Abstract
BACKGROUND
Few human papillomavirus (HPV)-positive head and neck squamous cell carcinoma (HNSCC) cell lines exist. We established UM-SCC-104, a new HPV(+) HNSCC cell linefrom a recurrent oral cavity tumor, and characterized it for the presence of cancer stem cells (CSC).
METHODS
Tumor cells were tested for biomarker expression by immunohistology and the presence of HPV was assessed by several methods.
RESULTS
UM-SCC-104 has a unique genotype, contains HPV-16 and expresses E6/E7. Inoculation of (Aldehyde Dehydrogenase) ALDH(+) and ALDH(−) cells in an immunocompromised mouse resulted in tumor growth from the ALDH(+) cells after 6 weeks that recapitulated the histology of the primary, while ALDH(−) cells did not produce tumors.
CONCLUSIONS
UM-SCC-104, a new HPV-16, CSC-containing HNSCC cell line will aid in studying recurrent HPV(+) tumors. The aggressive nature of this tumor is consistent with high uniform expression of EGFR and a functionally significant proportion of ALDH(+) CSC.
Keywords: HNSCC, HPV, cancer stem cells, head and neck cell lines, ALDH
Introduction
The establishment of cell lines from primary tumors is essential to head and neck squamous cell cancer (HNSCC) research. While many cell lines have been successfully established in the last thirty years(1–3), there are few that have been derived from cancers naturally infected with high-risk human papillomavirus (HR-HPV) in the head and neck. In recent years it has become clear that the overwhelming majority of oropharyngeal tumors are now associated with HR-HPV and there is strong evidence that the presence of HR-HPV in the tumor is an independent prognostic factor in overall survival for patients diagnosed with HNSCC(4, 5). However, not all patients with HPV-positive tumors respond well to therapy and the reasons for failure in some cases are not known(6, 7). The availability of HR-HPV-containing cell lines enhances our ability to study the role that HPV plays in HNSCC development and response or resistance to therapy.
The selective pressure on cells in the formation of an immortal cell line has been discussed previously(2, 8, 9). There are limitations in using cell line behavior to reflect that of the original tumor since it is speculative that cell lines truly reflect the behavior of primary tumors. However, despite this, it is well documented that cell lines remain the fundamental tools for preclinical investigations in the most efficient and cost-effective way. Other preclinical models, such as the use of xenografts, are equally valuable and important to investigate cancer cell behavior, however cell lines avoid high cost and ethical issues that inevitably arise when using animal models.
Our group and others have studied biomarker expression in HNSCC in HPV positive and negative tumors, with HPV and p16INK4a emerging as biomarkers associated with improved prognosis in oropharyngeal cancer(7, 10–12). EGFR, p53, and Bcl-xL are also relevant markers that modify the prognostic effect of HPV and may help guide the development of targeted therapy in HNSCC(6, 10). CDKN2a encodes the cyclin dependent kinase inhibitor p16INK4a, which is almost always overexpressed in HPV(+) oropharyngeal tumors, and is often lost or silenced in HPV(−) head and neck tumors(13) with only rare examples of p16INK4a positive/HPV- tumors(14). Overexpression of p16INK4a, like HPV, is an independent positive prognostic indicator for HNSCC(15). E-cadherin and nuclear β-catenin may also play a role in tumor progression in HPV-related tumors, and studies of these biomarkers may help to elucidate differences in tumor biology between HPV-positive and HPV-negative tumors(16). In our characterization, we chose to look at biomarkers that have been studied before and implicated in HNSCC prognosis.
Cancer stem cells (CSC) have been isolated in HNSCC using the cellular expression of aldehyde dehydrogenase (ALDH)(17–19). The ALDH+ CSC subpopulation (also called the tumor initiating cell population) is highly tumorigenic, can self re-new and has the capacity to recreate the initial tumor heterogeneity. Development of therapeutic strategies targeting CSC pathways necessitates identification of this important subpopulation in primary tumors and cell lines(20). In effect, distinguishing CSCs in new and existing cell lines is paramount to developing novel approaches in targeting these cancer-initiating cells. In this paper, we describe the development and characterization of a new HPV(+) HNSCC cell line that contains a tumorigenic CSC population.
Materials and Methods
Approvals for use of the animal model were obtained through the University of Michigan Committee for the Humane Care and Use of Animals. The University of Michigan’s Guide for the Care and Use of Laboratory Animals was followed.
Establishment of the cell line
Written informed consent was obtained from the patient for the study of his tissue and medical records after review and approval of the study and the consent form by the Medical School Institutional Review Board (IRB-Med). Surgically resected tumor tissue was carefully selected for culture by the surgeon, immediately transported to the lab and was washed extensively in Earle’s balanced salt solution containing penicillin, streptomycin, and amphotericin B. The tissue was then minced, placed in culture flasks and covered with complete Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum, L-glutamine, penicillin, streptomycin(2, 3). The tumor was heavily infected with yeast, therefore amphotericin B (20 µg/ml) was also added to the medium and frequent washing was employed to remove as much of the yeast as possible with each medium change. Eventually, sterile cultures were obtained and propagated. Trypsin with 0.125% EDTA was used for partial trypsinization to aid in fibroblast removal. When sufficient outgrowth of cells was observed, cells were detached using warm trypsin and plated into new culture flasks. The cells have been carried through multiple passages. Supernatants were tested for mycoplasma using Myco Alert Mycoplasma Testing Kit (Lonza, Rockland ME).
Analysis of Genetic Loci
DNA was extracted from UM-SCC-104 tumor cells and from fibroblasts separated from the UM-SCC-104 culture during cell line establishment using a modified Promega Wizard protocol (Promega, Madison WI). Briefly, the genotyping procedure is as follows: cell pellets are re-suspended in Nuclei Lysis Solution @ 55 degrees C. Proteins are precipitated, vortexed, then chilled on ice and separated by centrifugation at 13K RPM. DNA in the lysis supernatant is precipitated with isopropanol, diluted to 0.10 ng/µL and submitted to the University of Michigan Sequencing Core for analysis with the Profiler Plus PCR Amplification Kit (Applied Biosystems, Foster City, CA). Ten loci are analyzed: D3S1358, vWA, FGA, D8S1179, D21S11, D18S51, D5S818, D13S317, D7S820 and the amelogenin locus for identifying the presence of the X and Y chromosomes. PCR products are analyzed using the Applied Biosciences 3730 DNA sequencer and allele sizes are detected fluorescently.
Flow Cytometry
Aldehyde dehydrogenase (ALDH) expression was detected using the ALDEFLUOR kit (StemCell Technologies, Vancouver BC). Briefly, cells from the primary tumor were brought to single cell suspension in Aldefluor Assay Buffer (AAB), and incubated with ALDH substrate (BODIPY aminoacetaldehyde (BAAA), 5 µmol/l per 1 × 106 cells) for 45 minutes at 37° C. Concurrently, diethylaminobenzaldehyde (DEAB 50 mmol/l) was added to a separate sample also containing BAAA for an ALDH-inhibited control. Samples were washed and resuspended in AAB. 4’, 6-diamidino-2-phenylindole (DAPI; BD Pharmingen, San Diego CA) was used as a cell viability indicator. Fluorescence-activated cell sorting gates were established using the inhibited control (DEAB) and the cell viability sample (DAPI).
In Vivo Tumor Production from Cancer Stem Cells
ALDHHigh and ALDHLow fractions from UM-SCC-104 were collected into separate tubes and 5,000 cells from each fraction were suspended in 100 µl of DMEM with 100 µl of Matrigel (BD Biosciences, San Jose CA) for in vivo injections. The resulting 200 µl cell suspensions were injected subcutaneously into opposite flanks of a NOD/SCID (non-obese, diabetic/severe combined immunodeficiency) mouse (strain #001303, Jackson Laboratories, Bar Harbor ME). Tumor grown from the ALDHHigh inoculation was allowed to persist until the growth reached 1 cm in size (10 weeks) and subsequently harvested for sectioning. The tumor tissue was excised, washed in sterile PBS, transferred to 70% ethanol and delivered to the University of Michigan Research Histology and Immunoperoxidase Laboratory for automated vacuum infiltration tissue processing (Tissue Tek VIP 5, Sakura Finetek USA, Torrence, CA) in graded ethanol 70% 15 min, 95% 15 mins, 100% 15 times 2, followed by xylene and 4 cycles of paraffin infiltration and sectioning.
Immunohistochemistry
UM-SCC-104 cells were cultured on chamber slides until 50% confluent and then were fixed and permeabilized with 4% paraformaldehyde and 0.1% Triton-x for 15 minutes for each step (Sigma, St. Louis MO). Fixed slides were kept in PBS at 4°C until staining. Tissue slides from the surgical specimen and from the primary tumor murine xenograft were deparaffinized, rehydrated and peroxidase-quenched (Dako Cytomation, Glostrup Denmark). All slides except for one were incubated in Antigen Retrieval Solution (Dako Cytomation, Glostrup Denmark) for 40 minutes in 92°C water bath with a buffer change midway and allowed to cool to room temperature for 20 minutes. For Epidermal Growth Factor Receptor (EGFR), the antigen retrieval step was performed with pepsin incubation for 10 minutes at 37°C. Horse serum was used for blocking (30 minutes at room temperature). Sections were stained with primary antibodies: monoclonal mouse anti-EGFR/31G7 (Invitrogen, Camirillo CA), monoclonal mouse anti-Bcl-2/124 (Dako Cytomation, Glostrup Denmark), monoclonal mouse anti-p53/DO1 (Calbiochem, Darmstadt Germany), monoclonal rabbit anti-cyclin D1/SP4 and monoclonal mouse anti-Rb/51B7 (Neomarkers, Fremont CA). Primary antibodies were added and allowed to incubate overnight at 4°C. Staining for p16INK4a was performed per protocol supplied by the kit (CINtec p16INK4a Histology Kit; mtm Laboratories, Westborough MA). Slides were washed and incubated with corresponding biotinylated secondary antibodies (Vector Laboratories, Burlingame CA) at 1:200 dilution for 30 minutes. The slides were washed and incubated in a solution of coupled avidin/horseradish peroxidase (ABC Kit; Vector Laboratories, Burlingame CA), allowing linkage to the secondary antibodies via avidin-biotin binding. After washing, a DAB Peroxidase Substrate Kit (Vector Laboratories, Burlingame CA) was used to detect primary antibody binding. After a final washing step in tap water, the specimens were dehydrated and mounted. The stained slides were interpreted by a pathologist (J.B.M.).
Human Papillomavirus (HPV) Detection and HPV Type Identification
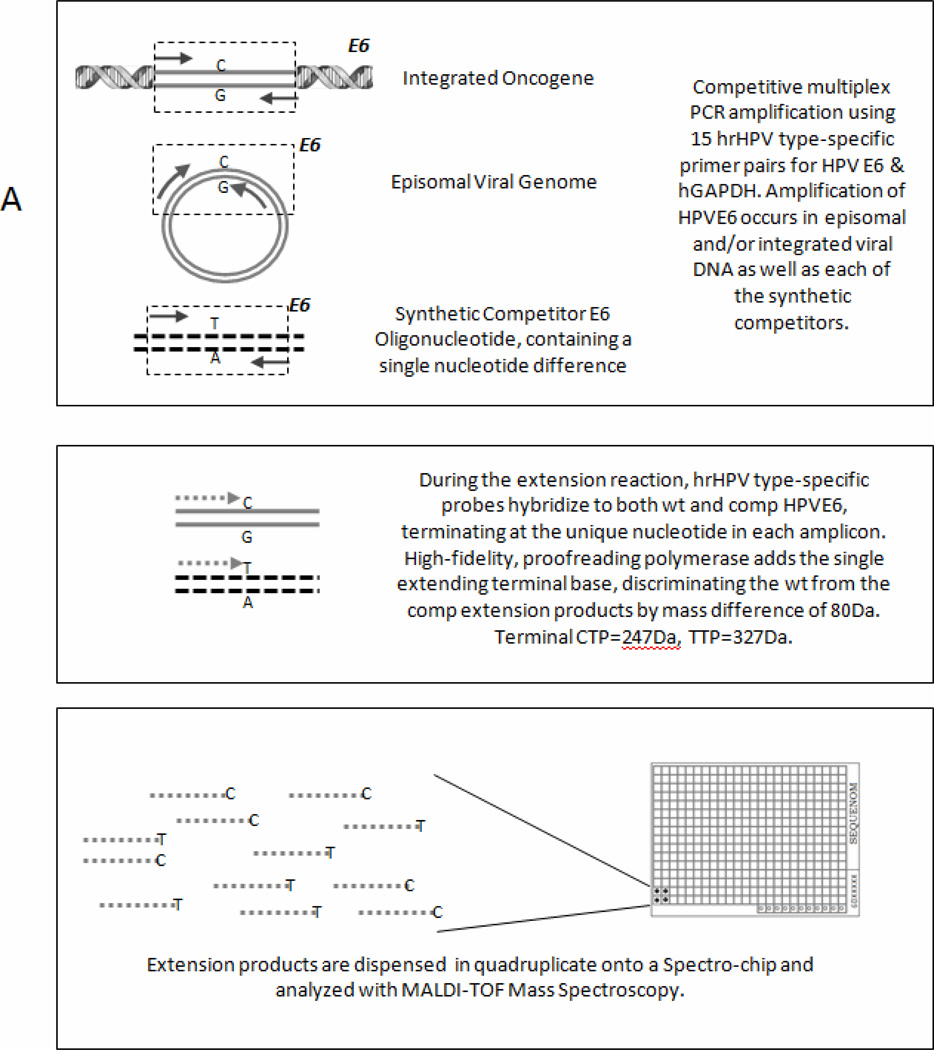
DNA was isolated from UM-SCC-104 and from the primary tumor tissue using DNeasy Blood and Tissue Kit (Qiagen). The presence and type of high risk HPV DNA was detected using the Mass Array technique developed by Dr. David Kurnit modified as described herein(21). Multiplex PCR amplification of the E6 region of 15 discrete high-risk HPV types (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68 and 73), as well as a human GAPDH control (See Table 1A for a list of primers) was run to saturation followed by shrimp alkaline phosphatase quenching. Amplification reactions included a competitor oligonucleotide identical to each natural amplicon except for a single nucleotide difference. Shrimp alkaline phosphatase removes phosphate from residual free dNTPs in the reaction mixture to prevent their incorporation in the next step, a single base extension reaction, which uses terminal bases. This multiplex single base extension reaction uses probes (Primer E, Table 1A) that identify unique sequences in the E6 region of each type, extending at the single base difference between wild-type and competitor HPV. The extension PCR product from each HPV type and its competitor can then be distinguished by mass (40 or 80 Dalton difference between wild type and competitor) when analyzed on the (Matrix Assisted Laser Desorption Ionization-Time of Flight) MALDI-TOF mass spectrometer(21, 22) (Figure 1). In situ hybridization was carried out in the Pathology Laboratory on formalin fixed paraffin embedded (FFPE) sections of the resected tumor using the INFORM HPVIII ISH assay (Ventana, Tucson AZ), which consists of a cocktail directed against 12 high-risk HPV genotypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 66).
Table 1.
Primer Sequences
| A. Primers used for identifying unique sequences in the E6 region for different subtypes of HPV | |
|---|---|
| Primer | Sequence |
| HPV16-F | ACGTTGGATGATGTTICAGGACCCACAGGA |
| HPV16-R | ACGTTGGATGCACGTCGCAGIAACTGTTGC |
| HPV16-E | TGCACAGAGCIGCAAACAA |
| HPV18-F | ACGTTGGATGATGCATGGACCIAAGGCAAC |
| HPV18-R | ACGTTGGATGGAAGGICAACCGGAATTTCA |
| HPV18-E | AGGCAACAITGCAAGAC |
| HPV31-F | ACGTTGGATGAAAGTGGTGAICCGAAAACG |
| HPV31-R | ACGTTGGATGTGCAATTICCGAGGTCTTTC |
| HPV31-E | /5dSp/GTGCAAACCIACAGACGC |
| HPV33-F | ACGTTGGATGCAAGACACIGAGGAAAAACCAC |
| HPV33-R | ACGTTGGATGCATTCCACGCACIGTAGTTC |
| HPV33-E | ATGATTIGTGCCAAGCATTGG |
| HPV35-F | ACGTTGGATGACATGTCAAIAACCGCTGTG |
| HPV35-R-2 | ACGTTGGATGAACAGGACAIACACCGACCT |
| HPV35-E-10 | CATCGGIGGACGGTGG |
| HPV39-F | ACGTTGGATGAATCCIGCAGAACGGCCATA |
| HPV39-R | ACGTTGGATGGGTTTGCTGIAGTGGTCGT |
| HPV39-E | CAGGACATTACAAIAGCCTGTGT |
| HPV45-F | ACGTTGGATGTTGTGGAAAAGIGCATTACAGG |
| HPV45-R | ACGTTGGATGTCTGTGCACAAAICTGGTAGC |
| HPV45-E | CAGGATGGCGCGCITTGACGATC |
| HPV51-F | ACGTTGGATGAAGGGTTAIGACCGAAAACG |
| HPV51-R | ACGTTGGATGTTCGTGGTCITTCCCTCTTG |
| HPV51-E | GTGCATAIAAAAGTGCAGTGGT |
| HPV52-F | ACGTTGGATGGAGGATCCIGCAACACGAC |
| HPV52-R | ACGTTGGATGTGCAGCCTIATTTCATGCAC |
| HPV52-E | GTGTGAGGIGCTGGAAGAAT |
| HPV56-F | ACGTTGGATGTTAACTCCGGIGGAAAAGC |
| HPV56-R | ACGTTGGATGAAACAIGACCCGGTCCAAC |
| HPV56-E | /5AmMC6T/AGGAAAAGCAAITGCATTGTGAC |
| HPV58-F | ACGTTGGATGACCACGGACAITGCATGATT |
| HPV58-R | ACGTTGGATGCAATTCGATTICATGCACAGA |
| HPV58-E | TTGCATGAITTGTGTCAGG |
| HPV59-F | ACGTTGGATGATTGCGAGCCTIACAGCA |
| HPV59-R | ACGTTGGATGCTGTACCTICCGAATCGG |
| HPV59-E | TGCAGCAAACCAGIAACCTG |
| HPV66-F | ACGTTGGATGCGTIAACACCGGAGGAAAAA |
| HPV66-R | ACGTTGGATGTGCATATGCTAIATAATGAAATCGTC |
| HPV66-E | GGAGGAAAAACAATIGCACTGTGAA |
| HPV68-F | ACGTTGGATGAATGGCGCIATTTCACAACC |
| HPV68-R | ACGTTGGATGACGTCAIGCAATGTGGTGTC |
| HPV68-E | CGCTATTICACAACCCTGAGG |
| HPV73-F | ACGTTGGATGTCCACTGGAIAAGCAAAAGC |
| HPV73-R | ACGTTGGATGCAGTTGCAGAIGGTCTCCAG |
| HPV73-E | GAAAAAAAACGGITTCATCAAATAG |
| GAPDH-F | ACGTTGGATGCAAGAAGGTGGTGAAGCAG |
| GAPDH-R | ACGTTGGATGTGAGCTTGACAAAGTGGTCG |
| GAPDH-E | GGTCTCCTCTGACTTCA |
| B. Primers used for RT- PCR of the E6 region for high risk HPV-16 | |
|---|---|
| HPV16-E6-F | ATGCACCAAAAGAGAACTG |
| HPV16-E6-R | TTACAGCTGGGTTTCTCTAC |
| HPV16-E7-F | ACCGGTCGATGTATGTCTTGTTG |
| HPV16-E7-R | CCGTACCCTCTTCCCCATTG |
| C. Primers used for sequencing p53 | |
|---|---|
| p53-1 F | GCGTGCTTTCCACGACG |
| p53-1 R | CCTTCCACTCGGATAAGATG |
| p53-2 F | TTGCATTCTGGGACAGCCAA |
| p53-2 R | GGCATCCTTGAGTTCCAAGG |
| p53-3 F | CACCATCATCACACTGGAAG |
| p53-3 R | CTGACGCACACCTATTGCAA |
Abbreviations: “F” represents Forward primer and “R” represents Reverse primer. The single base extension reaction uses a primer, “E” which represents Extension primer.
Fig 1.
PCR-MassARRAY using MALDI-TOF mass spectroscopy to detect and type human papillomavirus. Diagram depicting three-step process of HPV detection by competitive multiplex PCR amplification using 15 hrHPV type-specific primer pairs for HPV E6 & hGAPDH. Amplification of HPVE6 occurs in episomal and/or integrated viral DNA as well as each of the synthetic competitors.
RNA Extraction, Reverse Transcription Polymerase Chain Reaction, Gene Sequencing
RNA from UM-SCC-104 was isolated by phenol-chloroform extraction with TRIzol (Invitrogen, Carisbad CA). cDNA was synthesized using Reverse Transcription System Kit (Promega, Madison WI) according to the manufacturer’s protocol and detection of p53, E6 and E7 was performed using primers listed in Table 1B. The HPV16-containing CaSki cervical carcinoma cell line and the HPV16 (+) UM-SCC-47 oropharyngeal carcinoma cell line developed in our lab(23) were used as positive controls and the known HPV-negative cell line UM-SCC-38 was used as a negative control for E6 and E7 (amplicon sizes 477 base pairs and 400 base pairs, respectively) (See Table 1B for primers). The complete p53 cDNA was sequenced with three overlapping primer sets: amplicon 1 (652 base pairs), amplicon 2 (719 base pairs) and amplicon 3 (534 base pairs) (See Table 1C for primers). PCR products were submitted to the University of Michigan Sequencing Core for target gene sequencing. cDNA sequencing covers the entire p53 transcript including all exons and is the preferred method of sequencing as it can identify splicing variants as well as point mutations within the coding regions.
Results
Case Report
UM-SCC-104 (University of Michigan-Squamous Cell Carcinoma) was derived from a 56 year-old male with a recurrent floor of mouth squamous cell carcinoma. He was initially treated at a different hospital in 2008 with wide local excision for a posterior sublingual tumor that was described as moderately to poorly differentiated squamous cell carcinoma. Four months later he had a second wide local excision of a recurrent tumor of the floor of mouth and because of continuing suspicion of persistent disease in the floor of mouth-mandibular alveolus he was treated with chemoradiation (no details were available on the agents used, the doses or duration of treatment). Following the completion of radiation, the patient was referred to the University of Michigan where he presented with a painful erosive sublingual mass involving the mandibular alveolus and the lateral tongue, and was restaged as T4 N2b M0. Composite resection of the anterior mandible and anterior two thirds of the tongue, and reconstruction procedures with an osseus fibular and myocutaneous lattissimus dorsi flap were performed with bilateral selective neck dissections. The patient gave written informed consent for research on his tissue. Tissue from the resected oral cavity tumor was sent to the laboratory for culture and analysis. Three months after this surgery, there was an aggressive local recurrence deep to the reconstruction that was deemed unresectable. The patient returned home on palliative care. The patient’s social history was reported as 2 packs of cigarettes per day for twenty years and 2 alcoholic drinks per day. He was a non-smoker at the time of treatment for his cancer.
UM-SCC-104
Explanted tissue produced outgrowths of cells approximately on day 7 of culture (Figure 2A). At this point, multiple culture flasks were found to contain yeast and amphotericin B was added to the culture medium. Cultures were fed and rinsed with amphotericin B-containing medium until cultures no longer contained the contaminant. Differential trypsinization successfully removed fibroblast overgrowth leaving behind the epithelial cells. By day 21, the first passage of tumor cells to new flasks could be accomplished. The tumor cells were frozen at multiple low passages and were considered established as a cell line after the passage 20 milestone. UM-SCC-104 has a doubling time of 48 hours. Separate colony formation is typical of UM-SCC-104 and colonies will not become over-confluent (Fig 2B). This mirrors the histologic appearance of the grape like clusters of tumor cells in the patient’s anterior floor of mouth lesion tumor tissue, which was determined to be a moderately to poorly differentiated invasive squamous cell carcinoma (Figure 2C). This histologic appearance was recapitulated in subcutaneous tumors produced in the mouse after inoculation of ALDHHigh tumor cells (Fig 2D).
Fig 2.
Establishment of UMSCC-104 from a recurrent oral cavity squamous cell carcinoma. A, Outgrowths of squamous cells from explanted tissue placed in culture flasks. B, UM-SCC-104 at passage 8 growing in separate colonies which is its typical phenotype. C, Primary tissue stained with hematoxylin and eosin demonstrating infiltrating nests of tumor cells with abundant pink (keratinized) cytoplasm. Histology also showed intercellular bridges and focal squamous whorls typical of squamous cell carcinoma. D, Tumor from xenograft derived from cells expressing high ALDH expression. Similar to the primary tissue section, morphology consists of invasive poorly differentiated squamous cells.
UM-SCC-104 Genotype
The genotype of UM-SCC-104 is unique among the genotypes of our entire collection of cell lines(23). As shown in Table 2, the genotype of the UM-SCC-104 cell line and the fibroblasts that grew out from the normal stroma are nearly identical, except that the tumor cells show loss of one allele at each of two loci, D3S1358 and FGA.
Table 2.
Genotyping Results for UM-SCC-104
| Passage | AMEL | D3S1358 | vWA | FGA | D8S1179 | D21S11 | D18S51 | D5S818 | D13S317 | D7S820 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UM-SCC-104 Cell Line | 7 | X, Y | 17 | 16, 17 | 24 | 9, 13 | 30, 32.2 | 13, 14 | 12 | 8, 9 | 10 |
| UM-SCC-104 Fibroblasts |
4 | X, Y | 14, 17 | 16, 17 | 23, 24 | 9, 13 | 30, 32.2 | 13, 14 | 12 | 8, 9 | 10 |
Cancer Stem Cell Derived Xenograft
UM-SCC-104 cells sorted for ALDH expression revealed a 2.3% ALDHHigh population (Figure 3A, B). Cells selected for high and low ALDH activity were inoculated subcutaneously into opposite sides of NOD/SCID mice. Tumor growth resulted from the ALDHHigh injection after 6 weeks (Figure 3C, D) whereas no growth was observed from the ALDHLow injection even after 10 weeks of observation at which time the tumor from the ALDHHigh inoculation was harvested. Xenograft tissue morphology consisted of thick cords and clusters of poorly differentiated squamous cells similar to those in the tumor tissue taken from the patient (c.f. Figure 2C, D).
Fig 3.
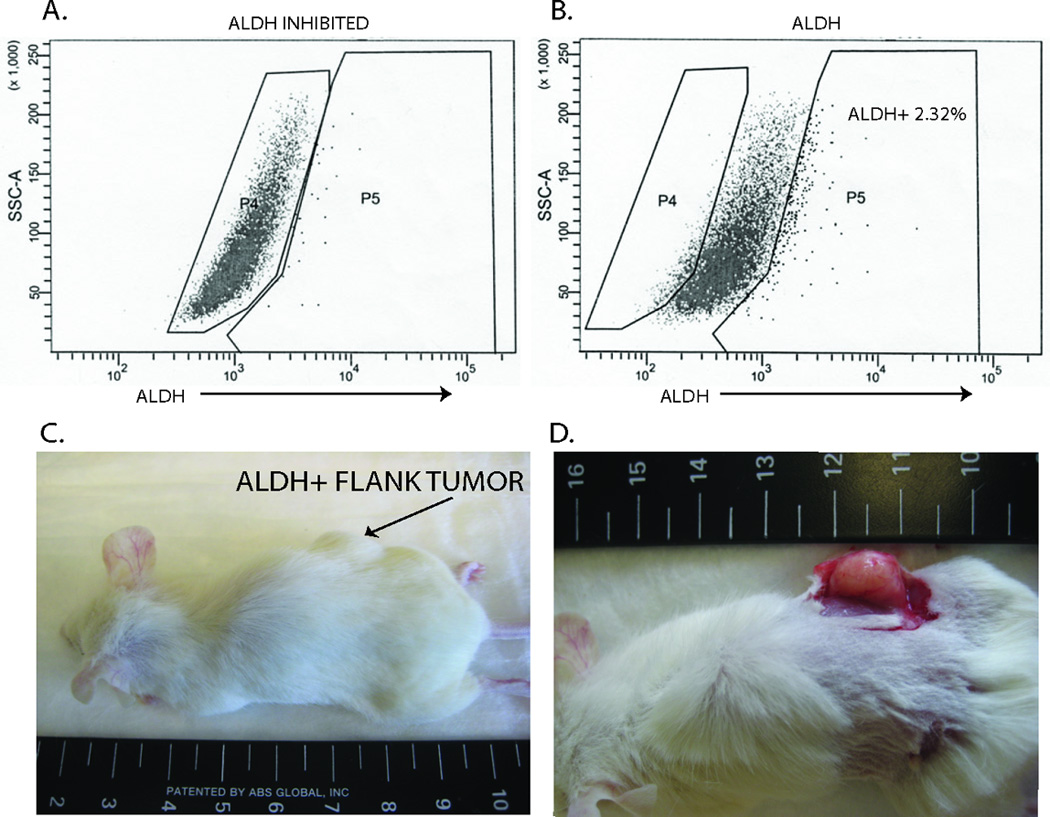
Identification of cancer stem cells from primary tissue. A, Flow cytometry analysis in which uptake of an ALDH substrate (BAAA) is inhibited with DEAB. This inhibition serves as a baseline for fluorescence activity. B, When BAAA is incubated with the cells without the inhibitor, an ALDH high population stands out (2.32%). Cells were collected from both the ALDH high population (P5) as well as the ALDH low population (P4). C, 5,000 cells were implanted in the flank of a NOD/SCID mouse. Cells collected in the ALDH high population grew tumor and cells from the ALDH low population did not produce tumor. D, Flank tumor of mouse derived from injection of ALDH high cells.
Head and Neck Cancer Biomarkers
Figure 4 shows a comparison of biomarker expression in the patient’s tumor tissue, the murine xenograft and the UM-SCC-104 cell line. Sections of the resected tumor tissue had weak expression of Rb and Bcl-2. In the xenograft tumor Rb was expressed only in mitotic cells but in the cell line all nuclei strongly express Rb. Bcl-2 also was not detected in the xenograft but is expressed in nuclei and cytoplasm of many cells in the UM-SCC-104 cell line, with the strongest expression in mitotic nuclei. p53 overexpression is present in a subset of the nuclei of the patient’s tumor, in a lower proportion of the xenograft cells and in a high proportion of nuclei in the cell line. Similarly, cyclin D expression is found in focal areas in the patient tumor and the xenograft, but is strongly expressed in nearly all of the cells in the cell line. All cells within the patient tumor, xenograft and the cell line strongly express diffuse nuclear and cytoplasmic p16INK4a. Similarly, all cells in the tumor, the xenograft and the cell line exhibit strong and diffuse membranous staining for EGFR..
Fig 4.
Immunohistologic staining of the patient’s resected anterior floor of mouth tumor (Primary Tumor), UM-SCC-104 cancer stem cell initiated murine xenograft tumor (CSC Derived Tumor), and UM-SCC-104 cells grown on glass slides (UM-SCC-104). Sections of the primary tumor tissue, the CSC derived tumor and the cell line all exhibited strong and diffuse nuclear and cytoplasmic staining for p16INK4a. Cell membrane staining was also strong and diffuse for EGFR in all three tumor cell samples. Approximately 10% of the resected tumor cells over-expressed p53, while staining was negative for RB, Bcl-2 and cyclin D. There was 10% focal staining of RB and cyclin D and 5% focal staining for p53, while no expression of Bcl-2 was seen on mouse tissue sections. UM-SCC-104 cultured on chamber slides uniformly over-expressed p16INK4a, EGFR and Rb. Focal staining of cyclin D1 (20%), Bcl-2 (20%) and p53 (30%) was presentin the cell line.
HPV and p53 status
In situ hybridization on the patient’s tumor demonstrated scattered cells with large punctate nuclear signals consistent with the presence of integrated HPV DNA (Figure 5A). In comparison an adjacent section cut from the same block at the same time exhibited strong p16 staining in all tumor cells (Figure 5B). The presence of HPV in DNA from the patient’s tumor and the UM-SCC-104 cell line was confirmed and identified as HPV16 by PCR-MassARRAY with MALDI-TOF (matrix assisted laser desorption-time of flight) mass spectroscopy (Figure 6). Expression of strong HPV16 E6 and E7 oncogenes were demonstrated by RT-PCR in UM-SCC-104 and in the HPV-16 positive controls, Caski and UM-SCC-47 cell lines (Figure 7). The HPV16 E6 primer set amplifies both the full length E6 (476 bp) and the alternately-spliced E6* variant (297 bp) in UM-SCC-47, Caski and UM-SCC-104 (Figure 7A). RT-PCR analysis of cDNA isolated from a small nidus of tumor cells in frozen tissue from the resected anterior floor of mouth tumor also demonstrated the presence of E6 and E7 message. The level of full length E6 message is barely detectable with a slightly stronger signal from the E6* alternate splice form and the E7 signal from the frozen tissue is comparatively strong. No E6 or E7 was detected in the HPV-negative UM-SCC-38 cell line. Sequencing p53 cDNA from UM-SCC-104 demonstrated that the tumor cells express wild-type p53 with the R72P polymorphism.
Fig 5.
In Situ Hybridization for high risk HPV and p16INK4a immunohistochemistry staining of the patient’s tumor tissue. A, In situ hybridization (blue stain) demonstrating rare punctate signals (arrows) within the nucleus indicative of integrated HPV DNA. B, p16INK4a immunohistochemistry demonstrating dark nuclear and diffuse cytoplasmic staining on an adjacent tumor tissue section to that used for in situ hybridization.
Fig 6.
MassARRAY analysis of HPV 16 DNA from UM-SCC-104 and the floor of mouth tumor tissue. In each case two peak areas representing the extension products of the target allele from the tumor cells and the corresponding competitive template allele separated by MALDI-TOF Mass Spectroscopy by the 80 Dalton mass difference resulting from the single base difference between wild-type HPV16 and the HPV16 competitor.
Fig. 7.

Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) detection of HPV16 E6 and E7 oncogenes in cDNA prepared from the UM-SCC-104 cell line, the patient’s anterior floor of mouth frozen tumor tissue and the HPV-positive control Caski and UM-SCC-47 cell lines. No bands were seen in HPV-negative UM-SCC-38 cell line. HPV16 E6 primer sets amplify the full length E6 (476 bp) and the alternately-spliced E6* variant (297 bp) in UM-SCC-47, Caski and UM-SCC-104. With the cDNA from the floor of mouth tumor tissue the E6 bands were too faint to show in the photograph, but the E7 amplicon was more abundant and easily visible.
Discussion
There are very few cell lines derived from HPV(+) HNSCC despite the increasing prevalence of HPV in oropharynx cancer. To our knowledge, there are only four published HPV(+) HNSCC cell lines (UM-SCC-47, established at the University of Michigan; UD-SCC-2, established at the University of Düsseldorf(24); 93-VU-147T, established at the Free University (VU) in Amsterdam; and UPCI-90, established at the University of Pittsburgh(23–27). Review of the literature indicates that all of the donors of HPV positive cell lines were smokers (Table 3). There are also two unpublished HPV+ HNSCC cell lines, UPCI-152 and UPCI 154, established from two different specimens from the same patient at the University of Pittsburgh (Personal Communication, Susanne Gollin), that are available for studying HPV oncogenesis in head and neck cancer. Herein, we present a new HPV-positive cell line derived from a persistent and multiply recurrent oral cavity tumor. The tumor and the cell line both contain high-risk HPV-16 with HPV persisting in the tumor cells in culture. This cell line, UM-SCC-104, is a new in vitro and in vivo resource for studying convergent viral and carcinogen-related pathways of carcinogenesis since the patient was a heavy smoker and used alcohol for many years. Unlike many HPV positive tumors this tumor was multiply recurrent, failed to respond to chemoradiation and thus provides a model for use in preclinical studies to identify potential targets for novel therapies. Tumors like UM-SCC-104 that represent the aggressive non-responsive tumors are those for which new therapies must be developed if we are to offer the afflicted individuals more effective tumor control.
Table 3.
Existing published HPV positive head and neck cancer cell lines
| Cell Line | Type | p53 | Smoking History |
Institute of Origin |
|---|---|---|---|---|
| UM-SCC-47 | HPV16 | Wild- type |
Positive | University of Michigan, Ann Arbor MI |
| 93-VU-147T | HPV16 | Wild- type |
Positive | VU University Medical Center, Amsterdam Netherlands |
| UD-SCC-2 | HPV16 | Wild- type |
Positive | University of Dusseldorf, Dusseldorf, Germany |
| UPCI-SCC90 | HPV16 | Wild- type |
Positive | University of Pittsburgh, Pittsburgh PA |
HPV(+) tumors of the head and neck are now viewed as a separate and biologically distinct entity from HNSCC linked to smoking and alcohol consumption(28–30). The presence of HPV is a powerful independent predictor of response to therapy and overall survival in patients with HNSCC(10, 11), furthermore patients with HPV positive tumors are significantly less likely to be tobacco users(10, 30, 31). Hafkamp et al. studying a group of tonsillar cancer patients that were mostly smokers (84%) observed that there was a marked difference in survival rate between smokers and non-smokers with HPV-associated tumors(7). Maxwell et al. reported that of the HPV(+) oropharynx cancer patient population, only about 1/3 were never smokers and 2/3 of HPV(+) patients were either current or former smokers(6). Furthermore, never smokers had a five-fold lower risk of cancer progression (recurrence, distant metastasis or second primary cancer of the head and neck) than that of current smokers with HPV(+) tumors. This observation was subsequently expanded upon in a large cooperative trial(4). Thus, a positive tobacco history in patients with HPV(+) tumors may represent a distinct group of head and neck cancers when all HNSCC are divided by etiologic factors (i.e. group 1: HPV(−) smokers, group 2: HPV(+) never smokers, group 3: and HPV(+) ever smokers). UM-SCC-104 belongs to the group 3 demographic which have a higher risk of recurrence than HPV(+) tumors from never smokers(6) supporting the need for representative cell lines for further studies.
The creation of an HPV(+) cell line in HNSCC is a rare event even though the majority of oropharynx tumors are now HPV-positive. For successful attachment and subsequent in vitro propagation, it is thought that the tumor cells must have acquired traits compatible with survival and immortality in the unnatural environment of in vitro cultivation(2, 8). HPV(+) head and neck cancer cell lines are generally from patients with aggressive tumors that fail to respond to initial therapy. From what has been published about the existing HPV(+) cell lines and from the history of UM-SCC-47, patients from which they were derived from were smokers(23, 27, 32, 33) (Table 3). Tumors from HPV(+) smokers may more readily accumulate the genetic differences necessary for in vitro survival, which may account for the aggressive tumor behavior seen in this demographic.
UM-SCC-104 was derived from a patient who was a welder and had an extensive smoking (20 pack/year) and consistent moderate alcohol history. Thus, this patient had the usual etiologic factors for head and neck cancer, but also had a tumor that is HPV positive. It was initially thought that his smoking and alcohol use and the floor of mouth primary site indicated a typical chemical carcinogen-induced tumor. However upon verification of his HPV(+) tumor status, it became clear that there was a more complex process involved in tumorigenesis. One large retrospective study stratified risk factors by tumor site and found the strongest risk for an oral cavity tumor site was alcohol, while smoking was most strongly associated with laryngeal cancer and HPV-16 with pharyngeal cancer(30). Had our patient tested negative for HPV, his tumor would have been attributed to carcinogens related to tobacco and alcohol usage and possibly environmental hazards from his occupation. Our patient’s high-risk HPV status most likely provided an additive and possibly synergistic effect with his other risk factors contributing to the development of the cancer(34). UM-SCC-104 is a cell line that will be an important resource to begin to investigate how these combined risk factors contribute to the formation of such a therapeutically non-responsive tumor.
The floor of mouth is an unusual site for HPV related tumors. We confirmed the presence of HPV in the surgical specimen and in UM-SCC-104 using multiple methods: the patient’s recurrent tumor demonstrated integrated HPV DNA (ISH); the tumor and the cell line exhibited uniform over-expression of p16INK4a (IHC); E6 and E7 expression (RT-PCR) and HPV type 16 (multiplex PCR/MALDI-TOF mass spectroscopy) were also observed in the tumor and the UM-SCC-104 cell line. There is currently no universal method for HPV detection in head and neck cancer although numerous techniques and algorithms have been proposed(35), and our data are unequivocal in demonstrating that this floor of mouth/oral cavity tumor is HPV-16 positive. Although the original site was described as posterior floor of mouth and designated as an oral cavity tumor, HPV positive tumors of the oral cavity are rare and it cannot be excluded that the tumor originated in a never discovered small base of tongue tumor but presented through local extension into the floor of mouth.
We characterized the primary tumor and UM-SCC-104 for additional HNSCC biomarkers that have been linked to both tumor behavior and response to therapy. The tumor and the cell line over-express EGFR. We previously reported the phenotype HPV(+)/EGFRHigh to be associated with poorer survival after chemotherapy and radiation than HPV(+)/EGFRLow tumors(10). Cyclin D1 is also over-expressed in a proportion of UM-SCC-104 cells. High cyclin D1 expression is linked to poor prognosis in head and neck cancer(36) and in a clinical trial of organ sparing therapy for larynx cancer, cyclin D1 over-expression was a single variable that was clearly indicative of poor survival (Bradford et al. Triologic Thesis, unpublished data). Reduced expression of cyclin D1 in HPV positive head and neck tumors has been reported when compared to HPV negative tumors induced by smoking and alcohol(12). This makes sense since cyclin D1 and HPV E7 oncoprotein both target Rb. E7 sequesters Rb allowing E2F mediated transcription of the genes involved in entry into the cell cycle to proceed unopposed whereas cyclin D1 activates cyclin dependent kinase that phosphorylates Rb achieving the same effect as E7, making both mechanisms unnecessary in the same tumor cell. We previously reported that smoking is associated with increased EGFR expression in oropharyngeal cancers, and that increased EGFR expression strongly reduced the beneficial effect on survival associated with HPV(10). The increased expression of cyclin D1 in UM-SCC-104 is consistent with the patient’s smoking history as well as increased EGFR expression, since EGFR signaling activates STAT3 which in turn up-regulates cyclin D1 expression(37) providing an alternative mechanism of Rb activation through cyclin dependent kinase activation. However, in UM-SCC-104 this activation may be countered to some degree by the E2F-driven high expression of p16INK4a, which acts as an inhibitor of cyclin-dependent kinases.
Curiously, the proportion of biomarker positive cells differed somewhat in the cell line and the tumor. Rb, p53, Bcl-2 and cyclin D1 all showed higher proportions of positive cells in the cell line compared to the original tumor specimen. Because cells with these characteristics may have greater growth potential, it is logical that the cell line might therefore be enriched in these types of cells particularly under conditions of proliferation in vitro. There have been conflicting data on p53 expression in HPV(+) HNSCC tumor cells. Some studies have observed high expression of nuclear p53 in some HPV-containing tumors with wild-type p53(14) and other studies demonstrating low p53 expression(38). The mechanism of over-expression of wild type p53 in the presence of the virus is unknown. The expectation in HPV positive cells, is that the E6 protein of high risk HPV types recruits E6-associated protein (E6AP), a cellular ubiquitin ligase to the E6-p53-E6AP complex, allowing ubiquitination of p53, which marks it for exportation from the nucleus and into the proteasome for degradation. Why some HPV-positive tumors show overexpressed nuclear p53 is not fully understood. In UM-SCC-104 cells the in situ hybridization signal for HPV was not uniform throughput the tumor suggesting that the viral genome may have been rearranged or might be in the process of being lost. We also noted in these cells higher expression of an alternate splice form of HPV E6 called E6* than of the full length E6 transcript. It is unknown whether the E6* is less effective in causing p53 degradation than the full length E6 protein, but it is interesting to speculate that this might be the case and thus provide an explanation for high p53 expression in these HPV transformed tumor cells.
In accordance with the cancer stem cell theory, a small subpopulation of high ALDH expressing cells(39) from the UM-SCC-104 cell line was capable of forming a new tumor in an immunocompromised mouse, whereas ALDH negative cells were unable to form a tumor. Xenotransplantation is considered the gold standard for identification of cancer stem cells. It allows demonstration of essential cancer stem cell characteristics including the ability for the cells to self-renew and differentiate into various cell progeny(40). The tumor derived from the cancer stem cells displayed the primary tumor phenotypic heterogeneity indicating that this selected cell population has inherent stem-like functions that other cells do not. Accordingly, we observed congruency in biomarker expression in the mouse tumor of EGFR, p16INK4a, cyclin D and p53 compared to the primary section.
Conclusions
The existence of HPV positive cell lines from head and neck tumors provides opportunities to study HPV related carcinogenesis in head and neck cancer. UM-SCC-104 is a new HPV positive cell line that also contains a small and functionally important cancer stem cell population. This cell line will be a valuable resource to further determine relevant molecular pathways in both HPV and cancer stem cell carcinogenesis.
Acknowledgements
This project was made possible by funding from the University of Michigan Comprehensive Cancer Center, the American Academy of Otolaryngology-HNS Percy Memorial Award, the Kirschstein National Research Service Award (NRSA) in Advanced Research Training in Otolaryngology: 5 T32 DC005356, and a gift from Patricia Korican of Korican Real Estate. Additional support was from NIDCR 1 R01-DE019126, NCI P50 CA97248 (Head and Neck SPORE), NIDCD P30 DC05188 (KHRI Research Center Core grant), NCI P30 CA46592 (Cancer Center Core Grant) and the DNA sequencing Core. None of the funders had any role in the design, conduct, or interpretation of the above-mentioned experiments. The principal investigators in this study had full access to all data and take responsibility for the integrity of the data and the accuracy of the data analysis. Thank you to the Flow Cytometry Core and the Department of Pathology at U-M for their help in acquiring this data.
Footnotes
Presented at the American Head and Neck Society 2010 Research Workshop Meeting in Arlington, VA on October 28–30, 2010
References
- 1.Lin CJ, Grandis JR, Carey TE, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29(2):163–188. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 2.Easty DM, Easty GC, Carter RL, Monaghan P, Butler LJ. Ten human carcinoma cell lines derived from squamous carcinomas of the head and neck. Br J Cancer. 1981;43(6):772–785. doi: 10.1038/bjc.1981.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krause CJ, Carey TE, Ott RW, Hurbis C, McClatchey KD, Regezi JA. Human squamous cell carcinoma. Establishment and characterization of new permanent cell lines. Arch Otolaryngol. 1981;107(11):703–710. doi: 10.1001/archotol.1981.00790470051012. [DOI] [PubMed] [Google Scholar]
- 4.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen CT, Lewis JS, Jr, El-Mofty SK, Haughey BH, Nussenbaum B. Human papillomavirus and oropharynx cancer: Biology, detection and clinical implications. Laryngoscope. 2010 doi: 10.1002/lary.20936. [DOI] [PubMed] [Google Scholar]
- 6.Maxwell JH, Kumar B, Feng FY, et al. Tobacco use in human papillomavirus-positive advanced oropharynx cancer patients related to increased risk of distant metastases and tumor recurrence. Clin Cancer Res. 2010;16(4):1226–1235. doi: 10.1158/1078-0432.CCR-09-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hafkamp HC, Manni JJ, Haesevoets A, et al. Marked differences in survival rate between smokers and nonsmokers with HPV 16-associated tonsillar carcinomas. Int J Cancer. 2008;122(12):2656–2664. doi: 10.1002/ijc.23458. [DOI] [PubMed] [Google Scholar]
- 8.Kim SY, Chu KC, Lee HR, Lee KS, Carey TE. Establishment and characterization of nine new head and neck cancer cell lines. Acta Otolaryngol. 1997;117(5):775–784. doi: 10.3109/00016489709113477. [DOI] [PubMed] [Google Scholar]
- 9.Sacks PG, Parnes SM, Gallick GE, et al. Establishment and characterization of two new squamous cell carcinoma cell lines derived from tumors of the head and neck. Cancer Res. 1988;48(10):2858–2866. [PubMed] [Google Scholar]
- 10.Kumar B, Cordell KG, Lee JS, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26(19):3128–3137. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 12.Hafkamp HC, Mooren JJ, Claessen SM, et al. P21 Cip1/WAF1 expression is strongly associated with HPV-positive tonsillar carcinoma and a favorable prognosis. Mod Pathol. 2009;22(5):686–698. doi: 10.1038/modpathol.2009.23. [DOI] [PubMed] [Google Scholar]
- 13.Reed AL, Califano J, Cairns P, et al. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56(16):3630–3633. [PubMed] [Google Scholar]
- 14.Hafkamp HC, Speel EJ, Haesevoets A, et al. A subset of head and neck squamous cell carcinomas exhibits integration of HPV 16/18 DNA and overexpression of p16INK4A and p53 in the absence of mutations in p53 exons 5–8. Int J Cancer. 2003;107(3):394–400. doi: 10.1002/ijc.11389. [DOI] [PubMed] [Google Scholar]
- 15.Harris SL, Thorne LB, Seaman WT, Neil Hayes D, Couch ME, Kimple RJ. Association of p16(INK4a) overexpression with improved outcomes in young patients with squamous cell cancers of the oral tongue. Head Neck. 2010 doi: 10.1002/hed.21650. [DOI] [PubMed] [Google Scholar]
- 16.Stenner M, Yosef B, Huebbers CU, et al. Nuclear translocation of beta-catenin and decreased expression of epithelial cadherin in human papillomavirus-positive tonsillar cancer: an early event in human papillomavirus-related tumour progression? Histopathology. 2011 doi: 10.1111/j.1365-2559.2011.03805.x. [DOI] [PubMed] [Google Scholar]
- 17.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prince ME, Ailles LE. Cancer stem cells in head and neck squamous cell cancer. J Clin Oncol. 2008;26(17):2871–2875. doi: 10.1200/JCO.2007.15.1613. [DOI] [PubMed] [Google Scholar]
- 19.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginestier C, Liu S, Diebel ME, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010;120(2):485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Yang K, Khafagi A, et al. Sensitive detection of human papillomavirus in cervical, head/neck, and schistosomiasis-associated bladder malignancies. Proc Natl Acad Sci U S A. 2005;102(21):7683–7688. doi: 10.1073/pnas.0406904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxwell JH, Kumar B, Feng FY, et al. HPV-positive/p16-positive/EBV-negative nasopharyngeal carcinoma in white North Americans. Head Neck. 2010;32(5):562–567. doi: 10.1002/hed.21216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner JC, Graham MP, Kumar B, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32(4):417–426. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann TK, Sonkoly E, Hauser U, et al. Alterations in the p53 pathway and their association with radio- and chemosensitivity in head and neck squamous cell carcinoma. Oral Oncol. 2008;44(12):1100–1109. doi: 10.1016/j.oraloncology.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Rampias T, Boutati E, Pectasides E, et al. Activation of Wnt signaling pathway by human papillomavirus E6 and E7 oncogenes in HPV16-positive oropharyngeal squamous carcinoma cells. Mol Cancer Res. 2010;8(3):433–443. doi: 10.1158/1541-7786.MCR-09-0345. [DOI] [PubMed] [Google Scholar]
- 26.Wald AI, Hoskins EE, Wells SI, Ferris RL, Khan SA. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck. 2010 doi: 10.1002/hed.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferris RL, Martinez I, Sirianni N, et al. Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): a natural disease model provides insights into viral carcinogenesis. Eur J Cancer. 2005;41(5):807–815. doi: 10.1016/j.ejca.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 28.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 29.Gillison ML. Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol. 2004;31(6):744–754. doi: 10.1053/j.seminoncol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Applebaum KM, Furniss CS, Zeka A, et al. Lack of association of alcohol and tobacco with HPV16-associated head and neck cancer. J Natl Cancer Inst. 2007;99(23):1801–1810. doi: 10.1093/jnci/djm233. [DOI] [PubMed] [Google Scholar]
- 31.Gillison ML, D'Souza G, Westra W, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100(6):407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 32.Gwosdz C, Balz V, Scheckenbach K, Bier H. p53, p63 and p73 expression in squamous cell carcinomas of the head and neck and their response to cisplatin exposure. Adv Otorhinolaryngol. 2005;62:58–71. doi: 10.1159/000082473. [DOI] [PubMed] [Google Scholar]
- 33.Steenbergen RD, Hermsen MA, Walboomers JM, et al. Integrated human papillomavirus type 16 and loss of heterozygosity at 11q22 and 18q21 in an oral carcinoma and its derivative cell line. Cancer Res. 1995;55(22):5465–5471. [PubMed] [Google Scholar]
- 34.Smith EM, Ritchie JM, Summersgill KF, et al. Human papillomavirus in oral exfoliated cells and risk of head and neck cancer. J Natl Cancer Inst. 2004;96(6):449–455. doi: 10.1093/jnci/djh074. [DOI] [PubMed] [Google Scholar]
- 35.Smeets SJ, Hesselink AT, Speel EJ, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121(11):2465–2472. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 36.Akervall J, Kurnit DM, Adams M, et al. Overexpression of cyclin D1 correlates with sensitivity to cisplatin in squamous cell carcinoma cell lines of the head and neck. Acta Otolaryngol. 2004;124(7):851–857. doi: 10.1080/00016480410017323. [DOI] [PubMed] [Google Scholar]
- 37.Masuda M, Suzui M, Yasumatu R, et al. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62(12):3351–3355. [PubMed] [Google Scholar]
- 38.Wilczynski SP, Lin BT, Xie Y, Paz IB. Detection of human papillomavirus DNA and oncoprotein overexpression are associated with distinct morphological patterns of tonsillar squamous cell carcinoma. Am J Pathol. 1998;152(1):145–156. [PMC free article] [PubMed] [Google Scholar]
- 39.Clay MR, Tabor M, Owen JH, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010 doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen ZG. The cancer stem cell concept in progression of head and neck cancer. J Oncol. 2009;2009:894064. doi: 10.1155/2009/894064. [DOI] [PMC free article] [PubMed] [Google Scholar]