Abstract
Phospholipid flippases in the type IV P-type ATPase family (P4-ATPases) are essential components of the Golgi, plasma membrane and endosomal system that play critical roles in membrane biogenesis. These pumps flip phospholipid across the bilayer to create an asymmetric membrane structure with substrate phospholipids, such as phosphatidylserine and phosphatidylethanolamine, enriched within the cytosolic leaflet. The P4-ATPases also help form transport vesicles that bud from Golgi and endosomal membranes, thereby impacting the sorting and localization of many different proteins in the secretory and endocytic pathways. At the organismal level, P4-ATPase deficiencies are linked to liver disease, obesity, diabetes, hearing loss, neurological deficits, immune deficiency and reduced fertility. Here, we review the biochemical, cellular and physiological functions of P4-ATPases, with an emphasis on their roles in vesicle-mediated protein transport.
1. Introduction
Newly synthesized secretory proteins travel from the endoplasmic reticulum (ER) through the Golgi complex to the trans-Golgi network (TGN) where they are packaged into exocytic carriers for delivery to the plasma membrane or extracellular space [1]. As a major sorting station in the secretory pathway, the TGN also segregates proteins destined for the endosomal/lysosomal system from exocytic cargo, thus making an essential contribution to the protein composition of lysosomes and endosomes. In fact, a remarkable number of distinct protein trafficking pathways, mediated by specific types of vesicles, transport proteins between the TGN, plasma membrane and endocytic/lysosomal compartments [2]. The small GTP-binding protein Arf and its ArfGEF and ArfGAP regulators, along with clathrin and its various adaptor proteins are critically important and highly conserved components of the protein trafficking machinery in this system. The Golgi also plays an important role in establishing the appropriate composition and organization of lipids in the plasma membrane and internal organelles. For example, sphingolipids and glycosphingolipids are synthesized in the lumenal leaflet of late Golgi elements and glycerophospholipids are translocated to the cytosolic leaflet by phospholipid flippases to establish an asymmetric membrane structure [3].
The type IV P-type ATPases (P4-ATPases) are phospholipid flippases that not only establish membrane phospholipid asymmetry, but are also tightly coupled to vesicle-mediated protein transport in the Golgi and endosomal systems. P4-ATPases were first implicated in vesicular transport through studies in Saccharomyces cerevisiae [4–6], but more recent studies in Arabidopsis thaliani, Caenorhabditis elegans and mammalian tissue culture cells indicate that this function is conserved [7–10]. Precisely how and why phospholipid flippases are coupled to vesicle budding events is uncertain and remains an active area of investigation.
Most flippases in the P4-ATPase family are comprised of a catalytic α-subunit (the P4-ATPase) and a noncatalytic β-subunit in the Cdc50 family of integral membrane proteins [3]. The budding yeast flippase Drs2, for example, associates with Cdc50 and the complex must be formed before the newly synthesized flippase is allowed to leave the ER [11]. This arrangement is well conserved though evolution and several metazoan P4-ATPases are known to have a functional requirement for association with a Cdc50 homolog [12, 13].
In this review, we will trace the lines of evidence supporting the contention that P4-ATPases are phospholipid flippases. The transverse flip of specific phospholipid species from the exofacial to the cytosolic leaflet is an unusual activity for a P-type ATPase as members of this protein family are more famous for their roles in pumping ions or heavy metals across membranes [14]. How the P4-ATPases evolved such a different transport substrate is unclear. Only a single human disease, familial intrahepatic cholestasis, is currently known to result from a P4-ATPase deficiency [15]. However, studies in mice are beginning to illuminate additional physiological roles for mammalian P4-ATPases that will be discussed. We will also describe specific protein transport pathways linked to P4-ATPase activity and a model for how flippases may help establish the membrane curvature required to bud vesicles from Golgi and endosomal membranes. Other emerging topics from studies in budding yeast that will be addressed are regulatory mechanisms controlling flippase activity with connections to sterol, sphingolipid and phosphoinositide metabolism. We will conclude with a discussion of future directions for the phospholipid flippase field.
2. The P4-ATPase and Cdc50 family of proteins
The first P4-ATPase sequence to appear in the literature was Drs2 from budding yeast, although it was initially thought to be a Ca++ ATPase because these pumps were the closest homologs known at the time [16]. Soon thereafter, Drs2 homologs with greater similarity appeared in the newly sequenced yeast genome and bovine ATPase II (now known as ATP8A1) was cloned and found to be nearly 50% identical to Drs2 [17]. As additional eukaryotic genomes were sequenced, it became apparent that Drs2 and ATPase II were the founding members of a very large subgroup in the P-type ATPase superfamily. Axelson and Palmgren formalized this relationship in their phylogenetic analysis of the P-type ATPases and the Drs2/ATP8A1-related subgroup was categorized as type IV P-type ATPases (P4-ATPases) [18]. While the Axelson and Palmgren nomenclature is most commonly used, an alternative Transporter Classification (TC) system places the Drs2/ATP8A1-related subgroup in P-type ATPase family 8 [19]. The P4-ATPases are not found in prokaryotes, but are typically represented by multiple members in every sequenced eukaryotic genome.
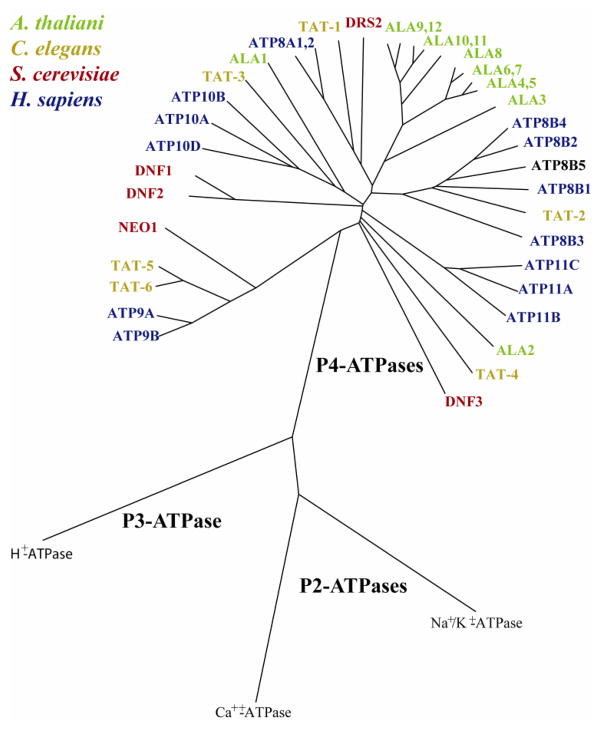
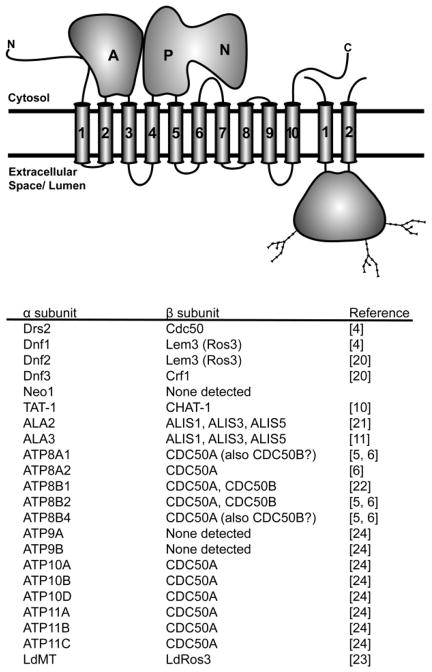
An example of the phylogenetic relationship of P4-ATPase catalytic subunits from yeast, plants and animals, for which functional studies have been performed, is shown in Figure 1. A unified nomenclature has been established for mammalian P4-ATPases, which are designated ATP8A1 through ATP11C. The human genome expresses 14 members of this family and the mouse genome contains a 15th member, ATP8B5, not found in the human genome [10]. The nomenclature used in model systems is less consistent with six Caenorhabditis elegans P4-ATPases named TAT-1 through TAT-6 for Transbilayer Amphipath Transporter, 12 Arabidopsis thaliani members named ALA1 through ALA12 for AminophosphoLipid ATPases, and five Saccharomyces cerevisiae P4-ATPases named Drs2 (Defect in Ribosome Synthesis), Neo1 (neomycin resistance) and Dnf1 through Dnf3 for Drs2 Neo1 Family ATPases. Only three noncatalytic subunits in the CDC50 family are encoded by mammalian (CDC50A, CDC50B and CDC50C), yeast (Cdc50, Lem3 and Crf1) and C. elegans (W03G11.2, R08C7.2a (CHAT-1) and F20C5.4) genomes while Arabidopsis expresses five (ALIS1 - ALIS5) (Fig 2). The relatively small number of β-subunits relative to α-subunits can be accommodated because not all P4-ATPases appear to require a β-subunit (e.g. Neo1) and several α-subunits can share the same β-subunit in independent heterodimer pairs (Fig 2) [7, 8, 11–13, 20–24].
Figure 1.
Phylogeny of P4-ATPases from Homo sapiens, Arabidopsis thaliana, Saccharomyces cerevisiae and Caenorhabditis elegans. An alignment was produced from amino acid sequences using EXPRESSO(3DCOFFEE) rooted with the P2- and P3-ATPases with known structures. Phylogenetic distances were calculated using PHYLIP 3.67 and presented using Kitsch drawtree. ATP8B5 is the Mus musculus sequence because Homo sapiens lacks this P4-ATPase. Accession numbers for sequences used are available upon request.
Figure 2.
P4-ATPases form an α–β heterodimer. The predicted topology of the P4-ATPase (α subunit) and the Cdc50 family protein (β subunit) along with the experimentally defined αβ pairs is shown. A question mark indicates a discrepancy in the cited literature for Cdc50B.
While no high-resolution structure for a P4-ATPase has been determined, the presence of well-conserved motifs found in all P-type ATPases suggests that they adopt the four-domain structure observed with the Ca++, Na+/K+ and H+ ATPases that have been crystallized [25]. These include the nucleotide-binding (N), phosphorylation (P), actuator (A) and membrane (M) domains (Fig. 2). Most of the P4-ATPases have relatively long N- and C-terminal cytosolic extensions that may form additional regulatory (R) domains and all of these pumps are predicted to have 10 transmembrane segments composing the membrane domain. The actuator domain folds from cytosolic sequences preceding the first transmembrane segment (TM1) and in the loop between TM2 and TM3, while the P and N domains are formed from the cytosolic loop between TM4 and TM5. The noncatalytic subunits are membrane proteins with two transmembrane segments separated by a large, glycosylated extracellular domain (Fig. 2).
For cation transporters, binding of ATP to the N-domain and the subsequent phosphorylation of an Asp residue in the P domain induces dramatic conformational transitions described as E1 to E2~P in the Post-Albers catalytic cycle [25]. Hydrolysis of the aspartyl-phosphate bond by sequences in the A domain allows the pump to relax back to the E1 conformation to complete this cycle. These conformational changes in the three cytosolic domains drive equally dramatic conformational changes in the substrate-binding membrane domain that pumps ions across the membrane against their electrochemical gradient. While it is likely that the P4-ATPases undergo similar E1–E2 conformational transitions, it is not known how these pumps couple ATP hydrolysis to translocation of phospholipid substrate across the membrane. Flipping of phospholipid has been a controversial activity for a P-type ATPase and so we provide below the lines of evidence that support this contention.
3. Evidence that the P4-ATPases are phospholipid flippases
The plasma membrane of living cells is an asymmetric structure with an uneven distribution of specific phospholipids between the inner (cytosolic) and outer (extracellular) leaflets. PS and PE, for example, are confined almost exclusively to the inner leaflet while PC and sphingolipids are enriched in the outer leaflet. This asymmetry creates different chemical environments on opposing membrane faces important for integral membrane protein function, interaction of soluble proteins with the membrane surface and cell-cell interactions [26]. In contrast, when bilayers are formed spontaneously from lipids suspended in water, the component phospholipids are evenly distributed between the two faces of the bilayer. The hydrophobic core of the membrane provides an energy barrier that prevents transverse movement of the hydrophilic headgroup, and so spontaneous flip/flop of component phospholipids like PC, PS or PE is a rare event in artificial membranes [27]. A low intrinsic rate of spontaneous phospholipid flip/flop allows plasma membrane asymmetry to be maintained with minimal energy input, but how is the ordered plasma membrane structure with PS and PE enriched in the cytosolic leaflet initially established?
The discovery of an energy-driven aminophospholipid translocase (APLT) activity in human erythrocytes provided the first mechanistic insight into how cells create an asymmetric plasma membrane structure [28]. Spin-labeled analogs of PS and PE incorporated into the extracellular leaflet are rapidly transported to the cytosolic leaflet of the erythrocytes, while spin-labeled PC remains in the extracellular leaflet. The selective transport of spin-labeled PS and PE to the cytosolic leaflet by the APLT suggested that this enzyme pumped endogenous PS and PE to establish asymmetry. The energy requirement for flipping the charged lipid headgroup across the hydrophobic membrane core is provided by ATP and vanadate inhibits translocation, providing the first hint that a member of the P-type ATPase family catalyzed this translocase activity. This study provided the first evidence for an APLT activity in human erythrocytes, but reports of an APLT activity in bovine chromaffin granules [29], platelets [30] and yeast [17, 31] followed.
The erythrocyte APLT is also able to flip unlabeled PS and PE to the inner leaflet as measured by changes in the shape of the cells [28, 32]. As described by the bilayer couple hypothesis, an unbalanced increase in the lipid number in one face of the membrane creates curvature in the direction of the leaflet with the greater surface area [33]. Incorporation of exogenous phospholipid into the extracellular face of the bilayer induces a dramatic outward bending of the membrane, and changes the erythrocyte shape from the normal discocyte shape to an echinocytic shape resembling a sea urchin. When a substrate of the APLT is incorporated into the membrane (PS or PE), erythrocytes rapidly revert from echinocytes back to discocytes, and then to crenated stomatocytes as phospholipid is pumped to the inner leaflet. However, if a non-substrate for the aminophospholipid translocase, such as PC, is incorporated into the outer leaflet, the cells retain their echinocyte shape [28, 32]. Importantly, these experiments demonstrated the potential of phospholipid flippases to induce curvature in biological membranes.
The identity of the APLT is still uncertain [34], although ATP8A1 remains the best candidate for the erythrocyte [35] and bovine chromaffin granule flippase [29]. This connection derived from the purification of a novel 115 kDa ATPase from bovine chromaffin granules, called ATPase II at the time, that is dramatically stimulated by PS (and PE to a minor extent) [36, 37]. A similar 110 – 120 kDa Mg++-dependent ATPase purified from human erythrocytes was suspected to be homologous to ATPase II and responsible for the aminophospholipid translocase activity of the erythrocyte plasma membrane [38]. This ATPase was partially purified from human erythrocytes and reconstituted into proteoliposomes, which displayed a PS translocase activity [39]. However, the protein responsible for the reconstituted APLT activity was not identified.
Validation that ATPase II (ATP8A1) is a P-type ATPase occurred when the bovine chromaffin granule enzyme was cloned, a discovery that nucleated a dramatic expansion of the field as additional homologs were uncovered [17]. ATPase II was initially found to be homologous to the yeast protein Drs2, and soon after to ATP8B1 (FIC1)(See section 4-Roles of P4-ATPases in disease)[40]. Disruption of DRS2 (drs2Δ) was reported to cause a defect in PS translocation at the plasma membrane of S. cerevisiae [17], although later reports suggested that Drs2 had a negligible impact on this activity [41, 42]. The discovery of protein trafficking defects in drs2Δ cells also raised concerns that the influence of Drs2 on the plasma membrane flippase activity was indirect (see section 5-Trafficking).
However, a number of subsequent genetic studies strongly supported a direct role for P4-ATPases in phospholipid translocation. In S. cerevisiae, Dnf1 and Dnf2 are required for the plasma membrane flippase activity towards NBD-labeled PC and PE [6]. Drs2 localizes to the TGN, and purified TGN membranes carrying a temperature-sensitive form of Drs2 contain an NBD-PS flippase activity that is inactivated at the nonpermissive temperature [43]. Secretory vesicles exiting the TGN contain a flippase activity that is Drs2 and Dnf3-dependent [44] and all of the DRS2 and DNF genes contribute to the establishment of membrane asymmetry [6, 45]. The Arabidopsis P4-ATPases ALA1, ALA2 and ALA3, and murine Atp8b5 (FetA) can support plasma membrane flippase activity when expressed in the yeast P4-ATPase mutants [8, 46]. In addition, overexpression of ATP8B1 in CHOK1 cells, results in an increased PS translocation at the plasma membrane [22, 47] (although see also [48]) and Atp8b1 deficiency in mice results in disruption of the membrane asymmetry in bile canalicular membranes [49, 50].
The P4-ATPase β subunits were independently identified in genetic screens for loss of membrane asymmetry or phospholipid translocase activity. For example, LEM3/ROS3 (encoding a member of the Cdc50 family [51]) was identified in a screen for mutants exhibiting a loss of phosphatidylethanolamine asymmetry in S. cerevisiae [52], and lem3Δ mutants were also found to be deficient in the uptake of edelfosine, a toxic PC analog [53]. More recently, a forward genetic screen for loss of PS asymmetry in C. elegans uncovered mutations in a Cdc50 homolog (CHAT-1) as well as TAT-1 [7]. The connection of the Cdc50 family to membrane asymmetry was not immediately clear because it seemed unlikely that these proteins could transport lipid directly as they have only two transmembrane segments and no ATPase domain. However, the Tanaka group linked a series of observations showing similar phenotypes of P4-ATPase mutants and Cdc50 family mutants to discover the functionally essential interaction between P4-ATPases (alpha subunits) and the Cdc50 family members (beta subunits) [11]. In sum, these genetic studies clearly demonstrate that the P4-ATPases and their β subunits are necessary for phospholipid flippase activities detected in several different membranes, and for the establishment of plasma membrane asymmetry. However, it still remained formally possible that these influences of P4-ATPase mutation or overexpression on flippase activity and membrane asymmetry were indirect.
Only recently has biochemical evidence emerged indicating that the P4-ATPases directly catalyze a phospholipid flippase activity. Working independently, the Graham and Molday groups were able to purify two P4-ATPases to near homogeneity, Drs2 and ATP8A2, respectively, reconstitute the P4-ATPases into proteoliposomes and demonstrate in vitro flippase activities [54, 55]. These studies provided the first evidence that a P4-ATPase is sufficient to catalyze an ATP-dependent phospholipid flippase activity. In the Drs2 proteoliposomes, a sub-stoichiometric amount of the Cdc50 beta subunit was present, and it is unclear if the flippase activity was catalyzed by the heterodimers in the preparation or if Drs2 can translocate substrate in the absence of Cdc50 [55]. ATP8A2 associates with CDC50A, and the complex was purified by a dual affinity approach using monoclonal antibodies that recognize each subunit. The purified heterodimer retained a robust PS-stimulated ATPase activity and NBD-PS flippase activity in proteoliposomes [56]. However, the ATP8A2 monomer was not assayed to determine if it is capable of flipping phospholipid in the absence of CDC50A.
It is generally accepted that most P4-ATPases associate with a beta subunit and this interaction is required for their mutual export from the ER [8, 11–13, 22, 24]. Whether or not the β-subunit has additional roles in the trafficking, substrate transport, or regulation of flippase activity is still uncertain. Recent evidence with Arabidopsis thaliana and human P4-ATPase complexes suggest the beta subunit is not responsible for the substrate specificity or localization of the P4-ATPase heterodimer [13, 21]. However, with both yeast and human complexes, the beta subunit appears to preferentially bind the E2~P form of the P4-ATPases, suggesting an important role in the catalytic cycle [12, 57]. In addition, conditional mutant forms of Cdc50 have been isolated that retain the ability to bind Drs2, but show a loss of function in vivo at the nonpermissive temperature. This implies an important role for the Cdc50-Drs2 interaction with the TGN-endosomal system [58]. By creating a series of chimeras between CDC50A and CDC50B, Coleman et al. found that the TM segments and ectoplasmic domain of CDC50A are critical for association with ATP8A2, and the N-terminal cytosolic domain had a significant influence on catalytic activity [56]. It appears that the β-subunit can modulate the activity of the α-subunit, but whether this is a regulatory interaction or an essential component of the pump is an important open question.
4. Influence of membrane asymmetry and flippases on metazoan physiology
The human diseases progressive familial intrahepatic cholestasis (FIC) and benign recurring intrahepatic cholestasis are caused by mutations in the ATP8B1 gene [15]. These remain the only known human diseases that are caused by a P4-ATPase deficiency. However, ATP8A2 has been linked to neurological defects of a single patient [59] and a recent genome-wide association (GWAS) study suggests a link between an ATP10D variant and increased risk of myocardial infarction [60]. In addition, studies in mice, Arabidopsis and C. elegans are indicating critical roles of P4-ATPases in diverse physiological processes.
4A. ATP8A P4-ATPases
By pumping PS to the inner leaflet of the plasma membrane, P4-ATPases create a concentration gradient intrinsic to the membrane that can be used for signal transduction in a manner analogous to how ion gradients established by cation-pumping P-type ATPases are used. Regulated disruption of membrane asymmetry and exposure of PS plays an important signaling role in apoptosis and blood clotting [61]. One of the first events in apoptosis is the Ca2+-dependent exposure of PS on the outer surface of dying cells. The exposed PS is an “eat-me” signal that is recognized by phagocytes such as macrophages, which engulf the cell corpse. In C. elegans, the ATP8A1 ortholog TAT-1 is critical for maintaining cell surface asymmetry of PS. In animals deficient for tat-1, PS is abnormally exposed on the cell surface of non-apoptotic cells, and these living cells are stochastically removed through a mechanism dependent on PSR-1, a PS-recognizing phagocyte receptor, and CED-1, which contributes to recognition and engulfment of apoptotic cells [62]. Thus, TAT-1 appears to function in preventing appearance of PS in the outer leaflet of plasma membrane, and inappropriate exposure of PS on the cell surface may result in removal of living cells by neighboring phagocytes. As described below (section 5A) tat-1 mutants also exhibit protein trafficking defects in the endocytic pathway.
ATP8A1 is the best candidate for the aminophospholipid translocase in erythrocytes and platelets. Restriction of PS to the inner leaflet in these blood cells may protect against thrombosis as exposure of PS is known to stimulate clotting reactions. A Ca2+ influx into activated platelets induces a “scramblase” activity that exposes PS on the extracellular leaflet [61]. Recently, a protein called TMEM16F was linked to a calcium-dependent scramblase activity in a mouse B-cell line [63]. Moreover, mutations in the TMEM16F gene have been found in patients with Scott syndrome, a rare bleeding disorder characterized by the lack of scramblase activity [63, 64].
ATP8A2 is disrupted by a molecularly balanced translocation between chromosomes 10 and 13 in a single patient with severe mental retardation, suggesting that haploinsufficiency of ATP8A2 caused the neurological condition [59]. ATP8A2 is highly expressed in brain, testes and retina, and was also identified as a membrane protein enriched in the disc membranes of photoreceptor outer segment preparations by a proteomic study [54, 59]. Although loss-of-function phenotypes have not been characterized for the visual system, the observation that ATP8A2 catalyzes ~50% of the total ATPase activity in the outer segment preparations suggests a high demand for PS translocation across the photoreceptor disc membranes [54]. This may be germane to a recent report showing opsin (or rhodopsin) can mediate energy-independent flip-flop of phospholipid when reconstituted into liposomes [65]. In order to maintain an enrichment of PS in the disc cytosolic leaflet, perhaps a robust ATP8A2 activity is needed to offset phospholipid “leak” caused by the high opsin concentration in this membrane.
4B. ATP8B P4-ATPases and cholestasis in humans
Mutations in ATP8B1 cause progressive familial intrahepatic cholestasis type 1 (PFIC1) or a less severe form of the disease called benign recurrent intrahepatic cholestasis type 1 (BRIC) [15]. Patients with PFIC1 suffer from chronic intrahepatic cholestasis that progresses to severe, end-stage liver disease, and often requires liver transplantation during the first or second decade of life [66]. Cholestasis results from a disruption of bile flow from the liver, which normally aids digestion and absorption of lipids in the small intestine. Bile salts are actively secreted into a canalicular lumen surrounded by the apical membranes of adjacent hepatocytes. ATP8B1 localizes to the apical membrane of epithelial cells, including hepatocytes and bile duct epithelial cells (cholangiocytes) [47].
In a mouse model for PFIC1, Atp8b1 deficiency disturbs canalicular membrane phospholipid asymmetry and decreases the resistance of the canalicular membrane to hydrophobic bile salts, as evidenced by enhanced biliary recovery of PS, cholesterol, and ectoenzymes. Atp8b1 deficiency also impairs the transport of hydrophobic bile salts into bile. The loss of phospholipid asymmetry of the canalicular membrane may lead to enhanced extraction of cholesterol from the canalicular membrane, which subsequently impairs hepatobiliary transport of bile salts and cause cholestasis [50]. Besides this critical function in the liver, Atp8b1 is abundantly expressed at the stereociliar membrane and Atp8b1 deficiency causes hearing loss associated with progressive degeneration of the cochlear hair cells [67].
Atp8b3 is exclusively expressed in the testis and localized in the acrosomal region of mouse sperm cells. Although Atp8b3−/− male mice are fertile, their litter sizes are slightly smaller than wild-type controls and in vitro fertilization rates with Atp8b3−/− sperm is significantly reduced. These sperm also display abnormal PS exposure on the outer leaflet of the plasma membrane [68]. Mouse Atp8b5 (also named as FetA) is also specifically expressed in testis and its intracellular localization implies a role in acrosome formation, a Golgi-derived organelle [46]. The co-expression of Atp8b3 and Atp8b5 in the sperm acrosome suggests that they redundantly function in the acrosome reaction.
The Arabidopsis ALA3 - ALA12 genes are closely related to the mammalian ATP8A and ATP8B P4-ATPases. Of this group, only ALA3 has been characterized and plants deficient for this P4-ATPase exhibit a defect in the growth of roots, pollen tubes and the patterning of trichomes [8, 69]. As described below (section 5), ALA3 is implicated in the formation of exocytic vesicles from the Golgi and this may underlie many of the mutant phenotypes. In C. elegans, TAT-2 has a strong influence on viability of worms starved for sterol or monomethyl branched chain fatty acids. The tat-2 mutations cause hypersensitivity to sterol deprivation [70], but suppress the growth arrest induced by depletion of the branched chain fatty acids [71].
4C. ATP9 P4-ATPases
The ATP9 subgroup contains the least well-understood members of the metazoan P4-ATPase family. Like Neo1 in yeast, ATP9A and 9B can exit the ER in the absence of CDC50-related β subunits [11, 24]. ATP9A and 9B localize to the TGN, with a portion of ATP9A also localizing to endosomes [24]. However, loss of function phenotypes have not been reported, nor is the substrate of these potential flippases known. The C. elegans TAT-5 gene appears to be ubiquitously expressed throughout the worm and RNAi knockdown is embryonically lethal [70]. NEO1 is also an essential gene in S. cerevisiae [5], implying that ATP9/NEO1/TAT-5 orthologs may be essential in all eukaryotes. Therefore, it is surprising that Arabidopsis appears to lack a member of this subgroup.
4D. Atp10 P4-ATPases and obesity
Atp10a (also called pfatp and Atp10c) has been implicated in type 2 diabetes associated with obesity. Mice heterozygous for a deletion of chromosome 7 that uncovers the pink-eyed dilution (p) locus and extends distally into Atp10a display diet-induced obesity, but only if the chromosomal deletion is inherited maternally [72]. Pups inheriting the chromosome deletion from the sire do not gain excessive weight. The Atp10a-deficient mice are hyperinsulinemic, insulin-resistant and have an altered insulin-stimulated response in peripheral tissues [73].
The mode of inheritance of diet-induced obesity suggests that Atp10a is only expressed from the maternally inherited chromosome. Both parental Atp10a transcripts are initially expressed in muscle and adipocytes, but after 12 weeks on a high fat diet, expression of the maternal allele in muscle tissue dramatically declines [74]. These studies suggest that Atp10a is subject to unique mode of imprinted gene silencing modulated by environmental conditions (high fat diet). The authors of this work suggested that a disruption in the trafficking or expression of Glut4, the insulin-responsive glucose transporter, may underlie the insulin resistance of peripheral tissues in Atp10a deficient mice [74].
ATP10D may also been linked to defects in lipid metabolism in mice and humans. A naturally occurring truncated variant of Atp10d might be linked to the fat-prone phenotype of C57BL/6 mice [75]. In addition, a recent genome-wide association study linked human ATP10D allele polymorphisms to variances in plasma levels of glucosylceramide and ceramide, which are associated with risk of myocardial infarction [60].
ALA1 from Arabidopsis appears to be most closely related to the ATP10 subgroup, although ectopic expression of ALA1 in Saccharomyces complements the cold-sensitive growth defect of a drs2Δ mutant (homologous to the ATP8A group) [76]. Remarkably, knockdown of ALA1 expression causes a cold-sensitive growth defect in plants [76]. These results imply a well-conserved role for the P4-ATPases in adaptation to growth at cold temperatures.
4E. Atp11 P4-ATPases and B-cell development
Recent work from two groups has uncovered a critical role for Atp11C in murine B cell development [77, 78]. Interestingly, the mutant mice also develop hepatocellular carcinoma, hyperbilirubinemia, anemia and cholestasis, suggesting important roles in liver homeostasis [77, 78]. In fact, dietary supplementation with cholic acid caused death of the Atp11c deficient mice within a few weeks, supporting an important role of Atp11c in bile export [79]. The extent of genetic overlap between Atp11c and Atp8b1 function has not yet been characterized, nor is the precise influence on B-cell development known.
5. Flippase influence on protein trafficking
P4-ATPases were first linked to protein trafficking when Drs2, and its chaperone, Cdc50, were discovered to genetically interact with Arf [4, 45, 80, 81]. Arf (ADP-ribosylation factor) is a small G-protein that cycles between GTP- and GDP-bound form through the action of several guanine nucleotide exchange factors (ArfGEFs) and GTPase activating proteins (ArfGAPs). In the GTP-bound form, Arf mediates the binding of adaptor proteins and coat proteins, such as COPI and clathrin, to sites of vesicle formation [1]. The synthetic lethal interaction between Drs2 and Cdc50 with Arf suggested that these proteins were important players in vesicle budding pathways. drs2Δ is also synthetically lethal with clathrin temperature-sensitive alleles, but not COPI or COPII mutations, which implicated Drs2 in clathrin-mediated protein trafficking pathways [4].
Drs2 primarily localizes to the trans-Golgi network (TGN) and this is one of the sites from which clathrin-coated vesicles bud. The phenotypes exhibited by drs2Δ cells are similar to the defects observed in cells deficient in clathrin. Both mutants accumulate enlarged Golgi cisternae and late Golgi enzymes involved in proteolytically processing pro-α-factor are mislocalized in both drs2Δ and clathrin mutants. Furthermore, the drs2Δ cells are markedly deficient in clathrin-coated vesicles (CCVs) that can be purified from these cells [4]. The screen for mutants defective in ribosome synthesis that first identified DRS2 may seem at odds with the roles described here in protein trafficking and membrane asymmetry. However, many mutants that exhibit defects in vesicle-mediated protein transport create membrane stress, which is sensed by the cell integrity pathway to downregulate ribosome production and attenuate translation [82, 83]. The defect in ribosome synthesis only appears after shifting drs2Δ cells to cold temperatures (below 20°C) for a few hours [16], while most of the Golgi defects are observed at any temperature [4, 84]. The localization of Drs2 to the TGN and observation that temperature-sensitive for function alleles of DRS2 (drs2-12 or drs2-31) cause a loss of vesicle formation within 30 minutes of temperature shift imply a direct role for Drs2 in vesicle budding [85].
There are many co-factors involved in the formation, budding and uncoating of clathrin-coated vesicles [86]. Most notably, adaptor proteins are essential in cargo selection and organizing the coat at sites of vesicle formation. In yeast, AP-1 (adaptor protein-1), a heterotetrameric complex composed of γ, β1, μ1, and σ1 adaptin subunits, is recruited by Arf and functions at the TGN and early endosomes. Another set of adaptor proteins, Gga1 and Gga2 (Golgi-localized, Gamma-ear containing, Arf-binding) bud clathrin-coated vesicles from the TGN that are targeted to the late endosome. The AP-3 tetrameric adaptor (composed of δ, β3, μ3 and σ3 adaptin subunits) also appears to bud from the TGN in an Arf-dependent, but clathrin-independent manner [3]. After a clathrin-coated vesicle buds, the yeast auxilin (Swa2) and Hsp70 (Ssa proteins) are required for clathrin disassembly and uncoating (Fig 3) [87, 88].
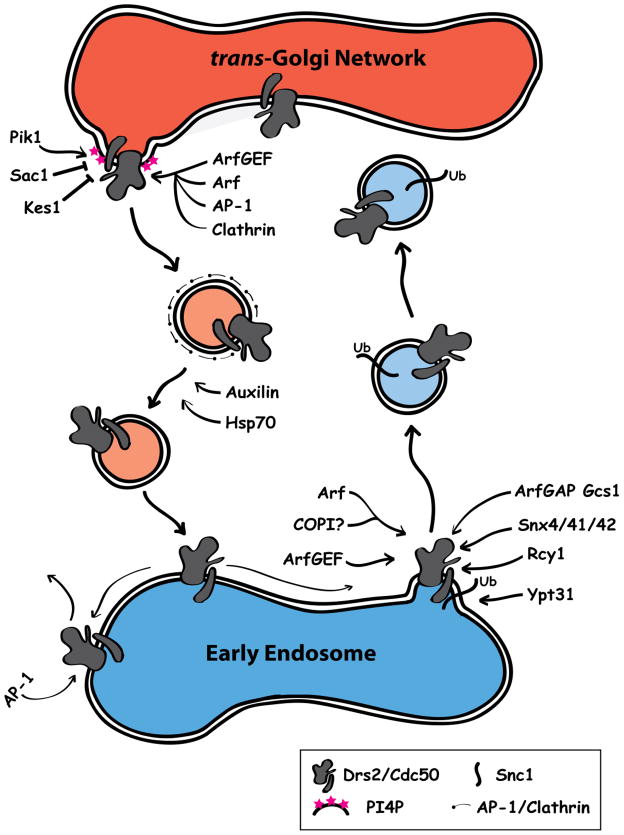
Figure 3.
The role of Drs2-Cdc50 and other factors in the formation and trafficking of transport vesicles between the late Golgi and the early endosome. The flippase activity of Drs2 is proposed to generate membrane curvature at sites of vesicle budding at the TGN. Arf, recruited to these sites by ArfGEF, mediates the recruitment of AP-1 and clathrin to the newly forming vesicle. Vesicle formation is controlled by various factors that regulate Drs2 flippase activity, including phosphatidylinositol-4-phosphate (PI4P), Sac1 (PI4P phosphatase), and Kes1 (oxysterol binding protein). At the early endosome, retrieval of Drs2-Cdc50 and the exocytic SNARE Snc1 back to the TGN is mediated by Rcy1 (F-box protein), an effector of the Ypt31 Rab protein, a sorting nexin complex (Snx4/41/42), and the Arf cycle, in particular the ArfGAP Gcs1.
Drs2 and Cdc50 are required for bi-directional transport between the TGN and the early endosome in pathways mediated by AP-1/clathrin and an F-box protein called Rcy1 (Fig. 3) [5, 20, 89]. The phenotypes associated with loss of Drs2 or AP-1 function within cells are very similar. For example, chitin synthase III (Chs3) is thought to cycle between the TGN and early endosome and is exported to the plasma membrane at certain times in the cell cycle [90]. Mutation of exomer subunit genes, such as CHS6, prevents transport of Chs3 to the plasma membrane. But, when chs6Δ is combined with either drs2Δ or AP-1 subunit mutations, Golgi-endosomal trafficking is disrupted and Chs3 redirects to the plasma membrane and late endosome or vacuole [89, 90]. These results suggest that Drs2, AP-1 and clathrin mediate both anterograde and retrograde transport of this cargo between the TGN and early endosome [89].
Drs2 also appears to cycle between the TGN and early endosomes (Fig 3). Occasionally, some Drs2 molecules find their way to the plasma membrane, but multiple endocytosis signals ensure a quick return to the early endosome and TGN system [91]. When AP-1 is disrupted, wholesale re-routing of Drs2 to the plasma membrane is observed and Drs2 can be held at this location with an endocytosis block. However, upon lifting the endocytosis block, Drs2 recycles through the endocytic pathway back to the TGN in the absence of AP-1 [89]. The role of AP-1, it seems, is to traffic Drs2 and Chs3 from the TGN to the early endosome, but unlike Chs3, Drs2 does not need AP-1 for the return trip. Because Drs2 is required to support Chs3 trafficking in AP-1 pathways, it seemed possible that the flippase activity may be required to support Drs2’s own transport by AP-1. In fact, inactivation of Drs2 activity (by shifting a GFP-tagged temperature sensitive form of Drs2 to the nonpermissive temperature) caused re-routing of Drs2 to the plasma membrane, comparably to what was observed by inactivation of AP-1 [89].
These observations support the idea that the formation of AP-1/clathrin-coated vesicles requires Drs2 flippase activity. Remarkably, however, the Arf-dependent recruitment of AP-1 and clathrin to the TGN is not perturbed in drs2Δ cells [89]. Moreover, the enlarged Golgi cisternae that accumulate in drs2Δ cells lack budding profiles or highly curved tubular elements [4, 85], even though the coat proteins are present on the membrane. Thus, a simple model that Drs2 facilitates coat recruitment to drive vesicle budding is incorrect. It seems more likely that Drs2 imparts curvature to the membrane by pumping phospholipid to the cytosolic leaflet, thereby producing a surface that the clathrin coat can more easily deform [92].
Drs2 and Cdc50 are also critical for directing cargo from the early endosome back to the TGN in the “Rcy1 pathway” [5, 20] (Fig 3). The F-box protein Rcy1 can form a Skp1-cullin-F-box (SCFRcy1) complex [93] and Rcy1 is required for the early endosome to TGN trafficking of Snc1, an exocytic SNARE protein that cycles between the early endosome, TGN and plasma membrane [94, 95]. The SCFRcy1 complex is an effector of the Rab proteins Ypt31 and Ypt32, and is proposed to ubiquitinate Snc1 to generate a signal for recycling out of the endosome [96]. Trafficking of Snc1 is unaffected by AP-1 disruption [89] and there is some indication that COPI might influence the Rcy1 pathway, although the vesicle coat acting in this pathway is unclear [97]. The sorting nexin complex Snx4/41/42 and the ArfGAP Gcs1 are also linked to this recycling pathway [81, 98]. The drs2Δ and cdc50Δ mutations disrupt retrograde trafficking of Snc1 from the early endosome to the TGN comparably to the rcy1Δ mutation. In addition, Cdc50 accumulates in the early endosomes of rcy1 mutants and so Drs2-Cdc50 also appears to travel the Rcy1 pathway back to the TGN [20]. In fact, growth defects associated with rcy1Δ can be suppressed by overexpression of Drs2/Cdc50 and Snc1, implying that these proteins are the critical cargos in the Rcy1 pathway [20]. While Rcy1 interacts with Drs2-Cdc50 [20], it is not known if Rcy1 ubiquitinates this flippase to facilitate retrieval or acts by another mechanism.
The C. elegans tat-1 and chat-1 mutants (potentially orthologous to Drs2-Cdc50) also show a strong defect in cargo recycling through endosomes [7, 9], indicating that this function in protein trafficking is well-conserved through evolution. TAT-1 and CHAT-1 co-localize within intestinal cells to the plasma membrane (apical and basolateral) along with early/recycling endosomes. In fact, CHAT-1 decorates tubules on recycling endosomes that require RAB-10 for formation, and these tubules are exaggerated in an rme-1 mutant (Eps15 homology domain (EHD) protein required for tubule scission) [7]. Intestinal cells of the tat-1 or chat-1 mutants accumulate large vacuolated structures bearing markers for late endosomes and lysosomes [7, 9]. In addition, several markers for the early and late endosomes, recycling endosomes and late Golgi show aberrant co-localization to aggregated vesicles. A number of proteins that normally recycle from the early endocytic pathway back to the plasma membrane, including human transferrin receptor, human IL-2 receptor a-chain and glucose transporter 1 (GLUT-1), are all trapped in abnormal endosomal intermediates in the mutants [7]. Moreover, the recycling endosome tubular extensions appear to be completely lost in the tat-1 and chat-1 mutants [7]. These data suggest that TAT-1 and CHAT-1 help drive the tubular membrane extensions from recycling endosomes that enrich cargo for delivery back to the plasma membrane.
In addition to bidirectional transport between the TGN and early endosome, and recycling from endosomes back to the plasma membrane, P4-ATPases may also contribute to budding of exocytic vesicles from the TGN [85]. Budding yeast produce at least two classes of exocytic vesicles distinguished by density and cargo [99]. Inactivation of either Drs2 or clathrin prevents formation of the denser class that accumulates when actin assembly is perturbed [85]. It remains unclear if these Drs2-dependent exocytic vesicles actually bud from the TGN or an early endosome, and it is possible that they are analogous to the tubular carriers produced by recycling endosomes in C. elegans.
However, phenotypes exhibited by Arabidopsis ala3 mutants suggest a defect in forming exocytic secretory vesicles directly from the TGN [8]. The peripheral columella cells in the root tip of wild-type plants display TGN cisternae with large, bulbous extensions filled with electron translucent slime polysaccharide. Secretory vesicles containing the slime polysaccharide derived from the Golgi are easily detected in the cytoplasm prior to their fusion to the plasma membrane. In contrast, the ala3 columella cells have a complete absence of the distended TGN cisternae and a marked deficiency of secretory vesicles in the cytoplasm [8]. ALA3 also plays a critical role in trichome formation on leaves, which is a wonderful model for directional membrane growth and complex cell patterning. The trichome branch elongation defect in ala3 plants may result from defects in membrane trafficking and/or regulation of the actin cytoskeleton. This is reminiscent of the polarized growth defects reported for lem3Δ mutants in budding yeast [100], and the loss of microvilli from the apical membrane of Caco-2 cells deficient for Atp8b1 [48].
Thus far, only Atp8b5 has been shown to have a role in protein trafficking in mammalian cells [46]. RNA interference of Atp8b5 in murine mastocytoma P815 cells causes the distension of Golgi cisternae and abrogates constitutive secretion. Interestingly, these secretory pathway defects are only observed at lower temperatures (33°C), perhaps because other P4-ATPases can compensate for the Atp8b5 deficiency at 37°C [46]. Alternatively, it is possible that Golgi membranes in Atp8b5 deficient cells are more sensitive to changes in membrane fluidity associated with reduced temperature.
In the budding yeast system, there is clear evidence for both overlapping and non-overlapping functions for the P4-ATPases [5]. The defects in bidirectional transport between the TGN and endosomes and exocytosis are observed in drs2Δ single mutants, in spite of the presence of four other P4-ATPases in these cells. Thus, Neo1 and the Dnf P4-ATPases cannot replace the critical function of Drs2 in these pathways. However, growth and protein trafficking defects associated with drs2Δ become more severe as additional DNF genes are knocked out. For example, neither a drs2Δ nor a dnf1Δ single mutant displays a defect in the AP-3-dependent transport of cargo to the vacuole. However, the drs2Δ dnf1Δ double mutant exhibits a substantial defect in sorting AP-3 cargo [5]. Thus, it appears that Drs2 and Dnf1 are interchangeable in this pathway. Neo1 and Dnf1 may also compensate for loss of Drs2 in delivering cargo from the Golgi to the late endosome, which is a GGA-dependent pathway [101–103]. In addition, the dnf1Δ dnf2Δ cells show a cold-sensitive endocytosis defect that exacerbated by addition of drs2Δ [6]. While cells are viable until all four DRS2 DNF genes are knocked out, the single neo1Δ mutation is lethal [5]. Thus, Neo1 has an essential role that cannot be replaced by Drs2 or Dnf proteins.
Neo1 appears to localize to both Golgi and endosomes and has been implicated in the COPI-dependent retrograde transport of cargo from the Golgi to the ER [101–103]. For example, the Rer1 protein rapidly cycles between the ER and Golgi, but COPI inactivation causes mislocalization of Rer1 to downstream compartments [104]. Similarly, Neo1 inactivation of causes the same Rer1 mislocalization phenotype [101]. Additionally, Neo1 has been implicated in GGA-dependent trafficking of cargo from the TGN to the late endosome. GGA is recruited to the TGN membrane by associations with both Arf and the Arf-like protein Arl1, which forms a complex with Mon2 (a large scaffold protein) and Neo1 [102].
6. Regulation of flippase activity
The activity of budding yeast P4-ATPases appears to be tightly regulated and responsive to changes in membrane composition. The first regulatory mechanism to be described came from a genetic screen designed to identify positive regulators of Dnf1 and Dnf2. This screen identified a closely related pair of kinases named flippase protein kinase 1 and 2 (Fpk1, Fpk2; orthologs of p70S6K) [105]. The fpk1Δ fpk2Δ double mutant exhibits many of the same phenotypes as the dnf1Δ dnf2Δ double mutant. Membrane asymmetry is perturbed and the fpk1Δ fpk2Δ cells are also deficient in flippase activity at the plasma membrane measured with fluorescent PC or PE derivatives. Protein trafficking defects observed in fpk1Δ fpk2Δ cells are consistent with a loss of Dnf1-Lem3 and Dnf2-Lem3 function. Moreover, Fpk1 directly phosphorylates the P4-ATPases in order of substrate preference: Dnf1=Dnf2 > Dnf3 > Drs2 > Neo1 [105]. But how does phosphorylation activate the flippases and what is being “sensed” by Fpk1 and Fpk2?
A recent study has uncovered a regulatory network of protein kinases controlling Fpk activity that is responsive to the sphingolipid composition of the membrane [106]. A nucleating discovery was that Ypk1 (ortholog of serum and glucocorticoid induced kinase (SGK)) directly phosphorylates Fpk1 and inhibits Fpk1 activity. Fpk1, in turn, phosphorylates Ypk1 and inhibits its kinase activity [106]. Several kinds of input into this tug of war between Ypk1 and Fpk1 can tip the balance in favor of one of the kinases. For example, sphingolipid long chain base stimulates Pkh1 (ortholog of human 3-phosphoinositide-dependent kinase (PDK1)) [107], which phosphorylates and activates Ypk1. Thus, conditions that increase sphingolipid long chain base (e.g. heat shock) should tip the balance in favor of Ypk1 and reduce flippase activity. On the other hand, mature glycosphingolipids stimulate Fpk1 activity by an unknown mechanism and should increase flippase activity [106]. These observations imply that a homeostatic mechanism is in place to ensure an appropriate organization of glycerophospholipid and sphingolipid in the plasma membrane.
The flippase activity of Drs2 is also subjected to complex regulatory mechanisms (Fig. 3). Genetic interactions suggested a relationship between Drs2 and phosphoinositol-4-phosphate (PI4P) produced by the Pik1 phosphatidylinositol 4-kinase [108]. Drs2 appears to have no influence on the production of PI4P, but ablation of PI4P by inactivation of Pik1 or destruction by a phosphoinositide phosphatase dramatically reduces Drs2 flippase activity in isolated TGN membranes [109]. A phosphoinositide binding site with preference for PI4P maps to a basic patch of residues within the C-terminal cytosolic tail of Drs2, and mutation of these residues abrogates Drs2 activity. In addition, the basic patch overlaps a binding site for Arf guanine nucleotide exchange factor (ArfGEF) and this interaction also stimulates Drs2 flippase activity [109]. By analogy to the plasma membrane calcium ATPase (PMCA1) regulation by calmodulin [110], the C-terminal tail of Drs2 is likely an autoinhibitory regulatory domain and binding interactions relieve the autoinhibition to stimulate activity. The C-terminal tail of Dnf1 and Dnf2 may also be a regulatory domain as Fpk1 phosphorylation sites (RXSLD) are found in this part of the protein [106].
Genetic studies also implied a relationship between yeast flippases, sterol metabolism and an oxysterol-binding protein called Kes1 (or Osh4)(Fig. 3) [84, 111]. Moreover, recombinant Kes1 can potently downregulate the flippase activity of Drs2 in isolated Golgi membranes [84]. A recent study suggests that oxysterol binding proteins regulate phosphoinositide metabolism by stimulating Sac1, a PI4P phosphatase [112]. Thus, it is possible that Kes1 downregulation of Drs2 activity is mediated by the destruction of PI4P and the loss of this positive activator. Interestingly, Drs2 is also capable of downregulating Kes1 function [84], perhaps by creating a membrane structure that restricts the ability of Kes1 to extract sterol from the membrane.
Another exciting development from the Tuck laboratory is the observation that the endocytic regulator Numb binds to the C-terminus of an intestine-specific spliced isoform of TAT-1 [113]. This interaction appears to inhibit TAT-1 activity and is responsible for the ability of overexpressed Numb to inhibit endocytic recycling in intestinal cells. Surprisingly, knockdown of phosphatidylserine synthase was able to suppress endocytic recycling defects caused by loss of tat-1 or overexpression of Numb. One interpretation of this observation is that inappropriate PS accumulation in the lumenal leaflet of the tat-1 mutant endosomes is inhibiting the budding of transport vesicles [113].
7. Concluding remarks and future directions
Based on what is currently known, the primary biochemical activity of the P4-ATPases is the unidirectional translocation of specific phospholipid species across the membrane bilayer. One major cell biological consequences of this activity is the generation of an asymmetric membrane structure with a high concentration of substrate lipids, such as PS and PE, in the cytosolic leaflet. The asymmetric membrane organization can influence the physical properties of the membrane, the binding of peripheral membrane proteins and likely impacts the activity of integral membrane proteins. Moreover, signals can be transduced by regulated disruption of the phospholipid concentration gradient allowing exposure of PS on the outer leaflet, an event linked to recognition of apoptotic cells by macrophages and blood clotting reactions.
A second important consequence of a unidirectional, ATP-dependent flippase activity is an increase in phospholipid number within the cytosolic leaflet relative to the exofacial leaflet. As first described by the bilayer couple hypothesis [114], this imbalance in surface area between the two leaflets should induce bending in the membrane towards the cytosol, a process that is essential to vesicle budding [115]. Indeed, flippases play a critical role in vesicle budding within the Golgi and endosomal systems of yeast, plants and animals. We favor a model whereby P4-ATPase flippase activity helps drive membrane curvature to support vesicle budding (see reference [92] for a more complete discussion of the potential influences of flippases on membrane curvature and vesicular trafficking). Many other types of transport proteins are also proposed to drive membrane curvature, including coat proteins (COPI, COPII and clathrin), small GTP binding proteins (Arf and Sar1), ENTH domain proteins, BAR domain proteins and lipid modifying enzymes [115, 116]. A significant challenge for the future is tease out the relative contributions of each of these membrane remodeling proteins on the vesicle budding process.
While the last decade has produced a dramatic expansion in the literature on P4-ATPases, many fundamental questions remain unanswered. On the biochemical front, how do P4-ATPases recognize and translocate phospholipid across the membrane? While atomic resolution mechanistic insight into substrate recognition and transport by cation transporting P-type ATPases is being achieved [25], it is not known if P4-ATPases transport phospholipid transport by the same mechanism. What is the precise role of the β-subunit in the catalytic cycle of the P4-ATPase? Does the β-subunit form part of the phospholipid translocation pathway or is it regulating the pump activity? It is becoming clear that P4-ATPases are tightly regulated by phosphorylation and/or binding partners, but how is this regulation achieved and why is it important to modulate flippase activity? On the cellular level, how are the membrane transformation events in the late Golgi system - translocation of glycerophospholipid to the cytosolic leaflet, sphingolipid biosynthesis in the lumenal leaflet, and cholesterol loading - integrated through the regulatory mechanisms described above? The hypothesis that flippases induce curvature in the membrane to support vesicle budding requires formal testing, as does the degree to which the role of flippases in vesicle trafficking is conserved in mammals. Physiological studies of P4-ATPase knockout mice are in their infancy and it seems very likely that genome-wide association studies will continue to identify additional human diseases linked to P4-ATPase deficiency. We have every reason to expect that the next decade will witness an even greater expansion of the P4-ATPase field.
Highlights.
Evidence that type IV P-type ATPases (P4-ATPases) are phospholipid flippases.
Phospholipid flippases create membrane phospholipid asymmetry.
Phospholipid flippases play critical roles in vesicle-mediated protein transport in the secretory and endocytic pathways.
P4-ATPases are linked to many disease states.
Mechanisms for regulating flippase activity are being uncovered.
Acknowledgments
Studies on the role of P4-ATPases on protein trafficking in our laboratory is supported by NIH grant RO1 GM62367 to T.R.G..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- 3.Muthusamy BP, Natarajan P, Zhou X, Graham TR. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim Biophys Acta. 2009;1791:612–619. doi: 10.1016/j.bbalip.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CY, Ingram MF, Rosal PH, Graham TR. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol. 1999;147:1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hua Z, Fatheddin P, Graham TR. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell. 2002;13:3162–3177. doi: 10.1091/mbc.E02-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomorski T, Lombardi R, Riezman H, Devaux PF, van Meer G, Holthuis JC. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen B, Jiang Y, Zeng S, Yan J, Li X, Zhang Y, Zou W, Wang X. Endocytic sorting and recycling require membrane phosphatidylserine asymmetry maintained by TAT-1/CHAT-1. PLoS Genet. 2010;6:e1001235. doi: 10.1371/journal.pgen.1001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulsen LR, Lopez-Marques RL, McDowell SC, Okkeri J, Licht D, Schulz A, Pomorski T, Harper JF, Palmgren MG. The Arabidopsis P4-ATPase ALA3 localizes to the golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell. 2008;20:658–676. doi: 10.1105/tpc.107.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruaud AF, Nilsson L, Richard F, Larsen MK, Bessereau JL, Tuck S. The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic. 2009;10:88–100. doi: 10.1111/j.1600-0854.2008.00844.x. [DOI] [PubMed] [Google Scholar]
- 10.Xu P, Ding X. Characterization and expression of mouse Cdc50c during spermatogenesis. Acta Biochim Biophys Sin (Shanghai) 2007;39:739–744. doi: 10.1111/j.1745-7270.2007.00342.x. [DOI] [PubMed] [Google Scholar]
- 11.Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryde S, Hennrich H, Verhulst PM, Devaux PF, Lenoir G, Holthuis JC. CDC50 proteins are critical components of the human class-1 P4-ATPase transport machinery. J Biol Chem. 2010;285:40562–40572. doi: 10.1074/jbc.M110.139543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Velden LM, Wichers CG, van Breevoort AE, Coleman JA, Molday RS, Berger R, Klomp LW, van de Graaf SF. Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J Biol Chem. 2010;285:40088–40096. doi: 10.1074/jbc.M110.139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 15.Bull LN, van Eijk MJ, Pawlikowska L, DeYoung JA, Juijn JA, Liao M, Klomp LW, Lomri N, Berger R, Scharschmidt BF, Knisely AS, Houwen RH, Freimer NB. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 16.Ripmaster TL, Vaughn GP, Woolford JL., Jr DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7901–7912. doi: 10.1128/mcb.13.12.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- 18.Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- 19.Thever MD, Saier MH., Jr Bioinformatic characterization of p-type ATPases encoded within the fully sequenced genomes of 26 eukaryotes. The Journal of membrane biology. 2009;229:115–130. doi: 10.1007/s00232-009-9176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Marques RL, Poulsen LR, Hanisch S, Meffert K, Buch-Pedersen MJ, Jakobsen MK, Pomorski TG, Palmgren MG. Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA alpha-subunit. Mol Biol Cell. 2010;21:791–801. doi: 10.1091/mbc.E09-08-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulusma CC, Folmer DE, Ho-Mok KS, de Waart DR, Hilarius PM, Verhoeven AJ, Oude Elferink RP. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology. 2008;47:268–278. doi: 10.1002/hep.21950. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Victoria FJ, Sanchez-Canete MP, Castanys S, Gamarro F. Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J Biol Chem. 2006;281:23766–23775. doi: 10.1074/jbc.M605214200. [DOI] [PubMed] [Google Scholar]
- 24.Takatsu H, Baba K, Shima T, Umino H, Kato U, Umeda M, Nakayama K, Shin HW. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50-independent manner. J Biol Chem. 2011 doi: 10.1074/jbc.M111.281006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmgren MG, Nissen P. P-type ATPases. Annual review of biophysics. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 26.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornberg RD, McConnell HM. Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry. 1971;10:1111–1120. doi: 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- 28.Seigneuret M, Devaux PF. ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: relation to shape changes. Proc Natl Acad Sci U S A. 1984;81:3751–3755. doi: 10.1073/pnas.81.12.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zachowski A, Henry JP, Devaux PF. Control of transmembrane lipid asymmetry in chromaffin granules by an ATP-dependent protein. Nature. 1989;340:75–76. doi: 10.1038/340075a0. [DOI] [PubMed] [Google Scholar]
- 30.Sune A, Bette-Bobillo P, Bienvenue A, Fellmann P, Devaux PF. Selective outside-inside translocation of aminophospholipids in human platelets. Biochemistry. 1987;26:2972–2978. doi: 10.1021/bi00385a003. [DOI] [PubMed] [Google Scholar]
- 31.Kean LS, Fuller RS, Nichols JW. Retrograde lipid traffic in yeast: identification of two distinct pathways for internalization of fluorescent-labeled phosphatidylcholine from the plasma membrane. J Cell Biol. 1993;123:1403–1419. doi: 10.1083/jcb.123.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daleke DL, Huestis WH. Incorporation and translocation of aminophospholipids in human erythrocytes. Biochemistry. 1985;24:5406–5416. doi: 10.1021/bi00341a019. [DOI] [PubMed] [Google Scholar]
- 33.Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc Natl Acad Sci U S A. 1974;71:4457–4461. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daleke DL. Phospholipid flippases. J Biol Chem. 2007;282:821–825. doi: 10.1074/jbc.R600035200. [DOI] [PubMed] [Google Scholar]
- 35.Soupene E, Kuypers FA. Identification of an erythroid ATP-dependent aminophospholipid transporter. Br J Haematol. 2006;133:436–438. doi: 10.1111/j.1365-2141.2006.06051.x. [DOI] [PubMed] [Google Scholar]
- 36.Xie XS, Stone DK, Racker E. Purification of a vanadate-sensitive ATPase from clathrin-coated vesicles of bovine brain. Journal of Biological Chemistry. 1989;264:1710–1714. [PubMed] [Google Scholar]
- 37.Moriyama Y, Nelson N. Purification and properties of a vanadate- and N-ethylmaleimide-sensitive ATPase from chromaffin granule membranes. Journal of Biological Chemistry. 1988;263:8521–8527. [PubMed] [Google Scholar]
- 38.Morrot G, Zachowski A, Devaux PF. Partial purification and characterization of the human erythrocyte Mg2+-ATPase A candidate aminophospholipid translocase. FEBS Letters. 1990;266:29–32. doi: 10.1016/0014-5793(90)81498-d. [DOI] [PubMed] [Google Scholar]
- 39.Auland ME, Roufogalis BD, Devaux PF, Zachowski A. Reconstitution of ATP-dependent aminophospholipid translocation in proteoliposomes. Proc Natl Acad Sci U S A. 1994;91:10938–10942. doi: 10.1073/pnas.91.23.10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bull LN, van Eijk MJ, Pawlikowska L, DeYoung JA, Juijn JA, Liao M, Klomp LW, Lomri N, Berger R, Scharschmidt BF, Knisely AS, Houwen RH, Freimer NB. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 41.Siegmund A, Grant A, Angeletti C, Malone L, Nichols JW, Rudolph HK. Loss of Drs2p does not abolish transfer of fluorescence-labeled phospholipids across the plasma membrane of Saccharomyces cerevisiae. Journal of Biological Chemistry. 1998;273:34399–34405. doi: 10.1074/jbc.273.51.34399. [DOI] [PubMed] [Google Scholar]
- 42.Marx U, Polakowski T, Pomorski T, Lang C, Nelson H, Nelson N, Herrmann A. Rapid transbilayer movement of fluorescent phospholipid analogues in the plasma membrane of endocytosis-deficient yeast cells does not require the Drs2 protein. European Journal of Biochemistry. 1999;263:254–264. doi: 10.1046/j.1432-1327.1999.00497.x. [DOI] [PubMed] [Google Scholar]
- 43.Natarajan P, Wang J, Hua Z, Graham TR. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc Natl Acad Sci USA. 2004;101:10614–10619. doi: 10.1073/pnas.0404146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alder-Baerens N, Lisman Q, Luong L, Pomorski T, Holthuis JCM. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol Biol Cell. 2006;17:1632–1642. doi: 10.1091/mbc.E05-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Wang J, Muthusamy BP, Liu K, Zare S, Andersen RJ, Graham TR. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 2006;7:1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- 46.Xu P, Okkeri J, Hanisch S, Hu RY, Xu Q, Pomorski TG, Ding XY. Identification of a novel mouse P4-ATPase family member highly expressed during spermatogenesis. J Cell Sci. 2009;122:2866–2876. doi: 10.1242/jcs.047423. [DOI] [PubMed] [Google Scholar]
- 47.Ujhazy P, Ortiz D, Misra S, Li S, Moseley J, Jones H, Arias IM. Familial intrahepatic cholestasis 1: studies of localization and function. Hepatology. 2001;34:768–775. doi: 10.1053/jhep.2001.27663. [DOI] [PubMed] [Google Scholar]
- 48.Verhulst PM, van der Velden LM, Oorschot V, van Faassen EE, Klumperman J, Houwen RH, Pomorski TG, Holthuis JC, Klomp LW. A flippase-independent function of ATP8B1, the protein affected in familial intrahepatic cholestasis type 1, is required for apical protein expression and microvillus formation in polarized epithelial cells. Hepatology. 2010;51:2049–2060. doi: 10.1002/hep.23586. [DOI] [PubMed] [Google Scholar]
- 49.Cai SY, Gautam S, Nguyen T, Soroka CJ, Rahner C, Boyer JL. ATP8B1 Deficiency Disrupts the Bile Canalicular Membrane Bilayer Structure in Hepatocytes, but FXR Expression and Activity Are Maintained. Gastroenterology. 2008 doi: 10.1053/j.gastro.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paulusma CC, Groen A, Kunne C, Ho-Mok KS, Spijkerboer AL, Rudi de Waart D, Hoek FJ, Vreeling H, Hoeben KA, van Marle J, Pawlikowska L, Bull LN, Hofmann AF, Knisely AS, Oude Elferink RP. Atp8b1 deficiency in mice reduces resistance of the canalicular membrane to hydrophobic bile salts and impairs bile salt transport. Hepatology. 2006;44:195–204. doi: 10.1002/hep.21212. [DOI] [PubMed] [Google Scholar]
- 51.Radji M, Kim J, Togan T, Yoshikawa H, Shirahige K. The cloning and characterization of the CDC50 gene family in Saccharomyces cerevisiae. Yeast. 2001;18:195–205. doi: 10.1002/1097-0061(200102)18:3<195::AID-YEA660>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 52.Kato U, Emoto K, Fredriksson C, Nakamura H, Ohta A, Kobayashi T, Murakami-Murofushi K, Kobayashi T, Umeda M. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2002;277:37855–37862. doi: 10.1074/jbc.M205564200. [DOI] [PubMed] [Google Scholar]
- 53.Hanson PK, Malone L, Birchmore JL, Nichols JW. Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J Biol Chem. 2003;278:36041–36050. doi: 10.1074/jbc.M305263200. [DOI] [PubMed] [Google Scholar]
- 54.Coleman JA, Kwok MC, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J Biol Chem. 2009;284:32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou X, Graham TR. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci U S A. 2009;106:16586–16591. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coleman JA, Molday RS. Critical role of the beta-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J Biol Chem. 2011;286:17205–17216. doi: 10.1074/jbc.M111.229419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lenoir G, Williamson P, Puts CF, Holthuis JC. Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholipid transporter Drs2p. J Biol Chem. 2009;284:17956–17967. doi: 10.1074/jbc.M109.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi Y, Fujimura-Kamada K, Kondo S, Tanaka K. Isolation and characterization of novel mutations in CDC50, the non-catalytic subunit of the Drs2p phospholipid flippase. J Biochem. 2011;149:423–432. doi: 10.1093/jb/mvq155. [DOI] [PubMed] [Google Scholar]
- 59.Cacciagli P, Haddad MR, Mignon-Ravix C, El-Waly B, Moncla A, Missirian C, Chabrol B, Villard L. Disruption of the ATP8A2 gene in a patient with a t(10;13) de novo balanced translocation and a severe neurological phenotype. European journal of human genetics: EJHG. 2010;18:1360–1363. doi: 10.1038/ejhg.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, Aulchenko Y, Franklin CS, Liebisch G, Erdmann J, Jonasson I, Zorkoltseva IV, Pattaro C, Hayward C, Isaacs A, Hengstenberg C, Campbell S, Gnewuch C, Janssens AC, Kirichenko AV, Konig IR, Marroni F, Polasek O, Demirkan A, Kolcic I, Schwienbacher C, Igl W, Biloglav Z, Witteman JC, Pichler I, Zaboli G, Axenovich TI, Peters A, Schreiber S, Wichmann HE, Schunkert H, Hastie N, Oostra BA, Wild SH, Meitinger T, Gyllensten U, van Duijn CM, Wilson JF, Wright A, Schmitz G, Campbell H. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009;5:e1000672. doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zwaal RF, Comfurius P, Bevers EM. Surface exposure of phosphatidylserine in pathological cells. Cell Mol Life Sci. 2005;62:971–988. doi: 10.1007/s00018-005-4527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darland-Ransom M, Wang X, Sun CL, Mapes J, Gengyo-Ando K, Mitani S, Xue D. Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science. 2008;320:528–531. doi: 10.1126/science.1155847. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 64.Castoldi E, Collins PW, Williamson PL, Bevers EM. Compound heterozygosity for 2 novel TMEM16F mutations in a patient with Scott syndrome. Blood. 2011;117:4399–4400. doi: 10.1182/blood-2011-01-332502. [DOI] [PubMed] [Google Scholar]
- 65.Menon I, Huber T, Sanyal S, Banerjee S, Barre P, Canis S, Warren JD, Hwa J, Sakmar TP, Menon AK. Opsin is a phospholipid flippase. Curr Biol. 2011;21:149–153. doi: 10.1016/j.cub.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klomp LW, Vargas JC, van Mil SW, Pawlikowska L, Strautnieks SS, van Eijk MJ, Juijn JA, Pabon-Pena C, Smith LB, DeYoung JA, Byrne JA, Gombert J, van der Brugge G, Berger R, Jankowska I, Pawlowska J, Villa E, Knisely AS, Thompson RJ, Freimer NB, Houwen RH, Bull LN. Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology. 2004;40:27–38. doi: 10.1002/hep.20285. [DOI] [PubMed] [Google Scholar]
- 67.Stapelbroek JM, Peters TA, van Beurden DH, Curfs JH, Joosten A, Beynon AJ, van Leeuwen BM, van der Velden LM, Bull L, Oude Elferink RP, van Zanten BA, Klomp LW, Houwen RH. ATP8B1 is essential for maintaining normal hearing. Proc Natl Acad Sci U S A. 2009;106:9709–9714. doi: 10.1073/pnas.0807919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Beserra C, Garbers DL. A novel aminophospholipid transporter exclusively expressed in spermatozoa is required for membrane lipid asymmetry and normal fertilization. Dev Biol. 2004;267:203–215. doi: 10.1016/j.ydbio.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 69.Zhang X, Oppenheimer DG. IRREGULAR TRICHOME BRANCH 2 (ITB2) encodes a putative aminophospholipid translocase that regulates trichome branch elongation in Arabidopsis. The Plant journal: for cell and molecular biology. 2009;60:195–206. doi: 10.1111/j.1365-313X.2009.03954.x. [DOI] [PubMed] [Google Scholar]
- 70.Lyssenko NN, Miteva Y, Gilroy S, Hanna-Rose W, Schlegel RA. An unexpectedly high degree of specialization and a widespread involvement in sterol metabolism among the C. elegans putative aminophospholipid translocases. BMC developmental biology. 2008;8:96. doi: 10.1186/1471-213X-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seamen E, Blanchette JM, Han M. P-type ATPase TAT-2 negatively regulates monomethyl branched-chain fatty acid mediated function in post-embryonic growth and development in C. elegans. PLoS Genet. 2009;5:e1000589. doi: 10.1371/journal.pgen.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dhar M, Webb LS, Smith L, Hauser L, Johnson D, West DB. A novel ATPase on mouse chromosome 7 is a candidate gene for increased body fat. Physiol Genomics. 2000;4:93–100. doi: 10.1152/physiolgenomics.2000.4.1.93. [DOI] [PubMed] [Google Scholar]
- 73.Dhar MS, Sommardahl CS, Kirkland T, Nelson S, Donnell R, Johnson DK, Castellani LW. Mice heterozygous for Atp10c, a putative amphipath, represent a novel model of obesity and type 2 diabetes. J Nutr. 2004;134:799–805. doi: 10.1093/jn/134.4.799. [DOI] [PubMed] [Google Scholar]
- 74.Dhar MS, Yuan JS, Elliott SB, Sommardahl C. A type IV P-type ATPase affects insulin-mediated glucose uptake in adipose tissue and skeletal muscle in mice. J Nutr Biochem. 2006;17:811–820. doi: 10.1016/j.jnutbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 75.Flamant S, Pescher P, Lemercier B, Clement-Ziza M, Kepes F, Fellous M, Milon G, Marchal G, Besmond C. Characterization of a putative type IV aminophospholipid transporter P-type ATPase. Mamm Genome. 2003;14:21–30. doi: 10.1007/s00335-002-3032-3. [DOI] [PubMed] [Google Scholar]
- 76.Gomes E, Jakobsen MK, Axelsen KB, Geisler M, Palmgren MG. Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell. 2000;12:2441–2454. [PMC free article] [PubMed] [Google Scholar]
- 77.Siggs OM, Arnold CN, Huber C, Pirie E, Xia Y, Lin P, Nemazee D, Beutler B. The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nature immunology. 2011;12:434–440. doi: 10.1038/ni.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yabas M, Teh CE, Frankenreiter S, Lal D, Roots CM, Whittle B, Andrews DT, Zhang Y, Teoh NC, Sprent J, Tze LE, Kucharska EM, Kofler J, Farell GC, Broer S, Goodnow CC, Enders A. ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nature immunology. 2011;12:441–449. doi: 10.1038/ni.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siggs OM, Schnabl B, Webb B, Beutler B. X-linked cholestasis in mouse due to mutations of the P4-ATPase ATP11C. Proc Natl Acad Sci U S A. 2011;108:7890–7895. doi: 10.1073/pnas.1104631108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen CY, Graham TR. An arf1Delta synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics. 1998;150:577–589. doi: 10.1093/genetics/150.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakane H, Yamamoto T, Tanaka K. The functional relationship between the Cdc50p-Drs2p putative aminophospholipid translocase and the Arf GAP Gcs1p in vesicle formation in the retrieval pathway from yeast early endosomes to the TGN. Cell Struct Funct. 2006;31:87–108. doi: 10.1247/csf.06021. [DOI] [PubMed] [Google Scholar]
- 82.Deloche O, de la Cruz J, Kressler D, Doere M, Linder P. A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae. Mol Cell. 2004;13:357–366. doi: 10.1016/s1097-2765(04)00008-5. [DOI] [PubMed] [Google Scholar]
- 83.Li Y, Moir RD, Sethy-Coraci IK, Warner JR, Willis IM. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol Cell Biol. 2000;20:3843–3851. doi: 10.1128/mcb.20.11.3843-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muthusamy BP, Raychaudhuri S, Natarajan P, Abe F, Liu K, Prinz WA, Graham TR. Control of protein and sterol trafficking by antagonistic activities of a type IV P-type ATPase and oxysterol binding protein homologue. Mol Biol Cell. 2009;20:2920–2931. doi: 10.1091/mbc.E08-10-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gall WE, Geething NC, Hua Z, Ingram MF, Liu K, Chen SI, Graham TR. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr Biol. 2002;12:1623–1627. doi: 10.1016/s0960-9822(02)01148-x. [DOI] [PubMed] [Google Scholar]
- 86.Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- 87.Gall WE, Higginbotham MA, Chen C, Ingram MF, Cyr DM, Graham TR. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr Biol. 2000;10:1349–1358. doi: 10.1016/s0960-9822(00)00771-5. [DOI] [PubMed] [Google Scholar]
- 88.Xiao J, Kim LS, Graham TR. Dissection of Swa2p/auxilin domain requirements for cochaperoning Hsp70 clathrin-uncoating activity in vivo. Mol Biol Cell. 2006;17:3281–3290. doi: 10.1091/mbc.E06-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu K, Surendhran K, Nothwehr SF, Graham TR. P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol Biol Cell. 2008;19:3526–3535. doi: 10.1091/mbc.E08-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- 91.Liu K, Hua Z, Nepute JA, Graham TR. Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol Biol Cell. 2007;18:487–500. doi: 10.1091/mbc.E06-07-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Graham TR. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 2004;14:670–677. doi: 10.1016/j.tcb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 93.Kus BM, Caldon CE, Andorn-Broza R, Edwards AM. Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins. 2004;54:455–467. doi: 10.1002/prot.10620. [DOI] [PubMed] [Google Scholar]
- 94.Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol Cell Biol. 2001;21:3105–3117. doi: 10.1128/MCB.21.9.3105-3117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen SH, Shah AH, Segev N. Ypt31/32 GTPases and their F-Box effector Rcy1 regulate ubiquitination of recycling proteins. Cellular logistics. 2011;1:21–31. doi: 10.4161/cl.1.1.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson M, Poon PP, Schindler C, Murray LE, Kama R, Gabriely G, Singer RA, Spang A, Johnston GC, Gerst JE. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol Biol Cell. 2006;17:1845–1858. doi: 10.1091/mbc.E05-09-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hettema EH, Lewis MJ, Black MW, Pelham HR. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. Embo J. 2003;22:548–557. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saito K, Fujimura-Kamada K, Hanamatsu H, Kato U, Umeda M, Kozminski KG, Tanaka K. Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev Cell. 2007;13:743–751. doi: 10.1016/j.devcel.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 101.Hua Z, Graham TR. Requirement for neo1p in retrograde transport from the Golgi complex to the endoplasmic reticulum. Mol Biol Cell. 2003;14:4971–4983. doi: 10.1091/mbc.E03-07-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singer-Kruger B, Lasic M, Burger AM, Hausser A, Pipkorn R, Wang Y. Yeast and human Ysl2p/hMon2 interact with Gga adaptors and mediate their subcellular distribution. Embo J. 2008;27:1423–1435. doi: 10.1038/emboj.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wicky S, Schwarz H, Singer-Kruger B. Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol Cell Biol. 2004;24:7402–7418. doi: 10.1128/MCB.24.17.7402-7418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sato K, Sato M, Nakano A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J Cell Biol. 2001;152:935–944. doi: 10.1083/jcb.152.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nakano K, Yamamoto T, Kishimoto T, Noji T, Tanaka K. Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol Biol Cell. 2008;19:1783–1797. doi: 10.1091/mbc.E07-07-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci U S A. 2010;107:34–39. doi: 10.1073/pnas.0912497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu K, Zhang X, Lester RL, Dickson RC. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J Biol Chem. 2005;280:22679–22687. doi: 10.1074/jbc.M502972200. [DOI] [PubMed] [Google Scholar]
- 108.Sciorra VA, Audhya A, Parsons AB, Segev N, Boone C, Emr SD. Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/rab-GTPase signaling pathway. Mol Biol Cell. 2005;16:776–793. doi: 10.1091/mbc.E04-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Natarajan P, Liu K, Patil DV, Sciorra VA, Jackson CL, Graham TR. Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat Cell Biol. 2009;11:1421–1426. doi: 10.1038/ncb1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Di Leva F, Domi T, Fedrizzi L, Lim D, Carafoli E. The plasma membrane Ca2+ ATPase of animal cells: structure, function and regulation. Archives of biochemistry and biophysics. 2008;476:65–74. doi: 10.1016/j.abb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 111.Kishimoto T, Yamamoto T, Tanaka K. Defects in structural integrity of ergosterol and the Cdc50p-Drs2p putative phospholipid translocase cause accumulation of endocytic membranes, onto which actin patches are assembled in yeast. Mol Biol Cell. 2005;16:5592–5609. doi: 10.1091/mbc.E05-05-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 113.Nilsson L, Jonsson E, Tuck S. Caenorhabditis elegans Numb Inhibits Endocytic Recycling by Binding TAT-1 Aminophospholipid Translocase. Traffic. 2011 doi: 10.1111/j.1600-0854.2011.01271.x. [DOI] [PubMed] [Google Scholar]
- 114.Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc Natl Acad Sci U S A. 1974;71:4457–4461. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]