Abstract
Synapses are specialized adhesive contacts characteristic of many types of cell-cell interactions involving neurons, immune cells, epithelial cells, and even pathogens and host cells. Cell-cell adhesion is mediated by structurally diverse classes of cell-surface glycoproteins, which form homophilic or heterophilic interactions across the intercellular space. Adhesion proteins bind to a cytoplasmic network of scaffolding proteins, regulators of the actin cytoskeleton, and signal transduction pathways that control the structural and functional organization of synapses. The themes of this review are to compare the organization of synapses in different cell types and to understand how different classes of cell adhesion proteins and cytoplasmic protein networks specify the assembly of functionally distinct synapses in different cell contexts.
Keywords: adhesion protein, cytoskeleton, epithelial cells, host-pathogen, immune cells, neuron, signaling
INTRODUCTION
Cell-cell adhesion is a fundamental characteristic of multicellular organisms. During development, specific adhesion between distinct cell types is required for the correct organization of cells into patterns that give rise to different organs and tissues. In the adult, cell-cell adhesion not only maintains the structural and functional integrity of those organs and tissues, but it must be sufficiently dynamic to allow the formation of new cell interactions and the remodeling of old ones.
In specialized cell contexts, cell-cell adhesion is synonymous with a “synapse,” a word coined in 1897 by the English physiologist and 1932 Nobel Laureate Sir Charles Sherrington from the Greek word “sunaptein” meaning to fasten together (“sun-” together, and “haptein” to fasten or bind) (for a historical perspective, and quotes see Reference 1). Sherrington and his colleagues used “synapse” to describe adhesions between neurons, although the etymology of the word implies a more general cellular context as discussed here.
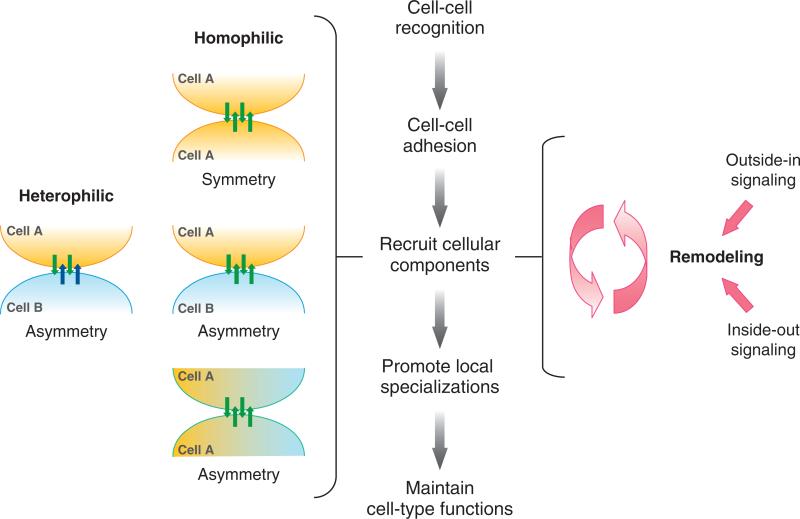
Correct interactions between cells to form synapses require cell-cell recognition by adhesion proteins, which leads to cell sorting as some cells adhere to each other and others are excluded. Because proper cell organizations arise from these specific cell interactions, cell adhesion proteins likely play instructive roles in promoting local specifications of the synapse that impact overall cellular functions (outside-in signaling) (Figure 1). Heterophilic adhesion on opposing cells potentially recruits different cytoplasmic signaling proteins to each membrane to generate structural and functional asymmetry at the synapse. Homophilic adhesion potentially recruits the same cytoplasmic signaling proteins to opposing membranes to generate a symmetric synapse, unless different sets of binding proteins were expressed in each of the opposing cells, resulting in an asymmetric synapse. In some instances, a synapse may establish cell polarity in a different cell axis (e.g., the apicobasal axis) (Figure 1). The recruitment of cytoplasmic signaling proteins provides an intracellular cue to modulate adhesive properties of the synapse (inside-out signaling) (Figure 1) and to propagate broader changes in cell structure and function. Finally, the formation of specific cell-cell interactions must be dynamic because many contacts are capable of undergoing reorganization in response to a variety of physiological stimuli (Figure 1).
Figure 1.
Schematic representation of sequential events following cell recognition and adhesion that promote local specialization of the synapse. (left) Heterophilic adhesion between different cells (A, B), expressing different adhesion proteins (blue and green arrows), leads to the formation of an asymmetric synapse; homophilic adhesion between the same cell types (A, A), expressing the same adhesion proteins (green arrows), leads to formation of a symmetric synapse and either a symmetric organization of cells (top), an asymmetric organization of the same cells (A, A) in the apicobasal cell axis (middle), or an asymmetric organization between two different cells (A, B). (right) The organization of adhesion proteins and cytoplasmic signaling complexes can be remodeled by either outside-in or inside-out signaling.
Here, we examine synapse organization in four major cell types as follows:
The classic neuronal synapse, an asymmetric structure that facilitates transduction of an action potential between cells;
The dynamic adhesion between leukocytes and endothelial cells in response to inflammation and the “immunologic synapse,” an asymmetric structure between T cells and antigen-presenting cells (APCs) that activates cytokine secretion from the T cell;
The epithelial synapse, a symmetric structure that nevertheless generates structural and functional asymmetry across the epithelium;
The pathogen-host cell synapse, an asymmetric structure between pathogen and host cell adhesion complexes.
The overall theme is to understand how similar classes of cell adhesion proteins and cytoplasmic protein networks assemble functionally distinct synapses.
THE NEURONAL SYNAPSE
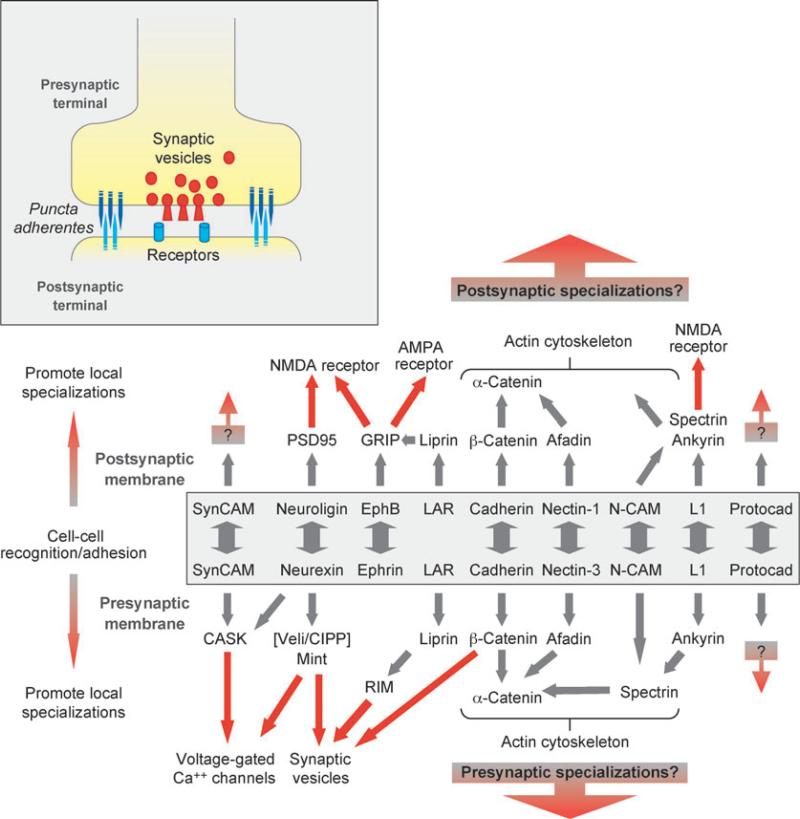
The neuronal synapse comprises an active zone formed between closely opposed membranes of the axon and dendrite, surrounded by an intercellular junctional complex termed the puncta adherentes (Figure 2, inset). The axon terminal (presynaptic membrane) is filled with neurotransmitter-containing synaptic vesicles, primed for fusion with the membrane in response to an action potential. The dendrite membrane (postsynaptic membrane) is enriched in neurotransmitter receptors, scaffolding, and signaling proteins (postsynaptic density), which respond to neurotransmitter release from the presynaptic membrane to initiate a new action potential (Figure 2, inset).
Figure 2.
Schematic representations of the neuronal synapse (inset) and protein interactions at the pre- and postsynaptic membranes. Different cell adhesion proteins form homophilic or heterophilic adhesions (boxed) and interact with downstream protein networks that describe functional specification of the presynaptic (recruit Ca2+ channels, synaptic vesicles) and postsynaptic membranes (recruit neurotransmitter receptors). Abbreviations: AMPA, α-amino-5-hydroxy-3-methyl-4-isoxazole propionic acid; CASK, calcium/calmodulin-dependent serine protein kinase; CIPP, channel-interacting PDZ domain protein; GRIP, glutamate receptor-interacting protein; LAR, leukocyte common antigen-related protein; Mint, Munc-18-interacting protein; N-CAM, neural cell adhesion molecule; NMDA, N-methyl-D-aspartic acid; PSD95, postsynaptic density 95; RIM, Rab3-interacting molecule; Veli, vertebrate LIN-7.
Proteins Involved in Cell-Cell Adhesion at the Neuronal Synapse
Several classes of adhesion proteins localize to the synapse: members of the immunoglobulin (Ig) superfamily (N-CAM, L1, nectin, SynCAM), neurexins and neuroligins, ephrin/Eph, and the cadherin family of Ca2+-dependent proteins. This subsection examines how each of these classes forms homophilic and heterophilic adhesions and how adhesion leads to cell sorting and the formation of neuronal circuits; the next subsection discusses the linkages between these adhesion proteins and cytoplasmic signaling networks that contribute to the functional organization of the pre- and postsynaptic membranes (Figure 2).
Ig superfamily
The founding member of the Ig superfamily of cell-cell adhesion proteins is N-CAM. Although earlier work focused on the role of N-CAM in cell-cell adhesion (2), it became clear that posttranslational addition of polysialic acid (PSA) to the extracellular domain of N-CAM dramatically reduced cell-cell adhesion, raising the possibility that PSA-modified N-CAM is (also) an anti-adhesion protein (3). Indeed, deletion of N-CAM or cleavage of PSA by topical addition of endosialidase N resulted in inhibition of cell migration and axon guidance as well as in disrupted synapse formation and plasticity (4, 5). It is likely that PSA addition to N-CAM physically increases the distance between opposed cells, hence sterically hindering binding between adhesion proteins and receptor-ligand complexes (3).
The L1 family (L1, neurofascin, NrCAM) forms homophilic adhesions and heterophilic adhesions with neurocan, integrins, TAG-1 and contactin (6). L1 proteins are expressed in the developing nervous system and are important in neuronal migration, axon growth, guidance and fasciculation, and synaptic plasticity (7). Genetic deletion of L1 in mice results in abnormalities in neuron morphology, axon guidance, and animal behavior (8, 9), and a number of X-linked forms of mental retardation may be caused by mutations in the L1 gene in humans (7).
SynCAMs (10) and nectins (11) are structurally related, and both are important in synapse formation and function. SynCAM forms homophilic adhesions at the synapse (10), although heterophilic interactions may also occur (12). Overexpression of SynCAM in cultured neurons increased synapse formation and function (10), but it is unclear whether these effects are due to increased recruitment of SynCAM-binding proteins (see next subsection) or SynCAM-facilitated clustering of other adhesion protein complexes. Nectins constitute a complex family of four proteins, each of which has multiple splice variants (13). Nectin subtypes are asymmetrically located at synapses (11), and deletion of one results in loss of the other from the synapse (14), indicating that heterophilic adhesion stabilizes nectin pairs (15). Deletion of either nectin-1 or -3 decreased the number of puncta adherentes at synapses (15). However, there was little effect of nectin deletion on the localization of synaptic marker proteins or the number and size of synapses (14), indicating that nectins may be important in synaptogenesis but not synapse maintenance and that other adhesion proteins are sufficient.
Neuroligin and neurexins
Neuroligin, encoded by five different genes in humans, is restricted to the postsynaptic membrane and forms heterophilic interactions with members of the neurexin family localized on the presynaptic membrane (16, 17) (Figure 2). The neurexin family is potentially large because it comprises hundreds of variants generated by alternate promoters and splicing (18). Overexpression of neuroligin increased synapse number and function (19, 20), whereas decreased expression reduced synaptic function and activity (21). The importance of neuroligin levels in synapse function may be due either to differences in the extent of cell-cell adhesion or the dosage of intracellular proteins recruited to the synapse by the neuroligin/neurexin complex (see next subsection). Significantly, contacts between neurons and heterologous cells (HEK293 cells) that expressed neuroligin or neurexin induced neuronal pre- and postsynaptic differentiation, respectively, indicating that the neuroligin/neurexin complex is instructive in generating structural and functional specializations on opposed membranes at the synapse (16, 22).
Classical cadherins
Members of the classical cadherin family of Ca2+-dependent homophilic cell adhesion proteins are single membrane-spanning proteins that contain five characteristic extracellular cadherin repeats (EC1-5) of which the EC1 domain regulates the specificity of cadherin-cadherin binding (23, 24). Cadherins are localized to puncta adherentes that surround the active zone of mature synapses (25, 26) (Figure 2, inset).
Many different cadherins are expressed in the nervous system (27–29), and they accumulate at the earliest stages of adhesion between neurons (26). Cadherin subtype expression in different neurons is gradually refined during development (30); for example, N-cadherin expression is initially uniform throughout the neuroepithelium (31) and then becomes restricted to a subset of brain nuclei, layers, and fibers and finally to specific neural circuits (32). In the adult, N-cadherin and E-cadherin are restricted to excitatory and inhibitory neurons, respectively (27), and loss of N-cadherin specifically suppresses synaptic activity of excitatory glutamatergic synapses (33).
Different cadherins appear to regulate cell sorting in the nervous system. For example, in the lateral motor column of the chick spinal cord, neurons are segregated into different motor pools, and misexpression of MN-cadherin, but not cadherin-6b, resulted in incorrect mixing of neurons in motor pools (34). Specificity for sorting appears to reside in the EC1 domain, as swapping this domain between MN-cadherin and cadherin-6b was sufficient to confer on cadherin-6b the ability to (mis-)sort neurons into motor pools similar to MN-cadherin (24). Cadherins may also facilitate sorting by regulating neuron navigation and pathfinding (35, 36). For example, N- and R-cadherin expression stimulates neurite outgrowth (37, 38), whereas cadherin-11 promotes axon elongation (39), and cadherin-13 acts as a repellant cue for growth cones (40). Because different cadherin subtypes are expressed within groups of cells in a neural circuit (25, 41), it has been suggested that each subtype may provide an “adhesion code” that recruits specific cells into each neural circuit (42).
Mutant cadherins have been expressed in neurons to test the role of cadherins in synapse assembly and function. Expression of a “dominant-interfering” mutant N-cadherin, lacking the extracellular domain, resulted in abnormal morphology and dynamics of dendritic spines as well as reduced presynaptic densities of both boutons and protein structures (43, 44). A point mutation at a site in EC1 (W2A), critical for cadherin-cadherin recognition and adhesion (23), had little effect on synapse adhesion, although there were abnormal structural responses of synapses to depolarization and repolarization (45). Function-blocking antibodies against the extracellular domain of N-cadherin (46) or cadherin-8 (47) increased the distance between membranes in the synaptic cleft and reduced synaptic function. Together, these results indicate that cadherins play roles in membrane (spine) dynamics, synaptic protein recruitment during synaptogenesis, and synaptic plasticity of mature synapses.
Protocadherins
Protocadherins (Pcdh) represent the largest subgroup of the cadherins, comprising more than 80 members arranged in three clusters in the genome termed Pcdhα, -β and -γ (30, 48). Each Pcdh protein has 6–7 extracellular cadherin (EC) domains and comprises variable and constant regions encoded by different exons (30). Pcdhγ proteins are expressed throughout development of the nervous system (49, 50). They are enriched at synapses and may be involved in synapse formation, specification, and maintenance (51, 52). Deletion of the entire Pcdhγ cluster resulted in a dramatic loss of interneurons from the spinal cord, but general alterations in axonal growth, adhesion, and migration were not observed (53); note that when the effect of Pcdhγ deficiency on apoptosis was circumvented, a decreased number and strength of both excitatory and inhibitory synapses was found (54). The lack of general effects of Pcdhγ deficiency on the nervous system may be due to compensation by other Pcdh family members. However, the localized effects of deletion of Pcdhγ on interneurons of the spinal cord indicates that expression of different members of the Pcdh family might specify both the survival and synaptic organization of neuronal subpopulations.
Other protein combinations (EphrinB/EphB, liprin/LAR)
EphrinB is a member of the Ephrin family of axon guidance proteins and binds to the EphB receptor (55). Ephrin and the EphB receptor are required for the maturation of dendritic spines, the structures that form the postsynaptic terminal; a triple knockout of EphB1, -2, and -3 resulted in defects in spine morphology (56), although the mice were viable, indicating that the effects might either be restricted to special neurons or that there is redundancy in adhesion proteins involved in synaptogenesis. Another protein that plays a central role in controlling synapse formation is liprin, a SAM domain-containing protein with a coiled-coil repeat that binds the receptor tyrosine phosphatase LAR (leukocyte common antigen-related) protein (57). Disruption of liprin function caused defects in active zone assembly and presynaptic bouton structure (58).
Linking Adhesion Proteins to the Cytoplasm and Cellular Specialization
How might combinations of these cell-cell adhesion proteins specify differences in the structural and functional organization of pre- and postsynaptic terminals? Either homophilic or heterophilic adhesion complexes could recruit protein complexes that specify various functions of the pre- and postsynaptic terminals (see Figure 1), and several adhesion proteins have been found to interact with different cytoplasmic scaffolding proteins, including the spectrin membrane skeleton, postsynaptic density 95-Discs large-ZO1 homologous domain (PDZ)-containing proteins, and a variety of proteins that interact directly or indirectly with the actin cytoskeleton (Figure 2).
The spectrin membrane skeleton is abundant in pre- and postsynaptic membranes (59). Spectrin is linked to cell-cell adhesion proteins either directly, in the case of N-CAM (60), or indirectly through ankyrinB, which binds to L1 (61) (Figure 2). Significantly, ankyrinB- and L1-deficient mice have a similar phenotype, and deletion of ankyrinB results in a concomitant loss of L1 (62). Also, in Drosophila, deletion of presynaptic, but not postsynaptic, spectrin resulted in loss of fasciclin (a member of the N-CAM family) and neuroglian (the Drosophila homolog of L1) from the synapse, and this deletion caused presynaptic membrane retraction and synapse elimination at the neuromuscular junction (63). These results indicate a role of the spectrin membrane skeleton in retention/stabilization of cell adhesion proteins at synaptic membranes. It is less clear whether ankyrin/spectrin specify different membrane organizations at the pre- and postsynaptic terminals because spectrin binds the ubiquitous actin cytoskeleton (Figure 2). However, spectrin has also been reported to interact with the C-terminal cytoplasmic domain of the NMDA-R (64), which would be a specialized interaction for the postsynaptic membrane.
The C terminus of several classes of adhesion proteins contains a PDZ-binding site for members of the PDZ/MAGUK family of scaffolding proteins, which forms links to structures (vesicles) and proteins (channels and receptors) specific to the pre- or postsynaptic membrane (65) (Figure 2). At the presynaptic membrane, the liprin/LAR complex binds RIM (66), which interacts with Rab3 bound to synaptic vesicles (67, 68), and the PDZ domain protein CASK binds SynCAM (10), neurexin (69), and voltage-gated Ca2+ channels (70) (Figure 2). At the postsynaptic membrane, the PDZ domain proteins, PSD95 (71) and GRIP (72), bind adhesion proteins and postsynaptic ion channels (Figure 3); PSD95 binds neuroligin (73, 74) and NMDA-R (75, 76); and GRIP binds ephrin receptors (77), the liprin/LAR complex (78) and AMPA receptors (72). Thus, structural and functional differences between the pre- and postsynaptic membranes correlate with the localization of various adhesion proteins, each with their own PDZ domain protein scaffold that recruits vesicles, ion channels, and receptors specific to those different membrane domains.
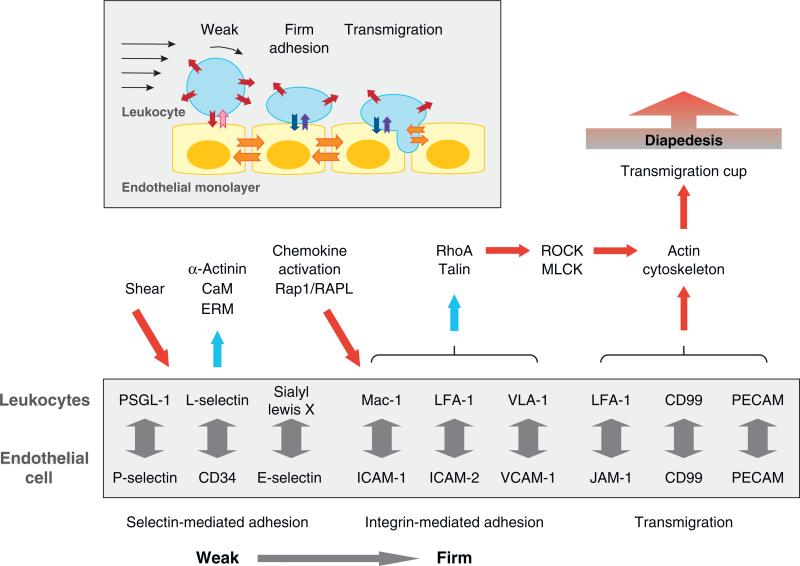
Figure 3.
Schematic representation of leukocyte-endothelial cell interactions during extravasation (inset). Selectin-mediated adhesion (red/pink arrows) initiates leukocyte rolling, and integrin-mediated adhesion (blue/purple arrows) stabilizes leukocyte adhesion to endothelial cells. Endothelial junction proteins (orange arrows) mediate leukocyte transmigration. Adhesion proteins (box) and protein networks assembled at the synapse between rolling leukocytes and endothelial cells (selectin- and integrin-mediated adhesion) as well as during leukocyte transmigration (JAM-1/LFA-1, CD99, and PECAM). Abbreviations: CaM, calmodulin; ERM, ezrin/radixin/moesin; MLCK, myosin light chain kinase; RAPL, regulator of adhesion and cell polarization enriched in lymphoid tissues; ROCK, Rho kinase.
The family of classical cadherins also binds specific cytoplasmic proteins, α- and β-catenin, which regulate cadherin function and the actin cytoskeleton (79). α-Catenin also binds afadin (80), a PDZ protein that binds nectins (81) (Figure 3). Functions of the catenins have been tested by genetic mutation and deletion and indicate involvement in the organization of the presynaptic, rather than the postsynaptic, terminal. Genetic deletion of neuronal α-catenin (α-Ncatenin) resulted in increased dendritic spine dynamics and filopodia formation but had little effect on the localization of synaptic vesicles (82). Interestingly, both the N terminus (dimerization and β-catenin-binding domain) and the C terminus (actin-binding domain) of α-Ncatenin were required to suppress spine filopodia formation. Studies in vitro showed that α-catenin homodimers inhibit actin-related protein 2 and 3 (Arp2/3)-mediated actin polymerization (83); thus deletion of α-catenin might result in activation of the Arp2/3 complex and an increase in actin dynamics and filopodia dynamics. In contrast, deletion of β-catenin had little effect on spine morphology but caused a decrease in the size of the recycling synaptic vesicle pool (84); deletion of the C-terminal PDZ-binding domain of β-catenin did not rescue the phenotype, indicating that β-catenin is required to bind PDZ scaffolding proteins rather than to link cadherin to α-catenin (84). Although it is assumed that the cadherin-catenin complex functions as a unit in cell-cell adhesion, these results indicate that each component plays a role in regulating different aspects of synapse organization and function, including dendritic spine dynamics (α-catenin), initial cell-cell adhesion (cadherin), and synaptic vesicle clustering (β-catenin). Clustering of the cadherincatenin complex upon cell-cell contacts may serve to recruit and concentrate each component for these functions at the synapse.
Changes in synaptic activity have also been shown to remodel synapse structure, which may involve the cadherin-catenin complex. Stimulation of the α-amino-5-hydroxy-3-methyl-4-isoxazole propionic acid receptor (AMPA-R) resulted in changes in spine dynamics and morphology during depolarization and repolarization (45). Changes in neuronal activity by a cAMP analog resulted in an increased number of N-cadherin puncta at synapses (85), and this affect is blocked by anti-N-cadherin antibodies (46). In general, KCl-mediated depolarization resulted in a re-distribution of N-cadherin along the expanding spine head and of β-catenin from dendritic shafts to spines as well as promotion of β-catenin binding to cadherin (86). Activation of NMDA-R (28) or addition of a Na-channel blocker (82) resulted in conversion of spines to dynamic filopodia, and this effect was inhibited by overexpression of α-catenin. Conversely, addition of the GABA-R antagonist bicuculline resulted in increased accumulation of N-cadherin, α-catenin, and β-catenin at synapses (82). These results indicate that remodeling of synaptic activity involves changes in the organization, and perhaps function, of the cadherin-catenin complex.
THE IMMUNOLOGICAL SYNAPSE
Immune cells travel throughout the blood and lymphatic systems and exit toward sites of inflammation and the lymph node (Figure 3) where APCs congregate (Figure 4). Immune cells express a wide variety of cell-surface receptors, some of which recognize antigens and, unlike other synapses, form only temporary asymmetric adhesions with target cells. Formation of these cell adhesions initiates the reorganization of membrane proteins and scaffolding networks that results in cell polarization. We discuss two examples of synapse formation in the immune system: first, a dynamic and asymmetric synapse between leukocytes and endothelial cells during extravasation (Figure 3) and, second, the formation of the immunologic synapse between T cells and APCs (Figure 4).
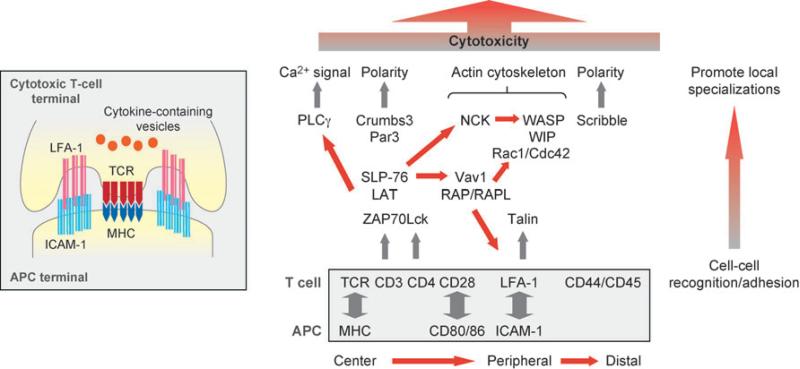
Figure 4.
Schematic representations of the immunologic synapse between T cells and antigen-presenting cells (APCs) (inset), adhesion proteins (boxed), and protein networks assembled at the synapse that remodel the T cell and initiate the immune response from the T cell. Abbreviations: ICAM-1, intercellular adhesion molecule 1; LAT, linker for activation of T cells; LFA-1, lymphocyte function-associated antigen 1; MHC, major histocompatibility complex; Par3, partition defective 3; PLCγ, phospholipase C γ; SLP-76, Src homology 2 domain-containing leukocyte-specific phosphoprotein of 76 kDa; TCR, T-cell receptor; WASP, Wiskott-Aldrich syndrome protein; WIP, WASP-interacting protein; ZAP70, zeta-chain-associated protein 70.
Crossing the Endothelial Barrier: The Leukocyte-Endothelial Synapse
In response to a foreign invasion, circulating leukocytes bind to and then migrate across the endothelium toward the site of inflammation (extravasation). This process requires sequential and dynamic cell-cell adhesion. Leukocytes initially attach to and then roll on the surface of endothelial cells via selectin-mediated adhesion and then form more firm, integrin-mediated adhesion to the apical surface of endothelial cells prior to diapedesis (87) (Figure 3, inset).
Selectins are calcium-dependent lectins that bind to glycoprotein ligands, and different subtypes are expressed on the surface of leukocytes (L-selectin) and endothelial cells (P- and E-selectin) (88). Selectin-mediated adhesion depends on the shear rate of blood flow. The number of leukocytes attached to, and rolling on, endothelial cells increases with shear force, but below a threshold rate, they no longer adhere (89). Two models have been proposed to explain shear flow-enhanced selectin adhesion. Shear flow may increase the lateral diffusion of adhesion molecules, which in turn increases the probability of bond formations that sustain the leukocyte rolling (valency enhancement) (90, 91). Alternatively, shear stress on adhesion molecules may induce conformational changes in adhesion proteins and thereby reduce their dissociation rate, leading to bond stabilization (catch bond) (92–94).
Membrane organization and cytoplasmic interactions of selectins are important for cell adhesion (Figure 3). L-selectin is concentrated at the tip of numerous microvilli on the surface of leukocytes (95, 96). A chimeric protein comprising the extracellular domain of L-selectin fused to the cytoplasmic domain of CD44, which is typically absent from microvilli, was excluded from microvilli (95) and reduced the tethering efficiency of cells (95, 97). The reduced adhesion of this L-selectin/CD44 chimera could also be due to the loss of cytoplasmic-binding partners, such as α-actinin, that interact with L-selectin (98). Indeed, a tailless L-selectin, despite its proper localization, led to reduced tethering at elevated shear stress (91, 99), indicating that binding of actin-associated proteins, and hence regulation of actin organization, are important in this process.
CD44 and integrins are also important in mediating adhesion between leukocytes and endothelial cells (Figure 3). CD44, together with selectins, mediates the initial attachment of leukocytes to the endothelial surface, which in turn promotes firm adhesion by α4β1 integrin (also termed, very late activation antigen-4, VLA-4) (100). This sequential series of adhesions is coordinated by the lateral interaction between CD44 and α4β1 integrin and requires functions provided by the CD44 cytoplasmic domain because a tailless CD44 abrogated firm adhesion of leukocytes to endothelial cells (101).
Firm adhesion between leukocytes and endothelial cells is achieved by integrin-mediated adhesion. Leukocyte integrins include αLβ2 (LFA-1), αMβ2 (Mac-1), αXβ2, α4β1 (VLA-4), and α4β7 (LPAM-1), and their ligands are immunoglobulin superfamily members (87) (Figure 3). Structural studies of integrin heterodimers have provided insights into mechanisms underlying affinity regulation of integrin adhesion (102). The predicted structure of αVβ3 integrin extra-cellular domain revealed highly flexible regions in both α- and β-subunits that form a V-shaped conformation (103). Furthermore, high-resolution electron micrographs showed that the active structure with high affinity toward ligands was extended, but the inactive structure was bent (104). Similar conformational changes were detected in live cells using fluorescence resonance energy transfer (FRET) (105). In addition, these structural studies showed close proximity of the C terminals of α- and β-subunits in the inactive state (103, 104, 106), which was also confirmed in live cells using FRET (107). Significantly, the interaction of the cytoplasmic domains could be disrupted by point mutations in the cytoplasmic domain of αIIb integrin or the talin-binding domain that activate integrins (108), and a greater separation of the cytoplasmic domains occurred upon activation of integrins by overexpressing talin or addition of chemokines (107). These results demonstrate the physiological importance in cell adhesion of inside-out and outside-in signaling mediated by conformational changes in integrins.
Leukocyte migration across the endothelial barrier, a process termed diapedesis, requires transient interactions between adhesion proteins on both cell types (Figure 3, inset). Immediately prior to diapedesis, LFA-1 integrin, on leukocytes (109) interacts with a transmigratory cup on the apical surface of endothelial cells comprised of microvilli-like projections that require RhoA activity (109, 110) and contain vascular cell adhesion molecule-1 (VCAM-1), ezrin, and intercellular adhesion molecule 1 (ICAM-1) (111). Significantly, transmigration of leukocytes does not disrupt the endothelial barrier (112), indicating that adhesion between leukocytes and endothelial cells must make a tight seal. Adhesion between endothelial cells is mediated by PECAM-1 and the junction-associated molecule (JAM) (members of Ig superfamily), VE-cadherin (a classical cadherin) and CD99 (an O-glycosylated transmembrane protein) (113). Homophilic interactions of PECAM-1, CD99, and possibly JAM between leukocytes and endothelial cells occur. Endothelial cell-cell adhesions contain numerous interdigitated membranes that are enriched with PECAM-1, which is constitutively recycled. During diapedesis, this recycling pool of PECAM-1 is targeted to the site of leukocyte-endothelial cell interactions (114). However, there appears to be redundancy in the requirement for all these adhesion proteins because leukocytes from PECAM-1-deficient mice were still recruited to the site of inflammation (115). Heterophilic interactions between the leukocyte integrin LFA-1 and the membrane proximal domain of JAM-1 on endothelial cells are also important (116). JAM-1 is dislocated from endothelial cell-cell adhesions as a part of the chemokine response (117), which may aid leukocyte diapedesis. CD99 may be involved in a late stage of diapedesis because addition of function-blocking antibodies to CD99 resulted in the arrest of monocytes between endothelial cells (118). Finally, recent studies revealed that interactions between leukocytes and the transmigratory cup can also trigger transcellular migration of lymphocytes, a process that requires caveolin and an ICAM-1-rich membrane domain that colocalizes with the pseudopodia of leukocytes (119).
T Cell–APC Adhesion: The Immunologic Synapse
When immune cells adhere to APCs or target cells, proteins on both cells orient themselves in a specific pattern toward the site of adhesion, termed the “immunological synapse” (Figure 4, inset). Although the reorganization and polarization of protein at the immunological synapse was first observed between T cells and APCs (120, 121), this process has also been demonstrated between natural killer cells and their target cells (122) as well as between B cells and APCs (123).
Detailed insight into the dynamic reorganization of proteins during assembly of the immunological synapse was provided by real-time analysis of T cells adhering to a surrogate lipid bilayer containing ICAM-1 and agonist peptides bound to a major histocompatibility complex (MHC) that mimics the surface of an APC (120). Upon T-cell attachment, LFA-1/ICAM-1 complexes cluster in the center of the adhesion, and the T-cell receptor (TCR)/MHC complexes are located at the periphery of the synapse (124). Later, TCR/MHC complexes become clustered in the center, whereas the LFA-1/ICAM-1 complexes are segregated to the periphery to form a ring around the TCR/MHC cluster (124) (Figure 4). Because the reorganization of receptors occurs between a T cell and proteins embedded in an artificial lipid bilayer, the formation and organization of the immunological synapse appears to be driven by the T cell and does not require direct input from the target cell.
Although the organization of different protein domains during immunological synapse formation is striking, the mechanisms involved are not known. The rapid recruitment of receptors to the immunological synapse is mediated by actin-driven membrane flow toward the contact site initiated by TCR binding to an agonist presented by the MHC (125). In addition, recycling endosomes, containing TCRs, are delivered to the site of immunological synapse in a soluble N-ethylmaleimide-sensitve factor attachment protein receptor (SNARE)-mediated process (126); how the SNARE complex is targeted to the immunological synapse is not known. The immunologic synapse also comprises a network of PDZ scaffolding proteins (127), including Crumbs, scribble, Dlg, and the Par complex, which could regulate vesicle trafficking and other signaling pathways important in the formation of the synapse (Figure 4). Disruption of this network prevents T-cell polarization and migration during antigen presentation (127).
Several mechanisms may be involved in partitioning these different protein complexes within the immunological synapse. Synapse formation can be viewed as a self-assembly process in which the physical properties of participating proteins drive their localization (128). Localization of the LFA-1/ICAM-1 complex at the center may occur because it is the first adhesion complex to form owing to the length of interacting extracellular domains (~42 nm). Ligation of LFA-1 to ICAM-1 might bring the two opposing membranes closer together, thereby increasing the probability of the TCR/MHC complex formation. The accumulation of shorter TCR/MHC complexes (~15 nm) may force the longer LFA-1/ICAM-1 complexes to migrate outward to avoid unfavorable bending of the membrane and of the LFA-1/ICAM-1 complexes (129, 130). Sorting of protein complexes might also depend on partitioning of proteins in the lipid microdomains, although there is conflicting evidence for the roles of lipid rafts (131, 132).
Formation of the immunological synapse results in the reorganization of cytoskeletal proteins at the site of cell-cell contact (Figure 4). The Wiskott-Aldrich syndrome protein (WASP), which localizes to the T cell–APC interface (133), activates the Arp2/3 complex to generate a branched actin network (134). Significantly, T cells derived from the WASP (135), or WASP-interacting protein, knockout mice (136) had a deficiency in activation, indicating an important role for actin polymerization during initial synapse formation and T-cell activation. WASP is regulated by the small GTPase Cdc42 (134), which is activated at the immunological synapse between the T cell and APC (137) and required to orient actin filaments and microtubules toward the immunological synapse (138). Cdc42 activation may be regulated by the synapse-localized guanine nucleotide exchange factor, Vav1, because Vav1-deficient thymocytes are less efficient in the polarization of microtubule-organizing center during the maturation of immunological synapse (139) (Figure 4).
What is the functional significance of the striking distributions of cell-surface and cytoplasmic proteins at the immunological synapse? Analysis of the minimum number of antigens required for induction of cytotoxic T lymphocyte (CTL) effector functions revealed that only three agonist/MHC complexes were required (i.e., not a mature synapse) (140, 141); small clusters of TCR were observed during the earliest stage of synapse formation (142, 143), which could play a part in activation of CTLs. However, the organization of the mature synapse might be important for amplifying signaling pathways. This possibility has been tested by geometrically constraining organization of proteins in the immunological synapse by plating T cells on micropatterned corrals containing a lipid bilayer with GPI-anchored MHC and ICAM-1, which resulted in prolonged TCR signaling and calcium mobilization (144).
THE EPITHELIAL SYNAPSE
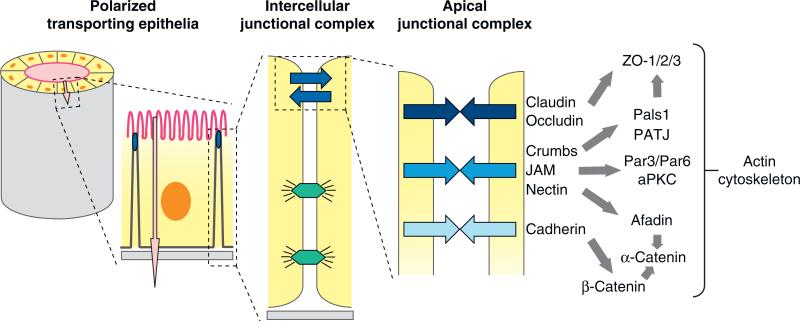
Epithelial cells form barriers that separate different biological compartments in the body and regulate the vectorial transport of ions and solutes between those compartments (Figure 5). Cell-cell adhesion and the barrier function are maintained by an apical junctional complex (AJC) localized at the boundary of the apical and lateral membrane domains (Figure 5). Each adhesion protein in the AJC has unique functions in regulating cell-cell interactions through specialized adhesion proteins, associated cytoplasmic proteins and cytoskeletal proteins, and signal transduction pathways. Although cell-cell adhesions formed between cells in the epithelium are symmetrical, the AJC plays a crucial role in regulating structural and functional asymmetry in the apicobasal axis between the apical and basal-lateral membrane domains. This asymmetry drives the vectorial transport of ions and solutes between compartments separated by the epithelium. Below, we discuss the structure, function, and protein interactions of each of the major AJC adhesion complexes.
Figure 5.
Schematic representations of the epithelial synapse. Polarized transporting epithelia comprise a closed monolayer of cells that surround a fluid-filed space (lumen) and vectorially transport ions and solutes (pink arrow) between the luminal space and the serosa; the plasma membrane domains facing the lumen space (apical) and serosa (basal lateral) are structurally and functionally different. Intercellular junctional complexes regulate cell-cell adhesion and the paracellular pathway (blue arrows) as well as maintain the structural integrity of the epithelium (desmosomes, lower junctions). The apical junctional complex comprises different cell adhesion proteins and downstream protein networks, and it is located at the boundary between the apical and basal-lateral membranes. Abbreviations: aPKC, atypical protein kinase C; JAM, junction-associated molecule; Pals1, protein-associated with Lin-7; Par3/Par6, partition defective 3/6; PATJ, Pals1-associated tight junction protein.
Tight Junction
The tight junction forms a continuous belt-like structure (zonula occludens) at the boundary between the apical and lateral membranes of polarized epithelial cells in vertebrates and regulates the flow of ions and solutes in the paracellular pathway between cells (145); in invertebrates, the septate junction may play a similar role (146). The membranes of opposed cells are very closely connected at the tight junction, and freeze-fraction electron microscopy has revealed a branched, interdigitating network of strands on opposing membranes, the complexity of which is directly proportional to the tightness of the tight junction barrier to ions and solutes (147).
The structural and functional organization of the tight junction is regulated by claudins (148). There are 24 mammalian claudins, some of which are expressed ubiquitously and others are expressed in a tissue- and a cell-type-specific manner (149), depending on the ion selectivity of the paracellular pathway (145). Claudins have four transmembrane helices, of which the first extracellular loop is highly variable between different claudins (150) and influences paracellular charge selectivity (151). The C-terminal domain has a PDZ-binding motif for binding PDZ domain proteins, including ZO-1, -2, -3 (152), PATJ (153), cingulin (154), and others (155) that are thought to form a protein network with the actin cytoskeleton (156) (Figure 5). Note, however, that claudins lacking the PDZ-binding domain are capable of forming a complex network of interdigitated strands between transfected fibroblasts, similar to that found in transporting epithelial cells (157), indicating that the organization of intercellular strands and intracellular signaling may be separate processes. Several other membrane proteins also contribute to tight junction organization, including the claudin-homology protein occludin (146) and the Coxsackie virus and adenovirus receptor (CAR), a member of the Ig superfamily (158).
In addition to these cytoplasmic structural proteins, the tight junction is a localization site for signaling proteins, including small GTPases and kinases, which may control tight junction assembly and function, and local actin dynamics (155). Several dual location proteins are also localized to the tight junction (159), including transcription factors of the Y-box family ZONAB (160, 161), huASH1 (162), and c-jun and c-fos (163), which regulate cell proliferation, gene expression, and cellular differentiation.
Adherens Junction
In vertebrates, the adherens junction is located immediately below the tight junction and forms a belt-like structure (zonula adherens) that circumscribes the cell (164) (Figure 5). The adherens junction is primarily involved in specifying adhesion between cells and, thereby, in sorting out different cell types during development (165). The adherens junction comprises adhesion proteins, including cadherins and nectins and associated cytoplasmic proteins that locally regulate the actin cytoskeleton.
The principal cell adhesion protein of the adherens junction is E-cadherin, a member of the family of classical cadherins (166). Cadherins are required for the formation of the earliest epithelial structures during embryogenesis (167), such as the trophectoderm in the preimplantation mouse blastocyst (168) and the ectoderm of the cellular blastoderm during early Drosophila development (169). During the initial stages of Ca2+-dependent cell-cell adhesion, cadherins rapidly concentrate at sites of cell-cell contacts (170). Subsequently, the contact expands laterally (also termed compaction), indicating an active process involving reorganization of the actin cytoskeleton (171), mediated by the small GTPases Rac1 and Cdc42, GTPases (172, 173), and actinomyosin contractility (174). Generally, formation of cadherin-mediated cell adhesion precedes assembly of other intercellular junctions (175, 176).
Cadherins bind directly to several cytoplasmic proteins, including β-catenin and p120 (165) (Figure 5). p120 may regulate the cadherin-actin cytoskeleton complex by locally controlling the activity of the Rho inhibitor, p190RhoGAP, and thereby activation of Rho and Rac (177), as well as the rate of cadherin endocytosis (178). β-catenin, which has an additional role as a transcriptional cofactor (179), plays a more direct role by binding to α-catenin (180), an actin filament-binding/bundling protein (181) that also forms binary interactions with other actin-binding proteins (182). Although these binary complexes with α-catenin were never shown to bind the cadherin/β-catenin complex, it has been generally assumed that α-catenin links the cadherin complex to the actin cytoskeleton (79). However, recent studies tested this assumption directly and found that α-catenin does not bind to β-catenin and actin simultaneously, suggesting that α-catenin behaves like an allosteric protein (83, 183). The molecular basis for α-catenin allostery appears to be the formation of either an α-catenin monomer, which preferentially binds β-catenin, or a homodimer, which preferentially binds and bundles actin filaments (83). Significantly, α-catenin homodimers also inhibit actin polymerization by the Arp2/3 complex (83). Because the Arp2/3 complex regulates actin branching in active lamellipodia (134), it has been proposed (83) that the increased local concentration of α-catenin during cadherin-mediated cell-cell contacts could locally suppress Arp2/3 activity and lamellipodia formation. This might result in strengthening adhesion and causing actin reorganization into bundles parallel to the adherens junction (184), perhaps with formins that polymerize unbranched actin filaments (185). Although α-catenin does not appear to bind directly to the actin cytoskeleton, it is possible that other interactions are important. α-Catenin binds afadin (80), a scaffolding protein that binds actin (186), and the nectin family of adhesion proteins (81) that cooperate with cadherins to initiate cell-cell adhesion (187).
Deletion of cadherin results in disruption of epithelial cell-cell adhesion and tissue integrity (168, 169). However, genetic deletion of α-catenin generates a constellation of defects that are different from those induced by E-cadherin deletion (188–190); cell-cell adhesion and aggregation were not severely affected per se, although cells dissociated from each other more easily than those expressing α-catenin. The most significant effects caused by the loss of α-catenin in vivo were increased cell migration, shortening of the cell cycle, increased proliferation (hyperplasia), and decreased apoptosis (189, 190). The effects of genetic deletion of β-catenin on cell-cell adhesion are more difficult to interpret because plakoglobin can substitute for β-catenin in the cadherin complex (191), and β-catenin functions in both cell-cell adhesion and gene transcription (179); however, whether β-catenin acts as a dual location protein or is located in separate pools at the adherens junction and nucleus is unclear (192, 193).
The adherens junction is also important in actomyosin-based contraction during invagination of epithelial sheets to form tubular structures in development (194–196). Although actin may not be linked directly to the cadherin/catenin complex (see above), several candidate proteins have been identified recently that localize to the adherens junction and bind the actomyosin contractile machinery: Shroom, a PDZ domain-containing actin-binding protein required for neural tube morphogenesis (197, 198), and bite-size, a synaptotagmin-like protein required for proper actin organization at adherens junctions during Drosophila cellularization (199).
Junction-Associated Molecule, Crumbs, and PAR Proteins
Several important signaling complexes are clustered in the AJC, including the membrane proteins JAM and Crumbs, with their associated cytoplasmic signaling complexes, which include the PAR complex (200, 201).
JAM is a member of the Ig superfamily of adhesion proteins (202). JAM forms mostly homophilic adhesions at contacts between epithelial or endothelial cells (203), but also forms heterophilic adhesions with the leukocyte integrin αLβ2 (116) (see above). JAM interacts with PDZ domain proteins, including cingulin and ZO-1 (204), CASK (205), and Par3 (206, 207)—some of which may link JAM to other adhesion proteins in the AJC and to the actin cytoskeleton to regulate tight junction assembly (202) (Figure 5).
The AJC is the location of two large interconnected protein scaffolds that are evolutionarily conserved and important in the generation of cell polarity in embryos, neurons, and epithelial cells (200, 201) (Figure 5). A complex of Par3, another PDZ domain protein Par6, and an atypical protein kinase C (aPKC) associate with the tight junction (208). Par6 is an effector of Cdc42, and binding of GTP-Cdc42 to the Par complex activates aPKC, which is also involved in junction assembly and cell polarization (209, 210). A second evolutionarily conserved signaling complex localized to the AJC comprises Crumbs and another PDZ domain protein scaffold of Pals1 and PATJ (200, 211). The Crumbs/Pals1/PATJ complex regulates ectoderm morphogenesis in Drosophila (212), tight junction formation in mammalian cells (213, 214), and cell polarization (214, 215). The Crumbs and Par complexes are not distinct, however, because there are a number of interconnections [e.g., Par6 binds Pals1 and Crumbs3 (213) (Figure 5), and aPKC phosphorylates and activates Crumbs (216), suggesting that there might be a hierarchical regulatory pathway of assembly and activation of these complexes as cells adhere and develop polarity].
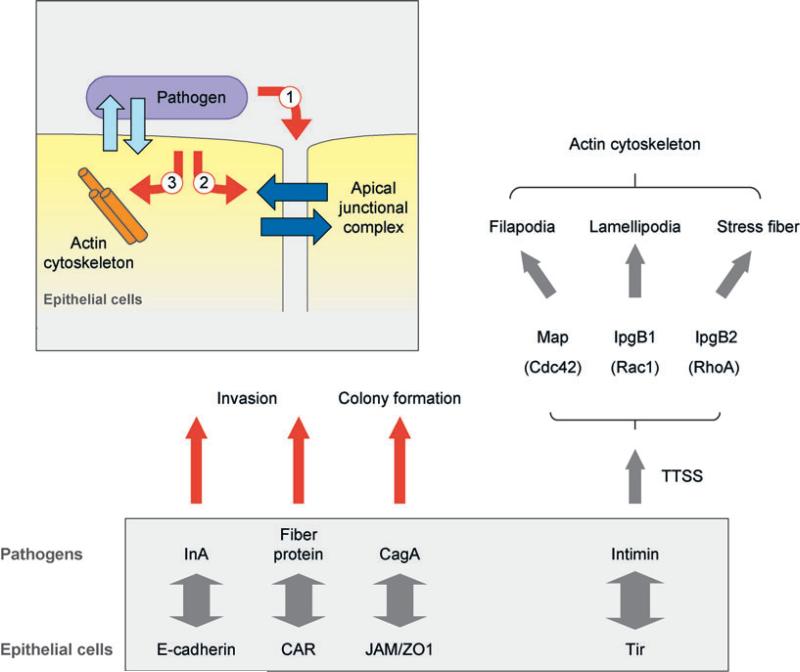
THE HOST-PATHOGEN SYNAPSE
Epithelial surfaces exposed to the outside, for example, in the gut, provide a physical barrier to invasion by bacterial and viral pathogens. Nevertheless, pathogens have evolved sophisticated molecular machineries to enable them to adhere to and, in some cases, disrupt cell-cell junctions to gain entry into the organism. The initial synapse between host cells and pathogens is via adhesion proteins on the apical surface of epithelial cells (Figure 6). This is followed by firm attachment and colonization of cells by pathogens and, in some instances, transit of pathogens across the epithelial barrier to enter the host and establish an infection (Figure 6, inset). Sometimes, pathogens target and disrupt cell-cell junctions by secreting proteases or toxins or by injecting factors into the host cytoplasm to induce remodeling of adhesion complexes.
Figure 6.
Schematic representations of novel synapses formed between pathogens and host cells. Pathogens form synapses on the luminal surface of epithelial cells (inset) and modify the host cell synapse from the outside (1) or inside (2), or through reorganization of the actin cytoskeleton (3). Pathogen surface proteins bind host cell adhesion proteins (left) or a pathogen protein inserted by a type III secretion system (TTSS) (right), and hijack host protein networks to modify the actin cytoskeleton and membrane dynamics. Abbreviations: CagA, cytotoxin-associated gene 1; CAR, Coxsackie virus and adenovirus receptor; InlA, internalin A; JAM, junction-associated molecule; Map, mitochondrial-associated protein; Tir, translocated intimin receptor; ZO1, zonula occludens-1.
The initial adhesion of pathogens to host epithelial cells in the gastrointestinal tract occurs in the context of the flow of mucus over the surface of the epithelium, a physical condition analogous to leukocytes’ endothelial adhesion (see above). To colonize under such conditions, many gram-negative bacteria are equipped with thin fibrous organelles, called pili (217), at the tip of which is localized the lectin-like adhesion molecule FimH that binds mannose presented on the apical surface of epithelial cells; FimH-mediated adhesion is enhanced by shear flow (218).
Some pathogens have surface receptors that specifically bind to host proteins localized on the lateral membrane. However, the tight junction barrier limits pathogen access to these receptors (Figure 6); for example, the ligand for internalin A (InlA) of Listeria monocytogenes is E-cadherin (219). It has been suggested that InlA gains access to E-cadherin through InlB-induced activation of the tyrosine kinase receptor c-Met, which then disrupts cell-cell adhesions allowing InlA/E-cadherin binding (220). However, recent studies in vivo show that L. monocytogenes invades the intestinal epithelium at the villus tip, where cell polarity is disrupted owing to apoptosis and cell sloughing, thereby exposing E-cadherin on the surrounding cells (221, 222).
Adenovirus binds via its fiber proteins to the CAR, a member of the Ig superfamily, which regulates tight junction assembly (158). Interestingly, the fiber protein expressed by adenovirus has higher affinity to CAR than that of CAR-CAR homodimers (223, 224); thus the fiber proteins can potentially compete with CAR homophilic adhesion to disrupt tight junctions. However, CAR is located below the tight junction barrier and is normally inaccessible to adenovirus and fiber protein in the airway lumen. However, aden-ovirus fiber protein produced in infected epithelial cells is secreted to the lateral membrane where it competitively binds CAR and disrupts the tight junction, thereby allowing adenovirus to escape into the airway (225).
Helicobacter pylori invades cells of the gastric mucosa and in chronic infections is located close to the tight junction (226). H. pylori manipulates tight junction permeability through the activity of cytotoxin-associated gene-1 (CagA). Upon adhering to host cells, CagA is injected into the host cytoplasm by a type IV secretion system (227) and forms a complex with JAM and ZO-1 (Figure 6), which is required for the firm attachment of H. pylori and results in loosening of the permeability barrier of the tight junction but not its overall structural organization (228). CagA can also activate c-Met, which, as noted above, disrupts cell-cell adhesion and induces cell scattering (229).
Some bacteria do not use host receptors but insert their own receptors to form firm adhesion with host membranes (Figure 6). These pathogens include enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) (230). Initial weak adhesion is mediated by intimin on the bacterial surface, host cell β1 integrin, (231) and nucleolin (232), although only nucleolin appears to be necessary for initial attachment (232). The bacterium then injects the intimin receptor Tir into the host plasma membrane by a type III secretion system (TTSS) (233). The interaction between intimin and Tir initiates actin reorganization and disassembly of microvilli on the host apical membrane and the formation in their place of pedestal-shaped structures, called attach and efface (A/E) lesions (234). A/E lesions allow strong adhesion between bacteria and host and provide EPEC and EHEC a competitive advantage over less-adherent microbes that are flushed away by the diarrheal response (230).
Pathogens also exploit epithelial cell adhesion complexes as an entry site into the host organism by secreting toxins or proteases that specifically degrade adhesion proteins. Staphylococcus aureus, which causes skin blistering and infections, produces an exfoliative toxin, which shares sequence homology with serine protease; exfoliative toxin specifically cleaves desmoglein 1, a desmosomal cadherin expressed in the superficial layers of the epidermis, resulting in loss of cell-cell adhesion in the epidermis and skin blistering (235). Vibrio cholerae and Bacteroides fragilis are diarrhea-causing pathogens that also target cell-cell adhesion proteins. V. cholerae secretes hemagglutinin/protease (HA/P), a zinc-containing metalloprotease, which specifically degrades occludin and alters ZO-1 localization at cell-cell junctions (236). B. fragilis secretes a zinc-dependent metalloprotease, B. fragilis enterotoxin, which cleaves E-cadherin (237).
Alternatively, pathogens disrupt the integrity of cell-cell adhesions by perturbing the actin cytoskeleton (Figure 6). Members of Rho GTPase family are important regulators of the actin cytoskeleton and cell-cell adhesion (238). Clostridium difficile secretes toxins A and B, which monoglucosylate and inactivate these small GTPases, thereby inducing actin reorganization and disruption of the tight junction (239). Cytotoxic necrotizing factor 1 is a toxin secreted by some E. coli that induces urinary and gastrointestinal tract infections (240). Unlike toxin A/B, the cytotoxic necrotizing factor 1 constitutively activates small Rho GTPase by deamidation of glutamine residues of RhoA, Rac1, or Cdc42, thereby inhibiting GTP hydrolysis, which leads to reorganization of the actin cytoskeleton and disruption of the tight junction (241). It is noteworthy that many virulence factors are being discovered that affect Rho GTPase activity (242), and their activity will likely also compromise the integrity of epithelial monolayer by disrupting the actin cytoskeleton and cell-cell adhesion complexes.
Other pathogen effector proteins seamlessly integrate into signaling and regulatory pathways that regulate the actin cytoskeleton (Figure 6). A recent study reveals that many virulence factors copy the activity of Rho GTPase. These effector proteins are delivered by TTSS and share a common Try-x-x-x-Glu (WxxxE) motif (243). For example, expression of Map, an effector protein from EPEC that contains the WxxxE motif, induced filopodia formation identical to that of activated Cdc42 signaling (243). This effect was not affected by expression of a dominant negative Cdc42 mutant, indicating that Map functionally mimicked Cdc42 rather than activating Cdc42 [see also (244)]. Interestingly, Map also disrupted tight junctions when injected by TTSS (245).
IpgB1 and IpgB2 from Shigella also induce increased stress fiber formation, even in the presence of several RhoA inhibitors, indicating that IpgB2 can replace the role of RhoA in mediating stress fiber formation (243) (Figure 6). Furthermore, IpgB2 directly interacts with downstream effectors of RhoA, ROCK, and mDia1 (243), which are required for actin stress fiber formation. This remarkable functional mimicry of bacterial factors emphasizes the importance of small GTPases for the regulation of actin cytoskeleton and cell-cell adhesion, and further studies are needed to understand how these effectors are used to invade host cells.
CONCLUSIONS AND PERSPECTIVES
We started with the proposition that many types of cell-cell interactions are synapses, even when synapses are defined as being asymmetric in the context of structural and functional differences between opposed cells. Interestingly, Sherrington preferred the word synapse to junction, although he was not accurate about the demise of the latter: “As to ‘junction’ I feel we are less easily reconcilable . . . The mere fact that junction implies passive union is alone enough to ruin the term . . . I think it does not want the gift of prophecy to fortell that it [the word junction] must become more and more obviously inapplicable as research progresses. Synapse, which implies a catching on, as e.g., by one wrestler of another—is really much closer to the mark.” [from a letter he wrote to his colleague Sharpy-Schafer in 1897, which was quoted in Reference 1]. From our survey of cell interactions by neurons, immune cells, epithelial cells, and even between pathogens and host cells, it is clear that in all cases these synapses are highly active, not passive, and that they generate asymmetry in the functional organization of cells at the synapse.
The neuronal synapse, as originally defined, is an asymmetric structure required to transmit an action potential from one cell to the other via the release of neurotransmitters from the presynaptic side that bind to, and activate, receptors on the postsynaptic side. Similarly, the immunological synapse is asymmetric because it forms between two functionally distinct cells, a T cell and an APC, and results in an asymmetric functional response by the T cell. Can we consider, in the same context, that a synapse is formed between simple epithelial cells? Because the adhesive contacts are symmetric between the same cell type, a superficial answer is no. However, the epithelial synapse is located at the boundary between structurally and functionally different membrane domains; the apical and basal-lateral domains generate asymmetry in the apicobasal axis of the cell to drive vectorial transport of ions and solutes from one side of the epithelium to the other. Other types of interactions between leukocytes, endothelial cells, and even pathogen and host cells are clearly asymmetric, involving different responses by the paired cells that mediate changes in the organization and fate of one or both of the cells.
How is structural and functional asymmetry generated at various synapses by these adhesion proteins (outside-in signaling)? Our survey of synapses revealed examples of adhesion between proteins of the same, or a different, class that were either homophilic or heterophilic. As suggested (see Figure 1), heterophilic adhesion potentially recruits different cytoplasmic scaffolding proteins to each membrane to generate structural and functional asymmetry at the synapse (e.g., neuroligin/neurexin) (Figure 2), whereas homophilic adhesion potentially recruits the same cytoplasmic scaffolding proteins and generates a symmetric synapse (e.g., cadherins) (Figure 5), unless different sets of binding proteins were expressed in each of the opposing cells and resulted in an asymmetric synapse (e.g., β-catenin at the presynaptic membrane) (Figure 2).
The recruitment of cytoplasmic signaling proteins to diverse adhesion proteins appears to play a role in modulating adhesive properties of the synapse (inside-out signaling), and to propagate changes in cell structure and function. Heterophilic adhesions between neurons or immune cells lead to the recruitment of scaffolding proteins and other membrane proteins that generate asymmetric signal transduction between opposed cells and ultimately different functions of the attached cells. Although the epithelial cell synapse forms symmetric adhesion between the same cell type, it recruits protein complexes that generate structural and functional asymmetry in the apicobasal axis of the cell by controlling the organization of the apical (Crumbs complex) and basal-lateral membrane domains (Par and Scribble complexes). Remarkably, pathogens have evolved machinery that hijacks host cell synapses to invade cells and organisms. Pathogens form novel synapses with host cell adhesion proteins and disrupt adhesion proteins or their downstream scaffolding and signaling networks to enable pathogen entry.
In summary, our survey of five diverse examples of cell-cell interactions revealed that cell adhesion proteins regulate the specificity of cell-cell interactions in terms of cell recognition and initial adhesion and that these proteins interact with networks of cytoplasmic scaffolding and signaling proteins to propagate changes in overall cell functions. What is surprising, however, is that many of the same classes of adhesion and cytoplasmic proteins are used at synapses in very different cellular contexts, with different functional outcomes. Although this is evidence of the evolutionary conservation of functions of these adhesion protein complexes, further studies are needed to understand how combinations of these adhesion protein complexes specify the assembly of functionally different synapses.
Synapse: a specialized adhesive junction formed between two closely opposed plasma membranes
Cell sorting and pattern formation: Cells interact in specific patterns to form functionally diverse tissues and organs. The specificity of sorting depends on cell adhesion proteins
APC: antigen-presenting cell
Cytoplasmic protein networks: a diverse number of cytoplasmic proteins that interact with adhesion proteins
Cell-cell adhesion proteins: a broad class of cell-surface membrane-attached glycoproteins, and its binding can be homophilic or heterophilic
PDZ: postsynaptic density 95-Discs large-ZO1 homologous domain
Extravasation: a complex series of stereotypical events by leukocytes in response to a foreign invader and inflammation
JAM: junction-associated molecule
MHC: major histocompatibility complex
TCR: T-cell receptor
AJC: apical junctional complex
CAR: Coxsackie virus and adenovirus receptor
Type III secretion system (TTSS): a complex multiprotein assembly used by certain pathogens to inject proteins into host cells
SUMMARY POINTS.
Cell-cell adhesion is a fundamental characteristic of multicellular organisms.
Cell-cell adhesion regulates sorting of cells into specific patterns during development.
Cell-cell adhesion proteins include members of the Ig superfamily, integrins and cadherins, that are expressed in diverse cell types, including neurons, epithelia, and cells of the immune system.
Host cell adhesion proteins are used by pathogens to gain entry into cells and the organism.
Different cell-cell adhesion proteins interact with networks of scaffolding proteins, the cytoskeleton, and signaling pathways to regulate synapse and cell function.
FUTURE ISSUES.
How are the repertoire and correct combination of cell-cell adhesion proteins regulated to achieve correct cell sorting and interactions in development?
How do cell adhesion proteins specify the structural and functional specialization of membrane domains that form different synapses in different cell types?
How are cell-surface levels of adhesion proteins determined and regulated?
How did pathogens evolve to hijack functions and downstream signaling by host cell adhesion proteins?
ACKNOWLEDGMENT
Work in the Nelson laboratory is supported by the National Institutes of Health grant GM35527 to W.J.N.
LITERATURE CITED
- 1.Bennett MR. Brain Res. Bull. 1999;50:95–118. doi: 10.1016/s0361-9230(99)00094-5. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham BA, Hemperly JJ, Murray BA, Prediger EA, Brackenbury R, Edelman GM. Science. 1987;236:799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- 3.Bruses JL, Rutishauser U. Biochimie. 2001;83:635–43. doi: 10.1016/s0300-9084(01)01293-7. [DOI] [PubMed] [Google Scholar]
- 4.Cremer H, Lange R, Christoph A, Plomann M, Vopper G, et al. Nature. 1994;367:455–59. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- 5.Hu H, Tomasiewicz H, Magnuson T, Rutishauser U. Neuron. 1996;16:735–43. doi: 10.1016/s0896-6273(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 6.Brummendorf T, Kenwrick S, Rathjen FG. Curr. Opin. Neurobiol. 1998;8:87–97. doi: 10.1016/s0959-4388(98)80012-3. [DOI] [PubMed] [Google Scholar]
- 7.Kamiguchi H, Hlavin ML, Yamasaki M, Lemmon V. Annu. Rev. Neurosci. 1998;21:97–125. doi: 10.1146/annurev.neuro.21.1.97. [DOI] [PubMed] [Google Scholar]
- 8.Dahme M, Bartsch U, Martini R, Anliker B, Schachner M, Mantei N. Nat. Genet. 1997;17:346–49. doi: 10.1038/ng1197-346. [DOI] [PubMed] [Google Scholar]
- 9.Cohen NR, Taylor JS, Scott LB, Guillery RW, Soriano P, Furley AJ. Curr. Biol. 1998;8:26–33. doi: 10.1016/s0960-9822(98)70017-x. [DOI] [PubMed] [Google Scholar]
- 10.Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu XR, et al. Science. 2002;297:1525–31. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 11.Mizoguchi A, Nakanishi H, Kimura K, Matsubara K, Ozaki-Kuroda K, et al. J. Cell Biol. 2002;156:555–65. doi: 10.1083/jcb.200103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shingai T, Ikeda W, Kakunaga S, Morimoto K, Takekuni K, et al. J. Biol. Chem. 2003;278:35421–27. doi: 10.1074/jbc.M305387200. [DOI] [PubMed] [Google Scholar]
- 13.Irie K, Shimizu K, Sakisaka T, Ikeda W, Takai Y. Semin. Cell Dev. Biol. 2004;15:643–56. doi: 10.1016/j.semcdb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Honda T, Sakisaka T, Yamada T, Kumazawa N, Hoshino T, et al. Mol. Cell. Neurosci. 2006;31:315–25. doi: 10.1016/j.mcn.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Togashi H, Miyoshi J, Honda T, Sakisaka T, Takai Y, Takeichi M. J. Cell Biol. 2006;174:141–51. doi: 10.1083/jcb.200601089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Cell. 2000;101:657–69. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 17.Dean C, Scholl FG, Choih J, DeMaria S, Berger J, et al. Nat. Neurosci. 2003;6:708–16. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Missler M, Fernandez-Chacon R, Sudhof TC. J. Neurochem. 1998;71:1339–47. doi: 10.1046/j.1471-4159.1998.71041339.x. [DOI] [PubMed] [Google Scholar]
- 19.Levinson JN, Chery N, Huang K, Wong TP, Gerrow K, et al. J. Biol. Chem. 2005;280:17312–19. doi: 10.1074/jbc.M413812200. [DOI] [PubMed] [Google Scholar]
- 20.Sara Y, Biederer T, Atasoy D, Chubykin A, Mozhayeva MG, et al. J. Neurosci. 2005;25:260–70. doi: 10.1523/JNEUROSCI.3165-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chih B, Engelman H, Scheiffele P. Science. 2005;307:1324–28. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- 22.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Cell. 2004;119:1013–26. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Proc. Natl. Acad. Sci. USA. 2005;102:8531–36. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, et al. Cell. 2006;124:1255–68. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 25.Fannon AM, Colman DR. Neuron. 1996;17:423–34. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- 26.Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. J. Cell Biol. 1996;135:767–79. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson DL, Tanaka H. J. Neurosci. 1998;18:6892–904. doi: 10.1523/JNEUROSCI.18-17-06892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka H, Shan W, Phillips GR, Arndt K, Bozdagi O, et al. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 29.Salinas PC, Price SR. Curr. Opin. Neurobiol. 2005;15:73–80. doi: 10.1016/j.conb.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Hirano S, Suzuki ST, Redies C. Front Biosci. 2003;8:d306–55. doi: 10.2741/972. [DOI] [PubMed] [Google Scholar]
- 31.Matsunaga M, Hatta K, Takeichi M. Neuron. 1988;1:289–95. doi: 10.1016/0896-6273(88)90077-3. [DOI] [PubMed] [Google Scholar]
- 32.Redies C, Engelhart K, Takeichi M. J. Comp. Neurol. 1993;333:398–416. doi: 10.1002/cne.903330307. [DOI] [PubMed] [Google Scholar]
- 33.Jungling K, Eulenburg V, Moore R, Kemler R, Lessmann V, Gottmann K. J. Neurosci. 2006;26:6968–78. doi: 10.1523/JNEUROSCI.1013-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Cell. 2002;109:205–16. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- 35.Honig MG, Petersen GG, Rutishauser US, Camilli SJ. Dev. Biol. 1998;204:317–26. doi: 10.1006/dbio.1998.9093. [DOI] [PubMed] [Google Scholar]
- 36.Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 37.Redies C, Inuzuka H, Takeichi M. J. Neurosci. 1992;12:3525–34. doi: 10.1523/JNEUROSCI.12-09-03525.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riehl R, Johnson K, Bradley R, Grunwald GB, Cornel E, et al. Neuron. 1996;17:837–48. doi: 10.1016/s0896-6273(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 39.Marthiens V, Gavard J, Padilla F, Monnet C, Castellani V, et al. Mol. Cell. Neurosci. 2005;28:715–26. doi: 10.1016/j.mcn.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Fredette BJ, Miller J, Ranscht B. Development. 1996;122:3163–71. doi: 10.1242/dev.122.10.3163. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki SC, Inoue T, Kimura Y, Tanaka T, Takeichi M. Mol. Cell. Neurosci. 1997;9:433–47. doi: 10.1006/mcne.1997.0626. [DOI] [PubMed] [Google Scholar]
- 42.Takeichi M, Uemura T, Iwai Y, Uchida N, Inoue T, et al. Cold Spring Harbor Symp. Quant. Biol. 1997;62:505–10. [PubMed] [Google Scholar]
- 43.Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 44.Bozdagi O, Valcin M, Poskanzer K, Tanaka H, Benson DL. Mol. Cell. Neurosci. 2004;27:509–21. doi: 10.1016/j.mcn.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamura K, Tanaka H, Yagita Y, Saeki Y, Taguchi A, et al. J. Cell Biol. 2004;167:961–72. doi: 10.1083/jcb.200406030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang L, Hung CP, Schuman EM. Neuron. 1998;20:1165–75. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- 47.Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, et al. J. Biol. Chem. 1999;274:19473–79. doi: 10.1074/jbc.274.27.19473. [DOI] [PubMed] [Google Scholar]
- 48.Junghans D, Haas IG, Kemler R. Curr. Opin. Cell Biol. 2005;17:446–52. doi: 10.1016/j.ceb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, et al. Neuron. 1998;20:1137–51. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 50.Frank M, Ebert M, Shan W, Phillips GR, Arndt K, et al. Mol. Cell. Neurosci. 2005;29:603–16. doi: 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Blank M, Triana-Baltzer GB, Richards CS, Berg DK. Mol. Cell. Neurosci. 2004;26:530–43. doi: 10.1016/j.mcn.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, et al. J. Neurosci. 2003;23:5096–104. doi: 10.1523/JNEUROSCI.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR. Neuron. 2002;36:843–54. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- 54.Weiner JA, Wang X, Tapia JC, Sanes JR. Proc. Natl. Acad. Sci. USA. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flanagan JG, Vanderhaeghen P. Annu. Rev. Neurosci. 1998;21:309–45. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- 56.Henkemeyer M, Itkis OS, Ngo M, Hickmott PW, Ethell IM. J. Cell Biol. 2003;163:1313–26. doi: 10.1083/jcb.200306033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serra-Pages C, Medley QG, Tang M, Hart A, Streuli M. J. Biol. Chem. 1998;273:15611–20. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- 58.Miller KE, DeProto J, Kaufmann N, Patel BN, Duckworth A, Van Vactor D. Curr. Biol. 2005;15:684–89. doi: 10.1016/j.cub.2005.02.061. [DOI] [PubMed] [Google Scholar]
- 59.Bennett V, Baines AJ. Physiol. Rev. 2001;81:1353–92. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 60.Leshchyns'ka I, Sytnyk V, Morrow JS, Schachner M. J. Cell Biol. 2003;161:625–39. doi: 10.1083/jcb.200303020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jenkins SM, Bennett V. J. Cell Biol. 2001;155:739–46. doi: 10.1083/jcb.200109026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scotland P, Zhou D, Benveniste H, Bennett V. J. Cell Biol. 1998;143:1305–15. doi: 10.1083/jcb.143.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pielage J, Fetter RD, Davis GW. Curr. Biol. 2005;15:918–28. doi: 10.1016/j.cub.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 64.Wechsler A, Teichberg VI. EMBO J. 1998;17:3931–39. doi: 10.1093/emboj/17.14.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheng M, Sala C. Annu. Rev. Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 66.Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, et al. Nature. 2002;415:321–26. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Okamoto M, Schmitz F, Hofmann K, Sudhof TC. Nature. 1997;388:593–98. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- 68.Dulubova I, Lou X, Lu J, Huryeva I, Alam A, et al. EMBO J. 2005;24:2839–50. doi: 10.1038/sj.emboj.7600753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hata Y, Butz S, Sudhof TC. J. Neurosci. 1996;16:2488–94. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maximov A, Sudhof TC, Bezprozvanny I. J. Biol. Chem. 1999;274:24453–56. doi: 10.1074/jbc.274.35.24453. [DOI] [PubMed] [Google Scholar]
- 71.Cho KO, Hunt CA, Kennedy MB. Neuron. 1992;9:929–42. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 72.Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. Nature. 1997;386:279–84. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 73.Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, et al. Science. 1997;277:1511–15. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 74.Song JY, Ichtchenko K, Sudhof TC, Brose N. Proc. Natl. Acad. Sci. USA. 1999;96:1100–5. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Science. 1995;269:1737–40. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 76.Muller BM, Kistner U, Kindler S, Chung WJ, Kuhlendahl S, et al. Neuron. 1996;17:255–65. doi: 10.1016/s0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- 77.Bruckner K, Pablo Labrador J, Scheiffele P, Herb A, Seeburg PH, Klein R. Neuron. 1999;22:511–24. doi: 10.1016/s0896-6273(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 78.Dunah AW, Hueske E, Wyszynski M, Hoogenraad CC, Jaworski J, et al. Nat. Neurosci. 2005;8:458–67. doi: 10.1038/nn1416. [DOI] [PubMed] [Google Scholar]
- 79.Gates J, Peifer M. Cell. 2005;123:769–72. doi: 10.1016/j.cell.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 80.Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. J. Biol. Chem. 2002;277:18868–74. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takai Y, Nakanishi H. J. Cell Sci. 2003;116:17–27. doi: 10.1242/jcs.00167. [DOI] [PubMed] [Google Scholar]
- 82.Abe K, Chisaka O, Van Roy F, Takeichi M. Nat. Neurosci. 2004;7:357–63. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- 83.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Cell. 2005;123:903–15. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bamji SX, Shimazu K, Kimes N, Huelsken J, Birchmeier W, et al. Neuron. 2003;40:719–31. doi: 10.1016/s0896-6273(03)00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bozdagi O, Shan W, Tanaka H, Benson DL, Huntley GW. Neuron. 2000;28:245–59. doi: 10.1016/s0896-6273(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 86.Murase S, Mosser E, Schuman EM. Neuron. 2002;35:91–105. doi: 10.1016/s0896-6273(02)00764-x. [DOI] [PubMed] [Google Scholar]
- 87.Springer TA. Cell. 1994;76:301–14. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 88.Simon SI, Green CE. Annu. Rev. Biomed. Eng. 2005;7:151–85. doi: 10.1146/annurev.bioeng.7.060804.100423. [DOI] [PubMed] [Google Scholar]
- 89.Finger EB, Puri KD, Alon R, Lawrence MB, von Andrian UH, Springer TA. Nature. 1996;379:266–69. doi: 10.1038/379266a0. [DOI] [PubMed] [Google Scholar]
- 90.Chang KC, Hammer DA. Biophys. J. 1999;76:1280–92. doi: 10.1016/S0006-3495(99)77291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dwir O, Solomon A, Mangan S, Kansas GS, Schwarz US, Alon R. J. Cell Biol. 2003;163:649–59. doi: 10.1083/jcb.200303134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barsegov V, Thirumalai D. Proc. Natl. Acad. Sci. USA. 2005;102:1835–39. doi: 10.1073/pnas.0406938102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sarangapani KK, Yago T, Klopocki AG, Lawrence MB, Fieger CB, et al. J. Biol. Chem. 2004;279:2291–98. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- 94.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Nature. 2003;423:190–93. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 95.von Andrian UH, Hasslen SR, Nelson RD, Erlandsen SL, Butcher EC. Cell. 1995;82:989–99. doi: 10.1016/0092-8674(95)90278-3. [DOI] [PubMed] [Google Scholar]
- 96.Berlin C, Bargatze RF, Campbell JJ, von Andrian UH, Szabo MC, et al. Cell. 1995;80:413–22. doi: 10.1016/0092-8674(95)90491-3. [DOI] [PubMed] [Google Scholar]
- 97.Stein JV, Cheng G, Stockton BM, Fors BP, Butcher EC, von Andrian UH. J. Exp. Med. 1999;189:37–50. doi: 10.1084/jem.189.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pavalko FM, Walker DM, Graham L, Goheen M, Doerschuk CM, Kansas GS. J. Cell Biol. 1995;129:1155–64. doi: 10.1083/jcb.129.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dwir O, Kansas GS, Alon R. J. Cell Biol. 2001;155:145–56. doi: 10.1083/jcb.200103042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Siegelman MH, DeGrendele HC, Estess P. J. Leukocyte Biol. 1999;66:315–21. doi: 10.1002/jlb.66.2.315. [DOI] [PubMed] [Google Scholar]
- 101.Nandi A, Estess P, Siegelman M. Immunity. 2004;20:455–65. doi: 10.1016/s1074-7613(04)00077-9. [DOI] [PubMed] [Google Scholar]
- 102.Dustin ML, Springer TA. Nature. 1989;341:619–24. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 103.Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, et al. Science. 2001;294:339–45. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takagi J, Petre BM, Walz T, Springer TA. Cell. 2002;110:599–11. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 105.Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. Biophys. J. 2003;85:3951–62. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Takagi J, Erickson HP, Springer TA. Nat. Struct. Biol. 2001;8:412–16. doi: 10.1038/87569. [DOI] [PubMed] [Google Scholar]
- 107.Kim M, Carman CV, Springer TA. Science. 2003;301:1720–25. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 108.Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas T, et al. Cell. 2002;110:587–97. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 109.Carman CV, Jun CD, Salas A, Springer TA. J. Immunol. 2003;171:6135–44. doi: 10.4049/jimmunol.171.11.6135. [DOI] [PubMed] [Google Scholar]
- 110.Carman CV, Springer TA. J. Cell Biol. 2004;167:377–88. doi: 10.1083/jcb.200404129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barreiro O, Yanez-Mo M, Serrador JM, Montoya MC, Vicente-Manzanares M, et al. J. Cell Biol. 2002;157:1233–45. doi: 10.1083/jcb.200112126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang AJ, Furie MB, Nicholson SC, Fischbarg J, Liebovitch LS, Silverstein SC. J. Cell Physiol. 1988;135:355–66. doi: 10.1002/jcp.1041350302. [DOI] [PubMed] [Google Scholar]
- 113.Dejana E. Nat. Rev. Mol. Cell Biol. 2004;5:261–70. doi: 10.1038/nrm1357. [DOI] [PubMed] [Google Scholar]
- 114.Mamdouh Z, Chen X, Pierini LM, Maxfield FR, Muller WA. Nature. 2003;421:748–53. doi: 10.1038/nature01300. [DOI] [PubMed] [Google Scholar]
- 115.Duncan GS, Andrew DP, Takimoto H, Kaufman SA, Yoshida H, et al. J. Immunol. 1999;162:3022–30. [PubMed] [Google Scholar]
- 116.Ostermann G, Weber KS, Zernecke A, Schroder A, Weber C. Nat. Immunol. 2002;3:151–58. doi: 10.1038/ni755. [DOI] [PubMed] [Google Scholar]
- 117.Ozaki H, Ishii K, Horiuchi H, Arai H, Kawamoto T, et al. J. Immunol. 1999;163:553–57. [PubMed] [Google Scholar]
- 118.Schenkel AR, Mamdouh Z, Chen X, Liebman RM, Muller WA. Nat. Immunol. 2002;3:143–50. doi: 10.1038/ni749. [DOI] [PubMed] [Google Scholar]
- 119.Millan J, Hewlett L, Glyn M, Toomre D, Clark P, Ridley AJ. Nat. Cell Biol. 2006;8:113–23. doi: 10.1038/ncb1356. [DOI] [PubMed] [Google Scholar]
- 120.Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, et al. Cell. 1998;94:667–77. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 121.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 122.Davis DM, Chiu I, Fassett M, Cohen GB, Mandelboim O, Strominger JL. Proc. Natl. Acad. Sci. USA. 1999;96:15062–67. doi: 10.1073/pnas.96.26.15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Batista FD, Iber D, Neuberger MS. Nature. 2001;411:489–94. doi: 10.1038/35078099. [DOI] [PubMed] [Google Scholar]
- 124.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, et al. Science. 1999;285:221–27. [Google Scholar]
- 125.Wulfing C, Davis MM. Science. 1998;282:2266–69. doi: 10.1126/science.282.5397.2266. [DOI] [PubMed] [Google Scholar]
- 126.Das V, Nal B, Dujeancourt A, Thoulouze MI, Galli T, et al. Immunity. 2004;20:577–88. doi: 10.1016/s1074-7613(04)00106-2. [DOI] [PubMed] [Google Scholar]
- 127.Ludford-Menting MJ, Oliaro J, Sacirbegovic F, Cheah ET, Pedersen N, et al. Immunity. 2005;22:737–48. doi: 10.1016/j.immuni.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 128.Chakraborty AK. Science STKE. 2002;2002:PE10. doi: 10.1126/stke.2002.122.pe10. [DOI] [PubMed] [Google Scholar]
- 129.Burroughs NJ, Wulfing C. Biophys. J. 2002;83:1784–96. doi: 10.1016/S0006-3495(02)73944-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Qi SY, Groves JT, Chakraborty AK. Proc. Natl. Acad. Sci. USA. 2001;98:6548–53. doi: 10.1073/pnas.111536798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Douglass AD, Vale RD. Cell. 2005;121:937–50. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Meiri KF. Philos. Trans. R. Soc. London Ser. B. 2005;360:1663–72. doi: 10.1098/rstb.2005.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Krause M, Sechi AS, Konradt M, Monner D, Gertler FB, Wehland J. J. Cell Biol. 2000;149:181–94. doi: 10.1083/jcb.149.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pollard TD, Borisy GG. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 135.Badour K, Zhang J, Shi F, McGavin MK, Rampersad V, et al. Immunity. 2003;18:141–54. doi: 10.1016/s1074-7613(02)00516-2. [DOI] [PubMed] [Google Scholar]
- 136.Anton IM, de la Fuente MA, Sims TN, Freeman S, Ramesh N, et al. Immunity. 2002;16:193–204. doi: 10.1016/s1074-7613(02)00268-6. [DOI] [PubMed] [Google Scholar]
- 137.Tskvitaria-Fuller I, Seth A, Mistry N, Gu H, Rosen MK, Wulfing C. J. Immunol. 2006;177:1708–20. doi: 10.4049/jimmunol.177.3.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stowers L, Yelon D, Berg LJ, Chant J. Proc. Natl. Acad. Sci. USA. 1995;92:5027–31. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ardouin L, Bracke M, Mathiot A, Pagakis SN, Norton T, et al. Eur. J. Immunol. 2003;33:790–97. doi: 10.1002/eji.200323858. [DOI] [PubMed] [Google Scholar]
- 140.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. Nat. Immunol. 2004;5:524–30. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 141.O'Keefe JP, Gajewski TF. J. Immunol. 2005;175:5581–85. doi: 10.4049/jimmunol.175.9.5581. [DOI] [PubMed] [Google Scholar]
- 142.Campi G, Varma R, Dustin ML. J. Exp. Med. 2005;202:1031–36. doi: 10.1084/jem.20051182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, et al. Nat. Immunol. 2005;6:1253–62. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- 144.Mossman KD, Campi G, Groves JT, Dustin ML. Science. 2005;310:1191–93. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 145.Van Itallie CM, Anderson JM. Annu. Rev. Physiol. 2006;68:403–29. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 146.Furuse M, Tsukita S. Trends Cell Biol. 2006;16:181–88. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 147.Claude P. J. Membr. Biol. 1978;39:219–32. doi: 10.1007/BF01870332. [DOI] [PubMed] [Google Scholar]
- 148.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. J. Cell Biol. 1998;141:1539–50. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tsukita S, Furuse M. Curr. Opin. Cell Biol. 2002;14:531–36. doi: 10.1016/s0955-0674(02)00362-9. [DOI] [PubMed] [Google Scholar]
- 150.Tsukita S, Furuse M, Itoh M. Nat. Rev. Mol. Cell Biol. 2001;2:285–93. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 151.Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Am. J. Physiol. Cell Physiol. 2002;283:C142–47. doi: 10.1152/ajpcell.00038.2002. [DOI] [PubMed] [Google Scholar]
- 152.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. J. Cell Biol. 1999;147:1351–63. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Roh MH, Liu CJ, Laurinec S, Margolis B. J. Biol. Chem. 2002;277:27501–9. doi: 10.1074/jbc.M201177200. [DOI] [PubMed] [Google Scholar]
- 154.Citi S, Sabanay H, Jakes R, Geiger B, Kendrick-Jones J. Nature. 1988;333:272–76. doi: 10.1038/333272a0. [DOI] [PubMed] [Google Scholar]
- 155.Aijaz S, Balda MS, Matter K. Int. Rev. Cytol. 2006;248:261–98. doi: 10.1016/S0074-7696(06)48005-0. [DOI] [PubMed] [Google Scholar]
- 156.Fanning AS, Jameson BJ, Jesaitis LA, Anderson JM. J. Biol. Chem. 1998;273:29745–53. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- 157.Sasaki H, Matsui C, Furuse K, Mimori-Kiyosue Y, Furuse M, Tsukita S. Proc. Natl. Acad. Sci. USA. 2003;100:3971–76. doi: 10.1073/pnas.0630649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cohen CJ, Shieh JT, Pickles RJ, Okegawa T, Hsieh JT, Bergelson JM. Proc. Natl. Acad. Sci. USA. 2001;98:15191–96. doi: 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Balda MS, Matter K. Trends Cell Biol. 2003;13:310–18. doi: 10.1016/s0962-8924(03)00105-3. [DOI] [PubMed] [Google Scholar]
- 160.Balda MS, Matter K. EMBO J. 2000;19:2024–33. doi: 10.1093/emboj/19.9.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Balda MS, Garrett MD, Matter K. J. Cell Biol. 2003;160:423–32. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nakamura T, Blechman J, Tada S, Rozovskaia T, Itoyama T, et al. Proc. Natl. Acad. Sci. USA. 2000;97:7284–89. doi: 10.1073/pnas.97.13.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Betanzos A, Huerta M, Lopez-Bayghen E, Azuara E, Amerena J, Gonzalez-Mariscal L. Exp. Cell Res. 2004;292:51–66. doi: 10.1016/j.yexcr.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 164.Farquhar MG, Palade GE. J. Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gumbiner BM. Nat. Rev. Mol. Cell Biol. 2005;6:622–34. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 166.Gumbiner BM. J. Cell Biol. 2000;148:399–404. doi: 10.1083/jcb.148.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Takeichi M. Development. 1988;102:639–55. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- 168.Larue L, Ohsugi M, Hirchenhain J, Kemler R. Proc. Natl. Acad. Sci. USA. 1994;91:8263–67. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang F, Dumstrei K, Haag T, Hartenstein V. Dev. Biol. 2004;270:350–63. doi: 10.1016/j.ydbio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 170.Adams CL, Chen YT, Smith SJ, Nelson WJ. J. Cell Biol. 1998;142:1105–19. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Adams CL, Nelson WJ. Curr. Opin. Cell Biol. 1998;10:572–77. doi: 10.1016/s0955-0674(98)80031-8. [DOI] [PubMed] [Google Scholar]
- 172.Ehrlich JS, Hansen MD, Nelson WJ. Dev. Cell. 2002;3:259–70. doi: 10.1016/s1534-5807(02)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Dev. Cell. 2002;3:367–81. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 174.Shewan AM, Maddugoda M, Kraemer A, Stehbens SJ, Verma S, et al. Mol. Biol. Cell. 2005;16:4531–42. doi: 10.1091/mbc.E05-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cereijido M, Robbins ES, Dolan WJ, Rotunno CA, Sabatini DD. J. Cell Biol. 1978;77:853–80. doi: 10.1083/jcb.77.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ando-Akatsuka Y, Yonemura S, Itoh M, Furuse M, Tsukita S. J. Cell Physiol. 1999;179:115–25. doi: 10.1002/(SICI)1097-4652(199905)179:2<115::AID-JCP1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 177.Anastasiadis PZ, Reynolds AB. Curr. Opin. Cell Biol. 2001;13:604–10. doi: 10.1016/s0955-0674(00)00258-1. [DOI] [PubMed] [Google Scholar]
- 178.Davis MA, Ireton RC, Reynolds AB. J. Cell Biol. 2003;163:525–34. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nelson WJ, Nusse R. Science. 2004;303:1483–87. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Aberle H, Schwartz H, Kemler R. J. Cell Biochem. 1996;61:514–23. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 181.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Proc. Natl. Acad. Sci. USA. 1995;92:8813–17. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kobielak A, Fuchs E. Nat. Rev. Mol. Cell Biol. 2004;5:614–25. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hirokawa N, Keller TC, Chasan R, Mooseker MS. J. Cell Biol. 1983;96:1325–36. doi: 10.1083/jcb.96.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kobielak A, Pasolli HA, Fuchs E. Nat. Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ikeda W, Nakanishi H, Miyoshi J, Mandai K, Ishizaki H, et al. J. Cell Biol. 1999;146:1117–32. doi: 10.1083/jcb.146.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sato T, Fujita N, Yamada A, Ooshio T, Okamoto R, et al. J. Biol. Chem. 2006;281:5288–99. doi: 10.1074/jbc.M510070200. [DOI] [PubMed] [Google Scholar]
- 188.Watabe-Uchida M, Uchida N, Imamura Y, Nagafuchi A, Fujimoto K, et al. J. Cell Biol. 1998;142:847–57. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Cell. 2001;104:605–17. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 190.Lien WH, Klezovitch O, Fernandez TE, Delrow J, Vasioukhin V. Science. 2006;311:1609–12. doi: 10.1126/science.1121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Development. 1995;121:3529–37. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 192.Brembeck FH, Rosario M, Birchmeier W. Curr. Opin. Genet. Dev. 2006;16:51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 193.Harris TJ, Peifer M. Trends Cell Biol. 2005;15:234–37. doi: 10.1016/j.tcb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 194.Wessells NK, Spooner BS, Ash JF, Bradley MO, Luduena MA, et al. Science. 1971;171:135–43. doi: 10.1126/science.171.3967.135. [DOI] [PubMed] [Google Scholar]
- 195.Young PE, Pesacreta TC, Kiehart DP. Development. 1991;111:1–14. doi: 10.1242/dev.111.1.1. [DOI] [PubMed] [Google Scholar]
- 196.Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, Wieschaus EF. Development. 2005;132:4165–78. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- 197.Haigo SL, Hildebrand JD, Harland RM, Wallingford JB. Curr. Biol. 2003;13:2125–37. doi: 10.1016/j.cub.2003.11.054. [DOI] [PubMed] [Google Scholar]
- 198.Hildebrand JD. J. Cell Sci. 2005;118:5191–203. doi: 10.1242/jcs.02626. [DOI] [PubMed] [Google Scholar]
- 199.Pilot F, Philippe JM, Lemmers C, Lecuit T. Nature. 2006;442:580–84. doi: 10.1038/nature04935. [DOI] [PubMed] [Google Scholar]
- 200.Knust E, Bossinger O. Science. 2002;298:1955–59. doi: 10.1126/science.1072161. [DOI] [PubMed] [Google Scholar]
- 201.Nelson WJ. Nature. 2003;422:766–74. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bazzoni G. Curr. Opin. Cell Biol. 2003;15:525–30. doi: 10.1016/s0955-0674(03)00104-2. [DOI] [PubMed] [Google Scholar]
- 203.Bazzoni G, Martinez-Estrada OM, Mueller F, Nelboeck P, Schmid G, et al. J. Biol. Chem. 2000;275:30970–76. doi: 10.1074/jbc.M003946200. [DOI] [PubMed] [Google Scholar]
- 204.Bazzoni G, Martinez-Estrada OM, Orsenigo F, Cordenonsi M, Citi S, Dejana E. J. Biol. Chem. 2000;275:20520–26. doi: 10.1074/jbc.M905251199. [DOI] [PubMed] [Google Scholar]
- 205.Martinez-Estrada OM, Villa A, Breviario F, Orsenigo F, Dejana E, Bazzoni G. J. Biol. Chem. 2001;276:9291–96. doi: 10.1074/jbc.M006991200. [DOI] [PubMed] [Google Scholar]
- 206.Ebnet K, Suzuki A, Horikoshi Y, Hirose T, Meyer zu Brickwedde MK, et al. EMBO J. 2001;20:3738–48. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, Tsukita S. J. Cell Biol. 2001;154:491–97. doi: 10.1083/jcb.200103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ohno S. Curr. Opin. Cell Biol. 2001;13:641–48. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- 209.Joberty G, Petersen C, Gao L, Macara IG. Nat. Cell Biol. 2000;2:531–39. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 210.Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, et al. Curr. Biol. 2004;14:1425–35. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 211.Roh MH, Margolis B. Am. J. Physiol. Renal. Physiol. 2003;285:F377–87. doi: 10.1152/ajprenal.00086.2003. [DOI] [PubMed] [Google Scholar]
- 212.Wodarz A, Hinz U, Engelbert M, Knust E. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 213.Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi MH, Medina E, et al. Mol. Biol. Cell. 2004;15:1324–33. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Shin K, Straight S, Margolis B. J. Cell Biol. 2005;168:705–11. doi: 10.1083/jcb.200408064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Straight SW, Shin K, Fogg VC, Fan SL, Liu CJ, et al. Mol. Biol. Cell. 2004;15:1981–90. doi: 10.1091/mbc.E03-08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sotillos S, Diaz-Meco MT, Caminero E, Moscat J, Campuzano S. J. Cell Biol. 2004;166:549–57. doi: 10.1083/jcb.200311031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Craig L, Pique ME, Tainer JA. Nat. Rev. Microbiol. 2004;2:363–78. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 218.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Cell. 2002;109:913–23. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 219.Mengaud J, Ohayon H, Gounon P, Mege RM, Cossart P. Cell. 1996;84:923–32. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 220.Pizarro-Cerda J, Cossart P. J. Pathol. 2006;208:215–23. doi: 10.1002/path.1888. [DOI] [PubMed] [Google Scholar]
- 221.Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, et al. Science. 2001;292:1722–25. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 222.Pentecost M, Otto G, Theriot JA, Amieva MR. PLoS Pathog. 2006;2:e3. doi: 10.1371/journal.ppat.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.van Raaij MJ, Chouin E, van der Zandt H, Bergelson JM, Cusack S. Structure. 2000;8:1147–55. doi: 10.1016/s0969-2126(00)00528-1. [DOI] [PubMed] [Google Scholar]
- 224.Freimuth P, Springer K, Berard C, Hainfeld J, Bewley M, Flanagan J. J. Virol. 1999;73:1392–98. doi: 10.1128/jvi.73.2.1392-1398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Walters RW, Freimuth P, Moninger TO, Ganske I, Zabner J, Welsh MJ. Cell. 2002;110:789–99. doi: 10.1016/s0092-8674(02)00912-1. [DOI] [PubMed] [Google Scholar]
- 226.Kusters JG, van Vliet AH, Kuipers EJ. Clin. Microbiol. Rev. 2006;19:449–90. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Backert S, Meyer TF. Curr. Opin. Microbiol. 2006;9:207–17. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 228.Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. Science. 2003;300:1430–34. doi: 10.1126/science.1081919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Churin Y, Al-Ghoul L, Kepp O, Meyer TF, Birchmeier W, Naumann M. J. Cell Biol. 2003;161:249–55. doi: 10.1083/jcb.200208039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Spears KJ, Roe AJ, Gally DL. FEMS Microbiol. Lett. 2006;255:187–202. doi: 10.1111/j.1574-6968.2006.00119.x. [DOI] [PubMed] [Google Scholar]
- 231.Liu H, Magoun L, Leong JM. Infect. Immun. 1999;67:2045–49. doi: 10.1128/iai.67.4.2045-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sinclair JF, O'Brien AD. J. Biol. Chem. 2002;277:2876–85. doi: 10.1074/jbc.M110230200. [DOI] [PubMed] [Google Scholar]
- 233.Nougayrede JP, Fernandes PJ, Donnenberg MS. Cell Microbiol. 2003;5:359–72. doi: 10.1046/j.1462-5822.2003.00281.x. [DOI] [PubMed] [Google Scholar]
- 234.Caron E, Crepin VF, Simpson N, Knutton S, Garmendia J, Frankel G. Curr. Opin. Microbiol. 2006;9:40–45. doi: 10.1016/j.mib.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 235.Amagai M, Matsuyoshi N, Wang ZH, Andl C, Stanley JR. Nat. Med. 2000;6:1275–77. doi: 10.1038/81385. [DOI] [PubMed] [Google Scholar]
- 236.Wu Z, Nybom P, Magnusson KE. Cell Microbiol. 2000;2:11–17. doi: 10.1046/j.1462-5822.2000.00025.x. [DOI] [PubMed] [Google Scholar]
- 237.Wu S, Lim KC, Huang J, Saidi RF, Sears CL. Proc. Natl. Acad. Sci. USA. 1998;95:14979–84. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Fukata M, Kaibuchi K. Nat. Rev. Mol. Cell Biol. 2001;2:887–97. doi: 10.1038/35103068. [DOI] [PubMed] [Google Scholar]
- 239.Nusrat A, von Eichel-Streiber C, Turner JR, Verkade P, Madara JL, Parkos CA. Infect. Immun. 2001;69:1329–36. doi: 10.1128/IAI.69.3.1329-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Landraud L, Pulcini C, Gounon P, Flatau G, Boquet P, Lemichez E. Int. J. Med. Microbiol. 2004;293:513–18. doi: 10.1078/1438-4221-00295. [DOI] [PubMed] [Google Scholar]
- 241.Hopkins AM, Walsh SV, Verkade P, Boquet P, Nusrat A. J. Cell Sci. 2003;116:725–42. doi: 10.1242/jcs.00300. [DOI] [PubMed] [Google Scholar]
- 242.Boquet P, Lemichez E. Trends Cell Biol. 2003;13:238–46. doi: 10.1016/s0962-8924(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 243.Alto NM, Shao F, Lazar CS, Brost RL, Chua G, et al. Cell. 2006;124:133–45. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 244.Kenny B, Jepson M. Cell Microbiol. 2000;2:579–90. doi: 10.1046/j.1462-5822.2000.00082.x. [DOI] [PubMed] [Google Scholar]
- 245.Dean P, Kenny B. Mol. Microbiol. 2004;54:665–75. doi: 10.1111/j.1365-2958.2004.04308.x. [DOI] [PubMed] [Google Scholar]