Abstract
Ubiquitination of proliferating cell nuclear antigen (PCNA) to ub-PCNA is essential for DNA replication across bulky template lesions caused by UV radiation and alkylating agents, as ub-PCNA orchestrates the recruitment and switching of translesion synthesis (TLS) polymerases with replication polymerases. This allows replication to proceed, leaving the DNA to be repaired subsequently. Defects in a TLS polymerase, Pol η, lead to a form of Xeroderma pigmentosum, a disease characterized by severe skin sensitivity to sunlight damage and an increased incidence of skin cancer. Structurally, however, information on how ub-PCNA orchestrates the switching of these two classes of polymerases is lacking. We have solved the structure of ub-PCNA and demonstrate that the ubiquitin molecules in ub-PCNA are radially extended away from the PCNA without structural contact aside from the isopeptide bond linkage. This unique orientation provides an open platform for the recruitment of TLS polymerases through ubiquitin-interacting domains. However, the ubiquitin moieties, to the side of the equatorial PCNA plane, can place spatial constraints on the conformational flexibility of proteins bound to ub-PCNA. We show that ub-PCNA is impaired in its ability to support the coordinated actions of Fen1 and Pol δ in assays mimicking Okazaki fragment processing. This provides evidence for the novel concept that ub-PCNA may modulate additional DNA transactions other than TLS polymerase recruitment and switching.
Keywords: FEN1, Okazaki fragment maturation, PCNA, polymerase delta, polymerase switching, translesion synthesis, Ubiquitination
Introduction
The maintenance of genomic integrity requires the faithful replication of genetic information during cell division by replication polymerases as well as an intricate network of cellular responses to DNA damage that lead to their repair.1 PCNA is a homotrimeric DNA sliding clamp that serves as an essential partner of DNA polymerase δ (Pol δ) and enables processive DNA synthesis. It also interacts with numerous other proteins involved in DNA replication and repair.2 Pol δ is suggested to be largely responsible for lagging-strand DNA synthesis in eukaryotic cells3 and also participates in the removal of RNA-DNA primers by strand displacement to create flaps that are removed by the flap endonucleases Fen1 or Dna2, creating nicks that can be joined by DNA ligase.4
Bulky lesions caused by UV radiation and alkylating agents can pose barriers to replication polymerases and, thus, progression of replication forks in S-phase cells. In response, eukaryotic cells activate a DNA damage bypass maneuver in which specialized translesion synthesis (TLS) polymerases extend the daughter strand across lesions.5 This DNA damage tolerance mechanism allows replication to proceed. The TLS polymerases Pol η, ι, κ and Rev1 are members of the Y family pols, while Pol ζ is a B family pol. The Y family TLS pols are not processive and generally error-prone; however, in some cases, a TLS pol can perform largely error-free translesion synthesis in vivo, as in the case of the bypass of UV-induced thymine dimers by Pol η.6 Because TLS pols are error-prone, their activities are tightly regulated to avoid interference with the normal processes of DNA replication. The seminal discovery by Hoege et al. was that PCNA is monoubiquitinated on K164 by RAD6 and RAD18 in budding yeast upon UV exposure.7 Consequently, ub-PCNA provides the key regulatory element in the recruitment of TLS pols to stalled replication forks and a polymerase switch between the TLS pols and the classical replication pols.8,9 Loss of any of the genes in the pathway for PCNA ubiquitination leads to increased cellular susceptibility to DNA damage.8,9 The monoubiquitination of PCNA mainly occurs during S phase, when DNA damaging agents are encountered and persist for a significant period of time.10 However, recent studies also indicate that PCNA ubiquitination can be separated from the cell cycle in certain genetic backgrounds; thus, post-replication repair may also be active in other phases of the cell cycle.11-14
There have been extensive studies at structural, functional and genetic levels to understand the roles of PCNA ubiquitination in the process of translesion synthesis. Pol η has been the most extensively studied in regard to both its recruitment to DNA repair foci and subsequent switching with Pol δ.6,15-18 The TLS pols possess PCNA binding peptide motifs (PIP boxes) as well as ubiquitin binding domains (UBZ/UBM) located in their extended C-terminal regions. These domains and motifs provide the means for recruiting the TLS polymerases to ub-PCNA as well as the means for competing with and displacing Pol δ. Those studies support a model in which ubiquitin binding greatly increases the affinity of TLS polymerases for ub-PCNA.19 In addition, it has been shown that PCNA must be loaded onto DNA for ubiquitination, and all three PCNA monomers are concurrently ubiquitinated.20,21
Despite the immense interest in understanding the functions of ub-PCNA, no structural information on native ub-PCNA is currently available due to the difficulties in obtaining adequate amounts of ub-PCNA. Here, we determined the structure of native human ub-PCNA in order to gain insights into the molecular basis for polymerase switching. We also show that the presence of ubiquitin may influence other aspects of DNA replication through modulating the binding of other PCNA-interacting proteins, specifically that of Fen1.
Results
PCNA monoubiquitination and crystallization.
Fully monoubiquitinated PCNA (ub-PCNA) was prepared by utilizing our observations that PCNA can be stoichiometrically ubiquitinated on K164 by the E3 ligase RNF8 and the E2 enzyme UbcH5 in vitro.22 We were able to purify PCNA ubiquitinated on all three monomers on a milligram scale to near-homogeneity (Fig. S1). We crystallized ub-PCNA with sodium citrate as precipitant. The crystals belong to space group P43212 with cell dimensions of a = 161.05, b = 161.05, c = 97.36 and diffract to 2.9 Å.
Structure of monoubiquitinated PCNA.
The structure was solved by molecular replacement with a PCNA monomer (1 vym) 23 as the initial search model. Three PCNA monomers in each asymmetric unit were identified by molecular replacement. Two of the three ubiquitin molecules were clearly identifiable in the initial (fo-fc) map (Fig. S2), whereas the density of the third ubiquitin was not sufficient for model building. The structure was refined to a nominal resolution of 2.9 Å (Table S1) with a trimeric ring of PCNA (chains A–C) and two molecules of ubiquitin (chains D and E) in each asymmetric unit.
Three PCNA monomers in each asymmetric unit form the prototypical trimeric ring structure.24 The PCNA trimer in ub-PCNA superimposes well on the PCNA portions of various PCNA complexes with minimal structural changes. The two ubiquitin molecules are projected radially away from the trimeric ring of PCNA at an angle of about 30° to the equatorial plane, toward the back face of PCNA (Fig. 1A and B). The overall orientation of the two ubiquitin molecules is similar, with one (E) extending further from the PCNA ring than the other (D) by about 2 Å. The position of the third ubiquitin molecule corresponding to that of the other two ubiquitins is occupied by a symmetry molecule in the crystal packing (Fig. S3). Unresolved electron densities indicate that the ubiquitin molecule is pushed into the groove on either side of the ridge within the PCNA plane (Fig. S3), in agreement with one of the conformations proposed in molecular simulation studies.25 Interestingly, ubiquitin D forms contacts with ubiquitin E from a neighboring symmetry unit with a buried surface area of 285 Å2, thus contributing to the defined density of both moieties (Fig. S4). Modeling of the third ubiquitin based on the structure of ub-D provides a view of the trimeric ub-PCNA structure (Fig. 1C and D).
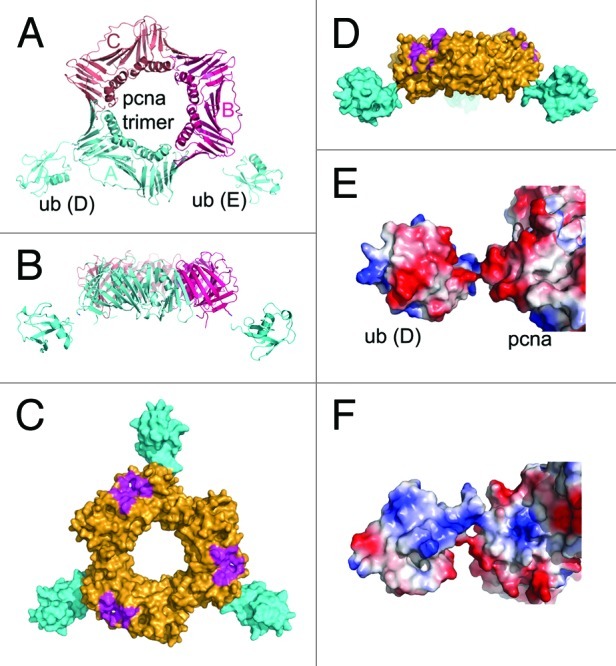
Figure 1. Overview of the ub-PCNA structure. (A) Structure of ub-PCNA in ribbon representation, viewed along the DNA axis. The two ubiquitins, ub-D and ub-E are shown in cyan. (B) Side view of ub-PCNA, at a 90° rotation from (A) looking into the plane of the PCNA structure. (C) Overview of ub-PCNA when the third ubiquitin molecule is modeled in by PCNA symmetry in surface representation. The surface in magenta represents PIP binding site. (D) Side view of ub-PCNA with all three ubquitin molecules modeled in. (E) The negative electrostatic surface of ubiquitin is adjacent to a negatively charged patch of PCNA. (F) The positive electrostatic surface of ubiquitin is adjacent to a positive electrostatic patch on PCNA.
In this structure, there are no other contacts between ubiquitin and PCNA aside from the isopeptide linkage. Ubiquitin has negatively charged and positively charged surfaces located on opposite sides of the molecule, oriented almost perpendicular to the plane of the PCNA trimer. The acidic face is adjacent to an acidic patch on PCNA (Fig. 1E), resulting in the formation of a continuous acidic surface, and the basic face is similarly adjacent to a basic patch on PCNA (Fig. 1F). These interactions contribute to the extended orientation of ubiquitin away from the PCNA surface. The overall structure of ub-PCNA displays high-temperature factors, with that of the ubiquitin moieties being even higher, indicating overall significant conformational flexibility.
PCNA-interacting proteins possess a short motif, the PIP-box, which binds to PCNA through a hydrophobic pocket next to the interdomain connector loop of PCNA, as shown for the PIP box of the p68 subunit of Pol δ (Fig. 2A).26 The ubiquitin molecules extend away from the PCNA ring and bend toward the back face, placing them far away from the PIP binding sites, which are on the front face (Fig. 2A). Thus, the ubiquitin moieties would not be expected to directly interfere with interactions of a PIP peptide with PCNA. We examined the abilities of PCNA and ub-PCNA to stimulate Pol δ activity, and found that they were equally effective (Fig. 2B–D). Similar findings have been reported for yeast ub-PCNA.20 Thus, the ubiquitin does not appear to affect the binding of Pol δ. Inspection of structures for the PCNA-RFC clamp-loader complex with PCNA shows that RFC resides exclusively on the front face of PCNA (Fig. S5),27 consistent with the fact that RFC can efficiently load ub-PCNA (data not shown). Also shown in Figure 2E and F are the locations of the UBZ binding surfaces of ubiquitin, which interact with the UBZ/UBM ubiquitin binding domains of Pol η and other TLS pols (Fig. S6).16
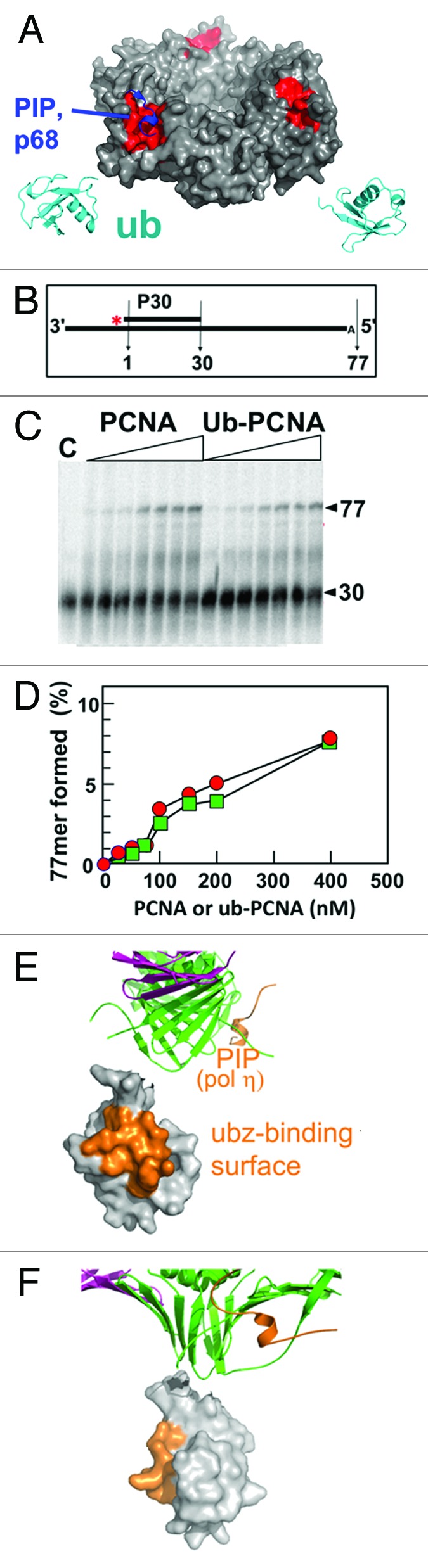
Figure 2. The orientation of ubiquitin molecules relative to PIP binding sites. (A) The ubiquitin molecules are shown in ribbon (cyan). The PCNA in surface representation is shown with the hydrophobic pocket for binding PIP boxes in red, with the PIP box of the p68 subunit of Pol δ in blue. Ubiquitin, on the back face of ub-PCNA, is far away from the PIP (red) binding sites on the front face. (B) Substrate design for polymerase assay. (C) Pol δ was incubated with increasing concentrations of ub-PCNA or PCNA (0, 25, 50, 75, 100, 150, 200, 400 nM) for 7.5 min, and the products examined by gel electrophoresis. Lane c is the control incubation without PCNA at 7.5 min. (D) Quantification of the products of the reaction from panel (C), values for PCNA are shown as solid circles and ub-PCNA as solid squares. (E and F) The relative orientations of the UBZ-binding surface (orange) on ubiquitin and the PIP box peptide of Pol η (orange) modeled by superimposing the structure of the PCNA-PIP peptide of Pol η (2ZVK).
Effect of monoubiquitination on Okazaki fragment maturation.
We considered the question of whether the ubiquitin moieties of ub-PCNA could interfere with the binding of other proteins involved in DNA replication or processing. Examination of the structure of Fen1, which plays an important role in replication of the lagging strand during the maturation of Okazaki fragments in coordination with Pol δ,28,29 shows that the ubiquitins could impose steric restrictions on Fen1 interactions with ub-PCNA. There are three Fen1 molecules bound to the PCNA trimer in the structure of the human Fen1-PCNA complex, each displaying a different conformation.30 In this structure, the N-terminal catalytic core of Fen1 is joined by a short linker sequence to the C-terminal PIP-box, which interacts with PCNA, allowing a wide range of movement of the catalytic core. The three conformations cover an angular range of ca. 100° from the front to the back face of PCNA. When we align our structure of ub-PCNA with that of the Fen1-PCNA complex, one of the three conformations of Fen1 is precluded due to steric hindrance from ubiquitin (Fig. 3A and B). The conformational change of PCNA-bound Fen1 from the engagement state (catalysis) to the idle state can be further illustrated with a morph movie (Video S1, 20 frames). The conversion of the two conformations involves a rotation around the hinge region.30 A snapshot of the conformation at the half-way point (frame 11) shows that the negatively charged surfaces on ubiquitin and Fen1 face each other, so that electrostatic repulsion would further limit the conformational flexibility of Fen1 bound to ub-PCNA (Fig. 3C). Thus, these structural analyses predict that ubiquitination of PCNA would restrict the conformational flexibility of Fen1. The coordination of Fen1 with Pol δ for flap removal requires that either Fen1 or Pol δ reside to the side of PCNA ring during Okazaki fragment maturation, as their access to the DNA primer/template is mutually exclusive.
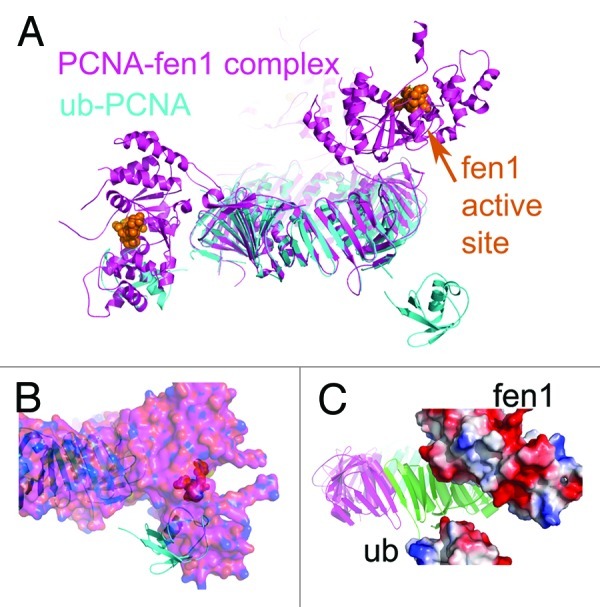
Figure 3. Structural alignment of ub-PCNA (cyan) with the PCNA-fen1 complex (magenta, 1UL1). (A) When the PCNA portions in the structures of ub-PCNA (cyan) and the PCNA-Fen1 complex are aligned, one of the three conformations (to the side of the PCNA ring) of Fen1 overlaps with ubiquitin. (B) Surface representation of the PCNA-Fen1 complex further illustrates the steric hindrance of ubiquitin molecules (cyan). When the PCNA portions are aligned as in (A), part of the ubiquitin molecule is buried in Fen1 (magenta). (C) Potential electrostatic repulsion between Fen1 and ubiquitin results from modeling the conformational changes among different conformations of Fen1. The conformational changes of Fen1 were simulated by a morph movie (Video S1) generated on the Molecular Movement Database morph server at Yale University at frame 20. When we take the middle stage (frame 11) of the conformational transition, the negatively charged electrostatic surface on ubiquitin is facing a negatively charged patch on Fen1.
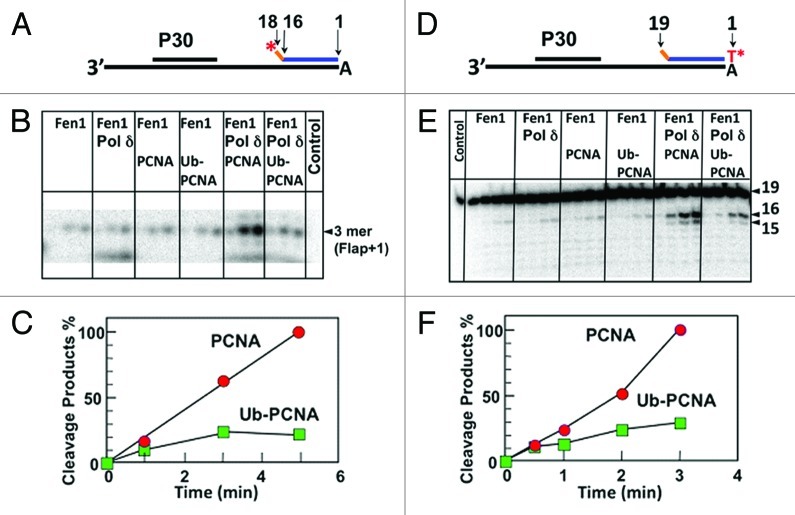
In order to test the hypothesis that ubiquitination creates a steric hindrance to a coordination of Fen1 and Pol δ, we compared the activity of Fen1 in the presence of Pol δ with either PCNA or ub-PCNA using gapped oligonucleotide substrates to mimic the Okazaki fragment maturation process.28 We also determined that ub-PCNA did not have a reduced affinity for Fen1 (not shown). The substrates consist of a 90mer oligonucleotide template annealed to a 30mer primer and a downstream blocking oligonucleotide with two non-complementary 5' nucleotides to provide a 2 nt flap structure (Fig. 4A and D). We labeled the blocking DNA with 32P, either on the 5'-end (Fig. 4A) or on the 3' end (Fig. 4D). The reactions were performed at near-physiological ionic strength. Pol δ filled the gap in the presence of either PCNA or ub-PCNA and dNTPs to produce the nicked substrates (Fig. S7). A representative experiment using the 5' end-labeled substrate is shown in Figure 4B; the major product is a 3 nt fragment, which is consistent with the flap + 1 specificity demonstrated with Fen1 flap substrates.31,32 The time course of formation of the 3 nt cleavage product is shown in Figure 4C. A low level of reaction was observed with Fen1 alone. In the presence of Fen1, Pol δ and PCNA, a marked stimulation of product formation is observed. When ub-PCNA was used instead of PCNA, the initial flap cleavage is significantly inhibited (Fig. 4B and C). The 3' end-labeled blocking oligonucleotide of 19 nt was used in order to observe the initial cleavage of the preformed flap as well as the subsequent cleavages, which reflect the combined actions of Pol δ and Fen1 (Fig. 4D). The initial cleavage of the preformed 2 nt flap gave rise to a 16 nt fragment, representing a flap + 1 cleavage. Subsequent cleavages produced a DNA ladder shortened by one nucleotide (Fig. 4E). These results reflect a cooperative strand displacement/flap removal performed by Pol δ and Fen1 in the presence of PCNA (Fig. S7b) in which Pol δ strand displacement is limited, so that a single nucleotide is cleaved by Fen1, rather than larger flaps of 2–10 nucleotides that are preferred by Fen1 alone. This has been previously demonstrated with yeast Pol δ and Fen133 but has not been previously demonstrated for human Pol δ and Fen1. The quantification of the 16 nt product confirmed the inhibition observed with the 3' end-labeled substrate (Fig. 4F). These results indicate that the coordinated actions of Pol δ/Fen1 are inhibited by the presence of the ubiquitin molecules in ub-PCNA. The reduced ability of ub-PCNA to support the actions of Pol δ and Fen1 is consistent with our structural analysis, which indicates that ubiquitination of PCNA would hinder the conformational flexibility of Fen1. Also noteworthy is that there is no qualitative change in the products formed, i.e., a selective inhibition of Fen1 that might result in longer flaps from Pol δ strand displacement is not observed. This is consistent with a tight coupling of the Pol δ and Fen1 actions.
Figure 4. Ubiquitination of PCNA impairs the coordinated processing of Okazaki fragments by Fen1 and Pol δ in vitro. (A) The oligonucleotide substrate consists of a 30 nucleotide primer and a 18 nucleotide blocking oligonucleotide containing a 2nt flap labeled with 32P at the 5′-end annealed to a template. (B) Phosphorimage of a sequencing gel of Fen1 activity assays showing the formation of the 3nt cleavage product from the 5′-end label substrate in the presence of different enzyme combinations with increasing time (1, 3 and 5 min). (The complete gel is shown in Fig. S8a). (C) Quantification of cleavage products in the preceding panel. The relative amounts of 3nt cleavage products formed was expressed as a percentage of highest amount of the product formed in the assay. Data in the presence of PCNA, Pol δ and Fen1 are shown as solid circles, in the presence of ub-PCNA, Pol δ and Fen1 as solid squares. (D) Template design when the 3′-end of the flap oligonucleotide is labeled (Fig. S7B). (E) Phosphorimage of Fen1 activity assays when the 3′-end is labeled showing the formation of the 16nt product from the initial flap cleavage and the subsequent limited strand displacement/flap cleavage (the complete gel is shown in Fig. S8b). (F) The relative amounts of cleavage products (16mer oligonucleotide).
Discussion
The ub-PCNA monomer is an unusual structure representing two independent globular proteins linked by a tether with limited direct surface contacts, in which the ubiquitin is radially extended away from PCNA. A structure of split yeast ub-PCNA has been reported, in which PCNA was split into two fragments between residues 163 and 164, and a ubiquitin molecule was fused via its C terminus and a linker (two glycines, for total of four glycine residues N-terminal to K164) of the C-terminal PCNA fragment.34 This structure differs from native ub-PCNA, in that the ubiquitin has extensive contacts with PCNA and is tucked under the plane of PCNA, with the UBZ domain interacting with PCNA (Fig. S9). In vivo, split ub-PCNA exhibits the ability to confer UV resistance in yeast strains in which it replaced PCNA.34 However, other fusions of ubiquitin to PCNA (N-terminal, C-terminal or split ub-PCNA) can substitute monoubiquitination PCNA in Saccharomyces cerevisiae to confer UV resistance as well.34,35
In solution, the ubiquitin moiety in our structure could be expected to display a limited range of motion governed by the length of the tether as well as interactions with the surface of PCNA. We cannot discount the possibility that the location of ubiquitin in split ub-PCNA is one that is possible with native ub-PCNA. However, our structure indicates that the electrostatic repulsions we presented in Figure 1 between ubiquitin and PCNA (which is conserved in yeast ub-PCNA when modeled) could contribute to the extension of the ubiquitin moiety. The ionic repulsions between PCNA and ubiquitin are present despite the fact that human ub-PCNA was crystallized in high ionic strength (> 1 M sodium citrate), indicating that the conformation adopted in the native ub-PCNA is preferred. A small angle X-ray scattering (SAXS) analysis of split PCNA in solution provided a predicted shape, in which a single ubiquitin was shown projecting away from the PCNA surface, indicating that ubiquitin did not occupy the position observed in the crystal structure.25 In the same study, computation modeling of ub-PCNA using tethered Brownian dynamics was used to generate a large number of conformations of a ubiquitin molecule tethered to a PCNA trimer. This showed a range of favored conformations that lie to the side of PCNA in the equatorial plane.25 Our structure is consistent with these findings for split ub-PCNA and suggests that the captured conformation in our crystals is a relevant conformation in solution.
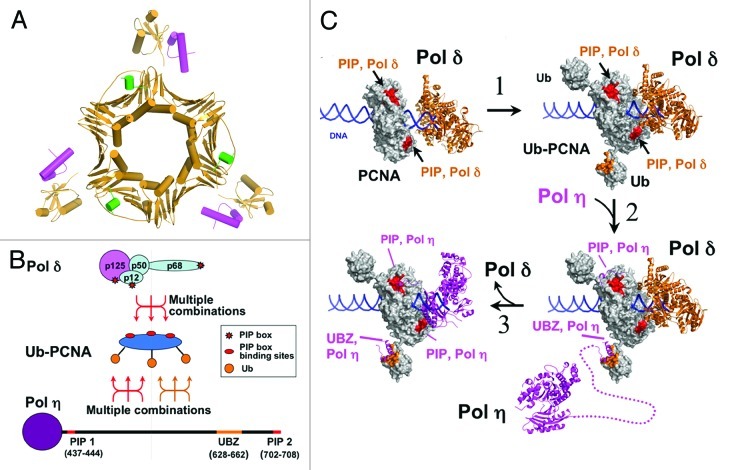
One of the key functions of ub-PCNA is to recruit translesion polymerases to replication forks stalled at sites of DNA damage. The recruitment of these polymerases depends on their possession of multiple ubiquitin binding domains (UBZ/UBM domains) and PIP boxes.36 These reside in their respective C termini, which range in length from 250–400 residues,16 much of which is unstructured.37 The conserved features of extended C-terminal regions as well as duplication of protein interaction domains can now be placed in context with the crystal structure of ub-PCNA. Multiple dynamic attachment modes utilizing different combinatorial interactions are possible, given that ub-PCNA is a hexavalent molecule (Fig. 5A and B). The distant location of the ubiquitins on the opposite face from the PIP binding surfaces on PCNA imposes spatial demands on the spacing of the UBZ/UBM and PIP-boxes in the C termini of the TLS pols. The length of these C termini, if present as unstructured regions, is unknown but could be quite long, given that they span several hundred residues.16,17 Thus, the structure of ub-PCNA now provides a rationale for the conserved features of the TLS pols viz., multiple protein interaction sites spaced along an extended C-terminus. The extension of the ubiquitins away from PCNA trimer also supports a model in which multiple TLS pols may be associated with ub-PCNA at the stalled replication fork,10 particularly when the extended nature of their C termini are considered. The combinatorial nature of TLS pol-ub-PCNA interactions could provide the versatility needed for arrangement of multiple TLS pols on ub-PCNA. This flexibility may account for the observations that various fusions of ubiquitin to PCNA can substitute monoubiquitinated PCNA in S. cerevisiae to confer UV resistance.34,35 However, it is not known if these substitutions are functional in mammalian cells.
Figure 5. Proposed model of polymerase recruitment and switching. (A) Structure of ub-PCNA with PIP motifs (cyan, from p21)64 and UBM domains (magenta)61 modeled in. (B) A diagrammatic view illustrates the potential for combinatorial interactions between ub-PCNA and a TLS polymerase such as Pol η. The upper figure shows ub-PCNA, emphasizing that ubiquitination converts PCNA from a trivalent to a hexavalent molecule, with three binding sites for UBZ on the side and three binding sites for PIP boxes on the front. This allows multiple modes of interaction with the C-terminus of Pol η. The bar shows the C-terminal region of Pol η to approximate scale, with the location of the UBZ and two PIP boxes as shown. Also shown is Pol δ, which has at least 3 PIP sequences. (C) The catalytic domain of human Pol δ (brown) is shown modeled onto PCNA (3K4X) and a DNA primer template based on the structure of yeast Pol δ (3IAY). The hydrophobic pockets on PCNA for binding of PIP motifs is shown in red. (1) PCNA is ubiquitinated when the replication fork stalls. The UBZ/UBM binding domains on ubiquitin are shown in brown. (2) Ubiquitin molecules on ub-PCNA then recruit Pol η (3MFH) through its UBZ domain. The dotted line indicates the extended C-terminus of Pol η. 3, Once bound to ub-PCNA, Pol η switching with Pol δ occurs via an intramolecular process as its PIP boxes compete with and displaces those of Pol δ.
The juxtaposition of these general features of the arrangement of protein-interaction sites in the C termini of TLS pols with the structure of ub-PCNA now allows the consideration and testing of models for their recruitment to the ub-PCNA/Pol δ complex at the replication fork and the switching mechanisms involved. One such model is shown in Figure 5C, which essentially envisages a two-stage process. Following ubiquitination of PCNA, the first stage is binding of the TLS pol (Pol η) via its UBZ domain to ubiquitin (Figs. 2 and 5C). Once bound to ubiquitin, the switching process is an intramolecular reaction (Figs. 3 and 5C) that favors the ability of the TLS pols to displace Pol δ. In this regard, it is noted that Pol δ has the potential for a trivalent interaction with PCNA, given that the p125,38 p68 39 and p12 40 subunits can interact with PCNA (Fig. 5B). Given that both Pol δ and the TLS polymerases have multiple PIP motifs, this could involve a stepwise process of displacement of the Pol δ binding interactions during switching.
The front face of PCNA acts as a platform on which several proteins can engage the primer terminus/template in a cooperative manner. The replication proteins of the hyperthermophilic archeon Sulfolobus solfataricus represent an orthologous system to that of the eukaryotes.41,42 In this system, PCNA consists of three non-identical subunits, each has a selective specificity for binding of the polymerase (PolB1), Fen1 and DNA ligase, respectively. Elegant structural and biochemical studies of the S. solfataricus replication proteins have provided experimental support for a model of a lagging-strand replication complex in which PolB1, Fen1 and DNA ligase are simultaneously bound to PCNA. In this model, a single PCNA molecule functions as a platform on which PolB1, Fen1 and DNA ligase are placed in a stereochemically defined order and act sequentially in Okazaki fragment processing as a physical complex.43-45 In order to permit their individual access to the primer terminus, all three enzymes have to adopt multiple conformations, such that they can switch positions at the primer terminus in a hand-off process.42,46 These studies provide an important perspective for eukaryotic Okazaki fragment processing. A quaternary structural complex of a polymerase, Fen1 and DNA ligase I in eukaryotes has not been established. However, there is functional evidence in that Pol δ and Fen1 act cooperatively in the presence of PCNA, since their combined actions lead to cleavage products of a single nucleotide, rather than the preferred flap cleavage of 2–10 nucleotides exhibited by Fen1.29,33,47 Structures of both bacterial46 and eukaryotic Fen1 32 and DNA ligase48 show that they have hinge regions, which allow large domains to move into different conformations that would facilitate their alternating access to the DNA primer terminus. Our interpretation of the inhibition of the cooperative activity of Pol δ/Fen1 is that Fen1 is impeded as it attempts to move to the side of PCNA, occupied by the ubiquitin molecules, to hand off the primer/template terminus to Pol δ and vise versa. This inhibition indirectly supports the possibility that the cooperative Pol δ and Fen1 reaction conforms to the S. solfataricus model, where the coupled reaction is based on the presence of a physical complex. Our findings differ significantly from earlier studies with proteins from S. cerevisiae, in which PCNA and ub-PCNA displayed no difference in stimulating Pol δ and Fen1 cooperation.20 These differences could be due to the fact that human Pol δ is a four-subunit protein, whereas S. cerevisiae Pol δ has three subunits.
The ability of ub-PCNA to alter the efficiency of Okazaki fragment processing raises the likelihood that this would lead to a reduction in the rate of replication fork progression in vivo, i.e., inhibition of DNA synthesis. The inhibition of DNA synthesis after DNA damage in response to UV in S phase cells is a well-established event, which is referred to as the intra-S phase checkpoint. Among the causes for inhibition of DNA synthesis is a reduced rate of replication fork progression (the elongation checkpoint).49,50 The molecular basis for this is still unclear, but the inhibition of Okazaki fragment processing by ub-PCNA could be a factor. Another possibility is the loss of the p12 subunit of Pol δ that occurs upon UV damage.51 Our findings that ubiquitination of PCNA inhibits the coupled Pol δ/Fen1 reaction also raises the question of whether ub-PCNA could act to regulate conformational flexibility of other proteins that use PCNA as a platform. The unique features of ub-PCNA structure will provide further impetus for elucidating the mechanism of polymerase switching and the construction of working models for eukaryotic DNA replication and repair.
Materials and Methods
Protein preparation and crystallization.
Purification of ub-PCNA and human Pol δ were as reported in references 22 and 52. Ub-PCNA was crystallized by the hanging-drop vapor diffusion method in a mixture of 1 μl protein (15 mg/ml) and 1 μl precipitant (0.9–1.2 M sodium citrate, pH 6.5). The crystals were frozen in liquid propane with paraffin oil as cryoprotectant.
Data collection and structure solution.
Diffraction data were collected on the X-ray Operations and Research beamline 19ID at the Advanced Photon Source, Argonne National Laboratory, and processed with the HKL-3000 system.53 The crystals belong to space group P43212 with cell dimensions of a = 161.05, b = 161.05, c = 97.36 and diffract to 2.9 Å. The structure was solved by molecular replacement with PCNA monomer (1 vym)23 as the initial search model with Phenix54 software packages. Model building was conducted in Coot.55 The ubiquitin moieties were identified by the difference Fourier map after refinement with the initial PCNA trimer model by the maximum entropy refinement (BUSTER).56 Two of the three ubiquitin molecules were clearly identifiable in the initial (fo-fc) map (molecule D and E), whereas the density of the third ubiquitin was not sufficient for model building. The structure was refined to a nominal resolution of 2.9 Å (Table S1) with a trimeric ring of PCNA (chain A, B and C) and two molecules of ubiquitin in each asymmetric unit with Phenix with R factors of 22.2% (Rwork) and 29.2% (Rfree). Figures were prepared with Pymol.
Oligonucleotide primer templates.
All oligonucleotides were purchased from Integrated DNA Technologies and purified by PAGE before use. For the DNA replication assay, the 90 nt template was annealed to a 5' 32P-labeled primer P30 (5'-AGG GAA GGG AGA GGG AGG AGA AGA AGG GAG-3'). For the Fen1 cleavage assay, the 90 nt template was annealed to cold P30 and the 5' 32P-labeled downstream blocking sequence, DF2 (5'-CCC CCA AAA CCA ACC CAC). For the Fen1 cleavage assay with a 3' labeled blocking sequence, the 90 nt template was annealed to DF2 (5'-CCC CCA AAA CCA ACC CAC) first and then labeled with [α-32P] dTTP by Klenow fragment. The template was subsequently annealed to unlabeled primer (P30) 28 before use.
Pol δ purification and assay.
Human Pol δ was prepared to near homogeneity, and its activities on oligonucleotide substrates were analyzed by electrophoresis of the reaction products on polyacrylamide gels; radioactive products were visualized by phosphorimaging and analyzed with ImageQuant software (Amersham Biosciences). The standard polymerase assay (10 μl) contained 50 mM TRIS-HCl (pH 8.0), 1 mg/ml BSA, 125 mM NaCl, 5 mM DTT, 0.5 mM ATP, 200 μM dNTP, 100 nM oligonucleotide substrate, 400 nM PCNA or ub-PCNA, 10 nM human Pol δ and 5 mM MgCl2. The reactions were initiated by the addition of MgCl2.
Fen1/Pol δ cleavage assay.
Fen1 activities were analyzed by sequencing gels using linear oligonucleotide primer-templates.28 The 10 μl assay mixture contained 50 mM TRIS-HCl (pH 8.0), 1 mg/ml BSA, 125 mM NaCl (final ionic strength of 0.15 M), 5 mM DTT, 0.5 mM ATP, 200 μM dNTP, 50 nM 3' or 5' end-labeled oligonucleotide substrate, 30 nM PCNA (trimer) or ub-PCNA (trimer), 30 nM human Pol δ and 30 nM Fen1. DNA synthesis and cleavage products were visualized by phosphorimaging.
Model building of Pol δ in complex with PCNA and Pol η in complex with ub-PCNA.
The duplex B DNA was located according to the structure of PCNA in complex with DNA (3K4X)57 after minimization of structural overlaps. The location of the PIP motif of the p68 subunit of Pol δ was identified after alignment of PCNA molecules (3K4X and 1U76).26 The relative localization of Pol δ catalytic domain was based on the structural alignment of the template (first 6 bases) from the yeast Pol δ in complex with substrate (3IAY).58 The localization of Pol η relative to ub-PCNA was identified through the alignment of the DNA template (3FMH).59 The localization of PIP motifs was based on the structure of PCNA in complex with the Pol η PIP motif peptide (2ZVK) or the Pol ι PIP motif peptide (2ZVM).60 The UBM and UBZ binding to ub-PCNA was based on the alignment of the ubiquitin molecules between ub-PCNA and UBM-ubiquitin complexes (2KWU and 2I5O).61-63
Acession number.
The coordinates and structure factors have been deposited at Protein Data Bank with accession code 3TBL.
Supplementary Material
Acknowledgments
Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02–06CH11357. This work was supported by grants from the United States Public Health Service GM31973 and ES14737.
Disclosure of Potential Conflicts of interest
No potential conflicts of interest were declared.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20595
References
- 1.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–19. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 2.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–7. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakrishnan L, Bambara RA. The changing view of Dna2. Cell Cycle. 2011;10:2620–1. doi: 10.4161/cc.10.16.16545. [DOI] [PubMed] [Google Scholar]
- 5.Kai M, Wang TS. Checkpoint responses to replication stalling: inducing tolerance and preventing mutagenesis. Mutat Res. 2003;532:59–73. doi: 10.1016/j.mrfmmm.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Biertümpfel C, Zhao Y, Kondo Y, Ramón-Maiques S, Gregory M, Lee JY, et al. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–8. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Bozza W, Zhuang Z. Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell Biochem Biophys. 2011;60:47–60. doi: 10.1007/s12013-011-9187-3. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann AR. Ubiquitin-family modifications in the replication of DNA damage. FEBS Lett. 2011;585:2772–9. doi: 10.1016/j.febslet.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–9. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Batters C, Zhu H, Sale JE. Visualisation of PCNA monoubiquitination in vivo by single pass spectral imaging FRET microscopy. PLoS ONE. 2010;5:e9008. doi: 10.1371/journal.pone.0009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–5. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16125–30. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulrich HD. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett. 2011;585:2861–7. doi: 10.1016/j.febslet.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann AR. Translesion synthesis in mammalian cells. Exp Cell Res. 2006;312:2673–6. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–54. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci USA. 2007;104:15591–8. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zahn KE, Wallace SS, Doublié S. DNA polymerases provide a canon of strategies for translesion synthesis past oxidatively generated lesions. Curr Opin Struct Biol. 2011;21:358–69. doi: 10.1016/j.sbi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaheen M, Shanmugam I, Hromas R. The Role of PCNA Posttranslational Modifications in Translesion Synthesis. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/761217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA. 2005;102:18361–6. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/S1097-2765(04)00259-X. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, Chea J, Meng X, Zhou Y, Lee EY, Lee MY. PCNA is ubiquitinated by RNF8. Cell Cycle. 2008;7:3399–404. doi: 10.4161/cc.7.21.6949. [DOI] [PubMed] [Google Scholar]
- 23.Kontopidis G, Wu SY, Zheleva DI, Taylor P, McInnes C, Lane DP, et al. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc Natl Acad Sci USA. 2005;102:1871–6. doi: 10.1073/pnas.0406540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–43. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 25.Tsutakawa SE, Van Wynsberghe AW, Freudenthal BD, Weinacht CP, Gakhar L, Washington MT, et al. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. Proc Natl Acad Sci USA. 2011;108:17672–7. doi: 10.1073/pnas.1110480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruning JB, Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12:2209–19. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–30. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 28.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278:1618–25. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 29.Balakrishnan L, Bambara RA. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J Biol Chem. 2011;286:6865–70. doi: 10.1074/jbc.R110.209502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–93. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–23. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–73. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freudenthal BD, Gakhar L, Ramaswamy S, Washington MT. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat Struct Mol Biol. 2010;17:479–84. doi: 10.1038/nsmb.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pastushok L, Hanna M, Xiao W. Constitutive fusion of ubiquitin to PCNA provides DNA damage tolerance independent of translesion polymerase activities. Nucleic Acids Res. 2010;38:5047–58. doi: 10.1093/nar/gkq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, et al. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol Cell. 2010;37:396–407. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 37.Ohmori H, Hanafusa T, Ohashi E, Vaziri C. Separate roles of structured and unstructured regions of Y-family DNA polymerases. Adv Protein Chem Struct Biol. 2009;78:99–146. doi: 10.1016/S1876-1623(08)78004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Mo JY, Perez A, Leon A, Liu L, Mazloum N, et al. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase delta. J Biol Chem. 1999;274:26647–53. doi: 10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]
- 39.Mo J, Liu L, Leon A, Mazloum N, Lee MY. Evidence that DNA polymerase delta isolated by immunoaffinity chromatography exhibits high-molecular weight characteristics and is associated with the KIAA0039 protein and RPA. Biochemistry. 2000;39:7245–54. doi: 10.1021/bi0000871. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Xie B, Zhou Y, Rahmeh A, Trusa S, Zhang S, et al. Functional roles of p12, the fourth subunit of human DNA polymerase delta. J Biol Chem. 2006;281:14748–55. doi: 10.1074/jbc.M600322200. [DOI] [PubMed] [Google Scholar]
- 41.Barry ER, Bell SD. DNA replication in the archaea. Microbiol Mol Biol Rev. 2006;70:876–87. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Cell. 2003;11:275–82. doi: 10.1016/S1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 43.Beattie TR, Bell SD. Molecular machines in archaeal DNA replication. Curr Opin Chem Biol. 2011;15:614–9. doi: 10.1016/j.cbpa.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 44.Beattie TR, Bell SD. The role of the DNA sliding clamp in Okazaki fragment maturation in archaea and eukaryotes. Biochem Soc Trans. 2011;39:70–6. doi: 10.1042/BST0390070. [DOI] [PubMed] [Google Scholar]
- 45.Beattie TR, Bell SD. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. EMBO J. 2012;31:1556–67. doi: 10.1038/emboj.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doré AS, Kilkenny ML, Jones SA, Oliver AW, Roe SM, Bell SD, et al. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. 2006;34:4515–26. doi: 10.1093/nar/gkl623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, et al. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol Cell. 2006;24:279–91. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 49.Kaufmann WK. The human intra-S checkpoint response to UVC-induced DNA damage. Carcinogenesis. 2010;31:751–65. doi: 10.1093/carcin/bgp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol Cell Biol. 2007;27:5806–18. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng X, Zhou Y, Lee EY, Lee MY, Frick DN. The p12 subunit of human polymerase delta modulates the rate and fidelity of DNA synthesis. Biochemistry. 2010;49:3545–54. doi: 10.1021/bi100042b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng X, Zhou Y, Zhang S, Lee EY, Frick DN, Lee MY. DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 2009;37:647–57. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 54.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–35. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–21. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 57.McNally R, Bowman GD, Goedken ER, O’Donnell M, Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16:979–86. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–43. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hishiki A, Hashimoto H, Hanafusa T, Kamei K, Ohashi E, Shimizu T, et al. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem. 2009;284:10552–60. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bomar MG, D’Souza S, Bienko M, Dikic I, Walker GC, Zhou P. Unconventional ubiquitin recognition by the ubiquitin-binding motif within the Y family DNA polymerases iota and Rev1. Mol Cell. 2010;37:408–17. doi: 10.1016/j.molcel.2009.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bomar MG, Pai MT, Tzeng SR, Li SS, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase eta. EMBO Rep. 2007;8:247–51. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burschowsky D, Rudolf F, Rabut G, Herrmann T, Peter M, Wider G. Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin. J Biol Chem. 2011;286:1364–73. doi: 10.1074/jbc.M110.135038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/S0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.