Abstract
The protein phosphatases PP2A and PP1 are major regulators of a variety of cellular processes in yeast and other eukaryotes. Here, we reveal that both enzymes are direct targets of glucose sensing. Addition of glucose to glucose-deprived yeast cells triggered rapid posttranslational activation of both PP2A and PP1. Glucose activation of PP2A is controlled by regulatory subunits Rts1, Cdc55, Rrd1 and Rrd2. It is associated with rapid carboxymethylation of the catalytic subunits, which is necessary but not sufficient for activation. Glucose activation of PP1 was fully dependent on regulatory subunits Reg1 and Shp1. Absence of Gac1, Glc8, Reg2 or Red1 partially reduced activation while Pig1 and Pig2 inhibited activation. Full activation of PP2A and PP1 was also dependent on subunits classically considered to belong to the other phosphatase. PP2A activation was dependent on PP1 subunits Reg1 and Shp1 while PP1 activation was dependent on PP2A subunit Rts1. Rts1 interacted with both Pph21 and Glc7 under different conditions and these interactions were Reg1 dependent. Reg1-Glc7 interaction is responsible for PP1 involvement in the main glucose repression pathway and we show that deletion of Shp1 also causes strong derepression of the invertase gene SUC2. Deletion of the PP2A subunits Pph21 and Pph22, Rrd1 and Rrd2, specifically enhanced the derepression level of SUC2, indicating that PP2A counteracts SUC2 derepression. Interestingly, the effect of the regulatory subunit Rts1 was consistent with its role as a subunit of both PP2A and PP1, affecting derepression and repression of SUC2, respectively. We also show that abolished phosphatase activation, except by reg1Δ, does not completely block Snf1 dephosphorylation after addition of glucose. Finally, we show that glucose activation of the cAMP-PKA (protein kinase A) pathway is required for glucose activation of both PP2A and PP1. Our results provide novel insight into the complex regulatory role of these two major protein phosphatases in glucose regulation.
Keywords: glucose sensing, yeast, protein phosphatases, PP1, PP2A
Introduction
Eukaryotic cells respond to nutrients with a variety of signaling pathways, many of which involve activation of protein kinases. Up to now, most attention in studying these pathways has been focused on the control of protein phosphorylation as triggered by activation of protein kinases. However, for an appropriate cellular response it appears likely that protein dephosphorylation must be also tightly controlled under the same conditions. This aspect of nutrient regulation has received much less attention.
In mammals, nutrient regulation has been studied intensively in cell types with a clear nutrient-sensing function, such as pancreatic β cells. Glucose-induced insulin secretion in these cells is likely affected by protein phosphorylation at different levels 1. In yeast, nutrient regulation has been studied in great detail and involves multiple protein kinase-mediated signaling pathways. Protein kinase A (PKA) can be activated by multiple nutrients in appropriately-starved cells, the Snf1 ortholog of mammalian AMP-activated protein kinase (AMPK) is activated by glucose addition and as in mammalian cells, the TOR protein kinase is activated by amino acid addition. These pathways exert pleiotropic effects on the yeast cells. PKA mediates nutrient control of fermentative growth, stationary-phase characteristics and developmental pathways, as shown by its effects on processes like fermentation, ribosomal protein gene expression, reserve carbohydrate accumulation, cell wall composition, stress tolerance, pseudohyphal growth and sporulation 2. The TOR protein kinase also influences several of these processes 3 and evidence for interaction with the cAMP-PKA pathway has been reported 4, 5. The Snf1 protein kinase functions in the main glucose repression pathway, which downregulates genes involved in respiration and the transport and metabolism of less-preferred carbon sources in the presence of high glucose 6. Its homolog, AMPK, is also subjected to glucose-induced inactivation in mammalian cells 7, 8.
The serine (Ser)/threonine (Thr) protein phosphatases PP2A and PP1 contribute > 90% of Ser/Thr phosphatase activity in eukaryotic cells and are major regulators of a variety of important cellular processes 9, 10. They are composed of catalytic subunits and multiple regulatory subunits, which direct the phosphatases to specific substrate proteins. The yeast Saccharomyces cerevisiae has two PP2A catalytic subunits, Pph21 and Pph22, which are 74% identical to the mammalian PP2Ac, and three PP2Ac-like catalytic subunits, Pph3, Ppg1 and Sit4 11, 12. The scaffolding A subunit is encoded by one gene, TPD3 13. In contrast to mammalian cells, only two types of regulatory subunits for PP2A are known, Cdc55 (class B) 14 and Rts1 (class B′) 15. The extreme C-terminus of Pph can be methylated, which greatly affects the heterotrimer formation between scaffolding, regulatory and catalytic subunits 16, 17, 18. Reversible methylation of PP2A is catalyzed by two conserved and PP2A-specific enzymes, leucine carboxyl methyltransferase (LCMT1) 19, 20 and PP2A methylesterase (PPME1) 21, and their yeast orthologs are called Ppm1 and Ppe1, respectively. It has been suggested that these posttranslational modifications of the C-terminal tail regulate the dynamic exchange of B-type regulatory subunits, thus affecting the specificity of PP2A 18. Moreover, activity of PP2A is also regulated by the phosphatase 2A phosphatase activator (PTPA) 22, 23, 24, for which yeast has two orthologs called as Rrd1/Ypa1 and Rrd2/Ypa2. Recently, a model integrating all these layers of regulation has been proposed, in which Rrd2 interaction with the complex appears to be a prerequisite for the formation of an active holoenzyme and Ppe1 plays a critical role by preventing premature generation of the active complex by counteracting the untimely methylation of the Pph subunits 25. PP2A has been implicated in a variety of cellular processes in yeast, including metabolic regulation, cell-cycle progression, DNA replication, transcription and translation 12, 26.
The catalytic subunit of PP1 in yeast is encoded by the GLC7 gene. Its amino-acid sequence is > 80% identical to that of mammalian PP1, suggesting strong functional conservation during evolution 12. Similar to PP2A, the substrate specificity of PP1 phosphatase action in yeast is determined by a number of regulatory subunits. Sip5 has been suggested to be involved in glucose repression 27. Reg1 controls glucose repression, growth and glycogen accumulation 28, 29, whereas Reg2 and Sds22 affect growth and cell-cycle progression 28, 30. Shp1, Gac1 and the Gac1-related proteins Gip2 and Pig1 appear to affect glycogen accumulation 31, 32, 33, 34. However, Gip1 is involved in meiosis and sporulation 35. Although Pig2 has been recognized as a PP1 subunit there is no clear evidence that links it with control of PP1 activity 32. Red1 and Glc8 are required for meiosis and chromosome segregation 36, 37. Bud14 plays a role in pheromone response and filamentous growth 38. Scd5 affects vesicular traffic in the secretion pathway 39, whereas Bni4 is involved in vesicle-driven recruitment of chitin synthase III at the bud neck 40. Finally, Ypi1 has also been reported as a PP1 regulator both as a phosphatase inhibitor 41 and as a positive regulator of nuclear PP1 activity 42.
Multiple mechanisms are involved in glucose sensing in yeast. The glucose sensors Snf3 and Rgt2, the nontransporting glucose carrier homologs, control expression of regular Hxt glucose transporters by low- and high-glucose levels, respectively 43, 44. Glucose activation of the cAMP-PKA pathway is mediated by a G-protein coupled receptor (GPCR) system, consisting of the Gpr1 receptor and the Gα protein Gpa2 45, 46, 47, and senses extracellular glucose. It activates adenylate cyclase in concert with an intracellular glucose sensing mechanism, which requires glucose phosphorylation by any of the glucose kinases Hxk1, Hxk2 and Glk2 48, and possibly acts through activation of the Ras proteins 49. The glucose sensing mechanism involved in activation of the Snf1 protein kinase in the main glucose repression pathway has remained unclear. Snf1 is phosphorylated on Thr210 by three upstream protein kinases, Sak1, Tos3 and Elm2 50, 51, 52. In addition, it has been shown that glycogen synthesis, which occurs in glucose-starvation conditions, inhibits Snf1 dephosphorylation on Thr210 by PP1 and Sit4 53, further shifting the balance towards Snf1 activation in glucose-deprived cells. As a consequence, the main repressor protein, transcription factor Mig1, becomes phosphorylated and is exported from the nucleus 54, 55, resulting in derepression of certain genes, such as SUC2. Addition of glucose causes inactivation of Snf1, resulting in Mig1 translocation to the nucleus and repression of these genes 55, 56. Evidence has been presented that Hxk2 also plays an important role in the phosphorylation state of Mig1, and as such regulates its nucleocytoplasmic distribution 57. Hxk2, however, may act at more than one level in the signaling pathway 58. Although HXK2 deletion enhances Snf1 phosphorylation, its role in the glucose sensing process remains unclear. No evidence has been found that the upstream protein kinases, Sak1, Tos3 and Elm2, are inactivated by glucose addition, which has led to the suggestion that Snf1 is deactivated through dephosphorylation by the PP1 protein phosphatase Glc7 and its regulatory subunit Reg1 59, 60. However, the precise glucose sensing mechanism for control of Snf1 has remained unclear. Interestingly, the mammalian Snf1 homolog, AMPK, is also inactivated by dephosphorylation of Thr172 (orthologous residue of Thr210 in yeast) following glucose addition and both PP1- and PP2A-type protein phosphatases have been proposed to be responsible for glucose-induced dephosphorylation 61, 62. Very recently it has been reported that in primary mouse adipocytes, PKA associates with and phosphorylates AMPKα1 at Ser173 to restrict Thr172 phosphorylation and thus activation of AMPKα1 by LKB1 kinase in response to lipolytic signals 63. However, it is not clear whether PKA is also involved in the regulation of glucose-induced dephosphorylation of AMPK. Taken together, the precise glucose-sensing mechanism in mammals has also remained unclear.
In the present paper, we report for the first time rapid posttranslational control of two major protein phosphatases PP2A and PP1 by a nutrient, glucose, in yeast. Among the many regulatory subunits known for these phosphatases, we identify specific components required for their glucose activation. Moreover, we show that certain so-called PP2A subunits also affect glucose-induced PP1 activation, and vice versa. In this respect, Rts1, Reg1 and Shp1 are essential for activation of both PP2A and PP1. In addition, we show that Rts1 physically interacts with both Pph21 and Glc7 under specific conditions, and that Reg1 is required for both interactions. For PP2A, glucose activation is associated with rapid C-terminal methylation of the Pph subunits. Glucose activation of both phosphatases requires activation of the cAMP-PKA pathway. We also show that activation of PP1 plays a role in the glucose sensing mechanism of the Snf1-dependent main glucose repression pathway. Moreover, we identify Shp1 as a new subunit of PP1 as well as a role for PP2A in the regulation of this pathway.
Results
Glucose triggers rapid activation of PP2A and PP1
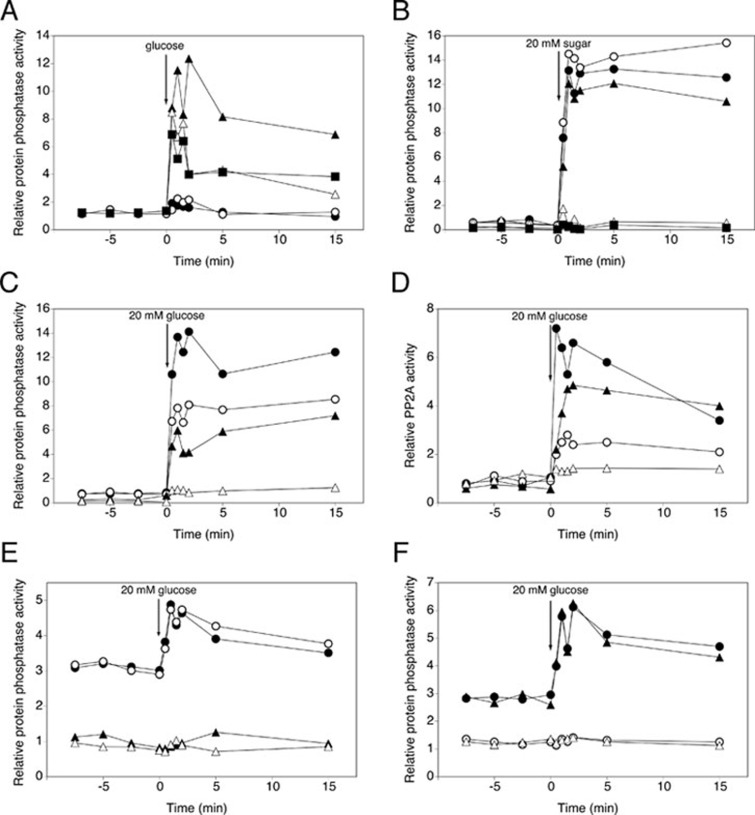
Addition of glucose to glucose-deprived (glucose-derepressed) yeast cells triggered rapid activation of protein phosphatase activity (within 1 min), as measured by the dephosphorylation of 32P-phosphorylated mammalian glycogen phosphorylase a (Figure 1A). Protein phosphatase activity was measured in cell extracts, protein concentration was determined for normalization and the measured relative phosphatase activity is shown as 'nmol phosphate released per min and per mg protein'. In spite of the variability in the basal level, the rapid increase in activity after addition of glucose could always be clearly recognized. For more details and comments concerning the reproducibility, please see Supplementary information, Data S1.
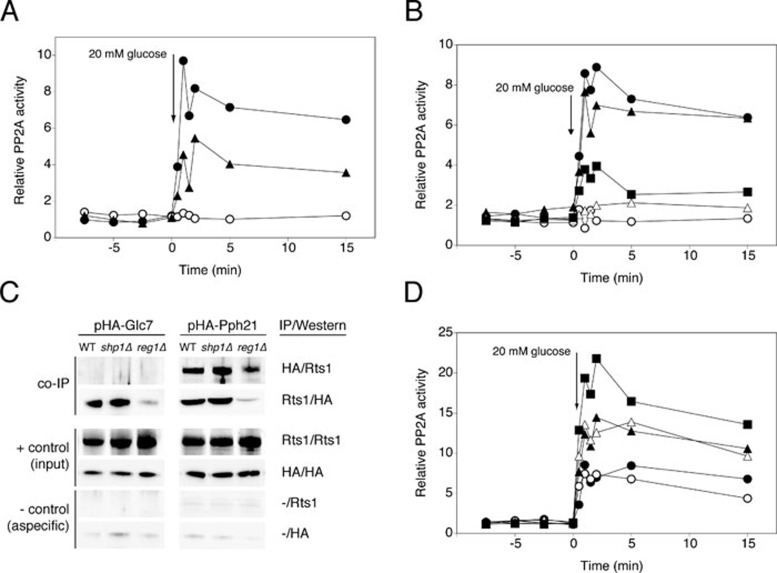
Figure 1.
Glucose activation of PP2A and PP1 protein phosphatase activity. At time 0 a given concentration of the indicated sugar was added to glucose-deprived (glycerol-grown) cells of BY-strains and cell extracts were used to measure protein phosphatase activity in the presence of the appropriate inhibitors: 0.2 μM inhibitor-2 to measure specific PP2A activity and 20 nM okadaic acid to measure specific PP1 activity. (A) Total phosphatase activity after addition of different glucose concentrations: 1 mM (•), 5 mM (○), 20 mM (▴), 60 mM (△) and 100 mM (▪). (B) Total phosphatase activity after addition of 20 mM of the indicated sugar: glucose (•), sucrose (○), fructose (▴), galactose (△) and sorbitol (▪). (C) Total (•) or specific PP2A (○) and PP1 (▴) phosphatase activity after addition of 20 mM glucose. Control: presence of both PP2A and PP1 inhibitor (△). (D) PP2A activity after addition of 20 mM glucose to WT strain (•), pph21Δ (○), pph22Δ (▴) and pph21Δ pph22Δ (△). (E) 20 mM glucose was added and activity was measured with HA-Pph21 immunoprecipitated from cell extracts: pph21Δ pph22Δ + pHA-Pph21 in the absence of inhibitors (•), in the presence of inhibitor-2 (○), in the presence of okadaic acid (▴) and pph21Δ pph22Δ + pEMPTY in the absence of inhibitors (△). (F) 20 mM glucose was added and activity was measured with HA-Glc7 immunoprecipitated from cell extracts: glc7Δ + pHA-Glc7 in the absence of inhibitors (•), in the presence of inhibitor-2 (○), in the presence of okadaic acid (▴) and WT + pEMPTY in the absence of inhibitors (△).
The use of different glucose concentrations revealed that 20 mM was an optimal concentration. Both lower and higher concentrations resulted in a reduced activation (Figure 1A). Although the precise reason for this is unclear, it might be due to stronger activation of a feedback inhibition mechanism at higher glucose concentrations. Addition of 20 mM sucrose or fructose triggered a similar increase as glucose, whereas galactose and sorbitol did not trigger activation (Figure 1B). All subsequent experiments were carried out with 20 mM glucose.
To discriminate between PP1 activity and PP2A activity, we used small-molecule inhibitors with a well-established specificity for the two phosphatases, which are standards in the literature and were confirmed in control experiments. For further details of the inhibitor optimization, we refer to Supplementary information, Data S1 and Figure S1. Using 0.2 μM inhibitor-2 to inhibit PP1 and 20 nM okadaic acid to inhibit PP2A in the protein phosphatase assay, we showed that glucose caused rapid activation of both PP1 and PP2A (Figure 1C). The observation that only combination of the two inhibitors completely suppressed activation of the phosphatases (Figure 1C) further supports the specificity of the inhibitors and indicates that PP2A and PP1 are the most important Ser/Thr phosphatases activated by glucose. This was further confirmed by the results that triple deletion of PPG1, PPH3 and SIT4, which encode PP2A-like phosphatases, did not affect the increase in PP2A activity (Supplementary information, Figure S2).
Glucose activation of PP2A requires Pph21 and Pph22, and PP2A and PP1 activation can be measured with immunopurified Pph21 and Glc7, respectively
We measured glucose activation of PP2A in strains lacking its catalytic subunits, either Pph21 or Pph22 or both (Figure 1D). Deletion of PPH21, and to a lesser extent PPH22, reduced the activation, while deletion of both largely abolished the activation (Figure 1D). This confirms that the measured phosphatase activity was due to PP2A. To further investigate the PP2A activation mechanism, we measured PP2A (Pph21) activity after immunopurification of N-terminally tagged Pph21 from cell extracts. Interestingly, compared to Pph21 in cell extracts (Figure 1D), a significant increase in activity was retained with Pph21 immunopurified from the extracts (Figure 1E). This indicates that at least part of the increase in activity is due to an inherent change in Pph21 or in a component tightly bound to Pph21. It has to be noted that in case of immunopurified Pph21 the basal phosphatase activity was higher, whereas the increase in activity after glucose addition was lower for protein phosphatase activity measured in whole cell extracts. Possible explanations are: (1) the huge increase in Pph21 protein used in the assay, which resulted from the immunopurification of the HA-tagged Pph21 subunit (Supplementary information, Figure S3A) and (2) the loss of small molecules or weakly interacting proteins during the purification step. The first hypothesis could explain both phenomena, due to the high amount of catalytic subunit itself and saturation of our assay, respectively. Using lower concentrations of cell extract for the immunopurification resulted in a decrease in both basal level and the magnitude of activation (Supplementary information, Figure S3B), suggesting that the hypothesis is valid for the basal level but not for the reduced activation. The second hypothesis could also explain both phenomena, because of the possible loss of inhibitory or stimulatory components, respectively. This possibility was investigated by mimicking the loss of small components by dialysis of the cell extracts. This did not affect the basal level but lowered the activation peak to a magnitude similar to that observed with the immunopurified samples (Supplementary information, Figure S3C). This indicates that part of the glucose-induced activation may be due to one or more stimulatory small molecules generated in the glucose metabolism. Addition of inhibitor-2 affected neither the high basal level nor the increase in immunopurified PP2A activity (Figure 1E), while addition of okadaic acid reduced the activity to the level in a pph21Δ pph22Δ strain with an empty plasmid (Figure 1E). This confirms that both the high basal level and the increase are solely due to PP2A.
Similarly, we measured glucose-induced activation with the catalytic subunit of PP1 immunopurified from cell extracts of a glc7Δ strain with a plasmid expressing N-terminally tagged Glc7 (Figure 1F). Since deletion of GLC7 is lethal, we used a wild-type (WT) strain in which an empty plasmid was present as a control. Figure 1F shows that the increase in PP1 activity was also clearly detected when the enzyme was immunopurified from the extracts, indicating that at least part of the activation is due to an inherent change in Glc7 or in a tightly bound component. Addition of inhibitor-2 abolished immunopurified Glc7 phosphatase activity completely, whereas okadaic acid had no effect (Figure 1F). This confirms that for immunopurified PP1 the measured phosphatase activity was also only due to PP1. Taken together, these data show that for both PP2A and PP1 at least a significant part of the increase in activity is not due to allosteric activation by a small molecule or weakly interacting protein present in the cell extracts.
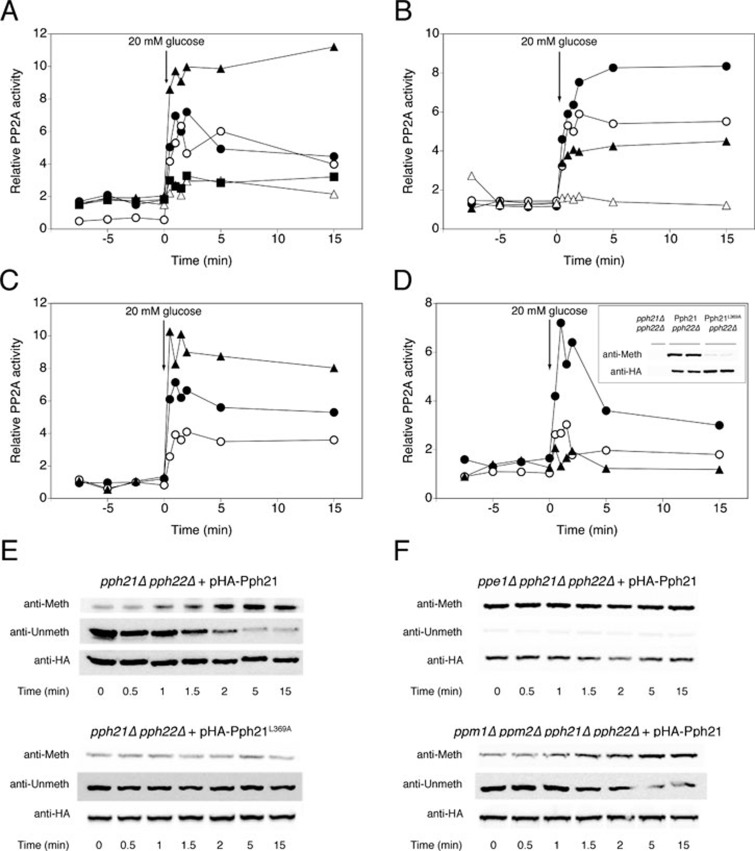
Glucose activation of PP2A is controlled by regulatory subunits Rts1 and Cdc55, and by the PP2A phosphatase activators Rrd1 and Rrd2, but not by scaffolding protein Tpd3
We tested glucose-induced activation in strains deleted for all the reported subunits of the PP2A complex: Rts1 and Cdc55, Tpd3, and Rrd1 and Rrd2. All strains used were from the ResGen/Invitrogen deletion collection, as listed in Supplementary information, Table S1, and were checked for the correct deletion by PCR (Supplementary information, Figure S4A). To our surprise, deletion of TPD3, encoding the scaffolding protein, did not affect the activation (Figure 2A). The absence of the genomic coding sequence, transcribed mRNA and translated protein from the tpd3Δ strain were confirmed by PCR (Supplementary information, Figure S4B), real-time quantitative PCR (RT qPCR) (Supplementary information, Figure S4C) and western blot (Supplementary information, Figure S4D), respectively. However, deletion of RTS1 and CDC55, encoding the regulatory subunits, both influenced the activation of PP2A. Rts1 appeared to be essential for activation, whereas Cdc55 seemed to be inhibitory (Figure 2A). The double deletion of these two genes abolished PP2A activation, indicating that the absence of Rts1 overrides the loss of Cdc55 inhibition (Figure 2A). This suggests an important role for Rts1 in the activation of PP2A. Since no detectable Rts1 could be co-immunoprecipitated with Pph21 in a tpd3Δ strain (Supplementary information, Figure S5), permanent binding of Rts1 to Pph does not seem to be important for the activation. In addition, the PP2A phosphatase activators, Rrd1 and Rrd2, appeared to be essential as well. The single deletions caused a partial reduction of PP2A activation, with a somewhat larger effect for Rrd2, while the activation was completely abolished in the rrd1Δ rrd2Δ strain (Figure 2B). We performed RT qPCR in all the deletion strains mentioned above and ruled out the possibility that the effects observed on PP2A phosphatase activation are due to changes in expression of the genes encoding the catalytic subunits Pph21 and/or Pph22 (Supplementary information, Figure S6A).
Figure 2.
Regulatory subunit Rts1, activators Rrd1 or Rrd2 and methylation at the C-terminus of Pph21 are required for glucose activation of PP2A, whereas regulatory subunit Cdc55 inhibits activation. At time 0, 20 mM glucose was added to glucose-deprived (glycerol-grown) cells of BY-strains with different deletions and cell extracts were used to measure specific PP2A activity (A, B, C and D), or to detect C-terminal methylation of catalytic subunit Pph21 (E and F). (A) WT strain (•), tpd3Δ (○), cdc55Δ (▴), rts1Δ (△) and rts1Δ cdc55Δ (▪). (B) WT strain (•), rrd1Δ (○), rrd2Δ (▴) and rrd1Δ rrd2Δ (△). (C) WT strain (•), ppm1Δ (○) and ppe1Δ (▴). (D) pph21Δ pph22Δ + pHA-Pph21 (•), pph21Δ pph22Δ + pHA-Pph21L369A (○) and pph21Δ pph22Δ + pEMPTY (▴). (E, F) Using antibodies specifically recognizing methylated or unmethylated PP2A catalytic subunits, we monitored changes in C-terminal methylation of pHA-Pph21 and pHA-Pph21L369A in a pph21Δ pph22Δ strain (E), and of pHA-Pph21 in strains ppe1Δ pph21Δ pph22Δ and ppm1Δ ppm2Δ pph21Δ pph22Δ (F). As a control for equal loading, the membranes were incubated with anti-HA after stripping.
Glucose addition causes rapid carboxymethylation of the catalytic subunits, which is necessary but not sufficient for full glucose-induced activation of PP2A
Addition of cycloheximide 15 min before glucose did not affect the response of either PP2A or PP1, indicating a posttranslational mechanism (Supplementary information, Figure S7A). For PP2A, methylation of the C-terminal leucine residue has been reported to be important for its activity and is controlled by the methyltransferase Ppm1 and the methylesterase Ppe1 16, 17, 25. Deletion of PPE1 resulted in increased activation, whereas deletion of PPM1 decreased but did not abolish PP2A activation (Figure 2C). Since Ppm1 has been regarded as the main methyltransferase for methylation of PP2A and the methylation is important for activity, the remaining activation in the ppm1Δ strain was unexpected. It has been stated that Ppm2 does not participate in methylation of PP2A 16, and we also found that deletion of PPM2 did not affect PP2A activation, neither alone nor in combination with the deletion of PPM1 (Supplementary information, Figure S7B). We next tested glucose-induced activation of PP2A in a pph21Δ pph22Δ strain transformed with a modified Pph21 in which the C-terminal leucine residue was mutated to an alanine (Pph21L369A), resulting in largely deficient methylation (Figure 2D inset). This showed that methylation of the C-terminal residue is necessary for full activation of PP2A (Figure 2D). Furthermore, we showed by western blot analysis that methylation of Pph21 rapidly increased within the first minute upon addition of glucose to glucose-starved cells, and that this process did not take place in a strain with only Pph21L369A as PP2A catalytic subunit (Figure 2E). In contrast, a strain lacking methyltransferases Ppm1 and Ppm2 still showed clear residual glucose-triggered methylation (Figure 2F), indicating that the existence of one or more alternative methyltransferases are able to methylate PP2A and respond to glucose activation.
When the PPE1 gene was deleted, the Pph21 subunit appeared constitutively hypermethylated and the signal did not increase after glucose addition (Figure 2F). Interestingly, the magnitude of glucose-induced activation of PP2A in a ppe1Δ strain was higher than in the WT strain, without altering the basal level of activity (Figure 2C). This suggests that methylation enhances activation of PP2A, but methylation in itself is not enough for activation.
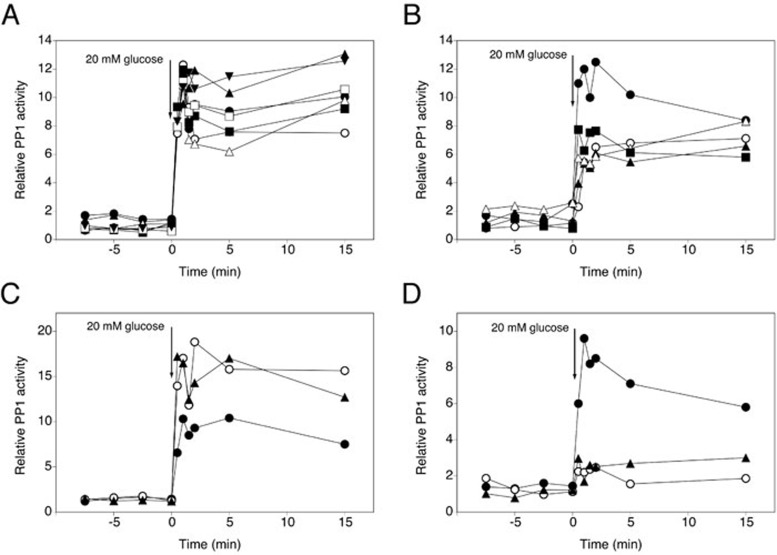
Glucose activation of PP1 is fully dependent on regulatory subunits Reg1 and Shp1
We investigated glucose activation of PP1 in strains containing a deletion of one of the genes encoding the most prominent and best-established regulatory subunits of PP1: Reg1, Reg2, Pig1, Pig2, Glc8, Red1, Gac1, Gip1, Gip2, Shp1, Bni4, Scd5, Sds22, Sip5 and Bud14. Since deletion of Sds22 is lethal, we have used a strain with a mutated Glc7 enzyme (Glc7G279S) that specifically lacks interaction with Sds22 64. Determination of glucose activation of PP1 showed that Bni4, Sip5, Gip1, Gip2, Bud14 and Sds22 are not required for activation (Figure 3A). In contrast, deletion of GAC1, GLC8, REG2 or RED1 resulted in a reduction of PP1 activation with ∼50% (Figure 3B). Deletion of PIG1 or PIG2 caused increased PP1 activation (Figure 3C), indicating that both Pig proteins inhibit PP1 activity. Interestingly, deletion of SHP1 or REG1 resulted in a complete loss of glucose-induced PP1 activation (Figure 3D), suggesting a crucial role for these two subunits in PP1 activation. It has to be noted that in the shp1Δ and pig2Δ strains GLC7 expression differed from WT levels, showing decreased and increased expression, respectively (Supplementary information, Figure S6B), which may contribute to the reduced and increased activation of PP1 in these strains.
Figure 3.
Shp1, Reg1 and Glc8 are required for glucose-induced activation of PP1, whereas absence of other subunits has only a partial effect on activation, or an inhibitory effect. At time 0, 20 mM glucose was added to glucose-deprived (glycerol-grown) cells of BY-strains with different deletions and cell extracts were used to measure specific PP1 activity. (A) WT (•), bni4Δ (○), sds22Δ (▴), sip5Δ (△), bud14Δ (▪), gip1Δ (□) and gip2Δ (▾). (B) WT (•), reg2Δ (○), glc8Δ (▴), red1Δ (△) and gac1Δ (▪). (C) WT (•), pig1Δ (○) and pig2Δ (▴). (D) WT (•), shp1Δ (○) and reg1Δ (▴).
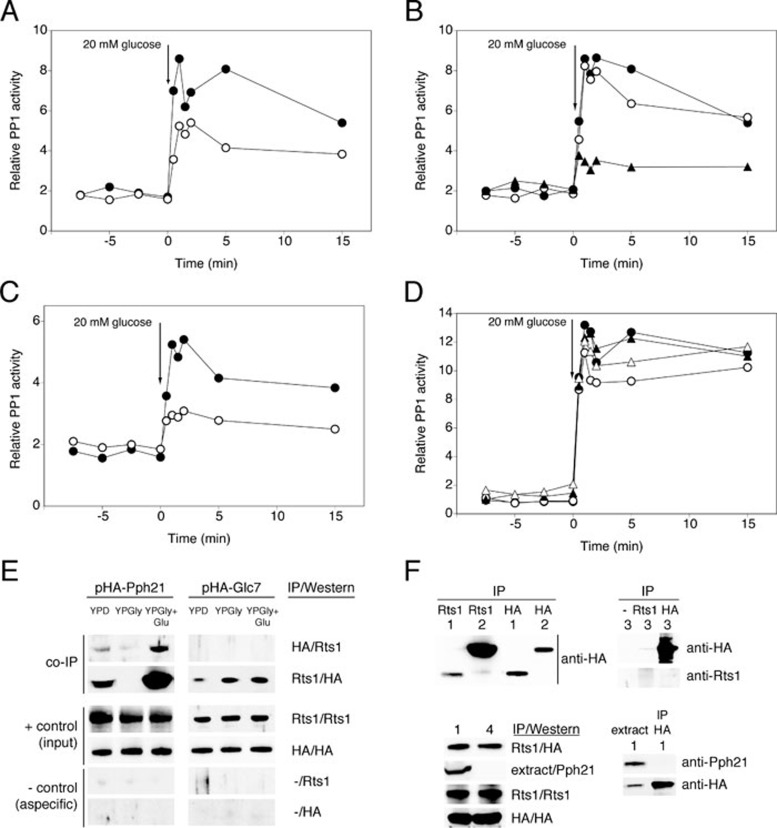
Full glucose activation of PP2A is also dependent on PP1 subunits Reg1 and Shp1, whereas full glucose activation of PP1 is dependent on PP2A subunit Rts1
We also investigated the role of so-called PP2A subunits in glucose activation of PP1 and vice versa. We focused on subunits shown to be either required or inhibitory for the activation of their phosphatases. First, we confirmed that the expression of the catalytic subunit(s) of the other phosphatase is unaffected in strains lacking these subunits (Supplementary information, Figure S6A and S6B). For PP1, we found that deletion of both catalytic subunits of PP2A resulted in decreased glucose-induced activation (Figure 4A). While Cdc55 (Figure 4B) and the PTPA orthologs Rrd1 and Rrd2 (Figure 4D) are not involved in PP1 activation, deletion of RTS1 strongly reduced activation of PP1 (Figure 4B). The large effect of rts1Δ (Figure 4B) compared to pph21Δ pph22Δ (Figure 4A) suggested that the effect of RTS1 deletion on PP1 activation is largely PP2A independent. This conclusion was further substantiated by the additional reduction in PP1 activation caused by deletion of RTS1 in the pph21Δ pph22Δ strain (Figure 4C). Moreover, we showed direct physical interaction of Rts1 with both Pph21 and Glc7 (Figure 4E). It has to be noted that we could show the Rts1-Glc7 interaction only in one direction. This is most probably due to the rather weak nature compared to the Rts1-Pph21 interaction (Figure 4F, upper left), making detection of Rts1 co-immunoprecipitated with the HA-tagged Glc7 impossible with our Rts1 antibody (Figure 4E). We ruled out the possibility that HA-Glc7 aspecifically bound to the beads (negative controls in Figure 4E) or the Rts1 antibody (Figure 4F, upper right). Moreover, since the Rts1-Glc7 interaction was also seen in a strain lacking both PP2A catalytic subunits (Figure 4F, lower left) and immunopurification of HA-Glc7 did not lead to any aspecific co-precipitation of Pph21 (Figure 4F, lower right), we ruled out the possibility that HA-Glc7 was indirectly immunopurified via an interaction with the Rts1-interacting PP2A subunits Pph21 and/or Pph22. The Rts1-Glc7 interaction is most prominent when cells are grown in glucose-deprived conditions (Figure 4E). Together with the weaker Rts1-Glc7 interaction compared to Rts1-Pph21, the requirement of a specific nutrient condition might explain why the interaction was not identified previously. For PP2A, on the other hand, Rts1 interacts with Pph21 during growth on glucose and not in the absence of glucose. Interestingly, the Rts1 subunit is rapidly, within 2 min, recruited to the complex upon glucose addition to glycerol-grown cells (Figure 4E).
Figure 4.
Requirement for Rts1, Rrd1 and Rrd2 in glucose-induced activation of PP1 and interaction of Rts1 with PP2A and PP1. At time 0, 20 mM glucose was added to glucose-deprived (glycerol-grown) cells of BY-strains with different deletions and cell extracts were used to measure specific PP1 activity. (A) WT (•) and pph21Δ pph22Δ (○). (B) WT strain (•), cdc55Δ (○) and rts1Δ (▴). (C) pph21Δ pph22Δ (•) and rts1Δ pph21Δ pph22Δ (○). (D) WT (•), rrd1Δ (○), rrd2Δ (▴) and rrd1Δ rrd2Δ (△). (E) WT cells expressing HA-tagged Pph21 or Glc7 were grown to exponential phase in either glucose (YPD) or glycerol (YPGly). A third sample was taken 2 min after addition of 20 mM glucose to the glycerol-grown cells (YPGly + Glu). Antibodies used for the immunopurification (IP) and immunodetection (western blot): HA: anti-HA; Rts1: anti-Rts1; -: no antibody. (F) Detection of Glc7 and Pph21 in total extract and in extract IP using the indicated antibodies by means of western blot with the antibodies indicated as follows: HA: anti-HA; Rts1: anti-Rts1; Pph21: anti-Pph21; -: no antibody. Strains used are: (1) WT + pHA-Glc7, (2) WT + pHA-Pph21, (3) rts1Δ + pHA-Pph21 and (4) pph21Δ pph22Δ + pHA-Glc7.
For PP2A, we found that deletion of REG1 or SHP1, up to now considered to encode PP1 subunits, resulted in abolished or partially reduced activation, respectively (Figure 5A). Hence, Reg1 and Shp1 seem to play a role in the activation of this phosphatase as well. Since GLC7 is an essential gene, we were unable to investigate the effect of reg1Δ or shp1Δ on PP2A activation in the absence of PP1. However, to check whether the effect of Reg1 on PP2A activation is an indirect effect of the absence of PP1 activation, we used specific mutant alleles of REG1. The Reg1Δ8 and Reg1I466M F468A mutant proteins, which are unable to interact with the PP1 catalytic subunit Glc7 65, caused either abolished or reduced activation of PP2A (Figure 5B). This suggests that the loss of PP2A activation due to malfunctioning of Reg1 is at least in part dependent on Reg1 interaction with PP1. The two Reg1 mutant proteins also showed the expected loss of PP1 activation upon addition of glucose (Supplementary information, Figure S8A, expression of these constructs was confirmed in Supplementary information, Figure S8B). The Reg1I466M F468A mutant protein still showed nearly half of the WT PP2A activation (Figure 5B) in spite of more strongly reduced PP1 activation (Supplementary information, Figure S8A), supporting that the effect on PP2A does not completely act through PP1. We were unable to demonstrate any direct Reg1-Pph21 interaction (data not shown), suggesting that the interaction may be weak or indirect. Interestingly, strains lacking Reg1 showed significantly reduced interaction of Rts1 with both Glc7 and Pph21 (Figure 5C), which supports the importance of Reg1 for PP2A activation and suggests that it may involve the Rts1 subunit. Strains lacking Shp1, on the other hand, were not affected in these interactions (Figure 5C).
Figure 5.
Requirement for Reg1 and Shp1 in glucose-induced activation of PP2A. (A, B and D) At time 0, 20 mM glucose was added to glucose-deprived (glycerol-grown) cells of different deletion strains and cell extracts were used to measure specific PP2A activity. (A) WT (•), reg1Δ (○) and shp1Δ (▴). (B) WT (•), reg1Δ (○), reg1Δ + pReg1 (▴), reg1Δ + pReg1Δ8 (△) and reg1Δ + pReg1I466M F468A (▪). (C) 20 mM glucose was added to glucose-deprived (glycerol-grown) cells of BY-strains with different deletions (WT, shp1Δ and reg1Δ) that are expressing HA-tagged Pph21 or Glc7, respectively. Antibodies used for the immunopurification (IP) and immunodetection (western): HA: anti-HA; Rts1: anti-Rts1; -: no antibody. (D) WT strain (•), pig1Δ (○), pig2Δ (▴), cdc55Δ (△) and pig2Δ cdc55Δ (▪).
Other proteins considered as PP1 subunits also affected PP2A activation. We found that Pig2 also inhibited glucose activation of PP2A, whereas Pig1 had no effect (Figure 5D). Moreover, Pig2 appeared to have a comparable effect as Cdc55 and combined deletion (pig2Δ cdc55Δ) increased the activation of PP2A even further (Figure 5D).
Several subunits involved in glucose activation of PP2A and PP1 affect the main glucose repression pathway
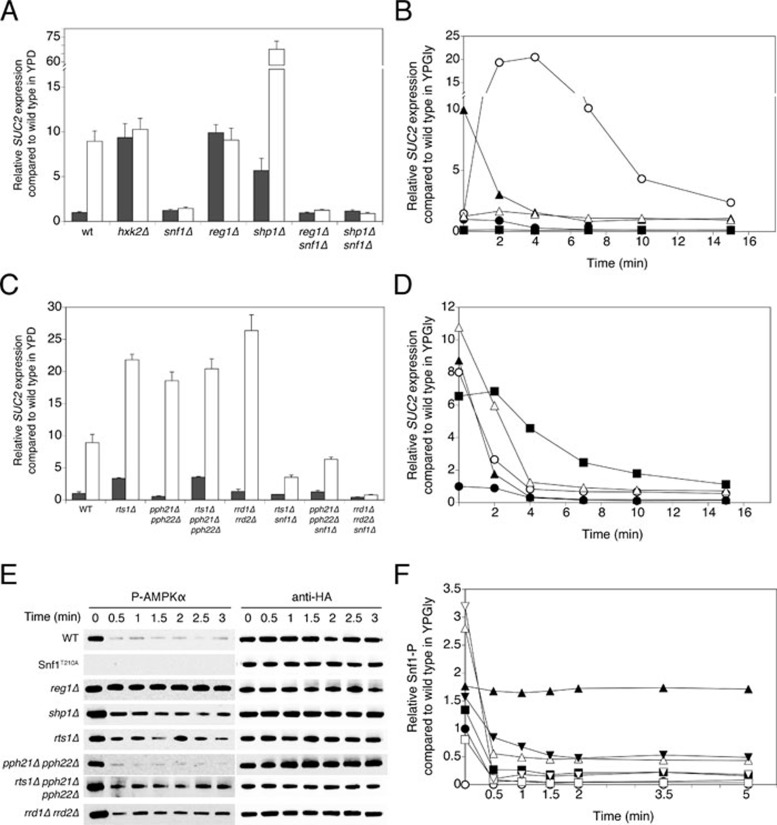
In order to investigate whether the subunits found to be of major importance for glucose-induced activation of PP2A and/or PP1 are also involved in the main glucose repression pathway, we studied the expression level of the invertase gene SUC2. We performed the analysis in two different ways: (1) we grew the cells in 2% glucose medium (repressed cells) and shifted them to medium with a low-glucose concentration of 0.05% for 3 h (derepressed cells); (2) we grew derepressed cells in glycerol medium and followed SUC2 repression upon addition of 20 mM glucose. Since it is generally accepted that PP1 is the phosphatase playing a key role in the main glucose repression pathway, we first studied the typical PP1 subunits together with appropriate controls, such as hxk2Δ and snf1Δ, which showed a constitutively derepressed and repressed phenotype, respectively (Figure 6A). Deletion of REG1 also resulted in a constitutively derepressed phenotype (Figure 6A). The Reg1 subunit of PP1 is known to play a crucial role in control of Snf1 phosphorylation. We noticed that addition of glucose to derepressed reg1Δ cells led to a rapid, transient increase in SUC2 expression (Figure 6B). Besides Reg1, we found that Shp1 is also involved in regulation of glucose repression. We detected higher SUC2 expression both in repressed and especially in derepressed shp1Δ cells compared to the corresponding WT levels (Figure 6A). Addition of glucose to shp1Δ cells caused a strong decrease in SUC2 expression, although it did not drop below the WT derepressed level (Figure 6B).
Figure 6.
Requirement for PP1 and PP2A subunits in regulation of glucose repression. (A, C) Cells were grown in YPD medium containing 2% glucose (repressed cells, dark bars) and shifted to YPGly medium with 0.05% glucose and left to grow for another 3 h (derepressed cells, white bars). Samples were taken for determination of SUC2 expression level. (B, D) Cells were grown in YPGly medium with 0.01% glucose, at time point 0, 20 mM glucose was added. Samples were taken for determination of SUC2 expression level. (B) WT (•), reg1Δ (○), shp1Δ (▴), hxk2Δ (△) and snf1Δ (▪). (D) WT (•), rts1Δ (○), pph21Δ pph22Δ (▴), rts1Δ pph21Δ pph22Δ (△) and rrd1Δ rrd2Δ (▪). (E) Snf1 phosphorylation (Snf1-P) was detected by P-AMPKα antibody at the indicated time points after addition of 20 mM glucose to glucose-deprived (glycerol-grown) cells of specific deletion strains. Anti-HA loading controls are shown. (F) Snf1 dephosphorylation as shown in E was quantified and normalized (Snf1-P signal divided by anti-HA signal and normalized to the t = 0 time point of the WT strain). WT (•), Snf1T210A (○), reg1Δ (▴), shp1Δ (△), rts1Δ (▪), pph21Δ pph22Δ (□), rts1Δ pph21Δ pph22Δ (▾) and rrd1Δ rrd2Δ (▽).
We also tested several subunits of PP2A for possible involvement in the main glucose repression pathway. We found that deletion of the catalytic subunits Pph21 and Pph22, or Rts1, or both (rts1Δ pph21Δ pph22Δ) enhanced the derepression level of SUC2 (Figure 6C and 6D). In addition, the strains lacking Rts1 showed a somewhat enhanced repression level of SUC2. Since the pph21Δ pph22Δ strain did not show higher repressed level and the rts1Δ pph21Δ pph22Δ strain showed the same level as rts1Δ, Rts1 does not act through PP2A (Figure 6C). In short-term repression, both rts1Δ and rts1Δ pph21Δ pph22Δ showed a higher residual SUC2 expression than the WT strain, even comparable with the WT derepressed level (Figure 6D). In all strains addition of glucose caused a rapid drop in the SUC2 messenger level (Figure 6D). Interestingly, deletion of the RRD1 and RRD2 genes caused a strong increase in the SUC2 derepression level, and in this case repression of SUC2 after addition of glucose was delayed (Figure 6C and 6D). Taken together, these results indicate that PP2A counteracts SUC2 derepression in the absence of glucose and that Rts1 may act on the derepression through PP2A and the repression through PP1. By studying strains in which each PP1 or PP2A subunit deletion was combined with a deletion of SNF1, we could conclude that all subunits affected SUC2 derepression largely or completely in an Snf1-dependent manner (Figure 6A and 6C).
Finally, we investigated Snf1 dephosphorylation of residue Thr210 upon addition of glucose to glucose-deprived cells in the deletion strains mentioned above (Figure 6E and 6F). As expected, the Snf1T210A allele showed no phosphorylation at all and the reg1Δ strain showed no dephosphorylation upon addition of glucose. Moreover, derepressed cells of the strain lacking Reg1 had an increased phosphorylation level compared to WT. shp1Δ, rrd1Δ rrd2Δ and rts1Δ pph21Δ pph22Δ showed an increased phosphorylation level in derepressed conditions, as could be expected from the SUC2 data. In agreement with these data, addition of glucose to shp1Δ and rts1Δ pph21Δ pph22Δ strains resulted in a 6-fold higher residual phosphorylation compared to WT repressed levels. More surprisingly, pph21Δ pph22Δ behaved more or less like WT, both in derepressed and repressed conditions, whereas deletion of RTS1 led to a small increase in phosphorylation in both conditions.
Glucose activation of PP1 and PP2A uses a similar glucose sensing mechanism as glucose activation of the cAMP-PKA pathway
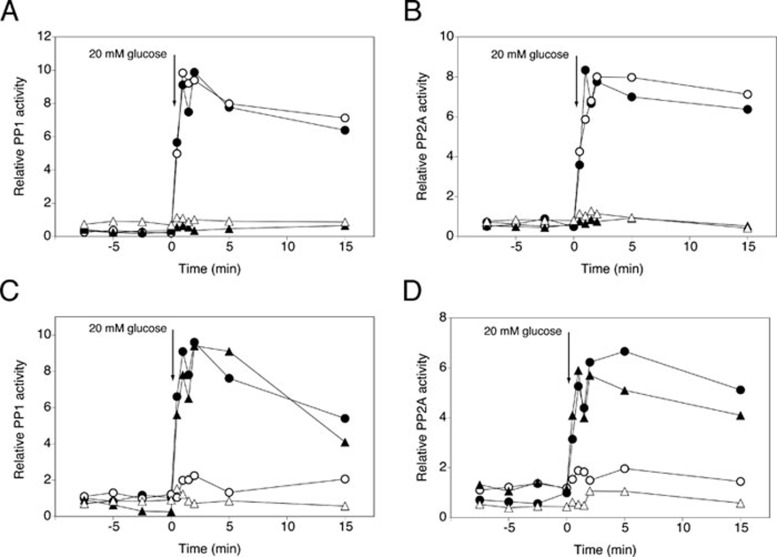
We next investigated which glucose-sensing system might be involved in activation of PP1 and PP2A. First, we ruled out the TOR pathway since addition of rapamycin did not affect glucose activation of either phosphatase (Supplementary information, Figure S9A). Also deletion of the glucose sensors Snf3 and Rgt2 43 had no effect (Supplementary information, Figure S9B and S9C). Next, we tested a possible connection with glucose activation of the cAMP-PKA pathway. For this purpose, we investigated glucose activation of PP1 and PP2A in yeast strains deficient in the glucose/sucrose-sensing G-protein coupled receptor, Gpr1, and in the three glucose kinases that can sustain activation of cAMP synthesis through the glucose phosphorylation-dependent mechanism. As opposed to rapid activation of cAMP synthesis, deletion of Gpr1 did not reduce glucose activation of PP1 or PP2A (Figure 7A and 7B). In contrast, deletion of the three glucose kinase genes, GLK1, HXK1 and HXK2, abolished glucose activation of PP1 and PP2A (Figure 7A and 7B), indicating that glucose phosphorylation is essential for activation of both phosphatases. To further investigate the possible involvement of the cAMP-PKA pathway in glucose activation of PP1 and PP2A, we used two strains with compromised PKA activity, tpk1Δ tpk2w2 tpk3Δ and tpk1Δ tpk2Δ tpk3Δ yak1Δ. In both strains, glucose activation of the two protein phosphatases was largely abolished (Figure 7C and 7D). Deletion of Yak1 only did not affect the activation (Figure 7C and 7D). Hence, our results suggest that rapid glucose activation of PP1 and PP2A may be triggered by rapid glucose activation of the cAMP-PKA pathway, with glucose phosphorylation-dependent glucose sensing being the most critical mechanism.
Figure 7.
Glucose activation of PP1 and PP2A phosphatases requires glucose phosphorylation and PKA. At time 0, 20 mM glucose was added to glucose-deprived (glycerol-grown) cells of BY-strains with different deletions and used to measure specific PP1 activity (A, C) or specific PP2A activity (B, D). (A, B) WT strain (•), gpr1Δ (○), glk1Δ hxk1Δ hxk2Δ (▴) and gpr1Δ glk1Δ hxk1Δ hxk2Δ (△). (C, D) WT strain (•), tpk1Δ tpk2W2 tpk3Δ (○), yak1Δ (▴) and tpk1Δ tpk2Δ tpk3Δ yak1Δ (△).
Discussion
Glucose activation of PP1 and PP2A
This work reveals for the first time that two crucial Ser/Thr protein phosphatases, PP2A and PP1, with broad pleiotropic roles in eukaryotic cell physiology, growth and development are rapidly activated in yeast by addition of glucose to glucose-deprived cells. By immunoisolation of the catalytic subunits of both PP2A and PP1, Pph21 and Glc7, respectively, we showed that glucose-induced phosphatase activation is to a significant extent due to an inherent increase in activity of the enzymes. The remaining part of the glucose-induced activation appears to be controlled by unknown allosteric activator(s), such as small molecule(s) or weakly interacting protein(s). Glucose activation of PP2A and PP1 is not inhibited by cycloheximide, which together with the rapidity of the response, within 1 min, indicates a posttranslational mechanism. Such a rapid change in protein phosphatase activity in response to an environmental stimulus has not previously been reported in yeast or in any other eukaryotic cell type 12. Our finding creates a direct link between nutrient regulation of cellular function and protein phosphatase activity.
Composition of the PP2A enzyme activated by glucose and its activation mechanism
Our results indicate that the Pph21 and Pph22 catalytic subunits (C subunits) form part of the PP2A enzyme that is involved in glucose activation. The other three known PP2Ac-like components, Pph3, Ppg1 and Sit4 are not involved. With respect to the regulatory B subunits, both Rts1 and Cdc55 are involved, in which Rts1 is required for the activation and Cdc55 appears to play a role as an inhibitor controlling the magnitude of activation. Although it has been reported that both the scaffolding A protein Tpd3 (to form the AC complex) and methyltransferase Ppm1 (to mediate C-terminal methylation of the C subunit resulting in efficient binding of the B regulatory subunits to the AC complex) are critical for PP2A activity 15, 16, 17, 25, our results indicate that neither of these is essential for its glucose-induced activation. Since Tpd3 is assumed to be essential for the function of regulatory subunit Rts1 in controlling PP2A function, it is striking that rts1Δ abolishes PP2A activation whereas tpd3Δ does not. Although it has been shown that Tpd3 is required for co-immunoisolation of Pph21 and Rts1 under normal growth conditions 15, other groups have reported PP2A-B′/C complex formation without A-scaffolding protein both in yeast 66 and in mammals 67. Nevertheless, we also could not show any significant Rts1-Pph21 association in a tpd3Δ strain under our growth conditions. Taken together, this suggests that, although Rts1 is required, its presence or weak association with Pph might be sufficient to support rapid activation by glucose signaling, whereas Tpd3 is only required for the strong interaction with Pph needed to assemble a stable heterotrimeric complex. Rrd1 and Rrd2, yeast orthologs of the mammalian PTPA protein, have been reported to play a crucial role in the regulation of PP2A activity 23, 24, 68. More recently, it has been shown that these Rrd proteins are able to re-activate an inactive protein phosphatase 2A population in which the association with methylesterase Ppe1 prevents it from premature methylation and subsequent holoenzyme assembly 25, 69. It has been proposed that in this process Rrd proteins interact physically and functionally with Tpd3 25. In spite of the lack of Tpd3 requirement for glucose activation, we showed that both Rrd1 and Rrd2 are required. This reveals a new regulatory function for the PTPA activator of PP2A.
The posttranslational nature of the glucose-induced activation of PP2A prompted us to study the C-terminal methylation of the catalytic subunit in more detail, which together with phosphorylation 70, 71 affects the process of assembly. We discovered that glucose activation of PP2A is associated with a rapid increase in methylation of the C-terminus and further work showed that methylation is important for glucose activation. Interestingly, it has been shown that PP2A methylation and activity significantly increases in bovine aortic endothelial cells when cultured in 30 mM glucose 72. Recently, it has been reported that in vitro activation of the PP2A active site stimulates methylation by LCMT-1, the human ortholog of Ppm1, suggesting a mechanism for efficient conversion of activated PP2A into substrate-specific holoenzymes 73. However, we show that hypermethylation of Pph subunits in glucose-starved cells of a ppe1Δ strain does not lead to PP2A activation prior to glucose addition, indicating that methylation is not sufficient to increase PP2A activity. Taken together, C-terminal methylation seems to prime the enzyme for another rapid modification that enhances inherent phosphatase activity, as previously suggested for steady-state conditions, such as growth on glucose-containing medium 25. Unexpectedly, we found that the ppm1Δ ppm2Δ strain did not only retain the C-terminal methylation of PP2A to some extent, but even showed a glucose-induced increase in C-terminal methylation. This indicates the existence of (an)other yet unidentified methyltransferase(s) subject to glucose signaling.
Composition of the PP1 enzyme activated by glucose and its role in glucose repression
With respect to the subunits required for glucose activation of PP1, they can be categorized into four groups: those not involved in the activation or at least showing redundancy (Bni4, Sds22, Sip5, Bud14, Gip1 and Gip2), those involved but not strictly required for activation (Reg2, Glc8, Red1 and Gac1), those that regulate the magnitude of the response by inhibiting the phosphatase (Pig1 and Pig2) and finally, those that are critical for PP1 activation (Shp1 and Reg1). It has to be noted that the effect of deletion of PIG2 and SHP1 may be partially due to altered expression of GLC7, which was found to be increased and decreased, respectively. Our work shows that glucose activation of PP1 most likely involves a posttranslational modification. Future work will have to show whether this modification is exerted on Glc7 itself or on one of its subunits. Our results also reveal for the first time that the PP1 phosphatase Glc7 could be a target of the glucose sensing process in the main glucose repression pathway. Up to now, it has remained unclear how glucose signaling causes dephosphorylation of Snf1 at residue Thr210, either by inhibition of the upstream protein kinases or by activation of the Glc7 protein phosphatase 6. The rapid posttranslational activation of PP1 that we have now discovered may constitute an important part of the link between glucose signaling and the rapid inhibition of Snf1 by stimulation of its dephosphorylation, resulting in rapid repression of downstream targets of the glucose repression pathway, such as SUC2. We found that deletion of Reg1 completely abolished glucose activation of PP1, dephosphorylation of Snf1 and glucose repression of SUC2. In addition, we have identified Shp1 as a new PP1 subunit involved in regulation of glucose repression. Deletion of SHP1 abolished PP1 activation and affected the glucose repression pathway in two ways, reducing repression by glucose and in particular enhancing the derepression level. Although a 6-fold higher SUC2 expression is observed in repressed conditions, significant Snf1 dephosphorylation still occurs in this strain upon addition of glucose. However, the residual phosphorylation of Snf1 (6-fold higher than the WT repressed level, but lower than the WT derepressed level) appears to be enough to sustain the highly elevated expression of SUC2 in the derepressed condition. A few possible explanations can be given for the significant glucose-induced dephosphorylation of Snf1 in a strain lacking PP1 activation. First, the glucose activation of the glucose repression pathway might be exerted at different points in the signaling pathway. This is consistent with previous results that hexokinase 2 is translocated into the nucleus and directly affects repression at the transcriptional level 57, while this enzyme normally would be considered to exert its glucose sensing function at the initiation of the glucose signaling pathway. Second, although we were unable to show any significant contribution of PP2A-like phosphatases in the glucose-induced activation of phosphatase activity, Sit4 appears to contribute to the dephosphorylation of Snf1 at Thr210 during growth in high glucose 53. Third, glucose-induced activation of PKA might result in phosphorylation of Snf1, as such affecting further phosphorylation of Snf1 by its upstream protein kinases, Sak1, Tos3 and Elm2 in glucose conditions (similar as shown for AMPKα1 in primary mouse adipocytes 63). However, although AMPK residues Thr172 and Ser173 are conserved in yeast Snf1 (Thr210 and Ser211), the latter is not embedded in a PKA recognition site.
Promiscuous use of PP2A and PP1 subunits in glucose activation
Although the subunit composition of PP2A and PP1 has been characterized in great detail, for other putative subunits the evidence linking them to either PP2A or PP1 is very limited. Still, our results that deletion of relatively well-established subunits of PP2A or PP1 affects glucose activation of the other phosphatase came as a surprise. They seem to suggest that some subunits may be common to both protein phosphatases or at least may also affect to some extent the other phosphatase. Our results reveal a more general role for Rts1, Shp1 and Reg1 in glucose-induced activation of PP1 and PP2A. Further investigation revealed that Rts1 interacts with the catalytic subunits of both PP1 and PP2A, although the Rts1-Pph21 interaction was clearly stronger than that between Rts1 and Glc7. In addition, Reg1 appears to play its role in PP2A and PP1 activation (at least partially) by enabling the recruitment of Rts1 to the phosphatases. In contrast to the loss of Rts1-Pph21 interaction in a strain lacking Tpd3, the loss of Rts1-Pph21 and Rts1-Glc7 in the reg1Δ strain correlates with abolished activation of both phosphatases. This suggests that Reg1 by itself is also directly required for activation of both phosphatases, whereas Tpd3 most likely has no role besides its scaffolding function. This adds another important regulatory role for Reg1 in glucose-induced signaling, in addition to the previously described recruitment of the PP1 subunit Glc7 to Snf1 58, 74, the inactivation of maltose permease 75, 76 and the degradation of fructose-1,6-bisphosphatase in the vacuole 77.
In terms of nutrient conditions, Rts1 is mainly present in the PP2A complex when glucose is available in the medium and the subunit is apparently rapidly recruited (within 2 min) when glucose is added to glucose-deprived cells. Hence, Rts1 recruitment correlates with the presence of C-terminal methylation on the catalytic subunits. In contrast, the Rts1-Glc7 interaction mainly occurs in glucose-deprived conditions and does not rapidly decrease upon glucose addition. This suggests that recruitment of Rts1 to PP1 under glucose-deprived conditions prepares it for a rapid response to the availability of glucose, triggered by a specific glucose-induced signaling mechanism resulting in activation of the Rts1-PP1 complex.
A role for Rts1 in particular and PP2A in general in glucose repression
Since Rts1 plays a direct role in the activation of both PP2A and PP1, we investigated the role of Rts1 and the other classical PP2A subunits, Pph21 and Pph22, and Rrd1 and Rrd2, in glucose repression. As already observed for the PP1 subunits, Reg1 and Shp1, strains lacking Rts1 (rts1Δ and rts1Δ pph21Δ pph22Δ) showed compromised glucose repression activity although to a more limited extent. Since both strains showed similar defects, Rts1 exerts its effect on the repression of SUC2 likely through PP1. In addition, a consistent effect was observed for PP2A in limiting the level of derepression of SUC2 in the absence of glucose. Since we showed that deletion of RTS1 or both PPH21 and PPH22 affects PP1 activation by glucose, the effects seen on glucose repression might act through PP1 and thus work indirectly. However, despite the fact that the Rrd proteins are not able to interact with Glc7 directly 23 and do not play any role in PP1 activation (our data), their deletion causes a higher derepressed level of SUC2, suggesting that their effect on derepression does not work through PP1. Taken together, our results reveal for the first time a complementary role of PP2A in glucose repression. While PP1 triggers repression in the presence of glucose, PP2A limits derepression in the absence of glucose.
For the regulatory subunits with a newly discovered function in glucose repression or derepression, Rts1, Shp1 and Rrd1, Rrd2, we observed a clear effect on the level of Snf1 phosphorylation in the deletion strains. However, for the Pph21 and Pph22 catalytic subunits this was not the case. In other cases, the correlation also did not always fit well. For instance, Snf1 phosphorylation in repressed conditions is rather low in strains lacking Rts1 in spite of the partial derepression of SUC2. However, the increase in residual phosphorylation after addition of glucose compared to WT might contribute to the 3-fold increase in SUC2 expression in the rts1Δ strain in repressed conditions. Similarly, the residual phosphorylation and the somewhat slower dephosphorylation seen in the rrd1Δ rrd2Δ strain might contribute to the slower repression of SUC2 upon addition of glucose. Compared to the persistently high level of Snf1 phosphorylation in the reg1Δ strain, a discrepancy is observed in derepressed conditions for the other strains, since the much higher SUC2 levels are not reflected in the rather low Snf phosphorylation of Thr210. In spite of this, we showed that the effects on SUC2 expression act through Snf1 and it has been shown that PP2A dephosphorylates the corresponding Thr172 of mammalian AMPK in vitro78. A possible explanation for the discrepancy between Snf1 phosphorylation and SUC2 expression might be that other residues in Snf1 also undergo phosphorylation/dephosphorylation and these are preferentially targeted by PP2A.
Signaling pathway for glucose activation of PP1 and PP2A
Glucose activation of PP2A and PP1 protein phosphatase activity is observed under the same conditions as glucose activation of the cAMP-PKA pathway and several other protein kinase-mediated signaling pathways 6, 79, 80. Our results show that glucose activation of PP2A and PP1 is independent of the TOR pathway and of the Snf3/Rgt2 sensor system. Surprisingly, the G-protein coupled receptor Gpr1 was also not essential. However, glucose activation of the phosphatases strictly requires the glucose phosphorylation-dependent system, similar to glucose activation of cAMP synthesis 81. The maximum of the glucose-induced cAMP increase occurs about 1 min after addition of glucose, which coincides with the initial activation of both PP2A and PP1. Hence, our data suggest that glucose activation of PP2A and PP1 is, at least partly, mediated by glucose activation of the cAMP-PKA pathway in yeast. The absence of PP2A and PP1 glucose activation in strains with compromised PKA activity provides further support for this concept. Interestingly, activation of D1-receptors in mammals leads to cAMP-dependent dephosphorylation of DARPP-32 through a mechanism in which PP2A is activated by a cAMP-PKA-dependent pathway 82. Moreover, very recently it has been shown that a C-terminal domain of mAKAP (muscle A-kinase anchoring protein, interacting with PKA by definition) binds PP2A, and PKA phosphorylation of mAKAP-bound B56δ (the mammalian ortholog of yeast Rts1) enhances the phosphatase activity in the complex 83. The involvement of the same glucose sensing systems in glucose activation of cAMP synthesis also explains why fructose and sucrose trigger the same activation as glucose and why galactose and sorbitol cannot trigger any activation. Fructose phosphorylation triggers a cAMP signal while sucrose activates the Gpr1 receptor, whereas galactose and sorbitol can neither be phosphorylated by Glk1 or Hxk1, Hxk2, nor do they activate Gpr1 48, 84.
Previous work has revealed cross talk between the cAMP-PKA pathway and the glucose repression pathway, but the precise connection between the two pathways has remained unclear 85, 86. It is also known that short-term and long-term glucose repression have different requirements 87, 88. While long-term glucose repression specifically requires hexokinase 2, short-term repression can be mediated by any one of the three glucose kinases 87. The latter is also true for glucose activation of cAMP synthesis 81 and rapid glucose activation of PP1.
Materials and Methods
Yeast strains and plasmids
The S. cerevisiae strains used in this work are shown in Supplementary information, Table S1. All experiments were carried out with isogenic WT and mutant strains. Since strains with deletion of both PPH21 and PPH22 are rather unstable, the strains were made several times independently and each set of experiments was started with freshly streaked strains (stored at −80 °C). Plasmids used are shown in Supplementary information, Table S2.
Recombinant DNA techniques
For all gene cloning experiments and PCR reactions, standard molecular biology techniques were used 89, 90.
Yeast culture conditions
Glucose-derepressed (glucose-deprived) yeast cells were cultured at 30 °C into exponential phase (OD600 nm = 1.5-2) in medium containing 1% w/v yeast extract, 2% w/v Bacto-peptone, 3% w/v glycerol and 0.01% w/v glucose. When selection for plasmids was required, cells were cultured in minimal medium containing 0.17% w/v Yeast Nitrogen Base without amino acids and ammonium sulfate, 0.5% w/v ammonium sulfate, auxotrophic supplements as required (pH 5.5), 3% w/v glycerol and 0.05% w/v glucose. Glucose-repressed cells were grown into exponential phase (OD600 nm = 1.5-2) on a YPD medium (1% w/v yeast extract, 2% w/v Bacto-peptone and 2% w/v glucose).
Protein phosphatase activity assay
Phosphatase activity was determined from the release of 32P-inorganic phosphate from 32P-phosphorylase a 91, which is in turn obtained from phosphorylation of glycogen phosphorylase b by glycogen phosphorylase kinase. The procedures for making radiolabeled phosphorylase a, for sampling of the cells, immunopurification of HA-tagged Pph21 and HA-tagged Glc7, and assay of protein phosphatase specific activity are described in detail in Supplementary information, Data S1. Relative protein phosphatase activity was calculated as nmol phosphate released per min and per mg protein.
Western blot analysis and real-time quantitative PCR
Detailed procedures for western blot analysis (the methylated and unmethylated fraction of PP2A catalytic subunits, the detection of Tpd3 and Pgk1 in extracts, Rts1 and Snf1 phosphorylation in immunoprecipitated samples) and for real-time quantitative PCR analysis of SUC2 expression are provided in Supplementary information, Data S1.
Reproducibility of the results
All experiments were repeated at least three times and in three independent experiments, showing consistent results and trends, one representative example is shown. For RT qPCR expression studies, SD are shown for comparisons between independent data points. Detailed procedures for standardization of the phosphatase studies are given in Supplementary information, Data S1.
Acknowledgments
We thank Stephen Castermans, Martine De Jonghe, Willy Verheyden and Evy Vanderheyden for excellent technical assistance and Nico Van Goethem for help with the preparation of the figures. We are also grateful to E Ogris (Medical University of Vienna, Austria), JR Broach (Princeton University, USA), JF Cannon (University of Missouri, USA), E Boles (Goethe-Universität, Germany), ET Young (University of Washington, USA), MC Schmidt (Children's Hospital of Pittsburgh, USA) and S Shenolikar (Duke-NUS Graduate Medical School Singapore) for the kind gift of strains, plasmids and/or antibodies. This work was supported by a predoctoral fellowship from the Institute for Scientific and Technological Research (IWT) to IS, by a Return Grant from the Belgian Federal Science Policy Office to MV and by grants from the Fund for Scientific Research - Flanders, Interuniversity Attraction Poles Network P5/30 and P6/14, and the Research Fund of the KULeuven (Concerted Research Actions) to JMT.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Supplementary Experimental Procedures
Optimization of okadaic concentration used for specific PP2A inhibition.
PP2Ac-like subunits are not required for glucose activation of PP2A.
High basal level of immunopurified Pph21 is most likely due to the high concentration of this catalytic subunit, and not to the loss of any interacting molecule/protein during the immunopurification.
Confirmation of all gene deletions, and in particular deletion of TPD3.
Rts1 does not interact with Pph21 in a tpd3Δ strain.
Expression of catalytic subunits of PP2A and PP1 is unaffected in strains showing altered glucose-induced phosphatase activation.
Both PP2A and PP1 are activated by glucose in a post-translational manner, and for PP2A, methylation by Ppm1 is partially required.
Reg1 mutant proteins unable to interact with Glc7 do not support glucose-induced PP1 activation.
Tor signalling and the Snf3/Rgt2 system are not involved in the activation of PP1 and PP2A.
List of yeast strains used in this study.
List of plasmids used in this study
References
- Guettier JM, Gorden P. Insulin secretion and insulin-producing tumors. Expert Rev Endocrinol Metab. 2010;5:217–227. doi: 10.1586/eem.09.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Martin DE, Hall MN. The expanding TOR signaling network. Curr Opin Cell Biol. 2005;17:158–166. doi: 10.1016/j.ceb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE, Hall MN. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 2004;24:338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez SA, Cardenas ME. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell. 2005;4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Xavier G, Leclerc I, Salt IP, et al. Role of AMP-activated protein kinase in the regulation by glucose of islet beta cell gene expression. Proc Natl Acad Sci USA. 2000;97:4023–4028. doi: 10.1073/pnas.97.8.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt IP, Johnson G, Ashcroft SJ, Hardie DG. AMP-activated protein kinase is activated by low glucose in cell lines derived from pancreatic beta cells, and may regulate insulin release. Biochem J. 1998;335 (Pt 3):533–539. doi: 10.1042/bj3350533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- Sneddon AA, Cohen PT, Stark MJ. Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J. 1990;9:4339–4346. doi: 10.1002/j.1460-2075.1990.tb07883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MJ. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast. 1996;12:1647–1675. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1647::AID-YEA71%3E3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- van Zyl W, Huang W, Sneddon AA, et al. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4946–4959. doi: 10.1128/mcb.12.11.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy AM, Zolnierowicz S, Stapleton AE, Goebl M, DePaoli-Roach AA, Pringle JR. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol Cell Biol. 1991;11:5767–5780. doi: 10.1128/mcb.11.11.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Yang H, Hallberg E, Hallberg R. Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol Cell Biol. 1997;17:3242–3253. doi: 10.1128/mcb.17.6.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Ashby DG, Moreno CS, et al. Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J Biol Chem. 2001;276:1570–1577. doi: 10.1074/jbc.M008694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Tolstykh T, Lee J, Boyd K, Stock JB, Broach JR. Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo. EMBO J. 2000;19:5672–5681. doi: 10.1093/emboj/19.21.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- De Baere I, Derua R, Janssens V, et al. Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the human homologue. Biochemistry. 1999;38:16539–16547. doi: 10.1021/bi991646a. [DOI] [PubMed] [Google Scholar]
- Lee J, Stock J. Protein phosphatase 2A catalytic subunit is methyl-esterified at its carboxyl terminus by a novel methyltransferase. J Biol Chem. 1993;268:19192–19195. [PubMed] [Google Scholar]
- Lee J, Chen Y, Tolstykh T, Stock J. A specific protein carboxyl methylesterase that demethylates phosphoprotein phosphatase 2A in bovine brain. Proc Natl Acad Sci USA. 1996;93:6043–6047. doi: 10.1073/pnas.93.12.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayla X, Van Hoof C, Bosch M, et al. Molecular cloning, expression, and characterization of PTPA, a protein that activates the tyrosyl phosphatase activity of protein phosphatase 2A. J Biol Chem. 1994;269:15668–15675. [PubMed] [Google Scholar]
- Fellner T, Lackner DH, Hombauer H, et al. A novel and essential mechanism determining specificity and activity of protein phosphatase 2A (PP2A) in vivo. Genes Dev. 2003;17:2138–2150. doi: 10.1101/gad.259903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof C, Martens E, Longin S, et al. Specific interactions of PP2A and PP2A-like phosphatases with the yeast PTPA homologues, Ypa1 and Ypa2. Biochem J. 2005;386:93–102. doi: 10.1042/BJ20040887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H, Weismann D, Mudrak I, et al. Generation of active protein phosphatase 2A is coupled to holoenzyme assembly. PLoS Biol. 2007;5:e155. doi: 10.1371/journal.pbio.0050155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabrocki P, Van Hoof C, Goris J, Thevelein JM, Winderickx J, Wera S. Protein phosphatase 2A on track for nutrient-induced signalling in yeast. Mol Microbiol. 2002;43:835–842. doi: 10.1046/j.1365-2958.2002.02786.x. [DOI] [PubMed] [Google Scholar]
- Sanz P, Ludin K, Carlson M. Sip5 interacts with both the Reg1/Glc7 protein phosphatase and the Snf1 protein kinase of Saccharomyces cerevisiae. Genetics. 2000;154:99–107. doi: 10.1093/genetics/154.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick DL, Tatchell K. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol Cell Biol. 1996;16:2922–2931. doi: 10.1128/mcb.16.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Chun KT, Goebl MG, Roach PJ. Genetic interactions between REG1/HEX2 and GLC7, the gene encoding the protein phosphatase type 1 catalytic subunit in Saccharomyces cerevisiae. Genetics. 1996;143:119–127. doi: 10.1093/genetics/143.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKelvie SH, Andrews PD, Stark MJ. The Saccharomyces cerevisiae gene SDS22 encodes a potential regulator of the mitotic function of yeast type 1 protein phosphatase. Mol Cell Biol. 1995;15:3777–3785. doi: 10.1128/mcb.15.7.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Guha S, Volkert FC. The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol Cell Biol. 1995;15:2037–2050. doi: 10.1128/mcb.15.4.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Huang D, Roach PJ. Yeast PIG genes: PIG1 encodes a putative type 1 phosphatase subunit that interacts with the yeast glycogen synthase Gsy2p. Yeast. 1997;13:1–8. doi: 10.1002/(SICI)1097-0061(199701)13:1<1::AID-YEA49>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Stuart JS, Frederick DL, Varner CM, Tatchell K. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol Cell Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Hart T, Wu X, Tatchell K. Protein phosphatase type 1 regulates ion homeostasis in Saccharomyces cerevisiae. Genetics. 2002;160:1423–1437. doi: 10.1093/genetics/160.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Song W, Carlson M. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4199–4206. doi: 10.1128/mcb.16.8.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EA, Roeder GS. Expression and DNA sequence of RED1, a gene required for meiosis I chromosome segregation in yeast. Mol Gen Genet. 1989;218:293–301. doi: 10.1007/BF00331281. [DOI] [PubMed] [Google Scholar]
- Tung HY, Wang W, Chan CS. Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol Cell Biol. 1995;15:6064–6074. doi: 10.1128/mcb.15.11.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF Jr. The Glc7p-interacting protein Bud14p attenuates polarized growth, pheromone response, and filamentous growth in Saccharomyces cerevisiae. Eukaryot Cell. 2002;1:884–894. doi: 10.1128/EC.1.6.884-894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KK, Holmer M, Lemmon SK. SCD5, a suppressor of clathrin deficiency, encodes a novel protein with a late secretory function in yeast. Mol Biol Cell. 1996;7:245–260. doi: 10.1091/mbc.7.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JR, Bharucha JP, Ceaser S, et al. Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:3040–3051. doi: 10.1091/mbc.E08-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gimeno MA, Munoz I, Arino J, Sanz P. Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type 1 protein phosphatase inhibitor. J Biol Chem. 2003;278:47744–47752. doi: 10.1074/jbc.M306157200. [DOI] [PubMed] [Google Scholar]
- Bharucha JP, Larson JR, Gao L, Daves LK, Tatchell K. Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1032–1045. doi: 10.1091/mbc.E07-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Pan X, Harashima T, et al. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, Ma P, Cauwenberg L, et al. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman L, Lemaire K, Ma P, et al. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- Rolland F, De Winde JH, Lemaire K, Boles E, Thevelein JM, Winderickx J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol Microbiol. 2000;38:348–358. doi: 10.1046/j.1365-2958.2000.02125.x. [DOI] [PubMed] [Google Scholar]
- Colombo S, Ronchetti D, Thevelein JM, Winderickx J, Martegani E. Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae. J Biol Chem. 2004;279:46715–46722. doi: 10.1074/jbc.M405136200. [DOI] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland CM, Hawley SA, McCartney RR, et al. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol. 2003;13:1299–1305. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- Nath N, McCartney RR, Schmidt MC. Yeast Pak1 kinase associates with and activates Snf1. Mol Cell Biol. 2003;23:3909–3917. doi: 10.1128/MCB.23.11.3909-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Xu X, Carlson M. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc Natl Acad Sci USA. 2011;108:6349–6354. doi: 10.1073/pnas.1102758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- Alms GR, Sanz P, Carlson M, Haystead TA. Reg1p targets protein phosphatase 1 to dephosphorylate hexokinase II in Saccharomyces cerevisiae: characterizing the effects of a phosphatase subunit on the yeast proteome. EMBO J. 1999;18:4157–4168. doi: 10.1093/emboj/18.15.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuatzi D, Riera A, Pelaez R, Herrero P, Moreno F. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J Biol Chem. 2007;282:4485–4493. doi: 10.1074/jbc.M606854200. [DOI] [PubMed] [Google Scholar]
- Sanz P, Alms GR, Haystead TA, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Carlson M. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6789–6796. doi: 10.1128/mcb.14.10.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnskjaer K, Boergesen M, Dalgaard LT, Mandrup S. Glucose-induced repression of PPARalpha gene expression in pancreatic beta-cells involves PP2A activation and AMPK inactivation. J Mol Endocrinol. 2006;36:289–299. doi: 10.1677/jme.1.01965. [DOI] [PubMed] [Google Scholar]
- Garcia-Haro L, Garcia-Gimeno MA, Neumann D, Beullens M, Bollen M, Sanz P. The PP1-R6 protein phosphatase holoenzyme is involved in the glucose-induced dephosphorylation and inactivation of AMP-activated protein kinase, a key regulator of insulin secretion, in MIN6 beta cells. FASEB J. 2010;24:5080–5091. doi: 10.1096/fj.10-166306. [DOI] [PubMed] [Google Scholar]
- Djouder N, Tuerk RD, Suter M, et al. PKA phosphorylates and inactivates AMPKalpha to promote efficient lipolysis. EMBO J. 2010;29:469–481. doi: 10.1038/emboj.2009.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy NT, Li L, Khalil M, Cannon JF. Regulation of yeast glycogen metabolism and sporulation by Glc7p protein phosphatase. Genetics. 1998;149:57–72. doi: 10.1093/genetics/149.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek KM, Voronkova V, Raney A, Young ET. Functional analysis of the yeast Glc7-binding protein Reg1 identifies a protein phosphatase type 1-binding motif as essential for repression of ADH2 expression. Mol Cell Biol. 1999;19:6029–6040. doi: 10.1128/mcb.19.9.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren R, Rainis L, Kleinberger T. The scaffolding A/Tpd3 subunit and high phosphatase activity are dispensable for Cdc55 function in the Saccharomyces cerevisiae spindle checkpoint and in cytokinesis. J Biol Chem. 2004;279:48598–48606. doi: 10.1074/jbc.M409359200. [DOI] [PubMed] [Google Scholar]
- Bennin DA, Don AS, Brake T, et al. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B' subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J Biol Chem. 2002;277:27449–27467. doi: 10.1074/jbc.M111693200. [DOI] [PubMed] [Google Scholar]
- Van Hoof C, Janssens V, De Baere I, et al. The Saccharomyces cerevisiae phosphotyrosyl phosphatase activator proteins are required for a subset of the functions disrupted by protein phosphatase 2A mutations. Exp Cell Res. 2001;264:372–387. doi: 10.1006/excr.2000.5144. [DOI] [PubMed] [Google Scholar]
- Longin S, Jordens J, Martens E, et al. An inactive protein phosphatase 2A population is associated with methylesterase and can be re-activated by the phosphotyrosyl phosphatase activator. Biochem J. 2004;380:111–119. doi: 10.1042/BJ20031643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Parsons S, Brautigan DL. Tyrosine phosphorylation of protein phosphatase 2A in response to growth stimulation and v-src transformation of fibroblasts. J Biol Chem. 1994;269:7957–7962. [PubMed] [Google Scholar]
- Guo H, Damuni Z. Autophosphorylation-activated protein kinase phosphorylates and inactivates protein phosphatase 2A. Proc Natl Acad Sci USA. 1993;90:2500–2504. doi: 10.1073/pnas.90.6.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Kowluru A, Kern TS. PP2A contributes to endothelial death in high glucose: inhibition by benfotiamine. Am J Physiol Regul Integr Comp Physiol. 2010;299:R1610–1617. doi: 10.1152/ajpregu.00676.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanevich V, Jiang L, Satyshur KA, et al. The structural basis for tight control of PP2A methylation and function by LCMT-1. Mol Cell. 2011;41:331–342. doi: 10.1016/j.molcel.2010.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabba S, Mangat S, McCartney R, Schmidt MC. PP1 phosphatase-binding motif in Reg1 protein of Saccharomyces cerevisiae is required for interaction with both the PP1 phosphatase Glc7 and the Snf1 protein kinase. Cell Signal. 2010;22:1013–1021. doi: 10.1016/j.cellsig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadura N, Robinson LC, Michels CA. Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics. 2006;172:1427–1439. doi: 10.1534/genetics.105.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayordomo I, Regelmann J, Horak J, Sanz P. Saccharomyces cerevisiae 14-3-3 proteins Bmh1 and Bmh2 participate in the process of catabolite inactivation of maltose permease. FEBS Lett. 2003;544:160–164. doi: 10.1016/s0014-5793(03)00498-8. [DOI] [PubMed] [Google Scholar]
- Cui DY, Brown CR, Chiang HL. The type 1 phosphatase Reg1p-Glc7p is required for the glucose-induced degradation of fructose-1,6-bisphosphatase in the vacuole. J Biol Chem. 2004;279:9713–9724. doi: 10.1074/jbc.M310793200. [DOI] [PubMed] [Google Scholar]
- Wu Y, Song P, Xu J, Zhang M, Zou MH. Activation of protein phosphatase 2A by palmitate inhibits AMP-activated protein kinase. J Biol Chem. 2007;282:9777–9788. doi: 10.1074/jbc.M608310200. [DOI] [PubMed] [Google Scholar]
- Versele M, de Winde JH, Thevelein JM. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- Beullens M, Mbonyi K, Geerts L, et al. Studies on the mechanism of the glucose-induced cAMP signal in glycolysis and glucose repression mutants of the yeast Saccharomyces cerevisiae. Eur J Biochem. 1988;172:227–231. doi: 10.1111/j.1432-1033.1988.tb13877.x. [DOI] [PubMed] [Google Scholar]
- Ahn JH, McAvoy T, Rakhilin SV, Nishi A, Greengard P, Nairn AC. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc Natl Acad Sci USA. 2007;104:2979–2984. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge-Kafka KL, Bauman A, Mayer N, et al. cAMP-stimulated protein phosphatase 2A activity associated with muscle A kinase-anchoring protein (mAKAP) signaling complexes inhibits the phosphorylation and activity of the cAMP-specific phosphodiesterase PDE4D3. J Biol Chem. 2010;285:11078–11086. doi: 10.1074/jbc.M109.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell. 2004;16:293–299. doi: 10.1016/j.molcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Townley R, Carlson M. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol Cell Biol. 2004;24:1836–1843. doi: 10.1128/MCB.24.5.1836-1843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonyi K, Van Aelst L, Arguelles JC, Jans AW, Thevelein JM. Glucose-induced hyperaccumulation of cyclic AMP and defective glucose repression in yeast strains with reduced activity of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1990;10:4518–4523. doi: 10.1128/mcb.10.9.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winde JH, Crauwels M, Hohmann S, Thevelein JM, Winderickx J. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur J Biochem. 1996;241:633–643. doi: 10.1111/j.1432-1033.1996.00633.x. [DOI] [PubMed] [Google Scholar]
- Sanz P, Nieto A, Prieto JA. Glucose repression may involve processes with different sugar kinase requirements. J Bacteriol. 1996;178:4721–4723. doi: 10.1128/jb.178.15.4721-4723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB.Methods in Yeast GeneticsNew York: Cold Spring Harbor, 1992
- Sambrook J, Fritsch EF, Maniatis T.Molecular Cloning: A Laboratory Manual2nd Edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 1989
- Wang Z, Lanner MC, Jin N, Swartz D, Li L, Rhoades RA. Hypoxia inhibits myosin phosphatase in pulmonary arterial smooth muscle cells: role of Rho-kinase. Am J Respir Cell Mol Biol. 2003;29:465–471. doi: 10.1165/rcmb.2002-0157OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Experimental Procedures
Optimization of okadaic concentration used for specific PP2A inhibition.
PP2Ac-like subunits are not required for glucose activation of PP2A.
High basal level of immunopurified Pph21 is most likely due to the high concentration of this catalytic subunit, and not to the loss of any interacting molecule/protein during the immunopurification.
Confirmation of all gene deletions, and in particular deletion of TPD3.
Rts1 does not interact with Pph21 in a tpd3Δ strain.
Expression of catalytic subunits of PP2A and PP1 is unaffected in strains showing altered glucose-induced phosphatase activation.
Both PP2A and PP1 are activated by glucose in a post-translational manner, and for PP2A, methylation by Ppm1 is partially required.
Reg1 mutant proteins unable to interact with Glc7 do not support glucose-induced PP1 activation.
Tor signalling and the Snf3/Rgt2 system are not involved in the activation of PP1 and PP2A.
List of yeast strains used in this study.
List of plasmids used in this study