Abstract
Chromatin modifiers regulate lifespan in several organisms, raising the question of whether changes in chromatin states in the parental generation could be incompletely reprogrammed in the next generation and thereby affect the lifespan of descendents. The histone H3 lysine 4 trimethylation (H3K4me3) complex composed of ASH-2, WDR-5, and the histone methyltransferase SET-2 regulates C. elegans lifespan. Here we show that deficiencies in the H3K4me3 chromatin modifiers ASH-2, WDR-5, or SET-2 in the parental generation extend the lifespan of descendents up until the third generation. The transgenerational inheritance of lifespan extension by members of the ASH-2 complex is dependent on the H3K4me3 demethylase RBR-2, and requires the presence of a functioning germline in the descendents. Transgenerational inheritance of lifespan is specific for the H3K4me3 methylation complex and is associated with epigenetic changes in gene expression. Thus, manipulation of specific chromatin modifiers only in parents can induce an epigenetic memory of longevity in descendents.
Results
Transgenerational epigenetic inheritance has been described for some traits, including flower symmetry and color in plants1–3, progeny production in worms4, heat stress response and eye color in drosophila5–7, and coat color in mammals8–10. However, the transgenerational epigenetic inheritance of longevity, and more generally of complex traits, is largely undefined. Chromatin modifiers have been shown to regulate longevity in several species11–18, raising the possibility that chromatin changes in parents might not be entirely reset between generations and thereby also regulate longevity in descendents. Deficiencies in the H3K4me3 regulatory complex composed of ASH-2, WDR-5, and SET-2 extend lifespan in C. elegans12. We asked if perturbation of members of the H3K4me3 regulatory complex (ASH-2, WDR-5, and SET-2) only in the parental generation could regulate the lifespan of descendents in subsequent generations in C. elegans.
Transgenerational inheritance of longevity
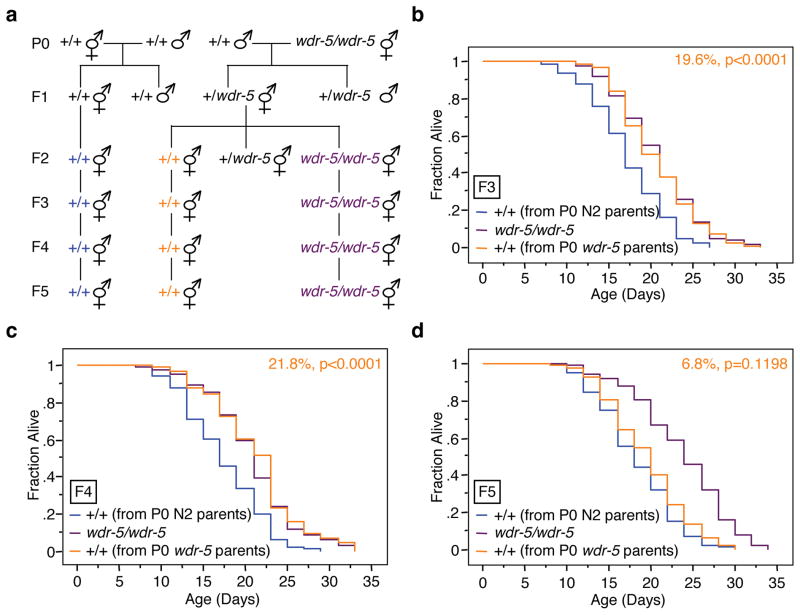
We first focused on WDR-5, a conserved regulatory component of the ASH-2 complex19 whose depletion decreases H3K4me3 levels12,20–22 and extends lifespan in worms12. To test if longevity could be inherited in a transgenerational epigenetic manner, we crossed wildtype males with wdr-5(ok1417) mutant hermaphrodites to generate F1 heterozygous hermaphrodites (Fig. 1a). These F1 heterozygous hermaphrodites were genotyped and then self-crossed to generate F2 hermaphrodites (wildtype, heterozygous, and homozygous at the wdr-5 locus), which were genotyped after they had laid F3 generation progeny. In parallel, we crossed a wildtype male with a wildtype hermaphrodite to generate pure wildtype descendents and control for any beneficial longevity effects that could come from crossing rather than self-mating (Fig. 1a). Longevity of the F3, F4, and F5 generations of worms was examined. Interestingly, genetically wildtype F3 descendents from P0 wdr-5 parents (+/+ from P0 wdr-5 parents) still exhibited a ~20% extension of lifespan (p<0.0001) compared to descendents from pure wildtype parents (+/+ from P0 N2 parents) (Fig. 1b). This 20% lifespan extension was similar in magnitude to the lifespan extension of pure F3 wdr-5(ok1417) mutants (wdr-5/wdr-5) (Fig. 1b). The lifespan of genetically wildtype descendents from wdr-5(ok1417) mutant parents (+/+ from P0 wdr-5 parents) was still extended in the F4 generation (Fig. 1c), but was no longer extended in the F5 generation (Fig. 1d). Thus, wdr-5 deficiency only in the parental generation can extend the lifespan of subsequent generations. Since the lifespan of F5 generation wildtype descendents from wdr-5 mutant parents is no longer extended, the lifespan extension observed in the F3 and F4 generations is unlikely to be due to extraneous mutations that might have been present in the parental wdr-5 mutant strain. Instead, the transgenerational inheritance of longevity may be due to epigenetic changes in H3K4me3 itself or in another molecule that can only be inherited for a limited number of generations.
Fig. 1. Genetically wildtype descendents from wdr-5 mutant parents have extended lifespan for several generations.
a, Scheme for generating wildtype descendents from wdr-5(ok1417) mutant worms. b–d, Lifespan of genetically wildtype F3 (b), F4 (c), F5 (d) descendents of wdr-5(ok1417) mutant worms (+/+ from P0 wdr-5 parents) compared to descendents of wildtype (N2) worms (+/+ from P0 N2 parents). Mean lifespan and statistics are presented in Supplementary Table 1.
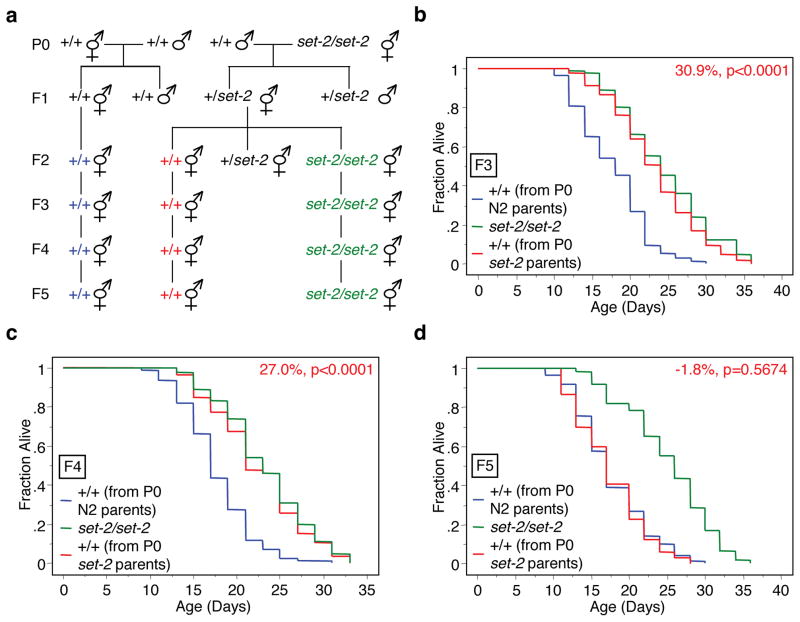
We next asked if a transgenerational epigenetic heritability of lifespan was also observed with SET-2, the H3K4me3 methyltransferase enzyme that functions together with ASH-2 and WDR-5 to regulate H3K4me3 levels12,20–22 and longevity in C. elegans12 (Fig. 2). Similar to what we observed for wdr-5, genetically wildtype descendents from set-2(ok952) mutants still exhibited a ~30% extension of lifespan (p<0.0001) in the F3 and F4 generations (Fig. 2b, c), but not in the F5 generation (Fig. 2d). Genetically wildtype F3 descendents from the reverse cross – P0 set-2(ok952) males crossed with wildtype hermaphrodites – were also long-lived (Supplementary Table 1), indicating that transgenerational inheritance of longevity is not linked to a particular gender in the parental generation.
Fig. 2. Genetically wildtype descendents from set-2 mutant parents have extended lifespan for several generations.
a, Scheme for generating wildtype descendents from set-2(ok952) mutant worms. b–d, Lifespan of genetically wildtype F3 (b), F4 (c), and F5 (d) descendents from set-2(ok952) mutant worms (+/+ from P0 set-2 parents) compared to descendents of wildtype (N2) worms (+/+ from P0 N2 parents). Mean lifespan and statistics are presented in Supplementary Table 1.
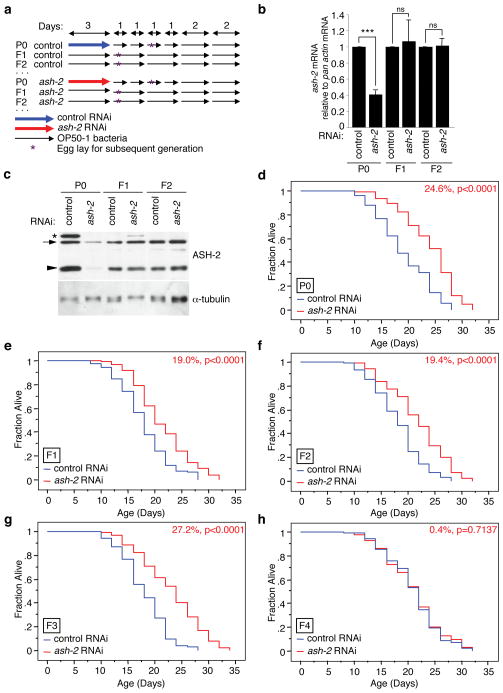
ASH-2 is important for the conversion of H3K4 dimethylation (H3K4me2) to H3K4me3 (ref. 23). ash-2 knock-down in worms decreases global H3K4me3 levels at the L3 stage12,22 and extends longevity12. We asked if ash-2 knock-down only in the parental generation affected the lifespan of several generations of descendents. Wildtype parent worms (P0) were placed on plates with bacteria expressing RNAi to ash-2 from birth to the larval stage L4, then switched every day for three days onto plates containing OP50-1 bacteria and streptomycin to selectively prevent the growth of RNAi-expressing bacteria (Fig. 3a). Endogenous ash-2 mRNA and ASH-2 protein levels were significantly decreased in the P0 generation, but returned to normal levels in subsequent generations (Fig. 3b, c), indicating that ash-2 RNAi is not itself inherited. The lifespan of worms from the F1, F2, and F3 generations in which ash-2 had been knocked-down only in the P0 parental generation was still significantly extended (19–27%, p<0.0001) compared to that of descendents of worms treated with empty vector control in the P0 parental generation (Fig. 3d–g). By contrast, F4 generation descendents no longer had extended lifespan (Fig. 3h). We obtained similar results after bleaching P0 worms to avoid potential carry over of RNAi-expressing bacteria (data not shown). Thus, alteration of the components of the H3K4me3 methyltransferase complex (ASH-2, WDR-5, SET-2) in parents affects the lifespan of descendents, supporting the possibility that transgenerational inheritance of longevity is due to epigenetic changes that may only be inherited for a limited number of generations.
Fig. 3. Knock-down of ash-2 only in the parental generation extends lifespan for several generations.
a, Scheme for generating wildtype descendents from RNAi treated parents. b, ash-2 mRNA levels at day 7 in different generations of worms treated with ash-2 RNAi or empty vector (control) only in the P0 generation. Mean ± s.e.m. of 3 independent experiments. ***p=0.0002 with paired t-test. c, ASH-2 protein levels at L3 stage in different generations of worms treated with ash-2 RNAi or empty vector (control) only in the P0 generation. Representative of 2 independent experiments. *: non-specific band; Arrow: ASH-2; Arrowhead: protein related to ASH-2, possibly a degradation product. d–h, Lifespan of P0 (d), F1 (e), F2 (f), F3 (g), and F4 (h) generations of worms with RNAi knock-down of ash-2 in parents only. Control RNAi: empty vector. Mean lifespan and statistics are presented in Supplementary Table 2.
Importance of H3K4me3 demethylase and germline
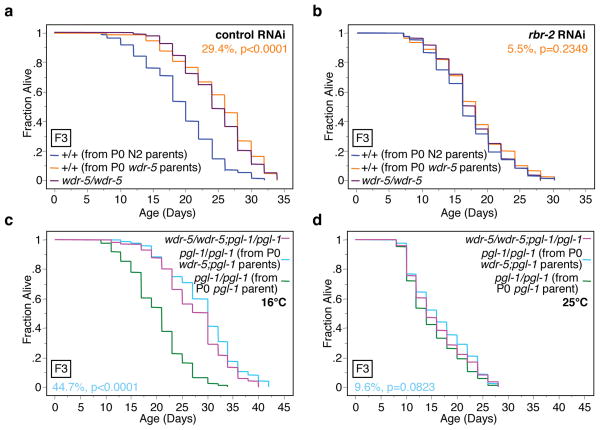
The H3K4me3 demethylase RBR-2 is necessary for the lifespan extension caused by deficiencies in members of the ASH-2 complex12. We asked if the transgenerational extension of longevity induced by deficiencies in members of the ASH-2 complex is dependent on RBR-2. The lifespan of genetically wildtype F3 descendents from P0 wdr-5 parents (+/+ from P0 wdr-5 parents) was no longer extended in the presence of rbr-2 RNAi (Fig. 4a, b). Similarly, F3 wildtype descendents from set-2;rbr-2 parents (+/+ from P0 set-2;rbr-2 parents) were no longer long-lived (Supplementary Fig. 1). Together, these data indicate that the transgenerational inheritance of longevity due to deficiencies in H3K4 trimethylation complex members is dependent on the H3K4me3 demethylase RBR-2. The fact that the longevity of wildtype descendents of wdr-5 and set-2 mutants is reverted by deficiencies in rbr-2 also suggests that this extended lifespan is unlikely to result from extraneous mutations in wdr-5 or set-2 strains. rbr-2 mutation or knock-down did not lead to a shortening of lifespan in descendents (Supplementary Fig. 2), indicating that by itself, RBR-2 deficiency does not affect longevity in a transgenerational manner.
Fig. 4. The transgenerational inheritance of longevity by deficiencies in ASH-2 complex members is dependent on the presence of the H3K4me3 demethylase RBR-2 and of an intact germline.
a–b, Lifespan of genetically wildtype F3 descendents from wdr-5(ok1417) mutant worms (+/+ from P0 wdr-5 parents) in the presence of empty vector (control RNAi) (a) or rbr-2 RNAi (b). c–d, Lifespan of pgl-1 F3 descendents from a wdr-5(ok1417);pgl-1(bn101) mutant worms (pgl-1/pgl-1 from P0 wdr-5;pgl-1 parents) compared to descendents from pgl-1(bn101) worms at the permissive temperature (16°C) (c) and at the restrictive temperature (25°C) (d). Mean lifespan and statistics are presented in Supplementary Tables 3 and 4.
Longevity due to modulation of the ASH-2 complex is dependent on a functioning germline12. To test if wildtype descendents of worms with deficiencies in ASH-2 complex members also require the presence of a functioning germline for lifespan extension, we used temperature-sensitive feminized fem-3(e2006) mutant worms, which do not produce mature eggs at the restrictive temperature24. Knock-down of ash-2 and wdr-5 only in parents extended the lifespan of the F1 generation in fem-3(e2006) mutant worms at the permissive temperature (16°C), but not at the restrictive temperature (25°C) (Supplementary Fig. 3). To independently examine if the germline is required for the longevity of wildtype descendents of mutants of ASH-2 complex members, we used pgl-1(bn101) temperature-sensitive mutants that cannot form a functioning germline at the restrictive temperature25 (Fig. 4c, d). F3 generation pgl-1 descendents from wdr-5;pgl-1 mutant parents no longer had an extended lifespan compared to pgl-1 descendents from pgl-1 parents at the restrictive temperature (25°C) (Fig. 4c, d). Thus, a functioning adult germline is necessary for the long lifespan of wildtype descendents of parents with deficiencies in members of the ASH-2 complex.
Specificity of epigenetic memory of lifespan
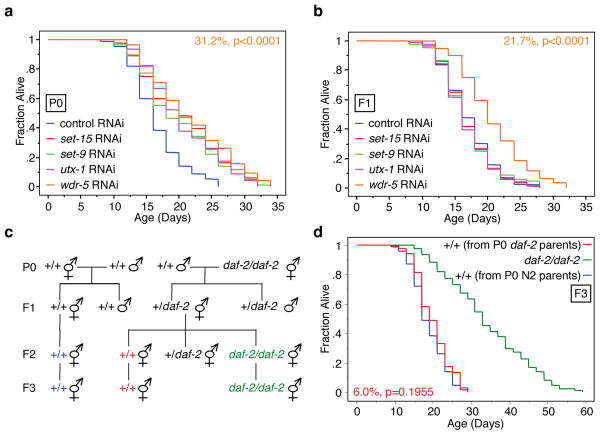
We then asked if the transgenerational inheritance of longevity is specific to H3K4me3 modifiers or if it is also observed with chromatin modifiers of other marks (set-9, set-15, and utx-1), and more generally with genes in known longevity pathways: insulin signaling (age-1 and dod-23), mitochondria (cco-1 and cyc-1), and stress resistance (asm-3)12,17,18,26–32. In contrast to what we observed for ash-2 and wdr-5, knock-down of set-9, set-15, utx-1, age-1, asm-3, cco-1, cyc-1, and dod-23 only in parents did not extend the lifespan of the F1 generation (Fig. 5a, b, Supplementary Fig. 5). Similarly, genetically wildtype F3 descendents from long-lived daf-2(e1370)33 mutant worms (+/+ from P0 daf-2 parents) had no significant extension of lifespan (6% p = 0.1955) (Fig. 5c, d). Collectively, these findings indicate that transgenerational extension of longevity is relatively specific to H3K4me3 chromatin modifiers, and further suggest that the H3K4me3 mark may be important for epigenetic memory of lifespan between generations. As SET-9, SET-15, and UTX-1, unlike members of the ASH-2 complex, regulate lifespan in a manner that is independent of the germline12,17,18, it is also possible that transgenerational inheritance of longevity is specific to chromatin regulators that act in the germline.
Fig. 5. Other longevity regulators do not have a transgenerational effect on lifespan.
a–b, Lifespan of P0 (a) and F1 (b) generation descendents from worms treated with set-9, set-15, utx-1, and wdr-5 RNAi only in the P0 generation. Control RNAi: empty vector. c, Scheme for generating wildtype progeny from daf-2(e1370) mutant worms. d, Lifespan of genetically wildtype F3 descendents from daf-2(e1370) mutant worms (+/+ from P0 daf-2 parents). Mean lifespan and statistics are presented in Supplementary Tables 2 and 4.
Transgenerational inheritance of gene expression
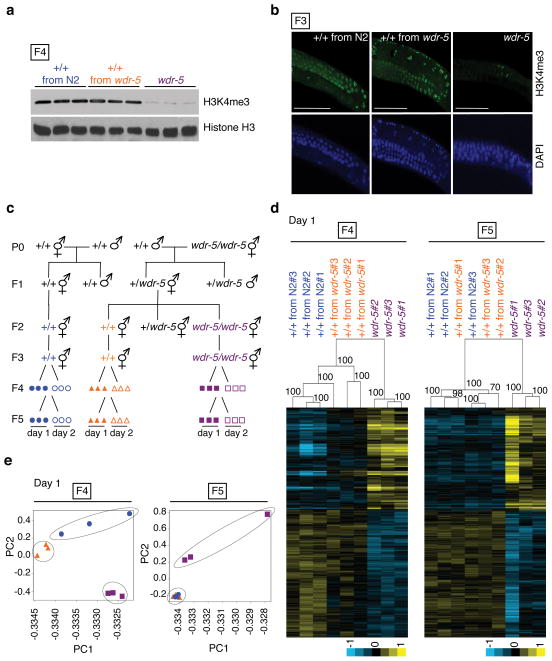
We next determined if transgenerational inheritance of lifespan was associated with heritable changes in H3K4me3. Western blot and immunocytochemistry revealed that global H3K4me3 levels were not decreased in F3 and F4 generation genetically wildtype descendents from wdr-5 and set-2 parents or in F1 and F2 generation descendents from ash-2 or wdr-5 knock-down only in parents (Fig. 6a, b, Supplementary Fig. 6, 7). Thus, transgenerational inheritance of lifespan is unlikely to be mediated by a heritable global decrease in H3K4me3 levels. Transgenerational inheritance of lifespan might be associated with heritable local changes of H3K4me3 at certain loci, which could affect expression of certain genes involved in longevity. To test this idea, we compared gene expression genome-wide in wildtype descendents from wdr-5 mutant and wildtype ancestors, and pure wdr-5 mutant descendents in the F4 and F5 generations (Fig. 6c). For each condition, we collected triplicates of L3 stage worms from the first or second day of egg-laying (Fig. 6c), with the first day of egg-laying corresponding to the samples used for lifespan assays. Statistical analysis of microarray (SAM) identified 759 genes that were differentially regulated in wdr-5 pure mutants compared to wildtype worms, regardless of the generation (Supplementary Table 7) and egg-laying day (Supplementary Fig. 8a) (p = 2.38×10−116, hypergeometric probability). These WDR-5 regulated genes are enriched for longevity, development, and growth Gene Ontology (GO) terms (Supplementary Fig. 8b), consistent with WDR-5’s reported functions12,21,22. As expected, WDR-5 regulated genes significantly overlap with ASH-2 regulated genes12 (p = 6.14×10−12, hypergeometric probability, Supplementary Fig. 8c) and are enriched for H3K4me3 (ref. 34,35) (p = 2.49×10−34, hypergeometric probability, Supplementary Fig. 8d). These observations suggest that WDR-5 functions together with ASH-2 to regulate a subset of genes by modulating H3K4me3 at these loci.
Fig. 6. Genetically wildtype descendents from wdr-5 mutant parents exhibit differences in gene expression, but not in global H3K4me3 levels, compared to descendents from wildtype parents.
a–b, Global H3K4me3 levels in the F4 generation by western blot (a) or in the F3 generation by immunocytochemistry (b) of L3 worms from genetically wildtype descendents from wdr-5 parents (+/+ from wdr-5) or wildtype parents (+/+ from N2), and wdr-5 mutants (wdr-5). Scale bars: 50 μm. c, Scheme for generating wildtype descendents from a cross between wdr-5(ok1417) null mutant worms and wildtype worms. Symbols represent RNA samples from L3 worms from 3 independent F2 ancestors on the first (closed symbols) or second (open symbols) day of egg-laying. d, Unbiased hierarchical clustering of WDR-5 regulated genes from the first day of egg-laying (Supplementary Table 9). Pvclust values are displayed on each node of the dendrogram. Values superior to 95 are considered significant. e, Principal component analysis (PCA) of the entire microarray datasets from the first day of egg-laying (Supplementary Table 5). PC: Principal component. Symbols represent gene expression data from L3 worms collected on the first day of egg-laying (Fig. 6c).
We asked if the expression of some WDR-5 regulated genes might be transgenerationally inherited. Interestingly, a significant subset of WDR-5 regulated genes was still differentially regulated in wildtype descendents from wdr-5 mutant worms in the F4 generation (Fig. 6d, Supplementary Fig. 10a), but not in the F5 generation (Fig. 6d, Supplementary Fig. 10a), consistent with the return to a normal lifespan in the F5 generation. Unbiased hierarchical clustering analysis revealed that WDR-5 regulated genes in wildtype descendents from wdr-5 mutant versus wildtype parents still clustered separately in the F4, but not the F5 generation (Fig. 6d, Supplementary Fig. 10a). Principal component analysis (PCA) confirmed that overall gene expression in wildtype descendents from wdr-5 parents versus wildtype parents is easily distinguishable in the F4, but not the F5 generation (Fig. 6e, Supplementary Fig. 10b). Genes with transgenerational inheritance of expression were slightly more enriched for H3K4me3 than expected by chance (p = 0.0123 and p = 0.0769 for the first and second day of egg-laying, respectively, hypergeometric probability), and may represent the genes that are the most affected by the loss of the H3K4me3 mark. A number of these genes are known longevity regulators and are expressed in the germline (Supplementary Table 7). GO analysis of genes with transgenerational inheritance of expression shows enrichment for different types of metabolic pathways (Supplementary Fig. 9, 10c), raising the possibility that changes in metabolism may play a role in the heritability of the phenotype. The genes with transgenerational inheritance of expression were different on the first versus second day of egg-laying, and were no longer identified when samples from different days of egg-laying were pooled (E.L.G. and A.B., data not shown). This could be because worms produced on the first day of egg-laying might be more susceptible to H3K4me3 depletion, because each collection day may represent a different snapshot in the rapidly changing L3 stage36, or because of inherent stochasticity in the transgenerational inheritance of gene expression. Overall, these results suggest that ancestral H3K4me3 status influences the gene expression of descendents for several generations.
Discussion
Our study provides the first example of epigenetic inheritance of longevity. Histone methylation marks and DNA methylation are generally, but not always, erased between generations with epigenetic reprogramming37,38. Our observations are consistent with the notion that H3K4me3 at specific loci may not be completely erased and replenished. Alternatively, the ASH-2/WDR-5/SET-2 complex could control the expression of the genes responsible for the erasure and replenishment of histone methylation marks between generations. Modulation of H3K4me3 modifiers in parents may also affect an unidentified protein or RNA that could in turn be inherited and cause lifespan changes. Interestingly, H3K4me3 regulators have been suggested to play a role in the inheritance of eye color in Drosophila5,6 and of active transcriptional states in Dictyostelium39. As the ASH-2 H3K4me3 regulatory complex is conserved from yeast to humans, manipulations of this complex in parents might have a heritable effect on longevity in mammals.
Supplementary Methods
Worm strains
wdr-5(ok1417) and set-2(ok952) strains were provided by the Caenorhabditis Genetics Center. Wildtype (N2), wdr-5(ok1417), and set-2(ok952) strains were genotyped for mut-16(mg461), a mutation that affects RNAi efficiency and that was found as an extraneous mutation in several laboratory strains. These strains did not contain the mut-16(mg461) mutation. wdr-5(ok1417) mutant worms were backcrossed four-nine times by crossing wildtype N2 males with wdr-5(ok1417) hermaphrodites. set-2(ok952) mutant worms were backcrossed four-six times by crossing wildtype N2 males with set-2(ok952) hermaphrodites. The transgenerational inheritance of longevity was similar, both in terms of magnitude and number of generation, whether wdr-5(ok1417) and set-2(ok952) worms were backcrossed four to nine times or four to six times, respectively (see Supplementary Table 1), arguing against a simple backcrossing effect to explain the increased lifespan of wildtype descendents from wdr-5 or set-2 parents. rbr-2(tm1231) mutant worms were backcrossed seven times, daf-2(e1370) mutant worms were backcrossed an additional two times by our lab, set-2(ok952) and rbr-2(tm1231) were backcrossed two times and six times respectively, before being crossed together to generate set-2(ok952);rbr-2(tm1231) and then crossed an additional time to wildtype N2 worms, wdr-5(ok1417) and pgl-1(bn101ts) were backcrossed four times each before being crossed together to generate wdr-5(ok1417);pgl-1(bn101ts) and then crossed an additional time to pgl-1(bn101ts) worms. For crosses involving set-2(ok952);rbr-2(tm1231) mutant worms, six F3 progeny were genotyped for each independent line to ensure the genotype of the second mutant loci. Temperature-sensitive fem-3(e2006) mutant worms were either maintained at 16°C or were switched to 25°C at birth and maintained at this temperature for the entirety of their lifespan. Temperature-sensitive pgl-1(bn101) and wdr-5(ok1417);pgl-1(bn101) mutant worms were either maintained at 16°C or were switched to 25°C at the L4 stage in F2 parents. F3 progeny from these worms was maintained at 16°C or 25°C for the entirety of their lifespan.
RNA interference
Adult worms were placed on NGM plates containing ampicillin (100 mg·ml−1) and IPTG (0.4mM) seeded with the respective bacteria and removed after 4–6 hours to obtain synchronized populations of worms. HT115 (DE3) bacteria transformed with vectors expressing dsRNA of the genes of interest were all obtained from the Ahringer library (a gift from M. W. Tan), except RNAi to rbr-2 that was from the Open Biosystems library (a gift from K. Shen). At the L4 stage, P0 worms were moved to NGM plates containing streptomycin (300 μg·ml−1) seeded with OP50-1 bacteria, which are streptomycin-resistant, to eliminate any potentially remaining RNAi HT115 (DE3) bacteria, which are streptomycin-sensitive. P0 worms were switched to fresh OP50-1 seeded plates every day until day 6 of life (day 2 of adulthood). Day 6 P0 worms were allowed to lay eggs for 4–6 hours and progeny from that stage were picked from these plates and their lifespan was examined. Subsequent generations were obtained by placing young adult F1, F2, F3, or F4 worms on fresh OP50-1 seeded plates for 4–6 hours. To perform RNAi in fem-3(e2006) mutant worms, one set of P0 worms was maintained at 16°C to allow them to lay eggs for the F1 generation, while a second set of P0 worms was analyzed in lifespan assays at both 16°C and 25°C. RNAi to rbr-2 was initiated at the eggs or L1 stage of F3 generation wdr-5(ok1417) mutant worms, wildtype descendents of wdr-5(ok1417) mutant worms, and wildtype descendents of wildtype parents.
Quantitative RT-PCR
Two hundred worms were picked to NGM plates with OP50-1 bacteria overnight two days in a row. Worms were then picked to NGM plates without bacteria and washed three times with M9 buffer (22 mM KH2PO4, 34 mM K2HPO4, 86 mM NaCl, 1 mM MgSO4). Worm pellets were resuspended in Trizol (Invitrogen), followed by six freeze-thaw cycles in liquid N2. One μg of total RNA was reverse transcribed with oligo dT primers using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Real time PCR was performed on a Bio-Rad iCycler or Roche LightCycler 480II using iQ SYBR green (Bio-Rad) or LightCycler480 SYBR green I master (Roche) with the following primers: pan-actin F: TCGGTATGGGACAGAAGGAC, pan-actin R: CATCCCAGTTGGTGACGATA, ash-2 F: CGATCGAAACACGGAACGA, ash-2 R: TGCCGGAATCTGCAGTTTTT, wdr-5 F: CCCTGAAACAATACACTGGACACG, wdr-5 R: AACTGGATGACAATCGGAGGC. The experiments were conducted in duplicate and the results were expressed as 2(−(target gene number of cycles pan – actin number of cycles)).
Protein analysis by western blot
Worms were synchronously grown to the L3 stage and washed off of plates with M9 buffer (22 mM KH2PO4, 34 mM K2HPO4, 86 mM NaCl, 1 mM MgSO4). Worms were washed several times in M9 buffer and snap frozen in liquid N2. Sample buffer (2.36% SDS, 9.43% glycerol, 5% β-mercaptoethanol, 0.0945 M Tris HCl pH 6.8, 0.001% bromophenol blue) was added to worm pellets and they were repeatedly snap frozen in liquid N2. Worm extracts were sonicated 3 times for 30 seconds at ~15W (VirSonic 600) and boiled for 2 minutes before being resolved on SDS-PAGE (10% or 14%) and transferred to nitrocellulose membranes. The membranes were incubated with primary antibodies (H3K4me3 (Abcam ab8580, Millipore 07–473), 1:500; H3 (Abcam ab1791), 1:1000; ASH-2 antibody50 (a gift from Dr. B. J. Meyer), 1:2000, alpha-tubulin (Sigma T9026), 1:1000), and the primary antibodies were visualized using horseradish peroxidase-conjugated anti-rabbit secondary antibody (Calbiochem 401393) and ECL Plus (Amersham Biosciences).
Whole-mount immunocytochemistry
Worms were washed several times to remove bacteria and resuspended in fixing solution (160 mM KCl, 100 mM Tris HCl pH 7.4, 40 mM NaCl, 20 mM Na2EGTA, 1 mM EDTA, 10 mM spermidine HCl, 30 mM Pipes pH 7.4, 1% Triton X-100, 50% methanol, 2% formaldehyde) and subjected to two rounds of snap freezing in liquid N2. The worms were fixed at 4°C for 30 minutes and washed briefly in T buffer (100 mM Tris HCl pH 7.4, 1 mM EDTA, 1% Triton X-100) before a 1 hour incubation in T buffer supplemented with 1% β-mercaptoethanol at 37°C. The worms were washed with borate buffer (25 mM H3BO3, 12.5 mM NaOH pH 9.5) and then incubated in borate buffer containing 10 mM DTT for 15 minutes. Worms were blocked in PBST (PBS pH 7.4, 0.5% Triton X-100, 1 mM EDTA) containing 1% BSA for 30 minutes and incubated overnight with H3K4me3 antibody (Millipore 07–473; 1:100) and with Alexa Fluor 594 secondary antibody (Invitrogen; 1:25–1:100). DAPI (2 mg/ml) was added to visualize nuclei. The worms were mounted on a microscope slide and visualized using a Leica SP2 confocal system or a Zeiss Axioskop2 plus fluorescence microscope.
Single-worm genotyping
Single worms were placed in 5 μl of worm lysis buffer (50 mM KCl, 10mM Tris pH 8.3, 2.5 mM MgCl2, 0.45% NP40, 0.45% Tween-20, 0.01% gelatin (w/v) and 60 mg/ml proteinase K), and incubated at −80°C for 1 hour, 60°C for 1 hour, and then 95°C for 15 minutes. PCR reactions were performed using the following primers: set-2 F: TGAAAGGATGATACTCGTGGGC, set-2 R: CGATGAGAGAAAGGGGATTTTGTAAC, wdr-5 F: TTGTGTGTTCGCTGTGCATG, wdr-5 R: GTATTTGCTCTCGGTCGATC, mut-16 F: AATATTCGATCGGCAAGCAG, mut-16 R: CCCGCCGATACAGAAACTAA, rbr-2 F: CAAGTGTCGTGTGATGCTGTGG, rbr-2 R: TGGCGATTGGAAACTCCGAG, pgl-1 F: TGATGTGATTGCCGAGGAACAC, pgl-1 R; GCTGAAGAAGACTGAAGACGCTAAG, daf-2 F: ACCTGGAGTCGCTCAAGTTTTG daf-2 R: TGCTTCGCTTTCATCGGTGTC PCR reactions were performed according to the manufacturer’s protocol (Qiagen) and PCR reactions were resolved on agarose gels. daf-2 PCR products were digested with BlpI to distinguish between wildtype and daf-2(e1370) genotypes. pgl-1 PCR products were digested with MseI to distinguish between wildtype and pgl-1(bn101) genotypes.
Microarray analysis
Total RNA was isolated using an RNAqueous kit (Ambion). Microarray hybridization was performed at the Stanford Protein and Nucleic Acid facility with oligonucleotide arrays (Affymetrix, GeneChip C. elegans Genome Arrays). The raw unfiltered microarray results are deposited at the Gene Expression Omnibus (GEO) under the Subseries entry GSE31043. Background adjustment and normalization was performed with RMA (Robust Multiarray Analysis). Two-class unpaired analysis in significance analysis of microarrays (SAM)51 was performed with 100 permutations and a 1×106 seed for the random number generator and a 5% false discovery rate (FDR) to compare gene expression in wdr-5 mutants and wildtype descendents from wildtype parents. To obtain a 5% FDR, a 1.06 delta value was used for samples collected at the first day of egg-laying (day 1) and a 0.93 delta value for samples collected at the second day of egg-laying (day 2). Significantly changed probes from these two lists were then used to compare wildtype descendents from wildtype parents to wildtype descendents from wdr-5 parents in each generation using a 5% FDR. To obtain a 5% FDR, a 0.66 delta value was used for the F4 generation at day 1, a 0.09 delta value was used for the F5 generation at day 1, a 0.92 delta value was used for the F4 generation at day 2, and a 0.61 delta value was used for the F5 generation at day 2. Similar results for wildtype descendents from wdr-5 parents compared to wildtype descendents from wildtype parents were observed when SAM was performed with the entire normalized lists of genes.
Hierarchical clustering
A complete linkage hierarchical clustering on the subset of WDR-5 regulated genes for each day (Supplementary Tables 9 and 10) was performed using Gene Cluster 3.0. Clustering results were analyzed further with Java Treeview. Further statistical analysis was performed using Pvclust52. For the clustering analysis, genes and then arrays were centered using the mean. The R package ‘Pvclust’ was used to apply complete linkage hierarchical clustering. As the data were centered, the uncentered Pearson correlation coefficient was used as a similarity measure, which was subsequently modified to dissimilarity by subtracting from 1. Experiments were conducted with 1000 bootstrap replications.
Principal component analysis
Principal component analysis (PCA)53 was conducted on the entire normalized lists of genes (Supplementary Tables 5 and 6). The data were scaled to obtain unit variance before conducting the PCA analysis. The Prcomp function in the R package ‘Stats’ was used. The first and the second principal components (PC1 and PC2) were plotted.
H3K4me3 ChIP-chip dataset from ModENCODE and comparison between datasets
The H3K4me3 ChIP-chip dataset was generated by the modENCODE consortium from worms at the L3 stage34,35. The data, protocols, and antibody information can be accessed at the modENCODE Data Coordination Center (http://intermine.modencode.org), accession ID 3550. Use of this dataset during the publication moratorium period was approved (S. Strome, personal communication). The raw unfiltered ChIP-chip data are deposited at GEO under the Subseries entry GSE30789. H3K4me3 ChIP intensity signals were divided by Input signals, log transformed, centered to mean zero, and scaled to standard deviation one. H3K4me3 enrichment peaks (4493) were called using the program ChIPOTle (54, http://sourceforge.net/projects/chipotle-2/) with a p-value cut-off of 10−20, window size 500 bp, step size 100 bp, and the Bonferroni p-value correction. A list of gene coordinates (transcript start-end) was obtained from WormBase WS170 (http://www.wormbase.org/). Peaks were mapped to 5062 genes by identifying the genes that had peaks overlap with their 5′ region (500 bp upstream and downstream from the transcript start site). For comparisons between different datasets, hypergeometric probabilities were calculated using http://stattrek.com/Tables/Hypergeometric.aspx.
Supplementary Material
Acknowledgments
We are grateful to J. Lieb, A. Rechtsteiner, and S. Strome for sharing their ModENCODE data pre-publication and for helpful discussion. We thank K. Shen, M.W. Tan, and T. Stiernagle and the Caenorhabditis Genetics Center for gifts of strains and reagents. We thank B. Meyer for her generous gift of the ASH-2 antibody. We thank A. Fire, S. Kim, J. Sage, S. Iwase, J. Lipsick, L. Pollina, A. Villeneuve, and members of the Brunet lab for helpful discussions and critical reading of the manuscript. We thank S. Han for screening different H3K4me3 antibodies for western blots in worm extracts. We thank R. Liefke and H. Tang for help with microarray analysis. This work was supported by NIH R01-AG31198 grant and by a generous gift from the Glenn Foundation for Medical Research to A.B.. E.L.G. was supported by an NSF graduate fellowship, by NIH ARRA-AG31198, by T32-CA009361, by a Helen Hay Whitney Post-Doctoral fellowship, and by a NIH R01-GM058012 (to Y.S.). T.J.M. was supported by NIH F32-AG037254. J.P.L. was supported by NIH T32-MH020016.
Footnotes
Author Contributions
E.L.G. conceived and planned the study with the help of A.B. E.L.G. performed the experiments and wrote the paper with the help of A.B.. E.L.G. performed some of the experiments in the lab of Y.S.. T.J.M. performed immunocytochemistry experiments (Fig. 6b, Supplementary Fig. 6c, 7c), D.U. performed Pvclust and PCA microarray analysis (Fig. 6d, e, Supplementary Fig. 10a, b). A.G.H. helped with Fig. 3b, 3c, Fig. 6a, Supplementary Fig. 6a, b, and Supplementary Fig. 7a, b. E.M. performed an independent repeat of the transgenerational wdr-5 RNAi longevity experiments (Supplementary Table 2). J.P.L. helped with Fig. 3c and Supplementary Fig. 7a, b. B.A.B. helped with bioinformatics analysis (Supplementary Table 7). All authors discussed the results and commented on the manuscript.
References
- 1.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–61. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 2.Brink RA. A Genetic Change Associated with the R Locus in Maize Which Is Directed and Potentially Reversible. Genetics. 1956;41:872–89. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodhouse MR, Freeling M, Lisch D. Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 2006;4:e339. doi: 10.1371/journal.pbio.0040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–20. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–18. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 6.Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–8. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- 7.Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of Stress-Induced, ATF-2-Dependent Epigenetic Change. Cell. 2011;145:1049–61. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 8.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–8. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 9.Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006;2:e49. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–74. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 11.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–7. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–7. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siebold AP, et al. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci U S A. 2010;107:169–74. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McColl G, et al. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem. 2008;283:350–7. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, et al. The conserved NAD(H)-dependent corepressor CTBP-1 regulates Caenorhabditis elegans life span. Proc Natl Acad Sci U S A. 2009;106:1496–501. doi: 10.1073/pnas.0802674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi Y, et al. Asymmetric arginine dimethylation determines life span in C. elegans by regulating forkhead transcription factor DAF-16. Cell Metab. 2011;13:505–16. doi: 10.1016/j.cmet.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00738.x. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin C, et al. Histone Demethylase UTX-1 Regulates C. elegans Life Span by Targeting the Insulin/IGF-1 Signaling Pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–4. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 20.Simonet T, Dulermo R, Schott S, Palladino F. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev Biol. 2007;312:367–83. doi: 10.1016/j.ydbio.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 21.Xiao Y, et al. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc Natl Acad Sci U S A. 2011;108:8305–10. doi: 10.1073/pnas.1019290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Kelly WG. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet. 2011;7:e1001349. doi: 10.1371/journal.pgen.1001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 24.Haag ES, Wang S, Kimble J. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr Biol. 2002;12:2035–41. doi: 10.1016/s0960-9822(02)01333-7. [DOI] [PubMed] [Google Scholar]
- 25.Kawasaki I, et al. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–45. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- 26.Fisher K, Southall SM, Wilson JR, Poulin GB. Methylation and demethylation activities of a C. elegans MLL-like complex attenuate RAS signalling. Dev Biol. 2010;341:142–53. doi: 10.1016/j.ydbio.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 27.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton B, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–55. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, Sun H. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell. 2007;6:489–503. doi: 10.1111/j.1474-9726.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–8. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 31.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 32.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–83. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 33.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 34.Liu T, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–36. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–87. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spencer WC, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–41. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–20. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–72. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Muramoto T, Muller I, Thomas G, Melvin A, Chubb JR. Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr Biol. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 40.Greer EL, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–56. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furuhashi H, et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–51. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 44.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–5. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 45.Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–68. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 46.Kaati G, Bygren LO, Pembrey M, Sjostrom M. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15:784–90. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- 47.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–96. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benyshek DC, Johnston CS, Martin JF. Glucose metabolism is altered in the adequately-nourished grand-offspring (F3 generation) of rats malnourished during gestation and perinatal life. Diabetologia. 2006;49:1117–9. doi: 10.1007/s00125-006-0196-5. [DOI] [PubMed] [Google Scholar]
- 49.Ng SF, et al. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–6. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 50.Pferdehirt RR, Kruesi WS, Meyer BJ. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 2011;25:499–515. doi: 10.1101/gad.2016011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 53.Pearson K. On lines and planes of closest fit to systems of points in space. Phil Mag Series. 1901;6:559–72. [Google Scholar]
- 54.Buck MJ, Nobel AB, Lieb JD. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6:R97. doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.