Abstract
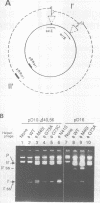
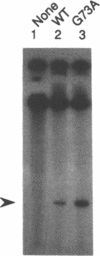
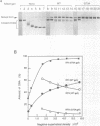
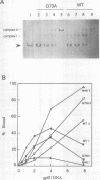
The origin of rolling circle replication in filamentous coliphage consists of a core origin that is absolutely required and an adjacent replication enhancer sequence that increases in vivo replication 30 to 100-fold. The core origin binds the initiator protein (gpII) which either nicks or relaxes negatively superhelical replicative form DNA (RFI). Nicking at the origin, but not relaxation, leads to initiation of DNA replication. Our results indicate that the ratio of nicking to relaxation (nicking-closing) in vitro depends on the superhelical density of the substrate. We have studied the effect of a single amino acid substitution in gpII, which allows wild-type levels of replication in the absence of the enhancer, on origin nicking and binding. The enhancer-independent mutation yields more nicking and less relaxation of RFI, compared to the wild-type protein. The mutant gpII also shows a reduced requirement for superhelicity of the substrate in the nicking reaction. At the same time, the mutant gpII increases the cooperativity of protein-protein interactions in origin binding. We propose that the relaxation activity of gpII negatively regulates replication initiation, and that both increase in the negative superhelicity of the substrate and action of the replication enhancer may antagonize the relaxation activity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Vovis G. F., Zinder N. D. Insertion mutant of bacteriophage f1 sensitive to EcoRI. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2699–2702. doi: 10.1073/pnas.76.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K. Replication of a plasmid containing two origins of bacteriophage. J Mol Biol. 1981 Nov 25;153(1):169–176. doi: 10.1016/0022-2836(81)90532-5. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. Initiation and termination of phage f1 plus-strand synthesis. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7122–7126. doi: 10.1073/pnas.79.23.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. The functional origin of bacteriophage f1 DNA replication. Its signals and domains. J Mol Biol. 1984 Feb 5;172(4):507–521. doi: 10.1016/s0022-2836(84)80020-0. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Zinder N. D. Increased intracellular concentration of an initiator protein markedly reduces the minimal sequence required for initiation of DNA synthesis. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1336–1340. doi: 10.1073/pnas.81.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Zinder N. D. Reduction of the minimal sequence for initiation of DNA synthesis by qualitative or quantitative changes of an initiator protein. Nature. 1984 Sep 20;311(5983):279–280. doi: 10.1038/311279a0. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Kornberg A. Purification and characterization of phiX174 gene A protein. A multifunctional enzyme of duplex DNA replication. J Biol Chem. 1979 Jun 25;254(12):5328–5332. [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Fulford W., Model P. Gene X of bacteriophage f1 is required for phage DNA synthesis. Mutagenesis of in-frame overlapping genes. J Mol Biol. 1984 Sep 15;178(2):137–153. doi: 10.1016/0022-2836(84)90136-0. [DOI] [PubMed] [Google Scholar]
- Geider K., Bäumel I., Meyer T. F. Intermediate stages in enzymatic replication of bacteriophage fd duplex DNA. J Biol Chem. 1982 Jun 10;257(11):6488–6493. [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Greenstein D., Horiuchi K. Interaction between the replication origin and the initiator protein of the filamentous phage f1. Binding occurs in two steps. J Mol Biol. 1987 Sep 20;197(2):157–174. doi: 10.1016/0022-2836(87)90115-x. [DOI] [PubMed] [Google Scholar]
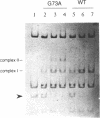
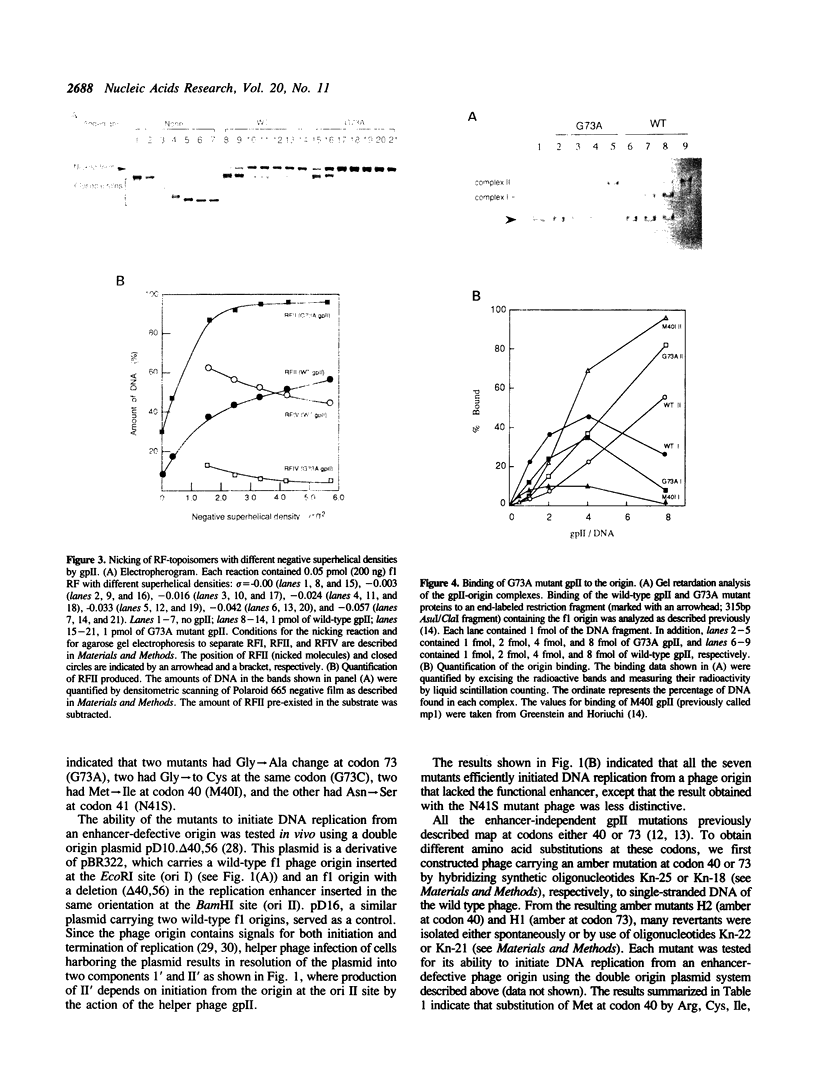
- Greenstein D., Horiuchi K. Replication enhancer-independent mutation increases the co-operativity with which an initiator protein binds its origin. J Mol Biol. 1990 Jan 5;211(1):91–101. doi: 10.1016/0022-2836(90)90013-C. [DOI] [PubMed] [Google Scholar]
- Greenstein D., Zinder N. D., Horiuchi K. Integration host factor interacts with the DNA replication enhancer of filamentous phage f1. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6262–6266. doi: 10.1073/pnas.85.17.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth G., Bäumel I., Meyer T. F., Geider K. Bacteriophage fd gene-2 protein. Processing of phage fd viral strands replicated by phage T7 enzymes. Eur J Biochem. 1981 Oct;119(3):663–668. doi: 10.1111/j.1432-1033.1981.tb05659.x. [DOI] [PubMed] [Google Scholar]
- Horiuchi K. Interaction between gene II protein and the DNA replication origin of bacteriophage f1. J Mol Biol. 1986 Mar 20;188(2):215–223. doi: 10.1016/0022-2836(86)90306-2. [DOI] [PubMed] [Google Scholar]
- Horiuchi K. Origin of DNA replication of bacteriophage f1 as the signal for termination. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5226–5229. doi: 10.1073/pnas.77.9.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Ravetch J. V., Zinder N. D. DNA replication of bacteriophage f1 in vivo. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):389–399. doi: 10.1101/sqb.1979.043.01.045. [DOI] [PubMed] [Google Scholar]
- Johnston S., Ray D. S. Interference between M13 and oriM13 plasmids is mediated by a replication enhancer sequence near the viral strand origin. J Mol Biol. 1984 Aug 25;177(4):685–700. doi: 10.1016/0022-2836(84)90044-5. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. H., Ray D. S. Mutational mechanisms by which an inactive replication origin of bacteriophage M13 is turned on are similar to mechanisms of activation of ras proto-oncogenes. J Virol. 1985 Mar;53(3):871–878. doi: 10.1128/jvi.53.3.871-878.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange-Gustafson B. J., Nash H. A. Purification and properties of Int-h, a variant protein involved in site-specific recombination of bacteriophage lambda. J Biol Chem. 1984 Oct 25;259(20):12724–12732. [PubMed] [Google Scholar]
- Lyons L. B., Zinder N. D. The genetic map of the filamentous bacteriophage f1. Virology. 1972 Jul;49(1):45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Geider K. Bacteriophage fd gene II-protein. II. Specific cleavage and relaxation of supercoiled RF from filamentous phages. J Biol Chem. 1979 Dec 25;254(24):12642–12646. [PubMed] [Google Scholar]
- Meyer T. F., Geider K. Enzymatic synthesis of bacteriophage fd viral DNA. Nature. 1982 Apr 29;296(5860):828–832. doi: 10.1038/296828a0. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Geider K., Kurz C., Schaller H. Cleavage site of bacteriophage fd gene II-protein in the origin of viral strand replication. Nature. 1979 Mar 22;278(5702):365–367. doi: 10.1038/278365a0. [DOI] [PubMed] [Google Scholar]
- Michel B., Zinder N. D. Translational repression in bacteriophage f1: characterization of the gene V protein target on the gene II mRNA. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4002–4006. doi: 10.1073/pnas.86.11.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991 Jul 12;66(1):129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A., Flamm E., Weisberg R. A., Miller H. I. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987 Sep;169(9):4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal Biochem. 1982 May 15;122(2):253–257. doi: 10.1016/0003-2697(82)90277-9. [DOI] [PubMed] [Google Scholar]
- Stark M. J. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene. 1987;51(2-3):255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Chaconas G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J Biol Chem. 1989 Feb 15;264(5):3028–3034. [PubMed] [Google Scholar]
- Tse Y. C., Kirkegaard K., Wang J. C. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J Biol Chem. 1980 Jun 25;255(12):5560–5565. [PubMed] [Google Scholar]