on the involvement of host proteins in viral RNA synthesis have revealed a number of proteins which are able to interact with the UTRs of viral genomes (29,30,32,40–43). Some of these proteins may also interact with other viral proteins as well (30,40). Our attempts to identify host proteins involved in this early process of the coronavirus life cycle yielded MADP1. This protein was shown to be localized to the nucleoplasm but excluded from the nucleolus, but its role in RNA splicing remains to be determined (37). MADP1 contains two conserved RNA-binding domains, the RNA recognition motif (RRM) 1 and universal minicircle sequence binding protein (UMSBP) domains (a zinc finger CCHC-type) (35). The former was determined to be the domain responsible for the interaction between MADP1 and IBV 5′-UTR. The MADP1 RRM 1 domain interacts with nucleic acid residues via aromatic and hydrophobic side chains at its active site, which in the case supplied by phenylalanine 55 and valine 53, respectively. Tyrosine 13 may have acted as an anchor for the phosphate backbone via electrostatic interactions.
In this study, interaction between MADP1 and the SARS-CoV and IBV 5′-UTR was initially identified by a yeast-based three hybrid screen and subsequently confirmed using an in vitro RNA pull-down assay with IBV 5′-UTR. A deeper look at the details of this interaction revealed that the RNA recognition motif, but not the zinc finger motif, of MADP1, is responsible for the interaction. This interaction is also shown to be specific and stem–loop I of IBV 5′-UTR is essential for the interaction to occur. Although MADP1 was reported to be a nuclear protein (35), it could be detected in the cytoplasm of IBV-infected cells and partially overlaps with the de novo synthesized viral RNA, which marks the location of the RTCs in infected cells in the presence of actinomycin D. Silencing of MADP1 resulted in a marked reduction in syncytium formation upon IBV infection. A closer examination revealed that the synthesis of both genome- (gRNA) and subgenome-length RNAs (sgRNA) was compromised, resulting in a drastic reduction of viral structural protein expression and release of viral progeny (titers), hence the overall reduction of viral infectivity in the cells.
Across different coronaviruses, the leader sequence situated in the extreme 5′ end of the genome, is composed of stem–loops I and II. Mutations introduced into either stem–loop I or II resulted in non-viable viruses, impaired (sense and anti-sense) sgRNA synthesis, but not the full-length gRNA synthesis (38,44). It was, however, observed in this study that silencing of MADP1 did render an impact on gRNA synthesis, although to a lesser extent compared to sgRNA synthesis. This might have been due to a secondary effect of decreased sgRNA synthesis, as proteins encoded by sgRNAs may enhance viral RNA synthesis (45). The predicted structure of stem–loop II indicated a strong secondary interaction, which is highly conserved across different groups of coronaviruses. The predicted stem–loop I structure, on the other hand, appears to fold into a hairpin of low thermodynamic stability, shows a wider sequence variation and is characterized by the presence of bulges, non-canonical base pairing as well as a prevalence of A-U base pairing (46). It has been shown in MHV that the structural liability of stem–loop I is a critical driving force in the 5′- and 3′-UTR interaction (44). Comparing the predicted stem–loop I structures of IBV to SARS-CoV and HCoV-OC43 (Figure 8B), it was noted that there exists a difference in the loop sequence. In addition, IBV stem–loop I has a shorter stem and the absence of bulges, although the structure may be as unstable thermodynamically as that of SARS-CoV and HCoV-OC43, due to the extremely high prevalence of weak base pairing between A and U as well as the presence of a non-canonical base pair at the base of the stem (46). Hence, sequence and structural differences may be one of the possible explanations for the observation of a weaker binding between MADP1 and SARS-CoV or HCoV-OC43 5′-UTR than with IBV 5′-UTR. In fact, the relatively weaker binding of MADP1 to the stem–loop I restoring mutant (5′-UTRΔ2M2) demonstrated in this study supports that primary sequences in the 5′-UTR may play a certain role in this interaction.
Most studies on host involvement in coronaviral RNA synthesis were so far performed using MHV (29–31,42,43). Identification of the interaction between MADP1 and 5′-UTR as well as its functional involvement in coronavirus replication, in this study, therefore may represent the first host protein identified to play a role in viral RNA synthesis by interacting with the 5′-UTR of the viral RNA in a gammacoronavirus. The functional implication of the interaction between MADP1 and IBV 5′-UTR may be extended to the rest of the members of the coronavirus family. In the case of hnRNP A1, it was initially reported to be functionally important for viral RNA synthesis for group II virus MHV (29,30). Subsequently, its involvement in viral RNA synthesis was also confirmed in TGEV, a group I coronavirus (32). In this study, we have shown that betacoronaviruses HCoV-OC43, SARS-CoV and gammacoronavirus IBV can bind to MADP1, albeit with different affinities. Due to the lack of a high containment facility, the functional implication of the relatively weaker interaction between SARS-CoV 5′-UTR and MADP1 was not further studied. It is, therefore, yet to be demonstrated if this weaker binding dictates less dependency on MADP1 in SARS-CoV RNA replication and infectivity.
Current evidence indicates that MADP1 is compartmentalized in the nuclei of cultured cells (35), markedly differing from the cytoplasmic, perinuclear localization of the coronavirus RTCs (47–49). As there was no report on the possibility of MADP1 shuttling between the nucleus and cytoplasm, our observation using indirect immunofluorescence that over-expressed MADP1 upon IBV infection became partially localized in the cytoplasm may represent a first report that MADP1 could be localized outside the nucleus. This could have been achieved with either an existing shuttling mechanism used by a nuclear protein or the assistance of viral factors. For example, IBV N protein is known to enter the nucleus while maintaining a predominantly cytoplasmic localization (50,51). Alternatively, binding of viral RNA may partially retain the newly synthesized MADP1 in the cytoplasm, as observed in this study.
It was observed that over-expression of Flag-tagged MADP1 was unable to fully restore IBV infection in MADP1-knockdown cells, even though the expression level of the introduced MADP1 construct far surpassed the endogenous level, as observed by western blot analysis. Considering the fact that only 30% of cells were transfected and over-expressed MADP1 protein despite the presence of a higher level of the protein in the transfected cells, it is understandable that the expression of viral proteins could not be restored after combining both transfected and untransfected cells. Interestingly, over-expression of silencing-sensitive MADP1 was unable to cause an increase in virus infection, comparing to that was observed for silencing-resistant MADP1 (FMmut) in shMadp1 cells, even though their expression levels were comparable. This lends further support to the conclusion that MADP1 is actively involved in the replication and infectivity of IBV.
Although the functional studies involving IBV, a chicken coronavirus, and a human protein, MADP1, were conducted using human and African green monkey cells, which were non-native. It is noteworthy that MADP1 (HomoloGene 12095) is conserved in humans (Homo sapiens), chimpanzees (Pan troglodytes), wolves (Canis lupus), cattle (Bos Taurus), mice (Mus musculus), rats (Rattus norvegicus) and chickens (Gallus gallus). The African green monkey genome is not available at NCBI, but an alignment search using basic local alignment search tool (BLAST) of the MADP1 amino acid sequence against the rhesus macaque (Macaca mulatta) RefSeq Protein library yields a 99% sequence similarity between the two species. The chicken homolog, on the other hand, bears 85% amino acid sequence similarity, but with an almost identical match in the N-terminal 120 amino acids, to the human MADP1 protein. As the predicted interaction domain lies in the N-terminus, it is highly likely that the homologs from other species could replace human MADP1 in the interaction studies.
In conclusion, the involvement of MADP1 in coronavirus RNA synthesis and its significance are demonstrated in this study in the tissue culture system. Further studies with an MADP1 knock-out animal system, which is currently not available, would be required to confirm further the involvement of MADP1 in coronavirus RNA synthesis.
FUNDING
A Biomedical Research Council grant (BMRC 08/1/22/19.589); and a grant from Nanyang Technological University (SUG M58080000), Singapore. Funding for open access charge: Agency for Science, Technology and Research (A*STAR), Singapore.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu K, Li W, Peng G, Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl Acad. Sci. USA. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada Y, Liu DX. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83:8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada Y, Liu XB, Fang SG, Tay FP, Liu DX. Acquisition of cell-cell fusion activity by amino acid substitutions in spike protein determines the infectivity of a coronavirus in cultured cells. PLoS One. 2009;4:e6130. doi: 10.1371/journal.pone.0006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brian DA, Baric RS. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plant EP, Dinman JD. The role of programmed-1 ribosomal frameshifting in coronavirus propagation. Front. Biosci. 2008;13:4873–4881. doi: 10.2741/3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Wong SM, Liu DX. Identification of hepta- and octo-uridine stretches as sole signals for programmed +1 and -1 ribosomal frameshifting during translation of SARS-CoV ORF 3a variants. Nucleic Acids Res. 2006;34:1250–1260. doi: 10.1093/nar/gkl017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 12.Fang S, Chen B, Tay FP, Ng BS, Liu DX. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology. 2007;358:136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang SG, Shen H, Wang J, Tay FP, Liu DX. Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells. Virology. 2008;379:175–180. doi: 10.1016/j.virol.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang S, Shen H, Wang J, Tay FP, Liu DX. Functional and genetic studies of the substrate specificity of coronavirus infectious bronchitis virus 3C-like proteinase. J. Virol. 2010;84:7325–7336. doi: 10.1128/JVI.02490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl Acad. Sci. USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim KP, Ng LF, Liu DX. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 2000;74:1674–1685. doi: 10.1128/jvi.74.4.1674-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan YJ, Lim SG, Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral. Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masters PS. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl Acad. Sci. USA. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brockway SM, Clay CT, Lu XT, Denison MR. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77:10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasternak AO, Spaan WJ, Snijder EJ. Nidovirus transcription: how to make sense…? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 24.Sawicki SG, Sawicki DL. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verheije MH, Hagemeijer MC, Ulasli M, Reggiori F, Rottier PJ, Masters PS, de Haan CA. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuniga S, Cruz JL, Sola I, Mateos-Gómez PA, Palacio L, Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi ST, Lai MM. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 2005;287:95–131. doi: 10.1007/3-540-26765-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi KS, Huang P, Lai MM. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology. 2002;303:58–68. doi: 10.1006/viro.2002.1675. [DOI] [PubMed] [Google Scholar]
- 29.Huang P, Lai MM. Heterogeneous nuclear ribonucleoprotein a1 binds to the 3′-untranslated region and mediates potential 5'-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 2001;75:5009–5017. doi: 10.1128/JVI.75.11.5009-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi ST, Huang P, Li HP, Lai MM. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 2000;19:4701–4711. doi: 10.1093/emboj/19.17.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi KS, Mizutani A, Lai MM. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J. Virol. 2004;78:13153–13162. doi: 10.1128/JVI.78.23.13153-13162.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galán C, Sola I, Nogales A, Thomas B, Akoulitchev A, Enjuanes L, Almazan F. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology. 2009;391:304–314. doi: 10.1016/j.virol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seay D, Hook B, Evans K, Wickens M. A three-hybrid screen identifies mRNAs controlled by a regulatory protein. RNA. 2006;12:1594–1600. doi: 10.1261/rna.145306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Gao MX, Li L, Wang B, Hori N, Sato K. Isolation, expression, and characterization of the human ZCRB1 gene mapped to 12q12. Genomics. 2007;89:59–69. doi: 10.1016/j.ygeno.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Tan YW, Fang S, Fan H, Lescar J, Liu DX. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Luhrmann R. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10:929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P, Li L, Millership JJ, Kang H, Leibowitz JL, Giedroc DP. A U-turn motif-containing stem-loop in the coronavirus 5' untranslated region plays a functional role in replication. RNA. 2007;13:763–780. doi: 10.1261/rna.261807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen H, Fang SG, Chen B, Chen G, Tay FP, Liu DX. Towards construction of viral vectors based on avian coronavirus infectious bronchitis virus for gene delivery and vaccine development. J. Virol. Methods. 2009;160:48–56. doi: 10.1016/j.jviromet.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo H, Chen Q, Chen J, Chen K, Shen X, Jiang H. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579:2623–2628. doi: 10.1016/j.febslet.2005.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spagnolo JF, Hogue BG. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nanda SK, Johnson RF, Liu Q, Leibowitz JL. Mitochondrial HSP70, HSP40, and HSP60 bind to the 3′ untranslated region of the Murine hepatitis virus genome. Arch. Virol. 2004;149:93–111. doi: 10.1007/s00705-003-0196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nanda SK, Leibowitz JL. Mitochondrial aconitase binds to the 3′ untranslated region of the mouse hepatitis virus genome. J. Virol. 2001;75:3352–3362. doi: 10.1128/JVI.75.7.3352-3362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Kang H, Liu P, Makkinje N, Williamson ST, Leibowitz JL, Giedroc DP. Structural lability in stem-loop 1 drives a 5′ UTR-3' UTR interaction in coronavirus replication. J. Mol. Biol. 2008;377:790–803. doi: 10.1016/j.jmb.2008.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen SC, Olsthoorn RC. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng LF, Liu DX. Membrane association and dimerization of a cysteine-rich, 16-kilodalton polypeptide released from the C-terminal region of the coronavirus infectious bronchitis virus 1a polyprotein. J. Virol. 2002;76:6257–6267. doi: 10.1128/JVI.76.12.6257-6267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu HY, Lim KP, Shen S, Liu DX. Further identification and characterization of novel intermediate and mature cleavage products released from the ORF 1b region of the avian coronavirus infectious bronchitis virus 1a/1b polyprotein. Virology. 2001;288:212–222. doi: 10.1006/viro.2001.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng LF, Liu DX. Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology. 2000;272:27–39. doi: 10.1006/viro.2000.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li FQ, Xiao H, Tam JP, Liu DX. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiscox JA, Wurm T, Wilson L, Britton P, Cavanagh D, Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]