Abstract
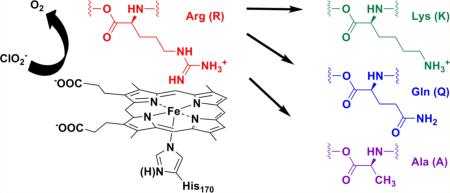
The chlorite dismutase from Dechloromonas aromatica (DaCld) catalyzes the highly efficient decomposition of chlorite to O2 and chloride. Spectroscopic, equilibrium thermodynamic, and kinetic measurements have indicated that Cld has two pH sensitive moieties; one is the heme, and Arg183 in the distal heme pocket has been hypothesized to be the second. This active site residue has been examined by site-directed mutagenesis to understand the roles of positive charge and hydrogen bonding in O–O bond formation. Three Cld mutants, Arg183 to Lys (R183K), Arg183 to Gln (R183Q), and Arg183 to Ala (R183A), were investigated to determine their respective contributions to the decomposition of chlorite ion, the spin state and coordination states of their ferric and ferrous forms, their cyanide and imidazole binding affinities, and their reduction potentials. UV–visible and resonance Raman spectroscopies showed that DaCld(R183A) contains five-coordinate high-spin (5cHS) heme, the DaCld(R183Q) heme is a mixture of five-coordinate and six-coordinate high spin (5c/6cHS) heme, and DaCld(R183K) contains six-coordinate low-spin (6cLS) heme. In contrast to wild-type (WT) Cld, which exhibits pKa values of 6.5 and 8.7, all three ferric mutants exhibited pH-independent spectroscopic signatures and kinetic behaviors. Steady state kinetic parameters of the chlorite decomposition reaction catalyzed by the mutants suggest that in WT DaCld the pKa of 6.5 corresponds to a change in the availability of positive charge from the guanidinium group of Arg183 to the heme site. This could be due to either direct acid–base chemistry at the Arg183 side chain or a flexible Arg183 side chain that can access various orientations. Current evidence is most consistent with a conformational adjustment of Arg183. A properly oriented Arg183 is critical for the stabilization of anions in the distal pocket and for efficient catalysis.
Heme proteins conduct a variety of chemical functions. These include the sensing and trafficking of small molecules, catalysis of one- and two-electron oxidations (peroxidases and peroxygenases), monoxygenation (cytochrome P450s), chlorination (chloroperoxidase), two- and four-electron reductions (N- and S-oxide reductases and cytochrome c oxidase), electron transfers (cytochromes), and heme trafficking (hemophores). The catalytic versatility of the heme is directed toward a particular function by the protein environment. A diverse body of literature has focused on reducing complex protein structures to the fewest possible elements responsible for controlling chemistry at heme-containing active sites. The proximal heme ligand is an important variable, ranging from a strong and polarizable thiolate to (neutral) imidazole, (anionic) imidazolate, or phenolate (Cys, His, or Tyr, respectively). The distal side of the heme often has either an open coordination site or a labile iron ligand, with surrounding residues controlling ligand access, the electrostatic environment, H-bonding, and, as a consequence of all these factors, the heme-mediated chemistry.
Chlorite dismutase (Cld) is a relatively recently described heme enzyme that is associated with respiratory perchlorate reduction in several Proteobacteria. Enzymes from perchlorate respirers detoxify chlorite (ClO2–), the end product of perchlorate (ClO4–) respiration, by converting it to Cl– and O2.1 This remarkable reaction is only the second well-described means of generating the O–O bond in biology.1,2 It is likewise difficult to reproduce efficiently using synthetic porphyrins.3–5 The enzyme from the perchlorate respiring organism Dechloromonas aromatica (DaCld) is fast, highly efficient [maximal kcat = (2.0 ± 0.6) × 105 s–1, and kcat/KM = (3.2 ± 0.4) × 107 M–1 s–1, at pH 5.2 and 4 °C], and highly specific, producing one molecule of O2 from each ClO2– even in the presence of large amounts of potential peroxidase (one-electron-donating) substrates.6 Because the majority of perchlorate is man-made, it has only recently become a significant environmental contaminant. When or how the chlorite decomposition reaction evolved is an open question. Interestingly, Cld homologues are found in a very broad array of bacterial and archaeal phyla, suggesting that the protein family is both ancient and necessary for reactions other than chlorite detoxification. Sequence and structural analyses indicate that the residues in the pocket above the open coordination position on the heme are likely diverse.7 We have therefore aimed to understand how the distal environment of wild-type (WT) and distally modified DaCld directs reactivity.
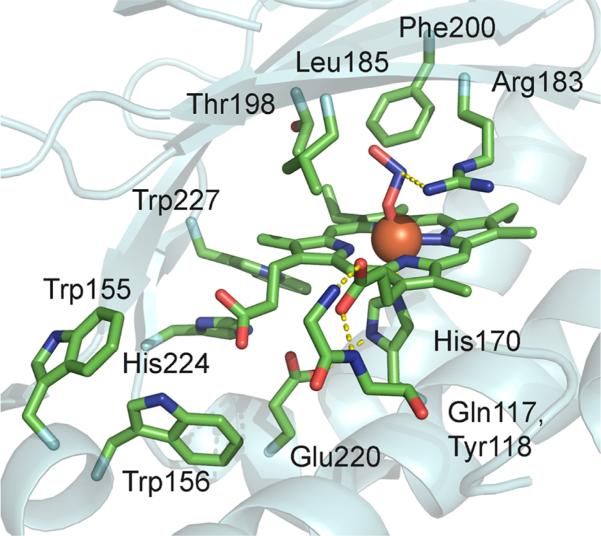
DaCld has been crystallographically and spectroscopically characterized and shown to have an unusual active site structure (Figure 1).8 It has a proximal His that is hydrogen bonded to the carboxylate side chain of Glu220. A similar histidine–aspartate hydrogen bonding pair is common in heme peroxidases,9 where it has been postulated to be important for the heterolytic cleavage of the coordinated hydroperoxy O–O bond. In spite of the His–Glu interaction, which might be expected to lend anionic (imidazolate) character to the ligating histidine, the resonance Raman (rR) spectrum of ferrous DaCld indicates a weak Fe–His bond relative to those of the peroxidases. This suggests that the His–Glu interaction in DaCld is in turn relatively weak, and that the ligating His is neutral in character.6 With the exception of an arginine residue (Arg183), the distal pocket contains only hydrophobic side chains, which form a sterically constrained pocket over the porphyrin plane. The guanidinium group of Arg183 hydrogen bonds to a heme-bound nitrito ligand in the DaCld crystal structure at pH 98 and to the heme-bound thiocyanate in the Cld from Azospira oryzae.10 At pH 6.5, the side chain in DaCld is nearly identically positioned but bridges the heme-bound nitrito ligand and a molecule of the buffer MES [2-(N-morpholino)-ethanesulfonic acid]. In the otherwise very similar structures of the imidazole and cyanide-bound Cld from Nitrospira defluvii, the guanidinium side chain turns away from the distal pocket and faces the protein surface. When this residue is mutated to lysine, its amine side chain forms a hydrogen bond to an Fe-bound water.11
Figure 1.
Active site of DaCld–nitrite complex (PDB entry 3Q08).
The various conformations assumed by the side chain in these structures suggest that it is mobile. Its hydrogen bonding patterns likewise suggest that it is potentially important for forming and stabilizing anion-bound forms of the protein, functions known to be served by the distal Arg in peroxidases.12 Evidence suggests that an analogously placed arginine plays a role in polarizing the heme-bound hydroperoxide ligand in peroxidases, promoting heterolytic cleavage of the O–O bond. It subsequently serves to stabilize the ferryl [FeIV=O] porphyrin cation radical species (Compound I) via donation of a hydrogen bond to the ferryl oxygen atom.13–16 Analogous functions for the DaCld distal arginine are possible. To further understand the role of Arg183 in the O2-generating ClO2– decomposition reaction, this residue has been substituted with a positively charged lysine, a neutral amide glutamine, and a small, hydrophobic alanine. The effects of these substitutions on solution structure, axial heme ligation, reactivity, and the reduction potential of these mutants were probed by UV–visible and resonance Raman (rR) spectroscopy, steady state kinetics of chlorite decomposition, and Fe3+/Fe2+ potential determination. The results are consistent with a conformationally mobile Arg183 side chain having roles in stabilizing the enzyme–substrate complex and promoting O–Cl bond cleavage.
EXPERIMENTAL PROCEDURES
Generation of DaCld(R183Q), DaCld(R183K), and DaCld-(R183A)
Site-directed mutagenesis of DaCld was conducted using a QuikChange polymerase chain reaction mutagenesis kit (Stratagene). The following primers and their complements were used to construct the three mutants (altered codon underlined): 5′-GTGAACGTCAAGCAGAAGTTGTACCAC-3′ (R183Q), 5′-CCTATCTTGTGAACGTCAAGAAGAAGTTGTACCACTCAACTGGC-3′ (R183K), and 5′-CCTATCTTGTGAACGTCAAGGCCAAGTTGTACCACTCAACT-3′ (R183A). The reaction products were digested with DpnI to isolate the nonmethylated (mutated) DNA and transformed into competent Escherichia coli cells (NovaBlue) by heat shock. Transformants were plated on kanamycin-supplemented LB agar. Sequencing was used to confirm the desired nucleotide mutations. The plasmids were transformed into E. coli TunerDE3 competent cells (EMD Biosciences) by heat shock.
Protein Purification
The mutant proteins were expressed in E. coli TunerDE3 cells and purified according to the procedure previously described for the WT protein.17 For DaCld(R183A), hemin was solubilized in DMSO and added during the sonication process (~60 μM).
Steady State Kinetics of Chlorite Decomposition
O2 production from chlorite decomposition was monitored continuously using a Clark electrode. The electrode was equilibrated to the set temperature for at least 1 h and calibrated to the expected O2 concentration in air-saturated, double-distilled Milli-Q water, adjusted for temperature and daily atmospheric pressure. The temperature was maintained at 4 °C by a circulating water bath (Thermo-Fisher Scientific) with a custom-built chamber jacketing the reaction vessel. Reactions were conducted in 1.5 mL volumes of 100 mM citrate-phosphate buffer (pH 6.8) with 0.080–500 mM sodium chlorite added from a stock made in the same buffer. Initial rates were determined by linear least-squares regression fits to the initial 5–10% of the progress of reaction curves (LoggerPro, VrTech).
Catalase Activity
For catalase activity measurements, the O2 concentration was continuously monitored via a Clark electrode. Equilibration and calibration were performed as described above, except calibration was conducted on an N2-saturated solution containing ditihionite. For each measurement, buffer alone was incubated for 5 min under N2 gas so that the initial O2 concentration was between 0 and 50 μM. The probe was then inserted into the solution and equilibrated. H2O2 was added to a final concentration of 5 or 50 mM via a gastight Hamilton syringe, and a baseline was measured. Enzyme (1–2 μM in the heme-containing DaCld monomer) was added via a gastight syringe, and the amount of O2 evolved was measured. Initial rates were determined by linear least-squares regression fits to the initial 5–10% of the progress of reaction curves (Kaleidagraph).
Peroxidase Activity
Oxidation of 2,2′-azinobis(3-ethyl-benzthiazoline-6-sulfonic acid) (ABTS) to the ABTS radical was monitored at 414 nm using an ε414 of 36 mM–1 cm–1 18 on a Varian Cary 50 spectrometer at 25 °C. Reactions were conducted in a quartz cuvette containing a saturating concentration of ABTS (79–84 μM) and 10 μL of the protein in 20 mM phosphate buffer (2.07 μM in heme-containing monomer) at the desired pH (final volume of 160 μL). The reaction was performed in 100 mM phosphate buffer (pH 6.8) at 25 °C. The reaction was initiated by the addition of 10 μL of a H2O2 stock prepared in the same buffer. Initial rates were determined by linear least-squares regression fits to the initial 5–10% of the progress of reaction curves (Kaleidagraph). The H2O2 concentration was varied from 0.04 to 116 mM.
UV–Visible pH Titrations
UV–visible spectra were recorded on a Varian Cary 50 spectrometer. Protein samples were titrated at 25 °C over three pH ranges. The following buffers were used: 50 mM citrate for pH 3.00–6.30, 100 mM phosphate for pH 6.70–10.9, and 100 mM disodium phosphate for pH 8.77–13.0. Each titration was conducted in an 8 mL reaction mixture with the protein at either ~1.8 or ~3.4 μM heme-containing monomer, where the heme concentration was determined via the pyridine hemochrome assay.19 The solutions were constantly stirred, and the pH was continuously monitored using a pH meter with a glass electrode (Corning pH 430). Small volumes of HCl (1 M) or NaOH (1 M) were added to the solution and the pH was measured, after which 200 μL aliquots were withdrawn and analyzed by UV–vis spectroscopy. The spectra were adjusted for dilution.
Resonance Raman Spectroscopy
Samples for rR experiments were prepared at 20–90 μM DaCld (based on heme-containing Cld monomers) in 100 mM citrate at pH 5.8, 100 mM sodium phosphate at pH 6.8 and 7.5, 100 mM Tris-HCl at pH 8.5, and 100 mM Ches at pH 10.0. Resonance Raman spectra were obtained with 406.7, 413.1, or 441.6 nm excitation from Kr+ and HeCd lasers using the 135° backscattering geometry. The spectrometer was calibrated against Raman frequencies of toluene, DMF, acetone, d6-dimethyl sulfoxide, and methylene bromide. Data were collected at ambient temperature from samples in spinning 5 mm NMR tubes. UV–visible spectra were recorded before and after rR experiments to probe whether the samples had been irreversibly altered in the laser beam. Laser power at ferric and ferrous samples ranged from 6 to 18 mW with 406.7 nm excitation and from 2 to 5 mW for ferrous–CO samples with 413.1 nm excitation; no spectral artifacts due to photoinduced chemistry were observed with these irradiation powers.
CO Complexes of WT and Mutant DaClds
Ferrous Cld was generated anaerobically at 20 °C by treatment of ferric Cld with an excess of buffered sodium dithionite stock solution. The corresponding CO complexes were prepared by flushing the ferrous proteins with 12CO or 13CO. Protein concentrations were typically 20–40 μM DaCld monomer in 100 mM Mes at pH 5.6, 100 mM citrate at pH 5.8, 100 mM sodium phosphate at pH 6.2, 6.8, and 7.5, 100 mM Tris-HCl at pH 8.0, 8.6, and 9.1, and 100 mM Ches at pH 10.0. The Cld–CO samples in D2O were prepared in a manner analogous to that of the samples in H2O except that the solution conditions were 100 mM Mes (pD 5.6) with an 88% D2O/12% H2O mixture.
Equilibrium Binding of Ligands
For each spectrophotometric titration, an ~8.0 μM protein sample was prepared in a 200 μL volume of the appropriate buffer in a quartz cuvette and its spectrum recorded. Solutions of the ligands (imidazole, azide, fluoride, and cyanide) were prepared in the same buffers at 0.002–200 mM and added in 5 μL aliquots. Spectra were repeatedly measured after each addition until the reaction mixtures had reached equilibrium. When the titration appeared complete, a ligand stock with a 10-fold higher concentration was added in 5 μL aliquots to be sure that a clear end point had been reached. Difference spectra were generated from spectra that had been corrected for sample dilution. The wavelength of the maximal absorbance change was used to construct a plot of ΔAbs versus [L]T (total concentration of added ligand) and fit by least-squares regression (Kaleidagraph) to an equilibrium isotherm of the form ΔAbs = ΔAbs∞[L]T/(KD + [L]T). The pH profiles were fit using two pKa equations or one pKa equation, as previously described.6 Here, c is a pH-independent measure of the parameter y (the KD for a given ligand), and Ka(1) and Ka(2) describe its dependence on the protonation state.
| (1) |
| (2) |
| (3) |
Redox Potentials
The reduction potentials of the WT and mutant DaClds were measured via the UV–visible titrimetric method of Massey (UV-2550 Shimadzu UV–vis spectrophotometer).20 WT DaCld (2 μM) was introduced into the body of an anaerobic cuvette with the reduction dye potassium indigo tetrasulfonate at ~20 μM (E = –46 mV21), 200 μM xanthine, and 1 μM benzyl viologen in 0.1 M potassium phosphate buffer at pH 7 and 25 °C. Benzyl viologen was used to ensure rapid equilibrium. An appropriate amount of xanthine oxidase was placed in the side arm of the cuvette, and several cycles of evacuation and flushing with argon gas were used to eliminate any oxygen. The spectrum of the solution of the enzyme and the dye was recorded. The reduction began as soon as the xanthine oxidase was mixed with the enzyme. Spectra were recorded every 2 min, and the reaction was stopped when the dye and the enzyme were fully reduced (minimum of 3 h). The extent of reduction of the dyes was monitored at 416 nm (an isosbestic point for the enzyme spectra as the enzyme was reduced). The extent of reduction of the enzyme was monitored at 432 nm. This is not an isosbestic point for the dye, so the contributions from the oxidized and reduced dye were subtracted. Reduction of the Cld mutants (R183A, R183K, and R183Q) was inhibited by the indicator dyes. Therefore, reduction potentials were determined by reducing the enzyme with the xanthine/xanthine oxidase system and then adding the indicator dyes. The indicator dyes reacted immediately with the enzymes, and the potentials were calculated on the basis of the extent of reactions. The same dye as WT DaCld was used for mutant R183Q, and the redox dyes pyocyanin (E = –34 mV) and 5-hydroxy-1,4-naphthoquinone (E = –3 mV) were used for mutants R183K and R183A, respectively.
RESULTS
Reactivity of DaCld R183 Mutants with Chlorite Ion
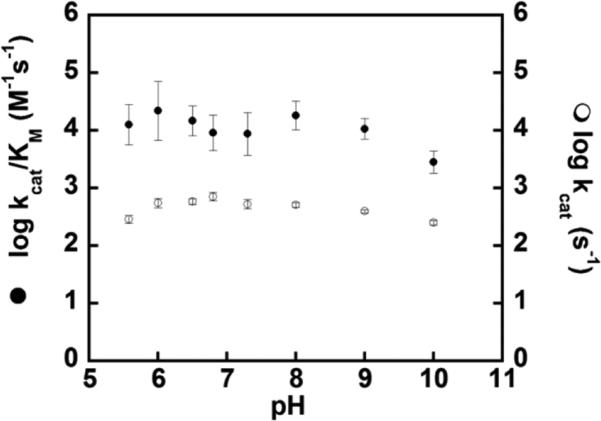
The steady state kinetic parameters for chlorite decomposition catalyzed by WT DaCld, DaCld(R183Q), DaCld(R183K), and DaCld(R183A) were determined at pH 6.8 and 4 °C (Table 1). The WT reaction has a kcat/KM of 2 × 107 M–1 s–1, which is 103-fold higher than that of DaCld(R183K) and 104-fold higher than that of either DaCld(R183Q) or DaCld(R183A). Because the Arg to Gln and Lys mutations have little effect on kcat, the changes in kcat/KM are primarily attributable to the sensitivity of KM to Arg183 mutations. This suggests that a positive charge and/or a polar, H-bond-donating residue (Q or K) is important for efficient formation of the Michaelis complex with chlorite. Both kcat and KM are significantly altered in DaCld(R183A). Plots of log(kcat) and log(kcat/KM) versus pH for DaCld-(R183Q) (Figure 2) show only small variations with pH. This is in contrast to the WT enzyme, for which bimodal pH profiles were observed for both log(kcat) and log(kcat/KM), indicating an optimally active low pH (pH ~6.5) and less active higher-pH forms of the enzyme.6
Table 1.
Steady State Kinetic Parameters for WT and Mutant DaClds for Their Chlorite Dismutase (top) and Peroxidase (bottom) Reactions at pH 6.8 and 4 °C
| KM (mM) | kcat/KM (M–1 s–1)a | kcat (s–1) | |
|---|---|---|---|
| WT Cld | 0.3 | 2 × 107 | 6 × 103 |
| R183K | 42 ± 12 | 2.5 × 104 | (1.0 ± 0.1) × 103 |
| R183Q | 50 ± 9 | 6.9 × 103 | (3.5 ± 0.2) × 102 |
| R183A | 14.6 ± 4.3 | 6.2 × 103 | 91 ± 6 |
| HRP-C14 | (11.5±1.1)×10-3 | 4.6 × 106 | 52.5 ± 3.9 |
| WT Cld | 1.6 ± 0.2 | (1.5 ± 0.2) × 103 | 2.5 ± 0.2 |
| R183K | 30.5 ± 5.1 | (2.3 ± 0.4) × 102 | 7.0 ± 0.4 |
| R183Q | 12.6 ± 3.0 | 5.6 ± 1.4 | 0.070 ± 0.005 |
| R183A | 9.7 ± 1.7 | 4.3 ± 0.8 | 0.042 ± 0.002 |
For the reactions in the top section, chlorite is the sole substrate. For reactions in the bottom section, the concentration of H2O2 was varied at a constant and saturating concentration of ABTS. As described in Experimental Procedures, initial rates were determined by monitoring the production of ABTS radical.
Figure 2.
pH–rate profiles of the R183Q mutant with chlorite. Plots of log(kcat) (○) and log(kcat/KM) (●) as a function of pH using chlorite at 4 °C in citrate/phosphate buffer.
Catalase and Peroxidase Activity of DaCld R183 Mutants
No catalase activity from either the wild type or the R183Q mutant (pH 6.8, 0.1 M phosphate, and 25 °C) was observed. In the presence of one-electron-reducing substrates, the WT enzyme exhibits modest peroxidase (Table 1) activity. Both kcat and kcat/KM(peroxide) for WT DaCld are roughly 4 orders of magnitude smaller than the same parameters measured for the chlorite decomposition reaction. Comparison of the mutants to the WT enzyme shows that kcat/KM(peroxide) is strongly diminished in the mutants while kcat either decreases by 1 order of magnitude for DaCld(R183Q) or slightly increases for DaCld(R183K). Similar to the chlorite reaction, having a polar H-bonding and/or charged residue at position 183 appears to be important for maintaining a high kcat/KM(oxidant).
pH Dependence of the Axial Coordination and Spin State of Ferric Heme in Mutant DaClds
UV–visible and rR characterization of WT DaCld showed that the enzyme undergoes a transition from its five-coordinate high-spin (5cHS) acidic form to its low-spin, ferric hydroxide form with a pKa of 8.7.6 UV–visible pH titrations for the three Arg183 mutant proteins were conducted over acidic (pH 3–6.8), neutral (pH 7–10), and basic (pH 8–12) ranges (Figures S1–S3 of the Supporting Information). In contrast to WT DaCld, the mutants do not exhibit a pH transition below pH ~10. Above pH 10, the dramatic loss of extinction in the Soret, α, and β bands is consistent with the loss of heme from the protein. Heme loss likewise occurs at low pH. On the basis of the heme spectrum, the mutant enzyme folds are stable from pH 5 to 9.9 (R183A) (Figure S2 of the Supporting Information), from pH 5.6 to 10.8 (R183K) (Figure S3 of the Supporting Information), and from pH 5.1 to 10.1 (R183Q) (Figure S1 of the Supporting Information).
DaCld(R183A)
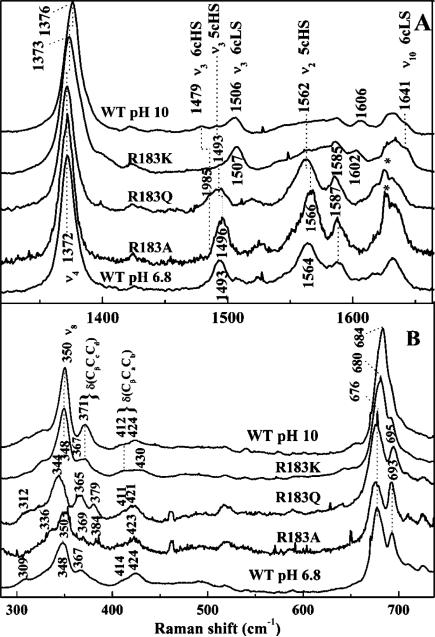
Like the acidic of the WT enzyme, the heme in DaCld(R183A) is 5cHS. At pH 6.8, the UV–visible spectrum of DaCld(R183A) (Figure 3) is similar to that of WT [λmax at 393 (Soret), 509 (Q), and 648 nm (CT)].6 The resonance Raman (rR) spectrum of the ferric protein at pH 6.8 is shown in Figure 4A with its pH dependence shown in Figure S4 of the Supporting Information. Over the pH range examined, those spectra exhibit ν4, ν3, and ν2 bands at 1372, 1496, and 1566 cm–1, respectively. These frequencies are indicative of a 5cHS heme. The alanine side chain in DaCld(R183A) cannot serve as a hydrogen bond donor to stabilize an anionic distal heme ligand. This likely accounts, at least in part, for the fact that this mutant does not form a hydroxide complex at alkaline pH values where the protein is stable. This conclusion is supported by the 5cHS rR signature of DaCld(R183A) at pH 10.0.
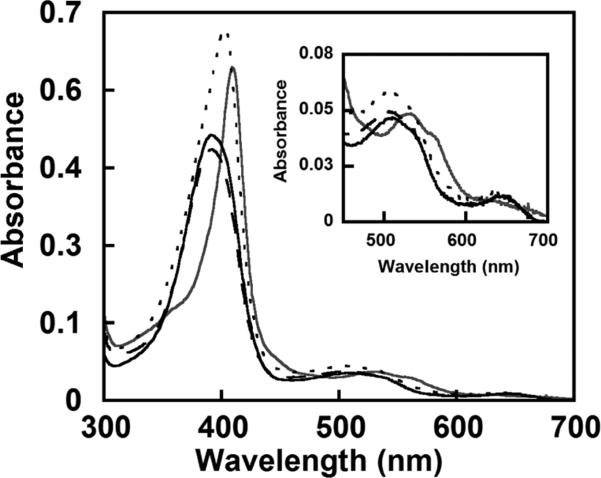
Figure 3.
UV–visible spectra for WT and DaCld mutants at pH 6.8 in 100 mM phosphate buffer. The spectrum of WT DaCld is represented with a thick black solid line with a Soret band at 393 nm. The R183A mutant (–––) has a spectrum similar to that of WT with a Soret band at 391 nm. The R183Q (---) and R183K (thick gray line) mutants have Soret bands at 403 and 410 nm, respectively.
Figure 4.
Soret-excited rR spectra of Cld mutants at pH 6.8. Samples were prepared in 100 mM sodium phosphate (pH 6.8): (A) high-frequency window and (B) low-frequency window. Spectra of WT Cld at pH 6.8 and 10.0 (100 mM Ches) are included for comparison to those of the mutants.
DaCld(R183Q)
The heme in DaCld(R183Q) exists as an equilibrium mixture of 5cHS and six-coordinate high-spin (6cHS) heme. Its UV–visible spectrum has a red-shifted Soret band maximum (403 nm) relative to that of WT, a shoulder at 380 nm, a broad α and β envelope at 509 nm, and a charge transfer (CT) maximum at 633 nm (Figure 3 and Table 2). Charge transfer bands are observed between 600 and 650 nm for HS but not LS ferric heme complexes, and their wavelengths are indicative of coordination number. Spectra of 5cHS species typically have a CT band at ≥640 nm, while the band is closer to 630 nm for 6cHS hemes. Additionally, a shoulder on the Soret band at 380 nm usually indicates the presence of a 5cHS species.22 The DaCld(R183Q) UV–visible spectrum with its shoulder at 380 nm and a CT band around 630 nm therefore suggests a mixture of 5cHS and 6cHS species. The ASoret/A380 absorptivity ratio (Table 2) reflects the relative abundance of 6cHS versus 5cHS species. The low ASoret/A380 ratio for WT and DaCld(R183A) is indicative of a primarily 5cHS species, while the higher ratio for DaCld(R183Q) suggests a mixture of 5c- and 6cHS heme.
Table 2.
UV–Visible Absorbance λmax Values (nanometers) for WT and Mutant DaClds at pH 6.8 and 10
| pH | Soret | CT 2 | β | α | CT 1 | spin sate | ν3 (cm–1) | ASoret/A380 | |
|---|---|---|---|---|---|---|---|---|---|
| WT Cld | 6.8 | 392 | 509 | ~536 | – | 644 | 5cHS | 1493 | 1.1 |
| R183K | 410 | – | ~530 | ~560 | – | 6cLS | 1507 | 2.6 | |
| R183Q | 403 | 509 | ~530 | – | 635 | 6c/5cHS | 1485/1493 | 1.5 | |
| R183A | 391 | 509 | ~530 | – | 644 | 5cHS | 1496 | 1.1 | |
| WT Cld | 10 | 408 | – | 533 | 576 | ~608 | 6cHS/LS | 1479/1506 | 2.05 |
| R183K | 410 | – | ~530 | ~560 | – | 6cLS | 1507 | 2.9 | |
| R183Q | 403 | 509 | 530 | – | 640 | 6c/5cHS | 1485/1493 | 1.4 | |
| R183A | 391 | – | ~530 | – | 625 | 5cHS | 1496 | 1.1 |
The rR spectrum of DaCld(R183Q) (Figure 4A) exhibits core size marker bands, ν4 and ν3, at frequencies that confirm the presence of HS ferric heme. The band in the ν3 region is broad, consistent with the presence of both 5c- and 6cHS hemes (ν3 at 1493 and 1485 cm–1, respectively). The rR spectra for this mutant, like the UV–visible spectra and kinetic parameters, are also insensitive to pH values between 6.8 and 10.0 (Figure S4 of the Supporting Information). The lack of a pH-dependent shift in the rR signature from a 5cHS or 6cHS species to a 6cLS species, which would be expected if a ferric hydroxide had formed, suggests that the sixth heme ligand of the 6cHS species is not hydroxide; the more likely possibility is that the proton-accepting Gln183 stabilizes a heme-bound water molecule that does not undergo proton loss.
DaCld(R183K)
Ferric DaCld(R183K) has a six-coordinate low-spin (6cLS) heme over the entire pH range examined. The UV–visible spectrum of DaCld(R183K) (Figure 3 and Table 2) has a sharp, red-shifted Soret peak (410 nm) relative to that of the WT Cld spectrum. The Soret wavelength together with the α and β bands at 530 and 560 nm, respectively, and the lack of a CT band in the visible region of the spectrum are characteristic of 6cLS heme complexes with bis-nitrogen axial ligation. The rR spectrum of DaCld(R183K) at pH 6.8 shown in Figure 4A has its core size marker bands, ν4, ν3, and ν10, at 1373, 1507, and 1641 cm–1, respectively. These frequencies confirm a ferric, 6cLS heme. The high-frequency, Soret-excited rR spectra of ferric DaCld(R183K) are pH-independent between pH 6.8 and 10.0 (Figure S4 of the Supporting Information). The lack of pH dependence in both the UV–visible and rR data argues against hydroxide being the sixth heme ligand. Instead, the data [the UV–visible features and detailed rR (see below)] support a His/Lys axial ligand pair in which the neutral form of Lys183 coordinates to the heme iron. The pKa of Lys183 (free lysine pKa = 10.53) must therefore be lower than 6.8 in the vicinity of the nearby Lewis acidic ferric iron center.
Effects of Mutations on Heme Conformation
The low-frequency region of the mutant rR spectra is shown in Figure 4B. Assignments for the HS DaCld(R183Q) and DaCld-(R183A) vibrations are made by analogy to HRP23 and Mb24 and indicated in Figure 4B. The δ(CβCaCb) bending modes for the vinyl groups [DaCld(R183Q), 411 and 421 cm–1; DaCld(R183A), 423 cm–1] are very similar to those observed for WT DaCld. The δ(CβCcCd) bending modes for the propionyl groups [DaCld(R183Q), 365 and 379 cm–1; DaCld-(R183A), 369 and 384 cm–1] are clearly resolved in the DaCld(R183Q) and DaCld(R183A) spectra but not in the WT spectrum. The latter has a broad shoulder at ~380 cm–1 on the propionate bending band (367 cm–1). These data suggest that the environment of at least one of the propionate groups in DaCld(R183Q) and DaCld(R183A) is perturbed relative to the WT heme. The LS DaCld(R183K) spectrum has multiple bands in the vinyl bending range [δ(CβCaCb), 412–430 cm–1] and a δ(CβCcCd) band at 367 cm–1. Compared to those of alkaline WT DaCld, which is also 6cLS, the frequencies of ν8 and the δ(CβCcCd) bands of DaCld(R183K) are shifted down by 2 and 4 cm–1, respectively. Arg183 therefore clearly influences the conformation of at least one of the peripheral heme propionate groups in solution. Although the crystal structure does not reveal any direct interaction, the distance between the guanidinium group of Arg183 and the carboxylate group of the 7-propionate could be bridged by a hydrogen-bonded water molecule. In the mutants, this interaction would range from less favorable to impossible, releasing the 7-propionate group to adopt a different conformation.
Axial Heme Coordination and Spin State of Ferrous DaCld Mutants
The visible spectra of ferrous DaCld(R183Q) (Soret band at 434 nm and α and β bands at 558 and 585 nm, respectively) and ferrous DaCld(R183A) (Soret band at 433 nm and α and β bands at 559 and 585 nm, respectively) are consistent with 5cHS ferrous heme. This is confirmed by the coordination and spin state marker bands, ν3 and ν4, for DaCld(R183Q) and DaCld(R183A) near 1473 and 1357 cm–1, respectively, in the high-frequency rR spectra (Figures S5 and S6 of the Supporting Information). This frequency signature is typical of 5cHS ferrous heme and is very similar to that observed for ferrous WT DaCld (1474 and 1359 cm–1).6 The UV–visible spectra of ferrous DaCld(R183K) indicate that it has an alkaline transition in the pH range of 6.8–10.0 (Figure S7 of the Supporting Information). The acidic form has its Soret band at 432 nm and α and β bands at 556 and 588 nm, respectively. This is very similar to the spectrum observed for ferrous WT DaCld (433, 556, and 587 nm) and for ferrous soybean seed coat peroxidase (434, 555, and 582 nm),23 and slightly blue-shifted relative to the spectra of ferrous HRP-C, CCP, BP,25 and CIP.26 Thus, the spectrum of acidic ferrous DaCld(R183K) is consistent with a 5cHS heme. When the pH is increased, the Soret band of DaCld(R183K) shifts to 423 nm and the α and β bands move to 528 and 558 nm, respectively. This suggests that the 5c heme is binding a sixth ligand under alkaline conditions. The acid–base transition of the heme is also observed in the pH dependence in the 406.7 nm excited rR of ferrous DaCld(R183K) (Figure 4). The acidic ferrous form has a ν3 (1474 cm–1) and a ν4 (1358 cm–1) consistent with a 5cHS heme and similar to those observed for ferrous WT DaCld. Between pH 7.8 and 8.8, a second ν3 band starts to appear at 1493 cm–1, consistent with the formation of 6cLS ferrous DaCld(R183K) under alkaline conditions. This behavior supports the conclusion that at low pH the amine side chain of Lys183 is protonated and unable to ligate the heme; in its alkaline form, Lys183 is depronotated and able to bind to the heme iron, generating a 6cLS heme. This behavior is in contrast to that of ferric DaCld(R183K), where the transition between 5c and 6c hemes is not observed, presumably because protonation of the coordinated amine would have to occur at a pH below the range of enzyme stability.
Proximal Heme Environment as a Function of Arg183 Mutation
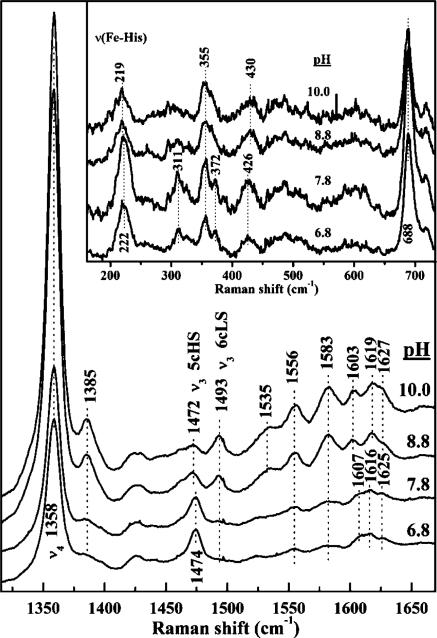
Resonance enhancement of the νFe–His mode increases considerably when the excitation is closer to the Soret band maximum for 5cHS ferrous hemes. In particular, 441.6 nm excitation elicits well-enhanced, low-frequency bands that can be attributed to νFe–His modes. The 441.6 nm-excited low-frequency spectrum of ferrous DaCld(R183A) is compared to its 413.1 nm-excited rR spectrum in Figure S6 of the Supporting Information. By virtue of its intensity and frequency, the 220 cm–1 band is tentatively assigned to the νFe–His mode of 5cHS ferrous DaCld(R183A). The νFe–His mode for ferrous DaCld(R183Q) is also tentatively assigned to a pH insensitive band at 220 cm–1 (Figure S5 of the Supporting Information). The pH dependence of the 441.6 nm excited low-frequency spectra of ferrous DaCld(R183K) is shown in the inset of Figure 5. The 222 cm–1 band is assigned to the νFe–His mode of 5cHS ferrous DaCld(R183K). This vibration is observed at the same frequency in WT DaCld under acidic conditions. The intensity of the νFe–His band decreases as the pH is increased; this is consistent with some of the 5cHS heme being converted to 6cLS heme as seen in the high-frequency rR spectra (Figure 5). The visible spectra of ferrous DaCld(R183K) at pH 6.8 and 10 are also consistent with this conversion (Figure S7 of the Supporting Information).
Figure 5.
pH dependence of the high-frequency rR spectrum of ferrous DaCld(R183K). Samples were prepared in 100 mM sodium phosphate at pH 6.8, 100 mM Tris-HCl at pH 7.8, 100 mM Tris-HCl at pH 8.8, or 100 mM Ches at pH 10.0 and reduced with sodium dithionite. Spectra were acquired with 406.7 nm excitation and 18 mW power at the sample. The inset shows the pH dependence of the low-frequency window of the rR spectrum of ferrous DaCld(R183K) obtained with 441.6 nm excitation.
The 220–222 cm–1 νFe–His frequency observed for WT and the Arg183 mutant enzymes is typical of a proximal His ligand in which the imidazole (ImH) side chain is H-bonded to a weak H-bond acceptor, such as an amide carbonyl group. Examples include α-Hb and Mb.27 If the proximal His ligand had imidazolate character, as in the peroxidases, the νFe–His frequency would be expected to be ≥230 cm–1.28 The Fe–His stretching band in HRP and CcP decreases 4 to 5 cm–1 in frequency when the distal arginine is replaced with a nonpolar amino acid [WT HRP, 243 cm–1; HRP(R38L), 238 cm–1; WT CcP, 233 and 246 cm–1; CcP(R48L), 242 cm–1]. The proximal side of the heme in DaCld, by contrast, is relatively uninfluenced by mutations in its distal pocket. When the arginine is replaced with lysine in HRP, a 3 cm–1 decrease in the νFe–His frequency is observed but no corresponding change is observed in the spectrum of CcP(R48K).12 The crystal structure of CcP(R48K) shows that Lys48 maintains the hydrogen bonding network present in WT CcP. At low pH, the same appears to be true for ferrous DaCld(R183K). The small effects of the distal mutation on the Fe–His stretching frequency of ferrous DaCld indicate that the differences in the enzymatic activity of the mutant DaClds can be attributed to structural and electronic changes within the distal pocket.
Distal Heme Environment Probed with Carbonyl Complexes of WT and Mutant DaClds
Two CO conformers, identified by isotopic substitution, have been reported for WT Cld–CO. The Fe–C and C–O stretching frequencies for these conformers have been reported at pH 5.8, 6.8, and 10.6 In the low-frequency window, the Fe–C stretching bands for these conformers overlap. The WT Cld–CO spectra were subjected to peak fitting so we could assign the frequencies for these isotope sensitive bands and ascertain the relative peak areas for each conformer (Figure S8 of the Supporting Information). Here we examined the pH dependence of WT Cld–CO in greater detail to determine if the FeCO vibrational signature of the heme carbonyl is sensitive to the pKa of 6.5 observed in the DaCld steady state reaction with chlorite. The high- and low-frequency Soret excited rR spectra of WT Cld–CO are independent of pH over the range of 5.8–9.1 (Figure S9 of the Supporting Information). The two conformers (νFe–C and νC–O for the so-called “open” form at 493 and 1956 cm–1, respectively and for the “closed” form at 518 and 1929 cm–1, respectively) are observed over the entire pH range examined here; their νFe–C and νC–O vibration frequencies and their relative populations (as judged by their relative peak areas) are independent of pH from 5.6 to 9.1 when the samples are prepared from fresh DaCld. If the protein is subjected to multiple freeze–thaw cycles or is stored frozen for several months, the νFe–C and νC–O bands assigned to the open conformer become sensitive to pH, shifting 5 and 15 cm–1, respectively, over the pH range of 5.6–10.0; further, the closed form ν(C–O) band is no longer observed at pH 10, as reported previously.6 Changes to spectroscopic heme signatures as a result of storage or sample handling have been reported for CcP and KatG.29–32
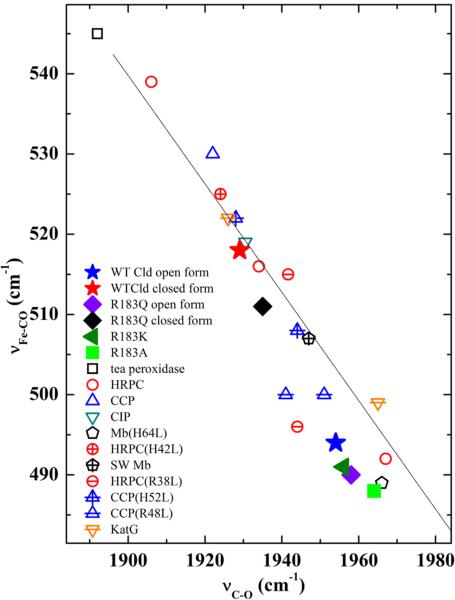
The inverse correlation between νFe–CO and νC–O frequencies is plotted for a number of heme–CO proteins and model complexes in Figure 7. The closed conformer of WT DaCld falls high on the νFe–C–νC–O correlation line, which was interpreted as being due to positive charge near or hydrogen bonding to the bound CO.6 Residues donating H-bonds to the CO on the distal side of the heme enhance π backbonding, weakening the C–O bond and strengthening the Fe–C interaction. This type of strong distal pocket–CO interaction results in points high on the imidazole line (i.e., νFe–C at 520 cm–1 and νC–O at 1935 cm–1).33 To assess hydrogen bonding to the CO ligand via the distal pocket Arg183, the rR spectra of WT DaCld–CO at pD 5.6 were examined for deuterium effects in both the νFe–C and νC–O regions of the spectra. No deuterium shift was observed for either conformer. Observation of a deuterium isotope effect on νC–O in D2O would confirm the presence of H-bonding. However, the deuterium isotope shifts for heme–CO complexes are very small, and their absence does not completely rule out the possibility of H-bonding. Therefore, by virtue of the WT closed conformer's position on the νFe–C–νC–O correlation line, H-bonding could contribute to the interaction between Arg183 and the heme-bound CO. Alternatively, its position on the νFe–C–νC–O correlation line could be due to electrostatic polarization of the heme HOMO by distal positive charge that resides on the side chain of Arg183, thereby concentrating electron density in the FeCO unit and increasing the structural manifestations of π backbonding. In the open conformer, this distal positive charge is distanced from the CO ligand by the Arg183 side chain reorienting toward the solvent and away from the heme pocket.
Figure 7.
νFe–C–νC–O correlation plot comparing CO complexes of WT and Arg183 Cld mutants with peroxidases and myoglobin. The data used to generate the plot are listed in Table S1 of the Supporting Information.
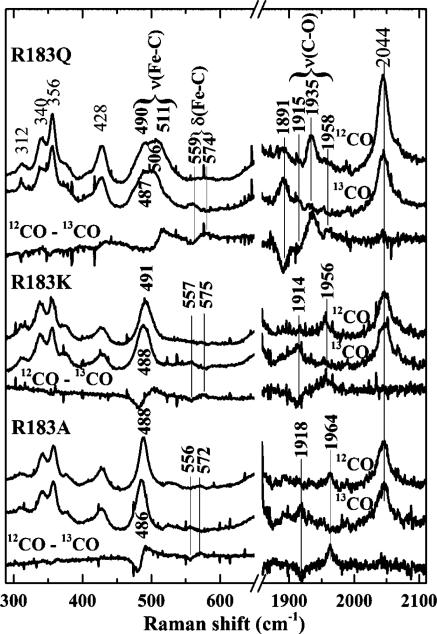
The isotope sensitive bands in the rR spectra of the 6c CO complexes of the Cld mutants have been identified by comparing their spectra obtained with 12CO and 13CO. Their rR spectra are shown in Figure 6, and their frequencies and 13C isotope shifts are summarized in Figure 7 and Table S1 of the Supporting Information. Spectra of DaCld(R183Q)–CO at pH 6.8 and 10.0 (Figure 7 and Figure S10 of the Supporting Information) reveal two isotope sensitive C–O stretching frequencies at 1935 and 1958 cm–1. The DaCld(R183Q)–CO spectra have a broad feature in the low-frequency region where iron–carbon stretching bands of heme–carbonyl complexes are expected. The 12CO–13CO difference spectrum in Figure 6 exhibits multiple features that can be attributed to isotope sensitivity. The frequencies of these bands and relative populations of the two conformers were determined by peak fitting (Figure S10 of the Supporting Information). Two νFe–C frequencies were assigned at 511 and 490 cm–1. Like WT, DaCld(R183Q)–CO exhibits open (490 and 1958 cm–1) and closed (511 and 1935 cm–1) conformers; the small variations with pH are attributed to small changes in the populations of these conformers (see Figure S10 of the Supporting Information). Further, the DaCld(R183Q)–CO conformers fall below their WT counterparts on the νFe–CO–νC–O correlation line. This suggests that the Gln183 is not as polarizing in its interactions with the bound CO, presumably because it lacks the distal positive charge supplied by Arg183 in the WT enzyme. The percentage of the total area of the νFe–C bands assigned to the open and closed conformers also differs between WT (79% open, 21% closed) and R183Q (65% open, 35% closed). Assuming that the relative resonance enhancements of this mode do not change substantially between the WT and R183Q proteins, the closed conformer is more favored in the mutant than in WT DaCld.
Figure 6.
Soret-excited rR spectra of the isotopomers of CO complexes of DaCld Arg183 mutants at pH 6.8. Spectra were acquired with 413.1 nm excitation and 2 mW power at the sample.
The Soret-excited rR spectra of DaCld(R183K)–CO (Figure 6) reveal three isotope sensitive bands at 491, 575, and 1956 cm–1. They are assigned to νFe–C, δFeCO, and νC–O modes, respectively. These vibrations are not sensitive to pH, as judged by comparison of the DaCld(R183K)–CO spectra at pH 5.5, 6.8, and 10.0. These data indicate that there is a single form of DaCld(R183K)–CO analogous to the open form of WT.
The isotopically sensitive bands in the DaCld(R183A)–CO complex are observed at 488, 572, and 1964 cm–1 and are assigned to the νFe–C, δFeCO, and νC–O vibrations, respectively (Figure 6). These features report a single form of DaCld-(R183A)–CO. The Fe–CO stretching frequency is the lowest observed for any of the DaCld–CO complexes that have been characterized to date, while the C–O stretching frequency is the highest, consistent with little or no nonbonded distal interaction with the bound CO ligand.
Similar to the WT DaCld, the locations of the Arg183 mutants on the νFe–C–νC–O correlation plot are consistent with 6c heme carbonyl complexes in which the heme is coordinated to a proximal histidine. All three Arg183 mutations exhibit a decrease in the observed νFe–C frequency and an increase in the νC–O frequency relative to that of WT. Their positions on the νFe–C–νC–O correlation line relative to WT are consistent with the decrease in dπ–π* backbonding expected with weakening polar interactions between the distal heme pocket and the bound CO ligand.
While the CO complexes of DaCld(R183Q) and WT are able to access two conformers, substitution of Arg183 with lysine or alanine eliminates the closed conformer: only the open form is observed in their rR spectra. For DaCld(R183A), the electrostatic interactions that stabilize the closed WT DaCld–CO conformer (H-bonding and/or charge interaction) are lost. DaCld(R183A)–CO falls very close to Mb(H64A)–CO and Mb(H64L)–CO on the νFe–C–νC–O correlation plot. In these Mb mutants, the distal histidine that normally donates H-bonds to the bound CO has been replaced with a hydrophobic amino acid residue. The diminished π back-bonding in these Mb mutants is reflected in a 17–18 cm–1 decrease in their Fe–C stretching frequencies and an increase of 19 cm–1 in the C–O stretching frequencies relative to those of WT Mb (Table S1 of the Supporting Information).34–36
The observation of a single DaCld(R183K)–CO conformer with little interaction between the distal pocket and the bound CO was unexpected. Because the charge and protonation state of lysine are similar to that of Arg183, one may predict that two CO conformers analogous to those of the WT enzyme would be formed. Between pH 5.8 and 10, DaCld(R183K) exhibits a single CO complex that falls slightly above the open form of DaCld(R183Q)–CO on the νFe–C–νC–O correlation plot. This corresponds to a weakly interacting distal pocket–CO pair. Lys183 appears flexible in the placement of its side chain. The crystal structure of ferric NdCld(R173K) showed the distal lysine hydrogen bonded to a sulfate ion well above the heme iron and not to the water molecule coordinated to the heme.11 The solution characterization of ferric DaCld(R183K) presented above clearly shows that the lysine side chain coordinates to the iron. The lack of a closed form of DaCld-(R183K)–CO suggests that, although the distal pocket structure can reorganize to allow coordination of the Lys183 side chain to the heme iron, the driving force for H-bond formation with bound CO is not sufficient to promote reorganization of the distal pocket. Finally, it is interesting to note that the open conformers of DaCld and its Arg183 mutants go down the correlation line in order of decreasing chlorite decomposition activity (WT > R183K > R183Q > R183A) and decreasing heme pocket polarity. This supports our current hypothesis that the positive charge in the pocket, rather than H-bonding, provides the most important structural basis for the enzyme's chlorite decomposition activity.
Resonance Raman data for the CO complexes of peroxidases and peroxidase mutants are summarized in Figure 7 and Table S1 of the Supporting Information for comparison with the DaCld–CO complexes reported here. Heme peroxidases typically have histidine and arginine residues in their distal heme pockets. The distal interactions in the peroxidases are stronger than those in DaCld, because their distal His and Arg residues act in concert. For example, comparison of WT DaCld–CO to the CO complexes of cytochrome c peroxidase (CcP) and horseradish peroxidase (HRP) shows that the Arg–CO interaction in the more strongly interacting closed DaCld conformer is still weaker than the distal CO interaction in these peroxidases. At neutral pH, the CO in the CcP–CO complex is hydrogen bonded to the distal arginine via a water molecule.37 This is a relatively strong interaction with its location on the νFe–C–νC–O correlation line well above the closed WT DaCld–CO. When the distal histidine in CcP (His52) is replaced with a nonpolar residue, its heme environment approaches that of closed WT DaCld–CO. Acidic and basic forms of CcP-(H52L)–CO are observed; the acidic form is slightly higher than the closed WT DaCld–CO form on the correlation line, while the basic form falls between open and closed WT DaCld.38 Replacement of the CcP distal arginine generating CcP(R48L) yields a CO complex that is well below closed WT DaCld–CO but still above open WT Cld–CO on the νFe–C–νC–O correlation plot. Two conformers of HRP–CO that fall on the imidazole νFe–C–νC–O correlation line have been reported.37 The form highest on the correlation line is attributed to direct interaction of the distal Arg with CO; the second conformer, with its weaker distal interaction, is attributed to interaction between CO and the distal histidine. When the distal histidine is mutated in HRP, its CO complex is still higher on the correlation line than WT DaCld. However, if the distal Arg is removed, the two HRP(R38L)–CO conformers fall between the open and closed conformers of WT DaCld–CO, reflecting the effect of the polar His remaining in the pocket.
Affinity of Ferric Mutant Clds for Exogenous Ligands
The equilibrium affinity of DaCld(R183K), DaCld(R183Q), and DaCld(R183A) for a series of ligands was measured as a function of pH from pH 5 to 10 to assess the effects of the R183 side chain on the stabilities of exogenous ligand complexes with ferric DaCld. The maximal measured values for KD are reported in Table 3.
Table 3.
Maximal KD (micromolar) Values for WT, R183K, R183Q and R183A with Ligands Demonstrating Their Varying Affinities at pH 7
| ligand | WT | R183K | R183Q | R183A |
|---|---|---|---|---|
| KCN | 4 | 300 ± 20 | 85 ± 6 | 11000 ± 2000 |
| imidazole | 9.6 ± 0.3 | 18 ± 1 | 27 ± 1 | 21± 1 |
| N3– | 8.3 ± 0.1 | NBa | NBa | NBa |
| F– | 15000 ± 1000 | NBa | NBa |
No binding observed optically up to 400 mM added ligand.
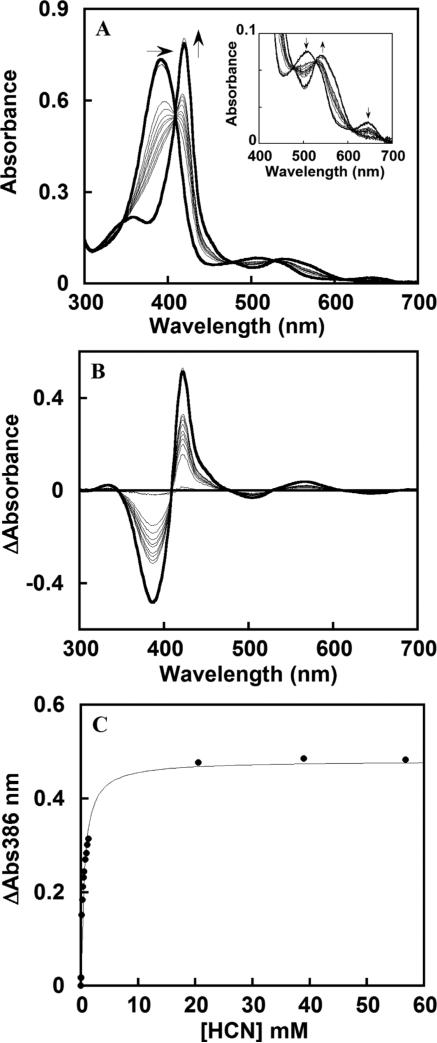
Hydrogen Cyanide
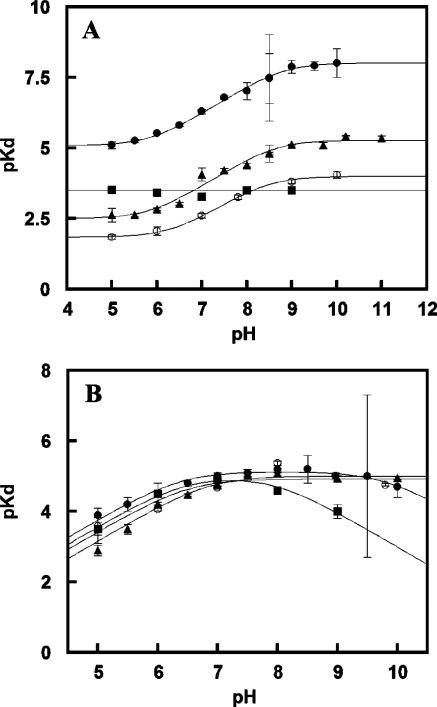
A representative titration of a mutant protein with hydrogen cyanide is shown for DaCld(R183A) in Figure 8A. The Soret band shifts from 391 to 420 nm with an isosbestic point at 411 nm. Values of δA were determined from the difference spectra in Figure 8B and used to generate the plot of δA versus total ligand concentration in Figure 8C. Hydrogen cyanide (HCN ⇆ H+ + CN–; pKa = 9.1) binds with high affinity to WT DaCld, with approximately 20- and 75-fold less affinity to DaCld(R183Q) and DaCld(R183K), respectively, and with substantially (close to 3000-fold) diminished affinity for DaCld(R183A) at pH 7. The sigmoidal shape of the DaCld(R183Q) and DaCld(R183A) pKD versus pH curves for HCN binding (Figure 9A) is similar to the shape of the curve for binding of HCN to WT Cld, generating similar pKa values [DaCld(R183Q), pKa1 = 5.8 ± 0.2 and pKa2 = 8.6 ± 0.2; DaCld(R183A), pKa1 = 6.3 ± 0.1 and pKa2 = 8.6 ± 0.1; WT, pKa1 = 6.0 ± 0.7 and pKa2 = 8.8 ± 0.76]. The higher pKa likely corresponds to the acid dissociation of HCN. The lower turning point was previously assigned to a protein-based deprotonation event. However, it seems more likely that it is not a true pKa but reflects the pH at which CN– is fully converted to HCN, particularly as the same turning point is observed for the WT and each mutant. The plateau in the lower-pH region of the plots indicates that neutral HCN enters the distal pocket with appreciable affinity, with binding likely facilitated in the active site by the Lewis acidity of Fe(III), which facilitates deprotonation of HCN. WT DaCld and the Gln and Ala mutants each have a stronger affinity for CN– than HCN, by 2–2.5 orders of magnitude, in spite of the greatly diminished affinity of the mutants for anionic azide or fluoride (see below). However, the pKD versus pH curves for DaCld(R183Q) and DaCld(R183A) are shifted ~2.5 and ~3 log units lower than that of WT DaCld, respectively. This indicates that the mutants have substantially lower affinity than WT for both CN– and HCN.
Figure 8.
Representative plots illustrating data quality and analysis for DaCld(R183A)–ligand titrations. (A) Titration of KCN at pH 7.8 in a 0.1 M citrate/phosphate buffer. Spectra were measured after addition of cyanide to final concentrations of 0, 24, 48, 163, 272, 377, 478, 574, 770, 959, 92, 1.1, 1.3, 20, 39, and 57 mM. The initial and final spectra are presented as thick black lines, and spectra are corrected for dilution. The inset shows an expanded view of the visible region of the spectrum. (B) Difference plot generated from the data in panel A. (C) Plot of δAbs386 as a function of ligand concentration. The data were fit to an equilibrium isotherm, as described in the text. The KD in this case was determined to be 553 ± 45 μM.
Figure 9.
pKD vs pH profiles describing the binding of (A) KCN and (B) imidazole to WT DaCld (●), DaCld(R183Q) (▲), DaCld-(R183K) (■), and DaCld(R183A) (○). Data are fit to the equations described in the text.
By contrast, the affinity of cyanide for DaCld(R183K) is essentially insensitive to pH (Figure 9A). Because lysine and hydrogen cyanide have somewhat similar pKa values (10.5 and 9.3, respectively) and spectroscopic data for DaCld(R183K) suggest that the pKa for Lys183 in the enzyme is lower than that for free lysine, both of these species could be deprotonated at similar pH values. Over the pH range examined and in the absence of HCN, Lys183 is deprotonated and forms a stable heme Fe–Lys species. This complex has to be dissociated for the formation of the heme–cyanide complex. At higher pH values, the concentration of cyanide ion in solution increases and the enzyme has a higher affinity for the anion than the hydrogen cyanide. However, the plot of pKD versus pH may be flat because the energy yield from the increased affinity of the enzyme for cyanide ion is offset by the higher free energy cost of displacing of the heme-bound Lys183 at alkaline pH.
Imidazole
The imidazole affinity for DaCld(R183Q) and DaCld(R183A) is substantial and relatively unchanged compared with that for WT DaCld. A curve with one pKa was found for the plots of pKD versus pH for imidazole and DaCld(R183Q) and DaCld(R183A) (Figure 9B). The pKa1 of 6.7 ± 0.1 and the pKa1 of 6.8 ± 0.1 appear to correspond to the pKa values of imidazolium and imidazole (pKa = 7). The amine side chain of DaCld(R183K) forms a strong complex with the ferric heme and must be displaced by any exogenous ligand. Imidazole binds DaCld(R183K) with an affinity similar to that for WT near neutral pH. Above pH 7, the affinity begins to decrease, again suggesting that the energy cost of displacing Lys183 becomes sufficiently large that imidazole cannot effectively compete for the heme iron. Clearly, the heme has a strong affinity for amine and imidazole ligands regardless of the residue at position 183.
Anions
The affinity of all of the mutants for the azide and fluoride anions is effectively lost (ligands added at ≤400 mM) (Table 3). Hence, with the exception of CN–, Arg183 is essential for stabilizing the binding of the anions examined in this study.
Reduction Potential
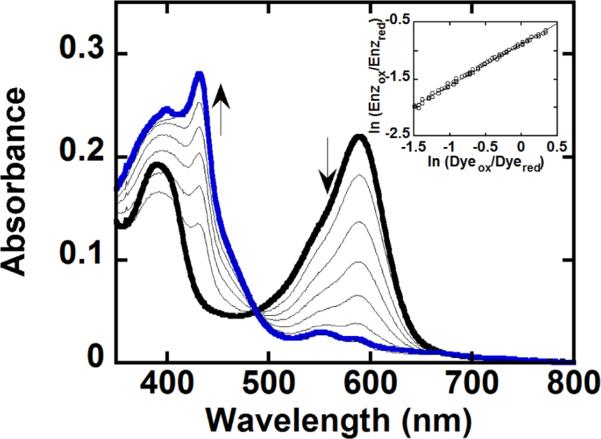
The reduction potential for WT DaCld was measured at –23 mV versus the standard hydrogen electrode (SHE), similar to the –21 mV potential of Cld from Ideonella dechloratans39 but very different from that reported for A. oryzae (Az) Cld (158 mV)10 (Table 4). A typical data set for the reduction potential measurement of the DaCld Arg183 mutants is shown in Figure 10 (the plot of the Nernst equation is represented in the inset); the values for the potentials for all three Arg183 mutants are given in Table 4. Several factors together control the reduction potential of heme proteins, including the electronic characteristics of the axial heme ligand(s), the polarity of the distal environment, and the bonding and non-bonding interactions between the heme and amino acid residues or solvent molecules in the heme vicinity.40
Table 4.
Redox Potentials of Several Heme Proteinsa
| enzyme | E′ (Fe3+/Fe2+) (V vs NHE) | ref |
|---|---|---|
| WT CcP from S. cerevisiae | –0.189 | 40 |
| CcP(R48K) from S. cerevisiae | –0.186 | 40 |
| WT APX from soybean | –0.206 | 40 |
| APX(R38K) from soybean | –0.214 | 40 |
| HRPC | –0.278 | 42 |
| Cld from A. oryzae | –0.158 | 10 |
| Cld from I. dechloratans | –0.021 | 39 |
| WT Cld from D. aromatica | –0.023 | this work |
| Cld(R183A) from D. aromatica | –0.004 | this work |
| Cld(R183K) from D. aromatica | –0.018 | this work |
| Cld(R183Q) from D. aromatica | –0.034 | this work |
All potentials are referenced to the SHE and measured at 25 °C and pH 7. In the case of DaClds, 100 mM phosphate buffer was used.
Figure 10.
Titration of DaCld and potassium indigo tetrasulfonate (dye) with xanthine oxidase used in the determination of its redox potential. An anaerobic solution of 2 μM Cld and 2 μM dye in 0.1 M potassium phosphate buffer (pH 7) with 1 μM benzyl viologen and 200 μM xanthine was reduced with an appropriate amount of xanthine oxidase. The spectra shown were recorded at 0, 50, 104, 158, 216, 266, 320, and 376 min. At time zero, the dye and DaCld are fully oxidized with maxima at 393 and 592 nm, respectively (—); at 376 min (blue line), they are fully reduced with a maximum at 432 nm for DaCld. The inset shows the logarithmic plot of the Nernst equation used to determine the reduction potential. The data were normalized to the concentration of Cld, and the wavelengths oft 432 and 416 nm were used to determine the reduction potential via the relationship ln(Enzox/Enzred) = 0.73 × ln(Dyeox/Dyered) – 0.88.
In the case of WT DaCld and the Arg183 mutants examined here, their proximal ligation is unperturbed, based on strong similarities among their Fe–His stretching frequencies (vide supra). Thus, differences in their reduction potentials likely reflect mutation-induced changes in the distal pocket. In the simplest analysis, the Fe(III)/Fe(II) potential is expected to be lowered (ferric species stabilized) by the introduction of a negative charge in the distal pocket and increased by a positive charge. At the same time, charged and polar residues can stabilize axial ligands to the iron or distal water molecules, electrostatically stabilizing the ferric state. Hence, the influence of distal residues on oxidation state is complex. Myoglobin has a distal histidine in an otherwise hydrophobic distal pocket, yielding a potential of 46 mV.41 Peroxidases by contrast have a much more polarizing distal environment with a neutral histidine and positively charged arginine. Accordingly, their reduction potentials are considerably lower.42 A wide range of reduction potentials have been reported for peroxidases (from –0.028 V for Mycobacterium tuberculosis KatG to –0.320 V for cucumber basic peroxidase) in part because of variations in their electronic structures.40 As it possesses just a distal arginine and no histidine, DaCld has a more hydrophobic pocket with a less electropositive character than typical heme peroxidases. This is in keeping with a measured reduction potential at the less negative end of the peroxidase range. In both DaCld and well-studied heme peroxidases, the arginine also serves to stabilize ligands bound to Fe(III) via hydrogen bonding. Hence, although it is positively charged, the arginine can contribute to the overall stability of the ferric state.
The reduction potential for DaCld(R183K) was measured at –18 mV (pH 7); this is 5 mV higher than the WT DaCld potential. Notably, at this pH, the ferrous and ferric forms of the protein are both 5cHS, so the change in oxidation state does not bring about a change in coordination. In DaCld-(R183A), which likewise remains 5cHS, the charge of the distal pocket is diminished by replacing the long and positively charged arginine side chain with the short and nonpolar methyl group of alanine. An increase in reduction potential of 19 mV relative to that of WT DaCld was observed, indicating that the ferrous form is relatively more stable in the mutant. The inability of the hydrophobic DaCld(R183A) pocket to stabilize anions, hydrogen-bonded water molecules, or hydroxide at higher pH values would cut off these possible avenues for electrostatically compensating for the higher charge of Fe(III). Finally, in the DaCld(R183Q) mutant, the reduction potential is 11 mV more negative than in the WT protein. DaCld(R183Q) is a mixture of 5cHS and 6cHS (presumably water-bound) heme, and coordination number affects reduction potential. The mixed coordination apparently supports a slightly more peroxidase-like potential, stabilizing the ferric state.
The reduction potentials for several peroxidase distal Arg mutants have been reported. For the ascorbate peroxidase mutant, APX(R38K) (ER38K – EWT = –8 mV) and CcP(R48K) (ER48K – Ewt = 3 mV),40 only small changes in the potential relative to that of WT are observed. On the basis of electrostatic considerations alone, no change in reduction potential is expected when the charged arginine is replaced with a similarly charged lysine. The effect of replacing the distal arginine with nonpolar residues in peroxidases is variable. In CcP(R48L), APX(R38A), and HRP-C(R38L), the mutations shift the reduction potentials to a substantial degree but in opposite directions: for CcP(R48L), ER48L – EWT = 25 mV; for APX(R38A), ER38A – EWT = –33 mV; and for HRP-C(R38L), ER38L – EWT = –35 mV.40 Loss of charged and polar residues in the distal pocket, its effect on the imidazolate character of the proximal histidine, and the effects of both on the potential all appear to depend on the peroxidase structure. Notably, the DaCld distal pocket does not contain hydrogen-bonding partners for the distal arginine, nor is this residue connected by hydrogen bonds to the proximal pocket.
DISCUSSION
The Clds from perchlorate-respiring bacteria catalyze a biologically unusual O–O bond-forming reaction via a heme cofactor with great efficiency in a structurally unique active site. We have sought to understand the role of the residue at the position of Arg183 in dictating the chemical, spectroscopic, and electrostatic properties of DaCld through its substitution with neutral (Gln), positively charged (Lys), and hydrophobic (Ala) residues and the use of CO as a probe. These data are important for understanding both the rapid O2 forming reaction catalyzed by DaCld and the likely reactivities of other proteins in the large and phylogenetically diverse Cld protein family, most of which are predicted to have a glutamine, serine, or alanine residue at the position corresponding to Arg183 of DaCld.7
Surprisingly, all of the mutants, including DaCld(R183A) that lacks any polar distal pocket residue, are catalytically active toward chlorite. The steady state turnover number (kcat) decreases only 6-fold in DaCld(R183K), close to 20-fold in DaCld(R183Q), and 65-fold in DaCld(R183A). The KM and kcat/KM values, by contrast, are much more strongly affected: kcat/KM decreases by 3–4 orders of magnitude in all three mutants. This suggests that Arg183 is particularly important for efficient formation of the Michaelis complex. Consistent with this observation, Arg183 is critical for stabilizing bound anions, including hydroxide. WT DaCld forms a hydroxide complex at moderately alkaline pH (pKa = 8.7).6 The three Arg183 mutants examined here, by contrast, exhibit no analogous alkaline transition. By the same token, with the exception of the π-acceptor CN–, none of the anionic ligands binds with measurable affinity to DaCld(R183Q), DaCld(R183K), or DaCld(R183A), and the affinity of the Fe(III) for CN– in the mutants is substantially diminished. The distal arginine forms hydrogen bonds with bound anions in several Cld crystal structures (vide infra). R183A offers no such hydrogen bonding capabilities, and R183Q is likely an insufficient H-bond donor to stabilize complexes with OH–, F–, and N –3. On the other hand, the primary amine of R183K, rather than acting as a hydrogen bond donor, itself binds to the ferric heme over the entire pH range for which the protein is stable. The Lewis acidity of the ferric iron and the hydrophobicity of the distal pocket likely assist in deprotonating R183K (pKa of free lysine = 10.53). Similar to the R183K side chain, imidazole binds avidly to the ferric heme. It does so regardless of the identity of the residue at position 183, and with a pKa in the plot of pKD versus pH corresponding to the imidazolium/imidazole deprotonation. This suggests that the WT and mutant distal pockets cannot accommodate exogenous cations, but the endogenous R183K can be deprotonated at the heme.
The WT enzyme has been shown to have several pH-dependent features in the pH range below the alkaline transition (pKa = 8.7). These could involve the polar-protic Arg183, either directly or indirectly, particularly as its side chain has a clear role in stabilizing Fe(III)-bound anions. This nonbonded interaction could be either a hydrogen bond or a salt bridge. The pH profiles of the steady state kinetic parameters for catalytic chlorite decomposition (kcat and kcat/KM) exhibit a number of turning points, indicating acid–base interconversions. Points at pH 4.8 and 10.3 define the limits of the enzyme's activity. Outside this range, irreversible inactivation of the enzyme occurs. The UV–visible spectra suggest that inactivation correlates with unfolding of the enzyme and dissociation of the heme from the protein. A pKa at pH 6.5 was also observed and was hypothesized to arise from acid dissociation of the Arg183 guanidinuim group, largely because it is the only active site residue with a protic side chain. By virtue of its proximity to the ferric iron center and its otherwise hydrophobic neighbors, we proposed that Arg183 could have a pKa of 6.5.6 Similarly low pKa values and acid–base roles for protein-bound arginine have been proposed in a number of other enzymes, including fumarate reductase,43 polysaccharide lyase,44 inosine 5′-monophosphate dehydrogenase,45 and photosystem II.46 Notably, mutation of Arg183 to Gln results in a loss of the transition in the plots of log kcat and log kcat/KM versus pH observed at pH 6.5 for WT DaCld. This suggests that this pKa indeed involves Arg183, either directly or indirectly.
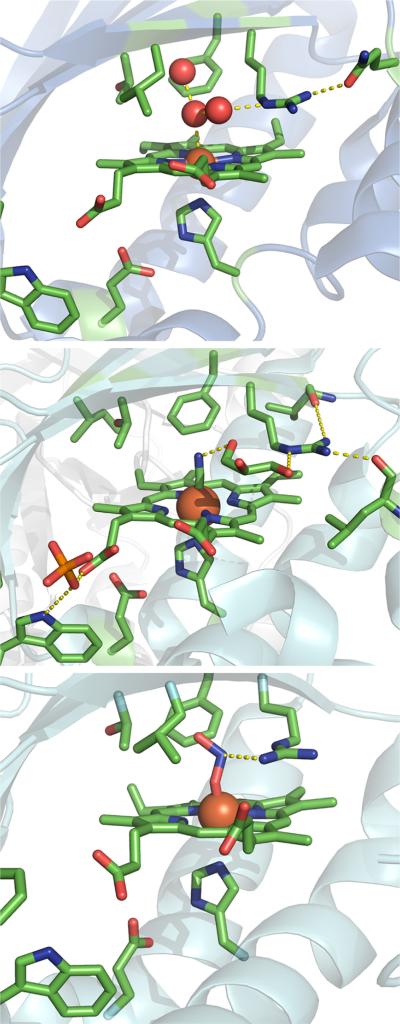
Support for an indirect effect of pH on Arg183 comes from the rR spectra of the DaCld(R183) mutants (discussed below) and structural data. Recent crystal structures of the homologous Clds from Candidatus Nitrospira defluvii (NdCld), Nitrosbacter winogradskyi (NwCld), and the DaCld enzyme show two accessible distal arginine conformers. In the structures of WT NdCld–CN and NdCld–ImH, the distal arginine is rotated away from the exogenous ligand such that it is not positioned over the porphyrin plane.11 In the structure of NwCld, the distal arginine is similarly rotated away from the porphyrin plane and outside of the distal pocket, forming a hydrogen bond to a nearby asparagine residue. The heme iron binds to a water molecule that forms part of a hydrogen-bonded water chain that appears to gain access to the active site when the arginine is rotated out. By contrast, in the DaCld–nitrito structure (pH 9), the Arg183 side chain is hydrogen bonded to the ligand and directly above the porphyrin plane.8 A similar arginine conformer was observed in the structure of the thiocyanate complex of the Cld from A. oryzae.10 The two types of side chain conformers are illustrated in Figure 11. The repositioning of the distal arginine side chain in the NwCld and NdCld structures clearly displaces positive charge from the heme pocket. On the basis of the steady state kinetics results for the WT and mutant DaClds described above, the displacement of charge from the distal pocket appears to reduce anion affinity and reaction efficiency.
Figure 11.
Comparison of open (top and middle) and closed (bottom) conformers hypothesized to modulate DaCld activity. The proteins are drawn as cyan cartoons with important residues depicted as sticks and elements designated by color: green for carbon, red for oxygen, blue for nitrogen, and orange for phosphorus. The iron atom is shown as a rust-colored sphere. In each case, hydrogen bonding interactions that are important for maintaining the distal arginine conformation are depicted as dotted lines. The top panel shows the active site of the Cld from N. winogradskyi (PDB entry 3QPI) with water as its axial ligand. This water molecule is at the center of a hydrogen bonding network that connects the iron to the distal arginine in its open conformation. The distal arginine is also hydrogen bonded to a glutamine. The middle panel shows the active site of the cyano–Cld complex from Ca. N. defluvii (PDB entry 3NN2). The Fe-bound cyanide ligand is connected to the distal arginine via a hydrogen bonding network containing a molecule of solvent (polyethylene glycol), again in the open conformation. The side chain is further hydrogen bonded to two backbone carbonyls. The bottom panel shows the DaCld–nitrito complex (PDB entry 3Q09). The distal arginine assumes its closed conformation. These figures were generated using PyMOL (http://www.pymol.org).
The observed pKa at 6.5 for WT DaCld could be due to an analogous structural shift in the flexible Arg183 side chain. In the closed conformer, the Arg183 side chain is over the porphyrin plane and points into the distal pocket, as in DaCld–ONO–. In the open conformer, the Arg183 side chain points away from the porphyrin plane and is oriented outside the distal pocket, as seen in NdCld–CN, NdCld–ImH, and NwCld–OH2. The mechanism by which pH could bias the populations of the open and closed conformers in DaCld is uncertain. However, a conformer model accounts for the pH profiles of kcat and kcat/KM via pH-dependent modulation of positive charge within the nonbonded interaction distance of the bound substrate. It also suggests a dynamic opening and closing of the heme pocket by a residue that can modulate access of the substrate to the heme, stabilize the enzyme–substrate complex, and possibly facilitate release of the product chloride ion.
The pH-dependent resonance Raman spectra of the ferric WT and mutant DaClds are consistent with a conformer model. The rR spectra of the WT ferric enzyme clearly showed formation of the heme–OH complex with a pKa of 8.7 but were independent of the pH 6.5 transition reported by the enzyme activity.6 The insensitivity of the rR features to the lower pKa is consistent with the associated acid–base chemistry occurring at a site remote from the heme iron. The FeCO vibrational signature of the heme carbonyl could be sensitive to the pKa at 6.5 if the transition results in conformational reorganization of the Arg183 side chain. The closed and open Arg183 conformations described above would be expected to impose distinct H-bonding and/or electrostatic environments on the terminal oxygen atom of coordinated CO. Under the influence of a positive charge or a H-bond donor, the closed conformer should fall high on the νFe–C–νC–O backbonding correlation line. By contrast, the open conformer would be expected to fall low and to the right, a position consistent with minimal nonbonded interaction between CO and the distal pocket. The two conformers seen in the rR spectra of WT DaCld–Co and DaCld(R183Q)–CO complexes are consistent with these expectations. Because glutamine does not contain a titratable proton, the observation of both closed and open active site forms for this mutant supports the conformer model rather than a distal acid/base model. The single conformer (open form) observed for the carbonyl complexes of DaCld(R183K) and DaCld(R183A) indicates that Lys and Ala cannot close the heme site in a manner analogous to that of Arg or Gln. In the case of Ala, the reason is most likely the lack of side chain bulk; for Lys, the reason is not apparent. Finally, the pH independence of the WT–CO complexes reported here is curious and suggests that the heme carbonyl is perhaps not an accurate mimic of the enzyme–substrate complex. Indeed, unlike the ferric–chlorite complex, the CO complex contains ferrous iron and a charge-neutral diatomic axial ligand. It is possible that, as a consequence, it cannot drive the pH-dependent reorientation of the Arg183 side chain in the same manner or to the same extent as the ferric–anion complex.6
Like Arg183 in the WT enzyme, Gln183 can access the open and closed conformers. We hypothesize that the closed form stabilizes coordination of a water molecule to the resting heme, giving rise to the observed mixed 5c/6cHS states. Given the lack of a clear LS heme signature in the spectra, even at high pH, we conclude that the pKa for formation of the heme hydroxide in this mutant is >10. This is consistent with the lack of an observed alkaline transition in the UV–vis titration of this mutant. The distribution of open and closed conformers in DaCld(R183Q)–CO is slightly pH-dependent, suggesting that the shorter Gln183 amide side chain is more sensitive than the guanidinium group of Arg183 to pH-driven reorientation.
The amine side chain of DaCld(R183K) does not interact with the distal pocket but instead binds to the heme Fe itself. Both the UV–visible and rR data support coordination of the primary amine side chain of Lys183 over the entire pH range in which the ferric enzyme is soluble. Consistent with this observation, the pH profiles of kcat and kcat/KM are featureless and offset to lower values than their WT counterparts (Figure 2). The ferrous enzyme contains a 5cHS heme under acidic conditions, but under basic conditions, a 6cLS ferrous complex is formed. The 6cLS ferrous Cld is probably the result of coordination of Lys183 to the Fe(II). The difference in pKa for coordination of the Lys to the heme iron from <6.8 for the ferric enzyme to ~9 for the ferrous enzyme can be attributed to the weaker Lewis acidity of Fe(II) relative to Fe(III). Binding of a distal lysine to the heme has precedence in the R48K distal pocket mutant of CcP. UV–visible and rR data indicate that ferric CcP(R48K) is 6cLS with a pKa for conversion to a 5cHS species below pH 5.2.47 The 6cLS species that predominates at neutral and alkaline pH values is attributed to coordination of the Lys side chain. Ferrous CcP(R48K) also undergoes a conversion from 5cHS to 6cLS ferrous heme between pH 7.0 and 8.533 in a manner similar to that of ferrous DaCld(R183K) (Figure 5).
Conformational mobility in the distal Arg likewise has precedence in heme peroxidases. In the 1.2 Å crystal structure of resting CcP, the distal arginine (Arg48) has two positions: one in which Arg48 is “out” toward the heme propionates and a second in which Arg48 is positioned “in” toward the heme iron.48 These out and in positions of Arg48 in the CcP crystal structure are proposed to be analogous to the open and closed conformers, respectively, observed here for ferric DaCld. In CcP Compound I, the in position of Arg48 is observed exclusively, allowing it to donate a hydrogen bond to the ferryl oxygen atom.50 The distal arginine is found in a similar in position in Compound I of HRP.49 Observation of the closed conformer of DaCld in solution under acidic conditions where the enzyme is most active is consistent with the closed conformer or in position of Arg183 stabilizing its substrate complex and possibly directing the reactivities of its intermediates, in both the reactions with chlorite and H2O2.
What do these results say about the likely roles of other enzymes in the chlorite dismutase family, given that they are known to have a diversity of residues at position 183, including Gln? The reactivity of the mutants is of note here. Arg183 could conceivably be necessary for polarizing and therefore promoting heterolytic cleavage of the Cl–O bond, or subsequently stabilizing the resulting Compound I–hypochlorite pair. If bond cleavage were instead homolytic, resulting in neutral chlorine monoxide and Compound II, roles for Arg in positioning the substrate and stabilizing Compound II could be proposed. However, all three mutants, including the alanine mutant, evolve O2. In other words, Cl–O bond cleavage and subsequent stabilization of intermediates and leaving groups can occur in DaCld containing a distal Gln, Lys, or Ala and no other polar residues. Many peroxidases are known to cleave the chlorite Cl–O bond when using chlorite as a “shunt” reagent for the generation of reactive Compound I. However, unlike DaCld and the R183 mutants described here, peroxidases do not efficiently promote the recombination of the hypochorite–Compound I or chlorine monoxide–Compound II pair. The hydrophobic and sterically confined nature of the Cld distal pocket appears to be important for protecting the reactive intermediates and promoting their reaction by rebound. (Notably, no uncoupling of chlorite consumption and O2 generation was observed in any of the mutants studied here.) It is therefore possible that other Cld proteins could have some O2 evolving capabilities even if they lack a distal Arg, though their efficiency is expected to be diminished. It was likewise interesting to observe no catalase and only weak peroxidase reactivity in any of the DaCld mutants. While such a lack of reactivity is consistent with the lack of a distal His or Asp base in the active site, it raises further questions about the likely biological and chemical roles of other Cld family proteins, which are often still described in genome annotations as peroxidases. In addition, the dye decoloring peroxidases (DyPs) and EfeB proteins, which share a monomer structure with the Clds and collectively make up the CDE (chlorite dismutase, dye decoloring peroxidase, EfeB) superfamily, possess both distal Asp and Arg residues.7 However, their hemes are flipped 180° relative to the heme in the Cld proteins, putting their distal arginine within close hydrogen bonding distance of one of the propionic acid side chains. Hence, the distal Arg in these proteins is conformationally constrained by this hydrogen bond. Moderate to substantial reactivities of DyP and EfeB proteins toward H2O2 have been reported.7,50
What do these results suggest about design elements that could be important in synthetic catalysts for O2 generation or, focusing specifically on the O–O bond-forming step, water reduction? Site isolation and the proximity of the two reactive O atoms would appear to be critical for promoting O–O bond formation. It is achieved in photosystem II by having both the high-valent metal–oxo species and the attacking nucleophilic or radical oxygen species bound to adjacent manganese and calcium sites on the manganese cluster. In DaCld, the high-valent metal–oxo species and the very reactive leaving group (which could be a nucleophilic hypochlorite or a chlorine monoxide) are generated concurrently in a confined and chemically inert pocket, promoting their recombination. Notably, efforts aimed at reacting a premade DaCld Compound I with exogenously added hypochlorite have to date not resulted in generation of O2.
CONCLUSIONS
R183 occupies a key position structurally and serves as the unique charged or polar entity in the DaCld distal pocket. Mutations at this position do not completely eliminate O2-evolving reactivity with chlorite. A positively charged residue at position 183 (Arg or Lys) is important for maintaining a high kcat, though the largest effect for all substitutions (Lys, Gln, and Ala) was a substantially increased value for KM. Consistent with this observation, R183 appears to be essential for maintaining the affinity of the ferric site for anions, including the hydroxide-bound alkaline form. Resonance Raman data are consistent with the R183 side chain accessing a closed conformation that maximizes positive charge in the vicinity of the heme iron, supporting the most efficient reactivity in the steady state. The ferric heme is somewhat less oxidizing than in a typical heme peroxidase, in either WT DaCld or the mutants, and the distal pocket less electrostatically polarizing. Rotation of the R183 side chain away from the porphyrin plane is expected to further reduce the amount of positive charge in the distal pocket. The fact that the WT and R183 mutants all successfully produce O2 suggests an especially important role for the pocket in promoting the rebound reaction between the high-valent Fe–O intermediate and the leaving group. It also suggests that Cld family proteins could still retain some chlorite decomposition activity, even though most appear to lack a distal Arg. This reactivity could be limited by their ability to stabilize a bound anionic substrate and/or leaving group.
Supplementary Material
ACKNOWLEDGMENTS
Garrett Moraski is thanked for helpful discussions.
Funding
Supported by National Institutes of Health Grants R01GM090260 (to J.L.D.) and R15GM094039 (to G.S.L.-R.).
ABBREVIATIONS
- DaCld
D. aromatica chlorite dismutase
- WT Cld
wild-type chlorite dismutase
- 5c
five-coordinate
- HS
high-spin
- 6c
six-coordinate
- LS
low-spin
- rR
resonance Raman
- UV–vis
UV–visible
- NdCld
Ca. N. defluvii chlorite dismutase
- NwCld
N. winogradskyi chlorite dismutase
- KatG
catalase-peroxidase
- CcP
cytochorne c peroxidase
- HRP
horseradish peroxidase
- PDB
Protein Data Bank
Footnotes
ASSOCIATED CONTENT
Supporting Information
UV–visible pH titrations over pH ranges (A) from 4 to 7, (B) from 7 to 11, and (C) from 8 to 12 for the R183Q, R183A, and R183K mutants (Figures S1–S3, respectively); high-frequency window rR spectra of Cld mutants as a function of pH (Figure S4); high-frequency rR spectra of ferrous Cld(R183Q) at pH 7.8 and 10.0 (Figure S5); high- and low-frequency regions of the rR spectrum of ferrous Cld(R183A) in 100 mM sodium phosphate (pH 6.8) (Figure S6); UV–visible spectra of ferrous Cld(R183K) as a function of pH (Figure S7); fit of WT DaCld–CO rR data (pH 5.8) for the determination of the Fe–C stretching frequency of the two conformers (Figure S8); Soret-excited rR spectra of WT Cld–CO as a function of pH (Figure S9); resonance Raman spectra and fits for the low-frequency data for the isotologs of Cld(R183Q)–CO at (A) pH 6.8 and (B) pH 10.0 (Figure S10); comparison of Fe–CO vibrational frequencies for several heme proteins that were used to construct Figure 7 (Table S1). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Coates JD, Achenbach LA. Microbial perchlorate reduction: Rocket-fuelled metabolism. Nat. Rev. Microbiol. 2004;2:569–580. doi: 10.1038/nrmicro926. [DOI] [PubMed] [Google Scholar]
- 2.Betley T, Surendranath Y, Childress M, Alliger G, Fu R. A ligand field chemistry of oxygen generation by the oxygen-evolving complex and synthetic active sites. Philos. Trans. R. Soc. London, Ser. B. 2008;363:1293–1303. doi: 10.1098/rstb.2007.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zdilla M, Lee A, Abu Omar M. Concerted dismutation of chlorite ion: Water-soluble iron-porphyrins as first generation model complexes for chlorite dismutase. Inorg. Chem. 2009;48:2260–2268. doi: 10.1021/ic801681n. [DOI] [PubMed] [Google Scholar]
- 4.Slaughter LM, Collman JP, Eberspacher TA, Brauman JI. Radical autoxidation and autogenous O2 evolution in manganese-porphyrin catalyzed alkane oxidations with chlorite. Inorg. Chem. 2004;43:5198–5204. doi: 10.1021/ic049922j. [DOI] [PubMed] [Google Scholar]
- 5.Zdilla M, Lee A, Abu Omar M. Bioinspired dismutation of chlorite to dioxygen and chloride catalyzed by a water-soluble iron porphyrin. Angew. Chem. 2008;47:7697–7700. doi: 10.1002/anie.200801521. [DOI] [PubMed] [Google Scholar]
- 6.Streit BR, Blanc B, Lukat-Rodgers GS, Rodgers KR, DuBois JL. How active-site protonation state influences the reactivity and ligation of the heme in chlorite dismutase. J. Am. Chem. Soc. 2010;132:5711–5724. doi: 10.1021/ja9082182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goblirsch B, Kurker R, Streit B, Wilmot C, DuBois J. Chlorite dismutases, DyPs, and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily. J. Mol. Biol. 2011;408:379–398. doi: 10.1016/j.jmb.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goblirsch B, Streit B, DuBois J, Wilmot C. Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase. J. Biol. Inorg. Chem. 2010;15:879–888. doi: 10.1007/s00775-010-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poulos TL, Fenna RE. Peroxidases: Structure, Function, and Engineering. Met. Ions Biol. Syst. 1994;30:25–75. [Google Scholar]
- 10.de Geus DC, Thomassen J, Hagedoorn P-L, Pannu NJ, van Duijn E, Abrahams JP. Crystal structure of chlorite dismutase, a detoxifying enzyme producing molecular oxygen. J. Mol. Biol. 2009;387:192–217. doi: 10.1016/j.jmb.2009.01.036. [DOI] [PubMed] [Google Scholar]
- 11.Kostan J, Sjoblom B, Maixner F, Mlynek G, Furtmuller P, Obinger C. Structural and functional characterisation of the chlorite dismuatse from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: Idientification of a catalytically important amino acid residue. J. Struct. Biol. 2010;172:331–342. doi: 10.1016/j.jsb.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Howes BD, Rodriguez-Lopez JN, Smith AT, Smulevich G. Mutation of distal residues of horseradish peroxidase: Influence on substrate binding and cavity properties. Biochemistry. 1997;36:1532–1543. doi: 10.1021/bi962502o. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Lopez J, Smith A, Thorneley R. Recombinant horseradish peroxidase isoenzyme C: The effect of distal haem cavity mutations (His42→Leu and Arg38→Leu) on compound I formation and substrate binding. J. Biol. Inorg. Chem. 1996;1:136–142. [Google Scholar]
- 14.Rodriguez-Lopez JN, Smith A, Thorneley R. Role of arginine 38 in horseradish peroxidase. A critical residue for substrate binding and catalysis. J. Biol. Chem. 1996;271:4023–4030. doi: 10.1074/jbc.271.8.4023. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Lopez JN, Lowe DJ, Hernandez-Ruiz J, Hiner ANP, Garcia-Canovas F, Thorneley RNF. Mechanism of reaction of hydrogen peroxide with horseradish peroxidase: Identification of intermediates in the catalytic cycle. J. Am. Chem. Soc. 2001;123:11838–11847. doi: 10.1021/ja011853+. [DOI] [PubMed] [Google Scholar]
- 16.Poulos T. Thirty years of heme peroxidase structural biology. Arch. Biochem. Biophys. 2010;500:3–12. doi: 10.1016/j.abb.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streit B, DuBois J. Chemical and steady-state kinetic analyses of a heterologously expressed heme dependent chlorite dismutase. Biochemistry. 2008;47:5271–5280. doi: 10.1021/bi800163x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savenkova MI, Ortiz de Montellano PR. Horseradish peroxidase: Partial rescue of the His-42 → Ala mutant by a concurrent Asn-70 → Asp mutation. Arch. Biochem. Biophys. 1998;351:286–293. doi: 10.1006/abbi.1997.0559. [DOI] [PubMed] [Google Scholar]
- 19.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemocrome spectra. Anal. Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 20.Massey V. A simple method for the determination of redox potentials. In: Curti B, Ronchi S, Zanetti G, editors. Flavins and Flavoproteins. Walter de Gruyter; Berlin: 1991. pp. 66–69. [Google Scholar]
- 21.Mansfield CW. Color chart of indicators. Williams & Wilkins Co.; Baltimore: 1921. [Google Scholar]
- 22.Ghiladi R, Medzihradszky K, Rusnak F, de Montellano P. Correlation between isoniazid resistance and superoxide reactivity in Mycobacterium tuberculosis KatG. J. Am. Chem. Soc. 2005;127:13428–13442. doi: 10.1021/ja054366t. [DOI] [PubMed] [Google Scholar]
- 23.Nissum M, Feis A, Smulevich G. Characterization of soybean seed coat peroxidase: Resonance Raman evidence for a structure-based classification of plant peroxidases. Biospectroscopy. 1998;4:355–364. doi: 10.1002/(sici)1520-6343(1998)4:6<355::aid-bspy1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 24.Hu SZ, Smith KM, Spiro TG. Assignment of protoheme Resonance Raman spectrum by heme labeling in myoglobin. J. Am. Chem. Soc. 1996;118:12638–12646. [Google Scholar]
- 25.Howes BD, Schiodt CB, Marzocchi MP, Ma J-G, Zhang J, Shelnutt JA, Smulevich G. The quantum mixed-spin heme stale of barley peroxidase: A paradigm for class III peroxidases. Biophys. J. 1999;77:478–492. doi: 10.1016/S0006-3495(99)76905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smulevich G, Feis A, Focardi C, Tams J, Welinder K. Resonance Raman study of the active site of Coprinus cinereus peroxidase. Biochemistry. 1994;33:15425–15432. doi: 10.1021/bi00255a024. [DOI] [PubMed] [Google Scholar]
- 27.Kitagawa T, Nagai K, Tsubaki M. Assignment of the Fe-Nε (His F8) stretching band in the resonance Raman spectra of deoxy myoglobin. FEBS Lett. 1979;104:376–378. doi: 10.1016/0014-5793(79)80856-x. [DOI] [PubMed] [Google Scholar]
- 28.Teraoka J, Kitagawa T. Structural implication of the heme-linked ionization of horseradish peroxidase probed by the Fe-histidine stretching Raman line. J. Biol. Chem. 1981;256:3969–3977. [PubMed] [Google Scholar]
- 29.Smulevich G, Evangelista-Kirkup R, English A, Spiro TG. Raman and infrared spectra of cytochrome c peroxidase-carbon monoxide adducts in alternative conformational states. Biochemistry. 1986;25:4426–4430. doi: 10.1021/bi00363a038. [DOI] [PubMed] [Google Scholar]
- 30.Yonetani T, Anni H. Yeast cytochrome c peroxidase. Coordination and spin states of heme prosthetic group. J. Biol. Chem. 1987;262:9547–9554. [PubMed] [Google Scholar]
- 31.Vitello LB, Huang M, Erman JE. pH-dependent spectral and kinetic properties of cytochrome c peroxidase: Comparison of freshly isolated and stored enzyme. Biochemistry. 1990;29:4283–4288. doi: 10.1021/bi00470a004. [DOI] [PubMed] [Google Scholar]
- 32.Kapetanaki SM, Chouchane S, Yu S, Zhao X, Magliozzo RS, Schelvis JPM. Mycobacterium tuberculosis KatG-(S315T) catalase-peroxidase retains all active site properties for proper catalytic function. Biochemistry. 2005;44:243–252. doi: 10.1021/bi048097f. [DOI] [PubMed] [Google Scholar]
- 33.Smulevich G, Mauro JM, Fishel LA, English AM, Kraut J, Spiro TG. Cytochrome c peroxidase mutant active site structures probed by resonance Raman and infrared signatures of the CO adducts. Biochemistry. 1988;27:5486–5492. doi: 10.1021/bi00415a015. [DOI] [PubMed] [Google Scholar]
- 34.Li T, Quillin ML, Phillips GN, Jr., Olson JS. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 35.Ling J, Li T, Olson JS, Bocian DF. Identification of the iron-carbonyl stretch in distal histidine mutants of carbonmonoxymyoglobin. Biochim. Biophys. Acta. 1994;1188:417–421. doi: 10.1016/0005-2728(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 36.Anderton CL, Hester RE, Moore JN. A chemometric analysis of the resonance Raman spectra of mutant carbonmonoxy-myoglobins reveals the effects of polarity. Biochim. Biophys. Acta. 1997;1338:107–120. doi: 10.1016/s0167-4838(96)00194-x. [DOI] [PubMed] [Google Scholar]
- 37.Smulevich G, Feis A, Howes BD. Fifteen years of Raman spectroscopy of engineered heme containing peroxidases: What have we learned? Acc. Chem. Res. 2005;38:433–440. doi: 10.1021/ar020112q. [DOI] [PubMed] [Google Scholar]
- 38.Smulevich G, Miller MA, Kraut J, Spiro TG. Conformational change and histidine control of heme chemistry in cytochrome c peroxidase: Resonance Raman evidence from Leu-52 and Gly-181 mutants of cytochrome c peroxidase. Biochemistry. 1991;30:9546–9558. doi: 10.1021/bi00103a023. [DOI] [PubMed] [Google Scholar]
- 39.Stenklo K, Thorell D, Bergius H, Aasa R, Nilsson T. Chlorite dismutase from Ideonella dechloratans. J. Biol. Inorg. Chem. 2001;6:601–607. doi: 10.1007/s007750100237. [DOI] [PubMed] [Google Scholar]
- 40.Battistuzzi G, Bellei M, Bortolotti C, Sola M. Redox properties of heme peroxidases. Arch. Biochem. Biophys. 2010;500:21–36. doi: 10.1016/j.abb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Taylor J, Morgan V. Oxidation-reduction potentials of the metmyoglobin-myoglobin system. J. Biol. Chem. 1942;144:15–20. [Google Scholar]
- 42.Makino R, Chiang R, Hagger L. Oxidation-reduction potential measurements on chloroperoxidase and its complexes. Biochemistry. 1976;15:4748–4754. doi: 10.1021/bi00666a033. [DOI] [PubMed] [Google Scholar]
- 43.Doherty MK, Pealing SL, Miles CS, Moysey R, Taylor P, Walkinshaw MD, Reid GA, Chapman SK. Identification of the active site acid/base catalyst in a bacterial fumarate reductase: A kinetic and crystallographic study. Biochemistry. 2000;39:10695–10701. doi: 10.1021/bi000871l. [DOI] [PubMed] [Google Scholar]
- 44.Charnock SJ, Brown IE, Turkenburg JP, Black GW, Davies GJ. Convergent evolution sheds light on the anti-β-elimination mechanism common to family 1 and 10 polysaccharide lyases. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12067–12072. doi: 10.1073/pnas.182431199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlippe YVG, Hedstrom L. Is Arg418 the catalytic base required for the hydrolysis step of the IMP dehydrogenase reaction? Biochemistry. 2005;44:11700–11707. doi: 10.1021/bi048342v. [DOI] [PubMed] [Google Scholar]
- 46.McEvoy JP, Brudvig GW. Water splitting. Chem. Rev. 2006;106:4455–4483. doi: 10.1021/cr0204294. [DOI] [PubMed] [Google Scholar]
- 47.Vitello L, Erman J, Miller M, Wang J, Kraut J. Effect of arginine-48 replacement on the reaction between cytochrome c peroxidase and hydrogen peroxide. Biochemistry. 1993;32:9807–9818. doi: 10.1021/bi00088a036. [DOI] [PubMed] [Google Scholar]
- 48.Bonagura CA, Bhaskar B, Shimizu H, Li HY, Sundaramoorthy M, McRee DE, Goodin DB, Poulos TL. High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase. Biochemistry. 2003;42:5600–5608. doi: 10.1021/bi034058c. [DOI] [PubMed] [Google Scholar]
- 49.Berglund G, Carlsson G, Smith A, Szoke H, Henriksen A. The catalytic pathway of horseradish peroxidase at high resolution. Nature. 2002;417:463–468. doi: 10.1038/417463a. [DOI] [PubMed] [Google Scholar]
- 50.Ahmad M, Roberts JN, Hardiman EM, Singh R, Eltis LD, Bugg TDH. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry. 2011;50:5096–5107. doi: 10.1021/bi101892z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.