Abstract
Members of the APOBEC protein family catalyze DNA cytosine deamination and underpin a variety of immune defenses. For instance, several family members, including APOBEC3B, elicit strong retrotransposon and retrovirus restriction activities. However, unlike the other proteins, APOBEC3B is the only family member with steady-state nuclear localization. Here we show that APOBEC3B nuclear import is an active process requiring at least one amino acid (Val54) within an N-terminal motif analogous to the nuclear localization determinant of the antibody gene diversification enzyme AID. Mechanistic conservation with AID is further suggested by APOBEC3B’s capacity to interact with the same subset of importin proteins. Despite these mechanistic similarities, enforced APOBEC3B expression cannot substitute for AID-dependent antibody gene diversification by class switch recombination. Regulatory differences between APOBEC3B and AID are also visible during cell cycle progression. Our studies suggest that the present day APOBEC3B enzyme retained the nuclear import mechanism of an ancestral AID protein during the expansion of the APOBEC3 locus in primates. Our studies also highlight the likelihood that, after nuclear import, specialized mechanisms exist to guide these enzymes to their respective physiological substrates and prevent gratuitous chromosomal DNA damage.
Keywords: antibody diversification, DNA cytosine deamination, retroelement restriction, active nuclear import
Introduction
Human cells can express up to eleven distinct polynucleotide cytosine deaminases (reviewed in1,2). AID, APOBEC1, APOBEC3A, APOBEC3B (A3B), APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G (A3G), and APOBEC3H have demonstrated DNA deaminase activity in one or more biochemical or biological assays (reviewed in1). Two other family members, APOBEC2 and APOBEC4, do not appear to have DNA editing activity3–5. Genomic cytosine to uracil deamination is potentially dangerous because uracil lesions can template the insertion of adenines and cause C/G-to-T/A transition mutations and, depending on DNA repair fidelity, other types of base substitutions, DNA breaks, and larger-scale chromosome aberrations (reviewed in6–8).
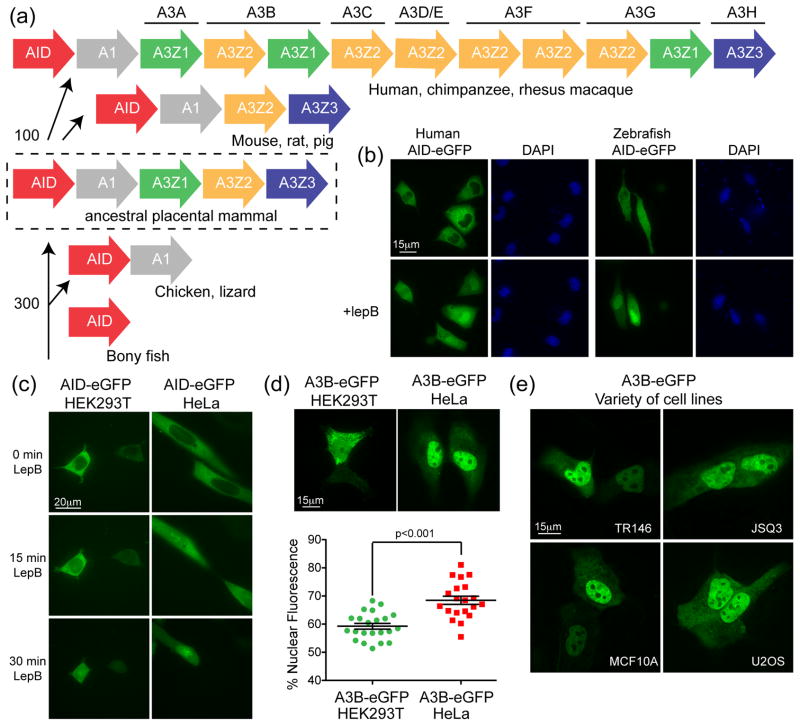
AID is arguably the most ancient family member because all vertebrates use it for adaptive immunity through antibody gene diversification (reviewed in1,9,10) (Fig. 1a). APOBEC1 appears to have evolved more recently, most likely from the duplication of an ancestral AID gene prior to the split of species such as birds and lizards from the vertebrate phylogenetic tree11,12 (Fig. 1a). In most species, the AID and APOBEC1 genes are located adjacent to one another on the same chromosome. Current models posit that these genes provided the substrate to root the APOBEC3 locus prior to the radiation of placental mammals, most likely by another ancestral gene duplication event2,13,14 (Fig. 1a). Thus, despite this considerable evolutionary gap, their common origin suggests that some present day APOBEC3 protein activities and/or regulatory programs may still be conserved with those of AID (in addition to their shared DNA deaminase activity). Indeed we and others have seen that localization through CRM1 mediated nuclear export is a conserved property of AID from a variety of different species15–19 (Fig. 1b).
Figure 1. Relationships between AID and A3B.
(a) Phylogenetic tree depicting the APOBEC loci in the indicated species. The split between fish and birds (~300 million years ago) and the divergence of the original placental mammal (~100 million years ago) are shown11,12,14. (b) Representative images of HeLa cells transfected with human or zebrafish AID-eGFP and treated with lepB or ethanol as a vehicle control. (c) Representative images of HEK293T or HeLa cells expressing human AID-eGFP after treatment for the indicated times with lepB. (d) Representative images of HEK293T or HeLa cells expressing A3B-eGFP (quantified below; mean and SD shown for >20 individual cell measurements). (e) Representative images of A3B-eGFP expressed in the buccal tumor epithelial line TR146, the squamous cell carcinoma line JSQ3, the breast epithelial line MCF10A, and the osteosarcoma line U2OS.
The physiological functions of AID in antibody diversification and APOBEC1 in APOB mRNA editing necessitate the transport of these enzymes to the nuclear compartment (reviewed in1,20). Despite <30 kDa protein sizes, which should allow for passive diffusion through nuclear pores, both enzymes are imported actively21–23. At least for AID, import is accomplished through a non-canonical nuclear localization signal (NLS) interacting with the adaptor importins α1, α3, and α5 in concert with the import factor β123. AID import may also be affected by interactions with other proteins such as GANP and CTNNBL1 (reviewed in24). Similarly, APOBEC1 binds with at least one other cellular protein, ACF, to enter the nuclear compartment and access APOB mRNA substrates25–28. However, at steady state, both AID and APOBEC1 appear cytoplasmic due to a strong C-terminal leucine-rich nuclear export signal (NES)15,16,29. This NES directs export through the CRM1 pathway, which can be blocked by incubating cells with leptomycin B (lepB) (Fig. 1b)30.
Curiously, A3B is the only human DNA deaminase family member with an apparent steady-state nuclear localization31–38. This property is also evident in rhesus macaque A3B38. Although human A3B has a putative positively charged NLS spanning residues 206–212, mutagenesis studies showed that these residues are dispensable36. However, individual domain as well as A3B/G chimera analyses were able to implicate an N-terminal region spanning residues 1–60 in nuclear import31,36,37.
Here we find that A3B actively imports into the nucleus through a non-canonical N-terminal NLS within the same region as the non-canonical NLS of AID. Additionally, both A3B and AID interact with adaptor importin proteins. Despite previous work showing that these enzymes can both inhibit retrotransposon replication31–33,39–41, other aspects of the regulation and function of A3B and AID have clearly bifurcated. For instance, we show that A3B and AID localize differently during mitosis. In addition, over-expression of A3B in primary B cells does not confer an ability to perform class switch recombination, and, unlike A3B, AID demonstrates little or no ability to restrict HIV-1 replication in single-cycle infectivity assays. Since A3B appears to have constitutive access to genomic DNA, and it is actively shuttled to the nuclear compartment, understanding additional levels of A3B regulation will be important for determining whether in certain circumstances it is able, like AID, to mutate genomic DNA and contribute to carcinogenesis.
Results
Relationship between A3B and AID
Consistent with a prior report23, we noticed that AID-eGFP has an import delay in HEK293T cells compared to HeLa cells after lepB treatment (see 15 minutes time-point in Fig. 1c). Similarly, we noticed less nuclear A3B-eGFP in HEK293T in comparison to HeLa cells, an average of 60% vs 70% of measurable fluorescent signal, respectively (Fig. 1d). A3B-eGFP displayed a HeLa-like nuclear concentration in a wide variety of other cell types (e.g., Fig 1e and additional data not shown). This activity was independent of the epitope tag and imaging procedure, as similar results were obtained for A3B-HA in fixed cells (Supp Fig. 1). These observations suggested that HEK293T cells have an import defect and provided correlative support for the hypothesis that A3B and AID share a common import mechanism.
A3B is actively imported into the nucleus of plasma membrane-permeabilized cells
The nuclear pore complex (NPC) allows objects smaller than 50 kDa to enter into the nucleus via passive diffusion, but passage is also influenced strongly by protein shape42,43. Native A3B is approximately 45.9 kDa, but the GFP tag brings the total mass to approximately 70 kDa. Since we see A3B-eGFP in the nuclear compartment, we would predict A3B to enter the nucleus through active import. However, occasionally proteins larger than 50 kDa are able to enter the nucleus through passive transport43,44. To address this possibility, we used a permeabilization assay in which the detergent digitonin permeabilizes the plasma membrane while leaving the nuclear membrane and pores intact44,45. Then, the original cytoplasm is washed away and the cellular import machinery can be added back with the addition of any cargos, such as a fluorescently tagged protein of interest.
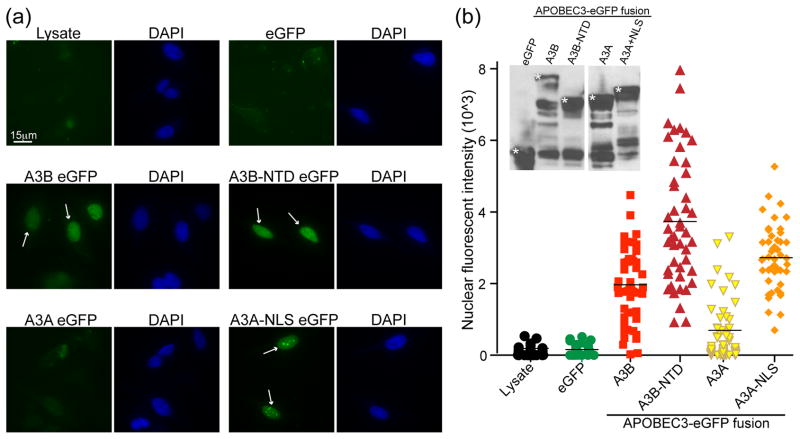
Using this assay, we found that full-length A3B-eGFP is imported within 20–30 minutes into the nuclear compartment, as is the N-terminal half of the protein (A3B NTD-eGFP) (Fig. 2a, quantified in 2b). In contrast, APOBEC3A-eGFP and eGFP, which are cell-wide31,46, were only found in nuclei at very low levels (barely distinguishable from non-fluorescent HeLa lysate) consistent with a passive diffusion mechanism (Fig. 2a, 2b). As a positive control, fusing the SV40 NLS45,47,48 to APOBEC3A-eGFP allowed the protein to concentrate in the nucleus (Fig. 2a, 2b). These data demonstrate an active nuclear import mechanism for A3B-eGFP.
Figure 2. A3B is actively imported into the nucleus.
(a) Representative images of digitonin treated HeLa cells incubated with lysates containing GFP, A3B-eGFP, A3B NTD-eGFP, A3A-eGFP, or A3A NLS-eGFP. White arrows highlight instances of active nuclear import. (b) Quantification of the results from (a) using the same exposure for all conditions (FITC=1 second; n≥30 cells were examined and the mean nuclear fluorescent intensity is indicated for each condition). The inset is an anti-GFP immunoblot of representative cell lysates with asterisks indicating the correct bands.
A3B and AID localize differently during the cell cycle
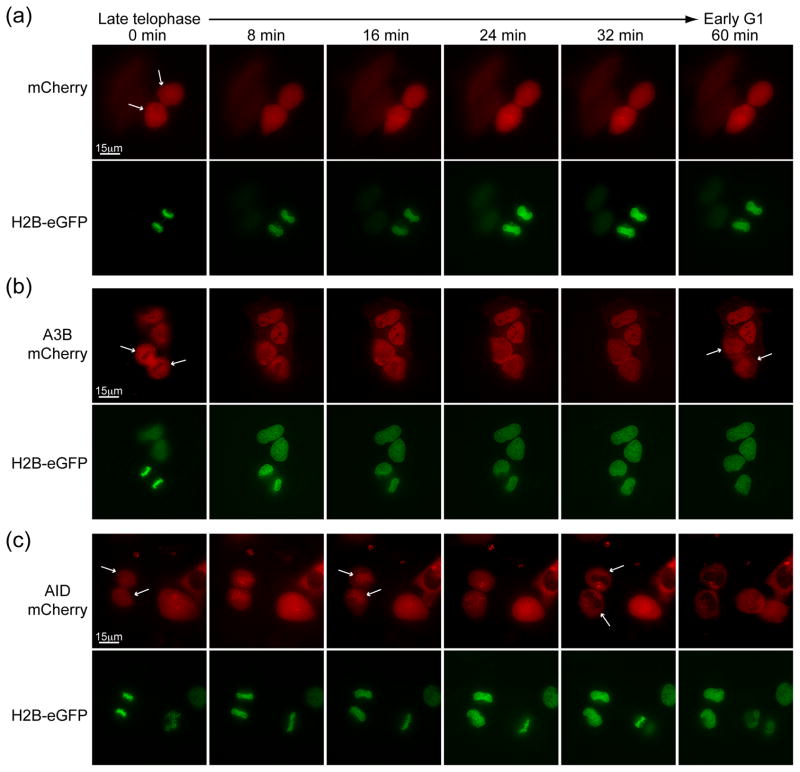
Human cells undergo open mitosis with nuclear envelope breakdown during prophase and reformation after telophase. We hypothesized that both A3B-mCherry and AID-mCherry would become cell wide upon breakdown of the nuclear envelope and return to their respective subcellular locations after telophase. Cell-wide proteins with no regulatory signals, such as mCherry, have an unaltered distribution throughout the cell cycle. In comparison, histone2B-eGFP (H2B-eGFP) elicits the expected strong but broad nuclear localization during G1 and a concentrated chromosomal distribution through mitosis (Fig. 3a and Supp. Movies 1–2).
Figure 3. A3B and AID localize differently during the mitosis.
Representative image frames of (a) mCherry, (b) A3B-mCherry, and (c) AID-mCherry localization during HeLa cell cycle progression from late telophase of mitosis to early interphase. H2B-eGFP images of the same cells are shown below each time series. The white arrows at 0 min highlight informative cells, and the arrows at other time-points indicate significant localization events discussed in the text.
Interestingly, during mitosis, although A3B-mCherry appears to shift as expected to a more cell-wide localization, it also seems to be excluded from metaphase chromosomes marked by H2B-eGFP. However, this localization phenotype only persists until ~10 minutes after telophase when A3B-mCherry rapidly (~20–30 minutes) returns to its characteristic nuclear localization (Fig. 3b and Supp. Movies 3–4). This rapid re-entry is further consistent with an active import mechanism. These data also demonstrate that A3B is not becoming trapped (and retained) with nuclear constituents as the nuclear membrane reforms after mitosis. AID-mCherry also shifts as expected to a more cell-wide localization at the early stages of mitosis. However, in contrast to A3B-mCherry, it appears to co-localize with H2B-eGFP labeled DNA during anaphase and telophase, and it remains concentrated there for ~20 minutes before rapidly shuttling to the cytoplasm (compare 24–32 minute images in Fig. 3c and Supp. Movies 5–6). These data support the conclusion that A3B is imported actively, and they also provide new visual evidence for differential regulation of A3B and AID during the cell cycle.
APOBEC3B and AID depend on the same predicted β2 region for nuclear import
Active nuclear import implies the existence of an NLS that directly or indirectly binds an importin. AID, APOBEC1, and A3B have putative classical localization signals that have been shown to be irrelevant for localization (although the putative NLS in AID is more controversial), implying that the localization determinant for each of these enzymes is non-classical or indirect15,16,22,23,36,49. Since the initial 60 residues are responsible for the nuclear localization of A3B36, we mutated residues in this region to those of the related, but cytoplasmic, A3G protein. However, no single mutational swap affected the localization of A3B NTD-eGFP (Supp. Fig. 2).
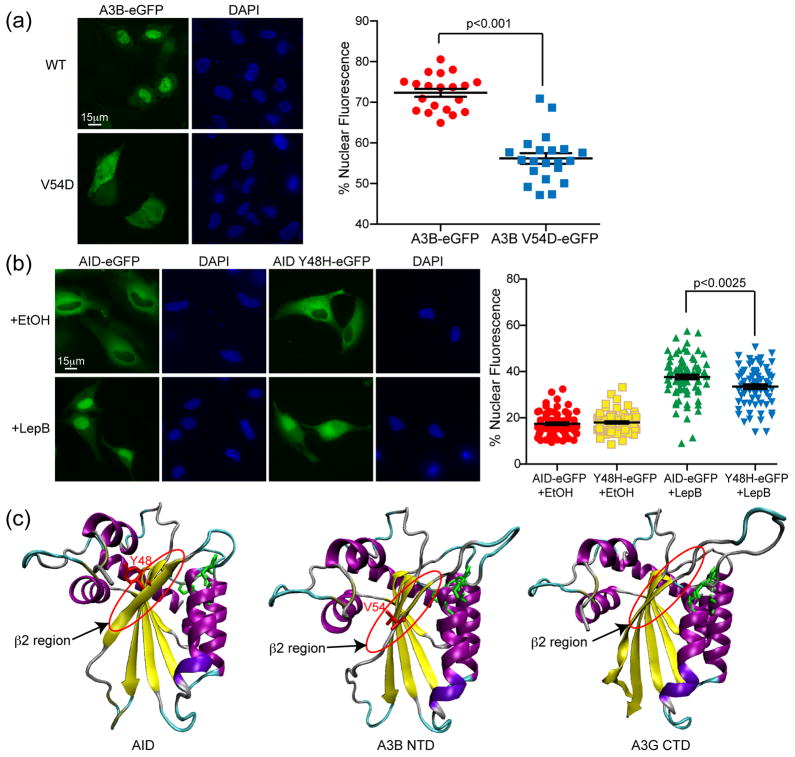
Our second strategy for identifying residues involved in A3B nuclear import was through comparison with AID. Amino acids 40–53 of AID have been implicated in nuclear import23. Focusing on the homologous region in A3B, we discovered that the single amino acid substitution mutant, valine (V) 54 to aspartic acid (D), caused full-length A3B-eGFP to become cell-wide (Fig. 4a). Mutation of A3B V54 to D also diminished the nuclear import of full-length A3B-eGFP in the digitonin import assay (Supp. Fig. 3). The homologous residue in AID, tyrosine 48, when mutated to histidine, modestly decreased the efficiency of nuclear import (Fig. 4b). This region of AID was predicted to form a β-strand (β2) when modeled using the recent A3G catalytic domain structure (pdb 3IR2)50–52 (Fig. 4c). Modeling of A3B using the same methodology predicted a similar β2 region on the external surface of the protein (Fig. 4c). The residue identified in A3B is consistent with previous reports on the importance of the N-terminal region for nuclear localization of A3B31,36 and complements what we know about the localization of AID. We conclude that the same structured region of AID and A3B, despite differences in primary amino acid sequence, is involved in nuclear import.
Figure 4. Single amino acid changes within the β2 region affects nuclear localization of A3B and AID.
(a) Representative images of A3B-eGFP and A3B V54D-eGFP in HeLa cells. The adjacent dot plot reports quantification of the nuclear to total fluorescent signal (n≥25 cells were analyzed for each condition with mean and SD shown). (b) Representative images of AID-eGFP and AID Y48H-eGFP in HeLa cells taken 3 hrs after ethanol or lepB treatment. The adjacent dot plot reports quantification of the nuclear to total fluorescent signal (n≥50 cells were analyzed for each condition with mean and SD shown). Model ribbon structures of AID and A3B NTD depicted adjacent to the actual structure of A3G C-terminal domain (CTD). The β2 region and key amino acids in AID and A3B are labeled in red and circled; the side chains of zinc-coordinating residues are illustrated in green.
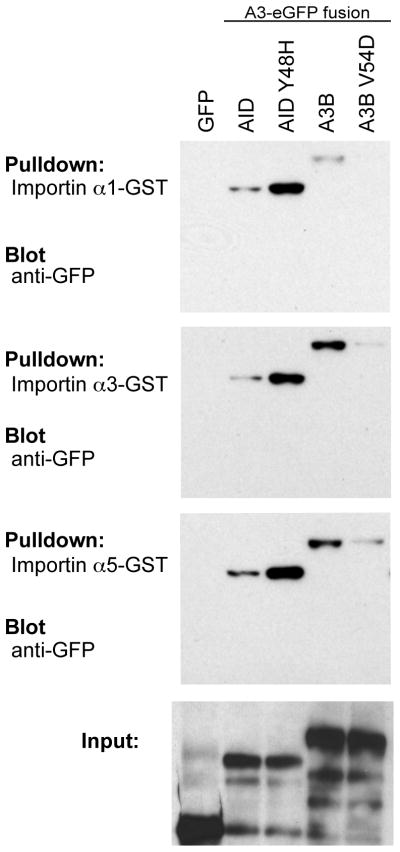
APOBEC3B can interact with the adaptor importins α1, α3 and α5
The classical pathway for nuclear import is driven by adaptor import proteins (reviewed in42,53–55). These proteins interact with the NLS as well as with karyopherin (importin β1) and the entire complex crosses through the nuclear pore using a GTP energy gradient. Because AID has been reported to interact with three of the adaptor importins, we tested these known interactors for interaction with A3B, with the hypothesis that A3B might have the same binding preferences. Indeed, pull-down experiments with GST-tagged adaptor importins indicate that A3B can also interact with importin α1, α3 and α5 (Fig. 5). As expected based on the localization defect described above, A3B V54D weakened these interactions (Fig. 5). As previously reported23, wild-type AID bound these same importins, but we were surprised to find that AID Y48H bound more strongly (Fig. 5). It is possible that the differential impact of the single amino acid substitution mutations may relate to interaction strength, with an optimal intermediate affinity being required for import and higher or lower affinities compromising this process. Nevertheless, despite this incongruence, these results show that both AID and A3B can form complexes with adaptor importins α1, α3 and/or α5.
Figure 5. Both AID and A3B interact with members of the adaptor importin family.
Immunoblots of input HEK293T protein lysates (bottom panel) and pulldown results for eGFP, AID-eGFP, AID Y48H-eGFP, A3B-eGFP and A3B V54D-eGFP with the indicated GST-importin substrates (top panels).
AID and A3B are functionally divergent for class switch recombination and HIV restriction
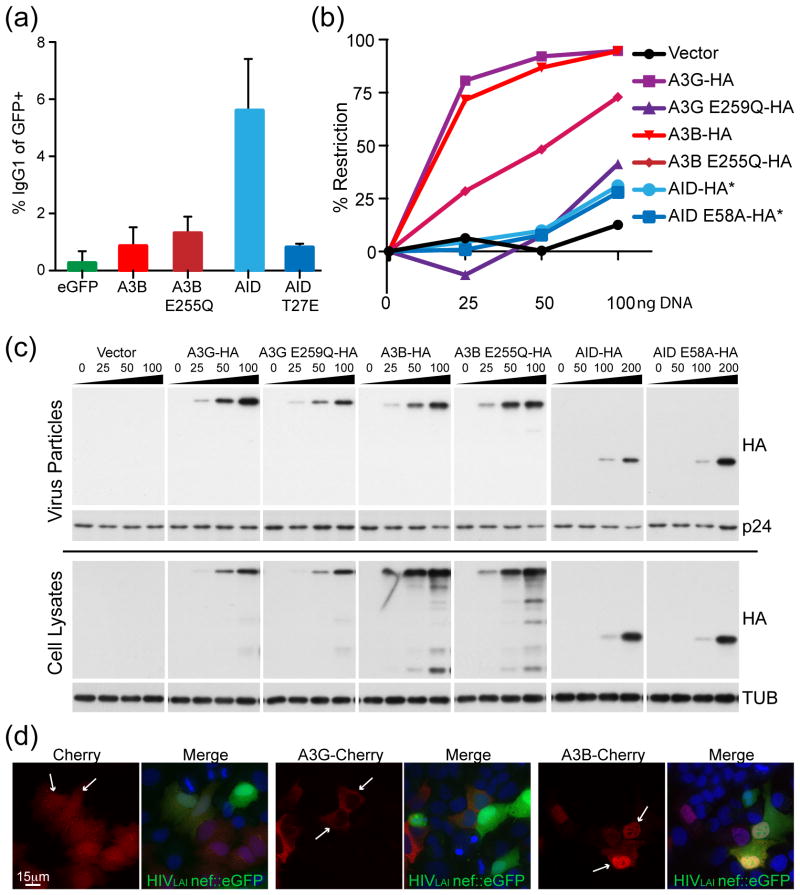
Since AID performs its predominant physiological function in the nucleus, we asked whether A3B could substitute for AID in promoting class switch recombination. Specifically, we tested whether AID-deficient mouse B cells, which are unable to switch antibody type, could be supplemented with A3B and convert from IgM to IgG56. In this assay, only the positive control, catalytically active AID, was able to affect the switch from IgM to IgG (Fig. 6a). Catalytically inactive AID (T27E), wild-type A3B and catalytically inactive A3B (E255Q) were all unable to promote isotype switching (Fig. 6a). We conclude that A3B’s ability to enter the nucleus, even via a similar pathway as AID, does not enable it to perform class switch recombination.
Figure 6. A3B does not perform class switch recombination and AID does not restrict HIV-1.
(a) Flow cytometry quantification of the isotype switch to IgG in AID-deficient murine B cells expressing AID, A3B or non-functional mutants (mean and SD are shown for duplicate samples). (b) Infectivity of HIV-1 produced in HeLa cells expressing HA tagged APOBEC3G (A3G), A3B, AID and their catalytic mutants. High levels of restriction correspond to lower levels of fluorescence in a reporter CEM-GFP cell line and a decrease in infectious virus. Asterisks indicate that twice as much DNA was required for adequate expression of AID (0, 50, 100 and 200 ng). (c) Immunoblots of viral particle proteins (top) or cell lysate proteins (bottom) probed for anti-HA (APOBEC/AID expression), anti-p24 for a virus loading control, or anti-tubulin for a cellular loading control. (d) Representative images of mCherry, A3B-mCherry or AID-mCherry in HeLa cells 24 hrs after infection with replication competent HIV-1LAI nef::eGFP. Cells expressing the protein of interest infected with HIV are indicated by white arrows.
To do the reciprocal experiment, we asked whether AID has the capacity to restrict HIV-1. AID has been previously tested for the capacity to restrict HIV57,58, but not in a dose-response study in which expression levels range. In comparison to A3B and A3G, which strongly restrict Vif-deficient HIV-1 in single-cycle infection assays, AID showed little activity despite having the capacity to package into viral particles (Fig. 6b & c). At low expression levels the catalytic glutamate of A3G (E259) was required, consistent with prior work59,60 (reviewed in61), whereas the analogous residue in A3B (E255) was only partly required (consistent with the latter enzyme having two active sites62). We also found that AID was a poor LINE1 (L1) inhibitor in comparison to A3B (Supp. Fig. 4). We conclude that AID does not possess a strong capacity to restrict Vif-deficient HIV-1 or L1, but note that this does not exclude it from possible roles in restricting the replication of other types of retroelement substrates, as suggested previously40,63,64.
One prior report indicated that HIV-1 infection, and specifically expression of the viral protein Vif, caused re-localization of several APOBEC3 proteins, including A3B65. In a reconstruction experiment, we infected A3B- or A3G-mCherry expressing cells with replication competent HIVLAI (Nef replaced by eGFP). No changes in localization were observed for mCherry, A3G-mCherry, or A3B-mCherry in HIV-1 infected (GFP-positive) cells (Fig. 6d and data not shown). Thus, HIV-1 does not appear to grossly alter the localization phenotypes described here, and we cannot readily explain why our studies diverge from the prior report that tried to address this question.
Discussion
AID is a well-studied enzyme with important functions in the adaptive immune response. A3B has not been as extensively studied because it is specific to primates (there are no exact homologs of any specific APOBEC3 in mice2), and it is more difficult to work with (it is lethal to E. coli33). To address the hypothesis that A3B and AID are regulated in the same way, we confirmed that A3B is generally nuclear, and we extended present knowledge by showing that A3B is actively imported into the nucleus through a predicted β2 region and that it has the capacity to interact with importins a1, a3, and a5. Why A3B is targeted specifically and constitutively to the nucleus is an avenue of current research.
While human AID mutations result in severe immune deficiencies 66–68, A3B is deleted in certain global populations with no obvious phenotypes (e.g., ~93% of Polynesians harbor homozygous A3B deletion alleles69). It is not surprising to find that A3B was unable to perform class switch recombination in AID deficient murine B cells. However, this was somewhat disappointing because we could envisage that forced expression of A3B in B cells might have resulted in some lesions in the antibody locus that allow for class switch recombination. The inability of A3B to perform class switch recombination echoes the failure of APOBEC1 to perform class switch recombination in AID-deficient B cells70,71 and it is unlikely that any family member, apart from AID itself, will have this capability.
We also found evidence for differential regulation of A3B and AID during the cell cycle. While A3B does not associate with H2B-labeled DNA during mitosis (particularly in late telophase), AID is cell-wide during division and appears to concentrate transiently with H2B-labeled DNA shortly after telophase and before being re-exported. We propose that A3B and AID interact with different proteins during mitosis. Several interactors of AID23,72–75 and A3B76,77 have been reported and one or more these proteins may contribute to the new localization phenotypes described here (Fig. 3 and Supp. Movies 1–6). In conclusion, our studies have demonstrated several similarities between A3B and AID with respect to nuclear import, but they have also highlighted several differences in regulation and function. Understanding the regulation of A3B, including its subcellular localization, is important for knowing how the cell regulates A3B’s cytosine deaminase activity - either for its benefit or detriment.
Materials and Methods
Constructs
Several constructs used in this paper have been reported previously including A3B-eGFP, N-terminal A3B-eGFP, AID-eGFP, zebrafish AID-eGFP, and MLV AID viral constructs 36,38,40,78. The pmCherry vectors used for time-lapse microscopy and the HIV localization experiment are direct derivatives of these constructs. The eGFP-tagged histone2B construct was a gift from J. Mueller. The GST-tagged importin α1, α3 and α5 constructs were gifts from J. Di Noia lab23. The HIVLAI nef::eGFP construct was a gift from M. Stevenson. The A3A NLS construct was made with an NLS encoding primer set that annealed to create a BsrGI site. This double-stranded oligo was inserted into the BsrGI site at the 3′ end the GFP gene of A3A-GFP (Fwd 5′-GTACAAGGATCCAAAAAAGAAGAGAAAGGTAGATCCAAAAAAGAAGAGAAAGGTA GATCCAAAAAAGAAGAGAAAGGTACT-3′ and Rev 5′-GTACAGTACCTTTCTCTTCTTTTTTGGATCTACCTTTCTCTTCTTTTTTGGATCTACCTT TCTCTTCTTTTTTGGATCCTT-3′). Site-directed mutagenesis was used to create Y48H in AID and V54D in A3B. (A3B V54D Fwd 5′-TGGGACACAGGGGACTTTCGAGGCCAG-3′ and Rev 5′-CTGGCCTCGAAAGTCCCCTGTGTCCCA-3′) (AID Y48H Fwd 5′-CTGGACTTTGGTCATCTTCGCAATAAG-3′ and Rev 5′-CTTATTGCGAAGATGACCAAAGTCCAG-3′). The A3B transducing virus was inserted into the AID MLV transducing virus backbone containing an IRES GFP and MLV long terminal repeats, but because A3B restricts production of its own virus, its coding sequence was disrupted with an inverted β-globin intron just before the second catalytic site (Fwd 5′-NNNNGTCGACATGAATCCACAGATCAGAA-3′ and Rev 5′-CCTTCTTCTCTTTCCTACAGgacccaggtgccattgtcc -3′, Fwd 5′-CTGTAGGAAAGAGAAGAA-3′, Rev 5′-GCCCATGTGCTGGTCCATCAGGTGAGTTTGGGGACCCTTG-3′ and Fwd 5′-GGGCTTTCTATGCAACGAGGCTAAGAATCTTCTCTG -3′, Rev 5′-NNNNACGCGTCTACTTGTACAGCTCGTCCAT-3′). The catalytic mutant was made by site directed mutagenesis (Fwd 5′-GCCGCCATGCGCAGCTGCGCTTC-3′, Rev 5′-GAAGCGCAGCTGCGCATGGCGGC-3′).
Cell culture
All cells except the MCF10A cell-line were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (HyClone). MCF10A cells were grown in MEBM base media with 100 ng/mL cholera toxin and MEGM additives except for gentamicin (Lonza). HeLa, HEK293T and MCF10A cells were obtained from the ATCC. U2OS cells were a kind gift from J. Mueller and the JSQ3 and TR146 lines were a gift from M. Herzberg.
Live cell fluorescence microscopy experiments
Microscopy experiments were performed as described36 with minor modifications. Approximately 20,000 cells of HeLa, HEK293T, U2OS, TR146, JSQ3 or MCF10A cells were plated on LabTek chambered cover glasses (Nunc), grown for 24 hours, transfected (Trans-IT LT1; Mirus) with 200 ng of DNA and incubated overnight. For live cell imaging, cells were incubated with clear DMEM containing 0.1% Hoechst dye to stain the nuclei and 10 mM HEPES, pH7.4 and kept at 37 degrees Celsius. Time lapse microscopy for cell cycle imaging was set up to take pictures at one-minute intervals using sealed chambered cover glasses with normal growth media. For nuclear import analysis of AID, AID-eGFP expressing live cells were treated with lepB (40 ng/ml) and followed over time78. Quantification of cells for lepB was performed by fixing the cells at the indicated time-points in 4% paraformaldehyde in PBS for 20 minutes before treating with 0.1% Hoechst dye in PBS. A DeltaVision deconvolution microscope (Applied Precision) at 60× or 40× magnification was used to collect all the images, and deconvolution was performed using DeltaVision softWoRx software (Applied Precision).
Quantification of imaging
Images were quantified for comparison with CellProfiler (Broad Institute)79,80. To quantify the nuclear localization of A3B-eGFP in HEK293T and HeLa cells, A3B-eGFP and A3B V54D-eGFP in HeLa cells, and AID-eGFP and AID Y48H-eGFP in HeLa cells, pipelines were set-up in CellProfiler to identify the nuclear compartment (based on Hoescht staining) and the cell, based on the GFP fluorescence. The intensity was measured in the defined nuclear and cellular compartments and the nuclear intensity divided by the total intensity. Data for analysis was collected from CellProfiler and assembled and statistics calculated with Prism 5.0 (GraphPad Software, Inc). The student t-test was used to calculate p-values where indicated. To quantify the import of eGFP tagged lysate into digitonin permeabilized cells we used CellProfiler to identify the nuclear compartment (based on Hoescht staining) and measured the intensity of fluorescence in the nucleus for each construct. This direct measurement was graphed with Prism 5.0 (GraphPad Software, Inc) and the Mann-Whitney test used to calculate statistics for A3B-eGFP and A3B V54D-eGFP.
Digitonin permeabilization import assays
Digitonin permeabilization has been previously described45 and modified as below. HeLa cells were plated onto 30 window slides at 50–150 cells/window and allowed to settle overnight (#63434-02; Electron Microscopy Sciences). The slides were incubated for five minutes on ice, washed five minutes in cold transport buffer (20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate), and then permeabilized in 70 ug/mL digitonin (EMD Chemicals, Inc) in transport buffer for five minutes. The digitonin solution was removed by washing with cold transport buffer then the import mixture was applied individually to each window. To make the lysates ~2.5 million HeLa cells were transfected with 5 ug of DNA (TransIT LT1; Mirus) and allowed to express protein for 48 hours before harvesting in 100 uL of transport buffer with protease inhibitors (Complete Protease Inhibitor Cocktail Tablets; Roche). To check for protein expression in the lysates, samples were run on acrylimide gels (BioRad), then transferred to PVDF membranes. After blocking for 1 hour (3% casein in PBS with 0.1% Tween), mouse anti-GFP (JL-8, BD Clontech 632 381) was used at a 1:2000 dilution. The membranes were incubated overnight 4 degrees C. The membranes were washed four times with PBS Tween before incubating with 1:5000 goat anti-mouse HRP (BioRad) secondary for 1 hour at room temperature. The total import mixture contained cell lysate, 0.5 mM GTP and 1 mM ATP. Each set of lysates was done in triplicate. The import mixture incubated on the slides for 20 minutes at room temperature and was washed away with cold transport buffer. The cells were fixed for 20 minutes with 4% paraformaldehyde on ice, washed with PBS, incubated for five minutes with PBS containing 0.1% Hoechst dye to stain the nuclei, and a coverslip was mounted with a 50% glycerol:PBS solution. A DeltaVision deconvolution microscope (Applied Precision) at 40× magnification was used to collect the images, using the same shutter exposure (1 second) for GFP fluorescence.
Protein modeling and alignments
The homology modeling server Swiss Model (http://swissmodel.expasy.org/) was used to generate structures of A3B and AID based off the crystal structure of the C-terminal domain of APOBEC3G (3IR2 Chain A)50–52. For A3B the QMEAN4 score was 0.52 and for AID the QMEAN4 score was 0.57 (range from 0–1, higher numbers characterize more accurate models)81. Models were loaded into the molecular visualization program, VMD82, and the selected residues highlighted. The chosen views were rendered for display. For alignment A3B sequence NCBI NP_004891.3, A3G sequence NCBI NP_068594.1, A3F sequence NCBI NP_660341.2 and AID sequence NCBI NP_065712.1 were used in a ClustalW alignment.
GST pulldown experiments
GST alone and GST-tagged importin α1, α3 and α523 were grown in BL21DE3 E. coli to OD600 0.6, induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 0.4 mM and expressed at 14 degrees for 16 hours. The cell pellet was resuspended in GST lysis buffer (25 mM HEPES pH 7.4, 10% glycerol, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM MgCl2, 1 mM ZnCl2) with protease inhibitors. The suspension was sonicated (Misonix XL-2000) three times for 2 minutes total (15 second pulses) with rests on ice. Debris was pelleted from the solution at 14,000 rpm and the supernatent was incubated with glutathione beads (Glutathione Sepharose 4 fast flow; GE Healthcare) overnight. The beads were washed four times with GST lysis buffer. HEK293T cells were transfected with 5 ug of DNA (Trans-IT LT1; Mirus) and allowed to express the GFP tagged proteins for 48 hours. These cells were harvested, lyzed and the insoluble fraction pelleted by centrifugation. Lysates were added to the glutathione beads and incubated overnight. The beads were washed six times with lysis buffer and the bound proteins eluted by boiling with SDS loading dye. Samples were run on acrylimide gels (BioRad), then transferred to PVDF membranes. After blocking for 1 hour (3% casein in PBS with 0.1% Tween), mouse anti-GFP (JL-8, BD Clontech 632 381) was used at a 1:2000 dilution. The membranes were incubated overnight 4 degrees C. The membranes were washed four times with PBS Tween before incubating with 1:5000 goat anti-mouse HRP (BioRad) secondary for 1 hour at room temperature. The blots were developed with chemiluminescent reagents A and B and film (Denville HyGlo A and B, HyBlot).
Class switch recombination assays
Both the class switch recombination assay and the AID deficient mice used to generate the B cells have been previously described56,78. All experiments followed the guidelines set forth by the University of Minnesota Animal Care and Use Committee. Briefly, B cells were purified from the mouse spleen using magnetic sorting (Miltenyi Biotech) and cultured in RPMI supplemented with 10% FBS, 50 ng/mL IL-4 and 50 ug/mL LPS. After 48 hours the cells were infected with GFP, AID, A3B and point mutant viruses, resuspended in fresh media and cultured for 4 days. Conversion of IgM to IgG was measured by flow cytometry (FACSCanto II, BD Biosciences) after staining with anti-IgG1-PE (BD Biosciences).
HIV single-cycle experiments
The single-cycle HIV assay and specific antibodies used for analysis have been described38 with the only modification being the use of HeLa cells. Briefly, either HEK293T or HeLa cells were transfected with 1 ug of the HIV 1IIIB A200C proviral construct and 0, 25, 50 or 100 ng of HA tagged APOBEC3 construct. Twice as much AID-HA (50, 100, 200 ng) was required for similar expression levels. The supernatents from the transfected cells were added to the reporter cell line, CEM GFP, and these cells were subsequently analyzed by flow cytometry to quantify viral titers. Cell and viral particle lysates were prepared and immunoblotted as previously reported38.
HIV Infection and fluorescent microscopy studies
HeLa cells transfected with mCherry, A3G-mCherry and A3B-mCherry were seeded into 6 well plates on coverslips and allowed to adhere overnight. HIVLAI nef::eGFP was added to half the samples and both treated and untreated cells were fixed with 4% PFA for 20 minutes in PBS at 0, 24 and 48 hours time-points. The nuclei were stained with Hoescht dye (0.1% in PBS). The coverslips were mounted in 50% glycerol in PBS and imaged on a DeltaVision deconvolution microscope (Applied Precision) at 40× magnification. Deconvolution was performed using DeltaVision softWoRx software (Applied Precision).
Supplemental Methods
Immunofluorescence experiments
Approximately 20,000 cells of HeLa, HEK293T, U2OS, TR146, JSQ3 or MCF10A cells were plated on LabTek chambered cover glasses (Nunc) and transfected with A3B-HA and incubated overnight. Cells were washed with PBS, then fixed with 4% paraformaldehyde in PBS for 20 minutes at room temperature. Cells were washed with PBS, incubated in blocking buffer for 1 hour (1% BSA, 1% goat serum and 0.2% Triton-X 100 in PBS) then stained with 1:200 mouse anti-HA 11 (Covance MMS-101P) overnight. Cells were washed with PBS five times then incubated with 1:200 goat anti-mouse FITC (Jackson 115095146) for 1 hour. After washing four times cells were incubated in PBS containing 0.1% Hoechst dye to stain the nuclei. A DeltaVision deconvolution microscope (Applied Precision) at 60× magnification was used to collect the images, and deconvolution was performed using DeltaVision softWoRx software (Applied Precision).
LINE1 studies
L1 assays were carried as previous described33. Briefly, a LINE1 construct containing eGFP with an inverted intron was transfected into HEK293T cells along with HA-tagged constructs of interest. All constructs were tested in triplicate. After 48 hours the cells were tested for eGFP expression by flow cytometry and then selected with 0.8 ug/mL of puromycin. Flow cytometry was performed at the indicated time-points. Western blots were performed from pooled cell lysates collected at 48 hours and probed with anti-HA and anti-tubulin antibodies.
Supplementary Material
Supplemental Figure 1. A3B-HA is localized to the nucleus. Representative images showing A3B-HA localization in HeLa, HEK293T, the head and neck cancer cell lines, TR146 and JSQ3, the breast epithelial cell line, MCF10A, and the osteosarcoma cell line, U2OS.
Supplemental Figure 2. Single point mutants in the N-terminal domain of A3B do not affect its localization. (a) Representative images of A3B NTD-eGFP and point mutants as indicated below the image. (b) Alignment of A3B with A3G, APOBEC3F (A3F) and AID. Both A3G and A3F are cytoplasmic proteins. Differences between A3B, A3G/A3F and AID are indicated with gray shading. The red stars indicate mutants shown in (a).
Supplemental Figure 3. A3B V54D-eGFP does not enter the nucleus after digitonin treatment. (a) Representative images of digitonin treated HeLa cells incubated with lysates containing eGFP, A3B-eGFP, or A3B V54D-eGFP. (b) Quantification of the results from (a) using the same exposure for all conditions (FITC=1 second; n≥25 cells were examined and the mean nuclear fluorescent intensity is indicated for each condition). (c) Anti-GFP immunoblot of cell lysates with asterisks indicating protein expression in the lysate concentration used for quantification.
Supplemental Figure 4. A3B restricts L1 transposition in a deaminase independent manner while AID is a poor restrictor. (a) Flow cytometry quantification of eGFP expressing cells as a marker for retrotransposition in APOBEC/AID expressing populations (mean and SD shown for triplicate experiments). Measurements are shown for vector (black, circles), A3A-HA (yellow, triangles), A3A E72A-HA (orange, triangles), A3B-HA (red, downward triangles), A3B E255Q-HA (maroon, diamonds), AID-HA (blue, squares), and AID E58A-HA (light blue, triangles).(b) Anti-HA immunoblot of cell lysates indicating expression of the APOBEC3/AID proteins with anti-tubulin as a loading control.
Movies showing mCherry fluorescence (a) or mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated. Frames from this series and a secondary series (not shown) were used to compose Fig. 3a.
Movies showing mCherry fluorescence (a) or mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated.
Movies showing A3B-mCherry fluorescence (a) or A3B-mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated. Frames from this series were used to compose Fig. 3b.
Movies showing A3B-mCherry fluorescence (a) or A3B-mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated.
Movies showing AID-mCherry fluorescence (a) or AID-mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated. Frames from this series and a secondary series (not shown) were used to compose Fig. 3c.
Movies showing AID-mCherry fluorescence (a) or AID-mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated.
Highlights.
APOBEC3B is imported actively into the nucleus.
APOBEC3B and AID can interact with several adaptor importin proteins.
APOBEC3B cannot substitute for AID in antibody gene diversification.
Despite some similarities, APOBEC3B and AID have distinct biological functions.
Acknowledgments
We thank J. Di Noia, M. Herzberg, J. Mueller and M. Stevenson for reagents, R. LaRue for the phylogenetic schematic, and J. Lee for technical assistance. This work was supported by NIH R01 AI064046 and P01 GM091743. L. Lackey was supported in part by an NSF Pre-doctoral Fellowship and subsequently by a position on the Institute for Molecular Virology Training Grant NIH T32 AI083196. Z.L. Demorest was supported in part by NIH T32 AI007313. A.M. Land was supported by a CIHR Postdoctoral Fellowship. J.F. Hultquist was supported by an NSF Predoctoral Fellowship.
Glossary The abbreviations used are
- AID
activation induced cytosine deaminase
- APOBEC
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like
- A3
APOBEC3
- A3B
APOBEC3B
- A3G
APOBEC3G
- CTD
C-terminal domain
- eGFP
enhanced green fluorescent protein
- D
aspartic acid
- H2B
histone 2B
- HIV-1
human immunodeficiency virus, type 1
- kDa
kilodalton
- lepB
leptomycin B
- L1
LINE1, long interspersed elements
- NTD
N-terminal domain
- NES
nuclear export signal
- NLS
nuclear localization signal
- mRNA
messenger RNA
- V
val, valine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaRue RS, Andrésdóttir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jónsson SR, Landau NR, Löchelt M, Malik HS, Malim MH, Münk C, O’Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–7. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao W, Hong SH, Chan BH, Rudolph FB, Clark SC, Chan L. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem Biophys Res Commun. 1999;260:398–404. doi: 10.1006/bbrc.1999.0925. [DOI] [PubMed] [Google Scholar]
- 4.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–5. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 5.Vonica A, Rosa A, Arduini BL, Brivanlou AH. APOBEC2, a selective inhibitor of TGFβ signaling, regulates left-right axis specification during early embryogenesis. Dev Biol. 2011;350:13–23. doi: 10.1016/j.ydbio.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–12. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 7.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–81. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Unniraman S, Schatz DG. AID and Igh switch region-Myc chromosomal translocations. DNA Repair (Amst) 2006;5:1259–64. doi: 10.1016/j.dnarep.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 9.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 10.Hirano M, Das S, Guo P, Cooper MD. The evolution of adaptive immunity in vertebrates. Adv Immunol. 2011;109:125–57. doi: 10.1016/B978-0-12-387664-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 11.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–77. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 12.Severi F, Chicca A, Conticello SG. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol Biol Evol. 2010;28:1125–9. doi: 10.1093/molbev/msq338. [DOI] [PubMed] [Google Scholar]
- 13.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 14.LaRue RS, Jónsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, Hötzel I, Andrésdóttir V, Smith TP, Harris RS. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–44. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–80. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichikawa HT, Sowden MP, Torelli AT, Bachl J, Huang P, Dance GS, Marr SH, Robert J, Wedekind JE, Smith HC, Bottaro A. Structural phylogenetic analysis of activation-induced deaminase function. J Immunol. 2006;177:355–61. doi: 10.4049/jimmunol.177.1.355. [DOI] [PubMed] [Google Scholar]
- 18.Wakae K, Magor BG, Saunders H, Nagaoka H, Kawamura A, Kinoshita K, Honjo T, Muramatsu M. Evolution of class switch recombination function in fish activation-induced cytidine deaminase, AID. Int Immunol. 2006;18:41–7. doi: 10.1093/intimm/dxh347. [DOI] [PubMed] [Google Scholar]
- 19.Barreto VM, Magor BG. Activation-induced cytidine deaminase structure and functions: a species comparative view. Dev Comp Immunol. 2011;35:991–1007. doi: 10.1016/j.dci.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2010;2:594–602. doi: 10.1002/wsbm.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Smith HC. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc Natl Acad Sci U S A. 1997;94:13075–80. doi: 10.1073/pnas.94.24.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Sowden MP, Smith HC. Intracellular trafficking determinants in APOBEC-1, the catalytic subunit for cytidine to uridine editing of apolipoprotein B mRNA. Exp Cell Res. 2001;267:153–64. doi: 10.1006/excr.2001.5255. [DOI] [PubMed] [Google Scholar]
- 23.Patenaude AM, Orthwein A, Hu Y, Campo VA, Kavli B, Buschiazzo A, Di Noia JM. Active nuclear import and cytoplasmic retention of activation- induced deaminase. Nat Struct Mol Biol. 2009;16:517–27. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 24.Storck S, Aoufouchi S, Weill JC, Reynaud CA. AID and partners: for better and (not) for worse. Curr Opin Immunol. 2011;23:337–44. doi: 10.1016/j.coi.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–9. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 26.Navaratnam N, Morrison JR, Bhattacharya S, Patel D, Funahashi T, Giannoni F, Teng BB, Davidson NO, Scott J. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993;268:20709–12. [PubMed] [Google Scholar]
- 27.Mehta A, Banerjee S, Driscoll DM. Apobec-1 interacts with a 65-kDa complementing protein to edit apolipoprotein-B mRNA in vitro. J Biol Chem. 1996;271:28294–9. doi: 10.1074/jbc.271.45.28294. [DOI] [PubMed] [Google Scholar]
- 28.Blanc V, Kennedy S, Davidson NO. A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor regulates nucleocytoplasmic import and shuttling. J Biol Chem. 2003;278:41198–204. doi: 10.1074/jbc.M302951200. [DOI] [PubMed] [Google Scholar]
- 29.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 30.Wolff B, Sanglier JJ, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–47. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 31.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–5. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–72. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 33.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–41. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 34.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, Candinas D, Sommer P, Wain-Hobson S, Vartanian JP, Greeve J. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–74. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 35.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–64. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J Virol. 2008;82:9591–9. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pak V, Heidecker G, Pathak VK, Derse D. The Role of Amino-Terminal Sequences in Cellular Localization and Antiviral Activity of APOBEC3B. J Virol. 2011;85:8538–47. doi: 10.1128/JVI.02645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–34. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacDuff DA, Demorest ZL, Harris RS. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 2009;37:1854–67. doi: 10.1093/nar/gkp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem. 2011 doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasiorowski JZ, Dean DA. Mechanisms of nuclear transport and interventions. Adv Drug Deliv Rev. 2003;55:703–16. doi: 10.1016/s0169-409x(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 43.Mohr D, Frey S, Fischer T, Güttler T, Görlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–53. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribbeck K, Görlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–30. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–16. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–5. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 47.Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Görlich D. Dominant-negative mutants of importinβ block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–63. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodward CL, Prakobwanakit S, Mosessian S, Chow SA. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J Virol. 2009;83:6522–33. doi: 10.1128/JVI.02061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoufouchi S, Faili A, Zober C, D’Orlando O, Weller S, Weill JC, Reynaud CA. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–68. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 51.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 52.Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, Kono T, Matsuo H, Harris RS, Somasundaran M, Schiffer CA. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–60. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 54.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–14. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 55.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–98. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 56.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 57.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–6. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 59.Schumacher AJ, Haché G, MacDuff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–60. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–21. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wissing S, Galloway NL, Greene WC. HIV-1 Vif versus the APOBEC3 cytidine deaminases: an intracellular duel between pathogen and host restriction factors. Mol Aspects Med. 2010;31:383–97. doi: 10.1016/j.mam.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bogerd HP, Wiegand HL, Doehle BP, Cullen BR. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364:486–93. doi: 10.1016/j.virol.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Macduff DA, Harris RS. Directed DNA deamination by AID/APOBEC3 in immunity. Curr Biol. 2006;16:R186–9. doi: 10.1016/j.cub.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 64.Rosenberg BR, Papavasiliou FN. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection. Adv Immunol. 2007;94:215–44. doi: 10.1016/S0065-2776(06)94007-3. [DOI] [PubMed] [Google Scholar]
- 65.Marin M, Golem S, Rose KM, Kozak SL, Kabat D. Human immunodeficiency virus type 1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J Virol. 2008;82:987–98. doi: 10.1128/JVI.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 67.Quartier P, Bustamante J, Sanal O, Plebani A, Debré M, Deville A, Litzman J, Levy J, Fermand JP, Lane P, Horneff G, Aksu G, Yalcin I, Davies G, Tezcan I, Ersoy F, Catalan N, Imai K, Fischer A, Durandy A. Clinical, immunologic and genetic analysis of 29 patients with autosomal recessive hyper-IgM syndrome due to Activation-Induced Cytidine Deaminase deficiency. Clin Immunol. 2004;110:22–9. doi: 10.1016/j.clim.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Imai K, Zhu Y, Revy P, Morio T, Mizutani S, Fischer A, Nonoyama S, Durandy A. Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2. Clin Immunol. 2005;115:277–85. doi: 10.1016/j.clim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 69.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eto T, Kinoshita K, Yoshikawa K, Muramatsu M, Honjo T. RNA-editing cytidine deaminase Apobec-1 is unable to induce somatic hypermutation in mammalian cells. Proc Natl Acad Sci U S A. 2003;100:12895–8. doi: 10.1073/pnas.2135587100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krause K, Marcu KB, Greeve J. The cytidine deaminases AID and APOBEC-1 exhibit distinct functional properties in a novel yeast selectable system. Mol Immunol. 2006;43:295–307. doi: 10.1016/j.molimm.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conticello SG, Ganesh K, Xue K, Lu M, Rada C, Neuberger MS. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell. 2008;31:474–84. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 73.Han L, Masani S, Yu K. Cutting edge: CTNNBL1 is dispensable for Ig class switch recombination. J Immunol. 2010;185:1379–81. doi: 10.4049/jimmunol.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ganesh K, Adam S, Taylor B, Simpson P, Rada C, Neuberger M. CTNNBL1 is a novel nuclear localization sequence-binding protein that recognizes RNA-splicing factors CDC5L and Prp31. J Biol Chem. 2011;286:17091–102. doi: 10.1074/jbc.M110.208769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maeda K, Singh SK, Eda K, Kitabatake M, Pham P, Goodman MF, Sakaguchi N. GANP-mediated recruitment of activation-induced cytidine deaminase to cell nuclei and to immunoglobulin variable region DNA. J Biol Chem. 2010;285:23945–53. doi: 10.1074/jbc.M110.131441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang W, Zhang X, Tian C, Wang T, Sarkis PT, Fang Y, Zheng S, Yu XF, Xu R. Cytidine deaminase APOBEC3B interacts with heterogeneous nuclear ribonucleoprotein K and suppresses hepatitis B virus expression. Cell Microbiol. 2008;10:112–21. doi: 10.1111/j.1462-5822.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 77.Chen Z, Eggerman TL, Bocharov AV, Baranova IN, Vishnyakova TG, Kurlander RJ, Csako G, Patterson AP. Hypermutation of ApoB mRNA by Rat APOBEC-1 Overexpression Mimics APOBEC-3 Hypermutation. J Mol Biol. 2012 Feb 9; doi: 10.1016/j.jmb.2012.02.005. http://dx.doi.org/10.1016/j.jmb.2012.02.005. [DOI] [PMC free article] [PubMed]
- 78.Demorest ZL, Li M, Harris RS. Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. J Biol Chem. 2011;286:26568–75. doi: 10.1074/jbc.M111.235721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–80. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–50. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. A3B-HA is localized to the nucleus. Representative images showing A3B-HA localization in HeLa, HEK293T, the head and neck cancer cell lines, TR146 and JSQ3, the breast epithelial cell line, MCF10A, and the osteosarcoma cell line, U2OS.
Supplemental Figure 2. Single point mutants in the N-terminal domain of A3B do not affect its localization. (a) Representative images of A3B NTD-eGFP and point mutants as indicated below the image. (b) Alignment of A3B with A3G, APOBEC3F (A3F) and AID. Both A3G and A3F are cytoplasmic proteins. Differences between A3B, A3G/A3F and AID are indicated with gray shading. The red stars indicate mutants shown in (a).
Supplemental Figure 3. A3B V54D-eGFP does not enter the nucleus after digitonin treatment. (a) Representative images of digitonin treated HeLa cells incubated with lysates containing eGFP, A3B-eGFP, or A3B V54D-eGFP. (b) Quantification of the results from (a) using the same exposure for all conditions (FITC=1 second; n≥25 cells were examined and the mean nuclear fluorescent intensity is indicated for each condition). (c) Anti-GFP immunoblot of cell lysates with asterisks indicating protein expression in the lysate concentration used for quantification.
Supplemental Figure 4. A3B restricts L1 transposition in a deaminase independent manner while AID is a poor restrictor. (a) Flow cytometry quantification of eGFP expressing cells as a marker for retrotransposition in APOBEC/AID expressing populations (mean and SD shown for triplicate experiments). Measurements are shown for vector (black, circles), A3A-HA (yellow, triangles), A3A E72A-HA (orange, triangles), A3B-HA (red, downward triangles), A3B E255Q-HA (maroon, diamonds), AID-HA (blue, squares), and AID E58A-HA (light blue, triangles).(b) Anti-HA immunoblot of cell lysates indicating expression of the APOBEC3/AID proteins with anti-tubulin as a loading control.
Movies showing mCherry fluorescence (a) or mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated. Frames from this series and a secondary series (not shown) were used to compose Fig. 3a.
Movies showing mCherry fluorescence (a) or mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated.
Movies showing A3B-mCherry fluorescence (a) or A3B-mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated. Frames from this series were used to compose Fig. 3b.
Movies showing A3B-mCherry fluorescence (a) or A3B-mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated.
Movies showing AID-mCherry fluorescence (a) or AID-mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated. Frames from this series and a secondary series (not shown) were used to compose Fig. 3c.
Movies showing AID-mCherry fluorescence (a) or AID-mCherry overlaid with H2B-eGFP fluorescence (b) during time-lapse microscopy. The time frames and scale are indicated.