Abstract
In vitro production of porcine embryos by means of in vitro fertilization (IVF) or somatic cell nuclear transfer (SCNT) is limited by great inefficienciy. The present study investigated chromatin and nucleolar dynamics in porcine embryos developed in vivo (IV) and compared this physiological standard to that of embryos produced by IVF, parthenogenetic activation (PA), or SCNT. In contrast to IV embryos, chromatin spatial and temporal dynamics in PA, IVF, and SCNT embryos were altered; starting with aberrant chromatin–nuclear envelope interactions at the two-cell stage, delayed chromatin decondensation and nucleolar development at the four-cell stage, and ultimately culminating in failure of proper first lineage segregation at the blastocyst stage, demonstrated by poorly defined inner cell mass. Interestingly, in vitro produced (IVP) embryos also lacked a heterochromatin halo around nucleolar precursors, indicating imperfections in global chromatin remodeling after fertilization/activation. Porcine IV-produced zygotes and embryos display a well-synchronized pattern of chromatin dynamics compatible with genome activation and regular nucleolar formation at the four-cell stage. Production of porcine embryos under in vitro conditions by IVF, PA, or SCNT is associated with altered chromatin remodeling, delayed nucleolar formation, and poorly defined lineage segregation at the blastocyst stage, which in turn may impair their developmental capacity.
Introduction
Fertilization represents a massive break-point in the status of the fully differentiated maternal and paternal genomes. During sequential morphological remodeling and functional reprogramming events, the differentiated state is reversed into the pluripotent state of the early embryo. In the last decade, several studies aimed on discovering the molecular mechanisms contributing to the functional reprogramming of the genome. The majority of the studies focused on epigenetic modifiers and processes enabling activation and/or silencing of developmentally important genes (Bourc'his and Voinnet 2010; Corry et al., 2009; Lorthongpanich et al., 2010). However, changes in selected epigenetic marks may not necessarily represent the ultimate marker of reprogramming. But genome-wide analysis is time consuming, expensive, and requires large amounts of biological material.
Besides functional reprogramming of the genome, the epigenetic changes are associated with morphological remodeling of the chromatin (Ahmed et al., 2010; Pichugin et al., 2010). Therefore, functional reprogramming is tightly linked to morphological chromatin remodeling and vice versa; changes in chromatin organization affect the expression profile of specific genes (Orkin and Hochedlinger, 2011; Pichugin et al., 2010; Thomas et al., 2011). Upon fertilization, the fully condensed chromatin of mammalian oocytes and spermatozoa undergoes rapid decondensation, and chromatin enclosure by nuclear envelope results in the formation of the maternal and paternal pronuclei (Laurincik et al., 1995, 1996). In subsequent cell divisions, the transcriptionally silent genome continues to be reprogrammed and chromatin is progressively rearranged. At the species-specific time point, the major portions of the newly formed genome becomes transcriptionally active indicating initiation of the embryonic developmental program (major embryonic genome activation, EGA) (Tomanek et al., 1989). Concomitantly, the heterochromatin decondenses and disperses throughout the nucleoplasm in mouse and cattle embryos, while a small fraction of condensed chromatin remains visible (Ahmed et al., 2010; Svarcova et al., 2007).
During differentiation, chromatin becomes organized into distinct territories characteristic for somatic cells. In mammals, the first differentiation results in establishment of two cell lineages, that is, the pluripotent inner cell mass (ICM) and unipotent trophectoderm (TE) of the blastocyst. The spatial organization of the nucleus in the TE is critically involved in regulating de novo gene expression by positioning the gene rich and decondensed euchromatin, from which genes are expressed, in the center and leaving the silent, highly condensed heterochromatin in the periphery along with the nuclear envelope (Cremer et al., 2006; Koehler et al., 2009; Mattout and Meshorer 2010; Pichugin et al., 2010). In contrast, in pluripotent mouse cells, that is, ICM and embryonic stem cells, the chromatin is uniformly dispersed throughout the nucleoplasm, mostly represented by euchromatin (Ahmed et al., 2010; Efroni et al., 2008).
In parallel with the chromatin, the nucleolus also dynamically evolves during early mammalian development. The nucleolus is the most prominent emerging nuclear structure around the ribosomal genes. Their transcription occurs in the periphery of fibrillar centers (FC) from where the primary transcripts localize to electron dense rims (dense-fibrillar component; DFC). After partial processing in DFC, the final assembly of ribosomal RNA into ribosomal subunits occurs in the granular component (GC) (Hernandez-Verdun et al., 2010). The functional nucleolus contains all three components organized in the reticulated manner. After fertilization, active nucleoli do not exist. Instead, the formation of the pronuclei is accompanied by formation of nucleolar precursor bodies (NPBs); electron dense spheres not capable of ribosome biogenesis (Hyttel et al., 2000a, 2001). The first functional nucleolar compartments develop during EGA consistent with the initiation of ribosomal genes transcription, rendering the nucleolus a useful marker of proper genome activation (Hyttel et al., 2001; Svarcova et al., 2008). However, the role of nucleolus in early chromatin reprogramming is largely unknown. Based on observations that the heterochromatin is tightly connected to NPBs it was previously suggested, that the NPBs may represent physical anchoring sites for α-satellite chromatin and other repetitive sequences during their redistribution within the nucleus (Martin et al., 2006a, 2006b). Interestingly, the presence or absence of perinucleolar heterochromatin rims in embryos developed in suboptimal conditions has not yet been characterized.
The domestic pig (Sus scrofa) represents a unique animal model for biomedicine. The genetic, physiological, and anatomical similarities of the domestic pig with humans render this species excellent for study of several human diseases, and as donor of functional xenografts to overcome the growing shortage of human organs (Cozzi et al., 2009; Gil et al., 2010; Kragh et al., 2009; Schmidt et al., 2010; Wilmut et al., 2009). However, after many years of intensive research, in vitro (IV) production of porcine embryos remains inefficient compared to mice (Gil et al., 2010; Petersen et al., 2008; Suzuki et al., 2006; Yamanaka and Blau 2010; Zhou et al., 2010; Zhu et al., 2003). The reduced embryonic development is most probably caused by subomptimal culture conditions, which in turn lead to insufficient epigenetic reprogramming as suggested by several studies (Ju et al., 2010; Miyamoto et al., 2011; Yamanaka et al., 2009; Zhao et al., 2010a, 2010b). In-depth analysis of chromatin changes during early porcine embryogenesis may be crucial for better understanding of reprogramming efficiency. Noteworthy, chromatin and nuclear dynamics in in vivo developed porcine embryos have not been studied yet. Thus, the field is still missing the standard required for comparison of developmental potentials of embryos of different origin.
Here we used transmission electron microscopy (TEM) to reveal chromatin changes in porcine embryos of different origins. The goal of the present study was designed to provide a detailed morphological description of chromatin dynamics in in vivo developed porcine embryos and compare this “physiological standard” with that observed in embryos produced under in vitro conditions, that is, parthenogenetic activation (PA), somatic cell nuclear transfer (SCNT), and in vitro fertilization (IVF) embryos. The data can be used as a guideline/model for the rapid evaluation of the reprogramming efficiency in early embryos and in reprogrammed somatic cells. Moreover, the results point out the role of nuclear envelope in proper chromatin remodeling and importance of the pre-EGA period for subsequent embryonic development in the pig.
Materials and Methods
Production of embryos
IV embryo collection
After insemination the sows were slaughtered at defined time points in a local slaughterhouse and uteri were transported to the laboratory at 37°C within 1 h. Uterine horns were flushed with prewarmed phosphate-buffered saline (PBS) enriched with 1% fetal bovine serum (FBS) to collect the embryos. The embryos were recovered from the flushed PBS at different time points: days 1–2 (zygotes), days 2–3 (two- and four-cell stages), days 3–4 (four- and eight-cell/morulae), days 5–6 (early blastocysts), and days 6–7 (late blastocysts).
In vitro fertilization (IVF)
The reagents were purchased from Sigma (Sigma-Aldrich, Vallensbæk, Denmark) and the plastic ware was from Nunclon (Nunc, VWR International, Albertslund, Denmark). Prewarmed (30 min) four-well plates containing 0.5 mLof the appropriate medium covered with 0.4 mL mineral oil (M 8410) were used for IV maturation of oocytes (IVM), IVF, and embryo culture (IVC). Incubations were performed at 38.5°C in 5% CO2 [95% humidified air (IVM and IVF), or in 5% O2, 90% N2 (IVC)]. Cumulus–oocyte complexes (COCs) were isolated from abattoir Danish Landrace×Yorkshire gilt ovaries, which were maintained at 30–33°C until follicle aspiration. Only 3–8-mm follicles were aspirated, and COCs where at least three layers of cumulus cells were selected for the experiments. IVM was performed for 42–44 h in TCM-199 (M 2154) supplemented with 50 ng/mL epidermal growth factor (EGF; E4127), 10 IU/mL equine chorionic gonadotropic (eCG), 5 IU/mL human chorionic gonadotropic (hCG; Suigonan Vet., Intervet Scandinavia, Skovlunde, Denmark), 50 μg/mL gentamicin (G 1264), 100 μM 2-mercaptoethanol, 5 μL/ml Insulin-Transferrin-Selenium, L-Glutamine–Penicillin–Streptomycin (G6784), and 4 mg/mL bovine serum albumin (BSA, A 3311).
Mixed Duroc semen in extender was purchased from a boar station (Hatting-KS, Ringsted, Denmark) and kept at 16–18°C until use. Semen was washed in noncapacitating medium (113 mM NaCl, 5 mM KCl, 5.56 mM glucose, 1.2 mM KH2PO4, 1.2 mM MgSO4, 22 mM Na-lactate, 5 μg/mL phenol red, 50 μg/mL gentamicin), and then resuspended in capacitating IVF medium (90 mM NaCl, 12 mM KCl, 0.5 mM NaH2PO4, 25 mM NaHCO3, 0.5 mM MgSO4, 2 mM Na-pyruvate, 8 mM CaCl2, 1.9 mM caffeine, L-Glutamine–Penicillin–Streptomycin (G6784), 5 μg/mL phenol red, 4 mg/mL BSA), and finally preincubated for 10–15 min. IV insemination was performed with a final sperm concentration of 1×106/mL. At 24 h postinsemination, excess sperm and cumulus cells were removed from presumptive zygotes and were cultured in porcine zygote medium (PZM) supplemented with 5% FBS (Yoshioka et al., 2002) and collected at 26 and 34 h postinsemination. Embryos were collected on day 2 (two-cell stages), day 3 (four-cell stages), day 4 (eight-cell/morulae), days 5–6 (early blastocysts), and days 6–7 (late blastocysts) post insemination. The blastocyst rate in the system varied between 1 and 10% with little variation between experimental replicates.
Parthenogenetic activation (PA)
Slaughterhouse derived (25–35°C) ovarian follicles (3–8 mm) were used for COCs aspiration and, selected COCs were cultured in 400 μL bicarbonate-buffered TCM-199 (M-2154; Sigma) supplemented with 10% (v/v) cattle serum (BS; Danish Veterinary Institute, Copenhagen, Denmark), 10% (v/v) pig follicular fluid (pFF), 15 IU/mL Suigonan, 0.8 mM glutamine and 0.05 mg/mL gentamicin (Suigonan Vet; Intervet, Skovlunde, Denmark), at 38.5°C in four-well plates with 5% CO2 and 100% humidity air using Submarine Incubation System (SIS) (Vajta et al., 2004). The oocytes were equilibrated in the activation medium [0.3 M mannitol, 0.1 mM MgSO4, 0.1 mM CaCl2, supplemented with 0.01% (w/v) PVA] after removal of the excess cumulus cells from the COCs. The equilibrated oocytes were aligned on the wire of the fusion chamber using the same AC (0.006 kV/cm and 700 kHz) and a single DC pulse (0.126 kV/ cm for 80 μs) was used for the activation. The pairs were carefully transferred and activated in the NUNC four-well plates containing PZM-3 medium (Yoshioka et al., 2002) supplemented with 5 μg/mL CB and 10 μg/mL CX at 38.5°C in 5% CO2, 5% O2, and 90% N2 with maximum humidity for 4 h. Activated zona-intact embryos were cultured in the four-well plate, with 400 μL PZM-3 medium at 38.5°C in 5% CO2, 5% O2, and 90% N2 with maximum humidity. Embryos were collected at different time points after activation: 4–6 h (early one-cell stages), 16–20 h (late one-cell stages), day 2 (two-cell stages), day 3 (four-cell stages), day 4 (eight-cell/morulae), day 5 (early blastocysts), and day 6 (late blastocysts). The blastocyst rate in the system varied between 50 and 60%.
SCNT
Slaughterhouse derived ovarian follicles (2–6 mm) from prepubertal gilts were aspirated and IVM was performed in NCSU-37 maturation medium supplemented with 0.1 mg/mL cysteine, 10 ng/mL EGF, 10% pFF, 50 M 2-mercaptoethanol, 0.5 mg/mL dibutyryl cyclic AMP (dbcAMP, Sigma-Aldrich, Germany), 10 IU eCG (Intergonan®, Intervet, Germany), and 10 IU hCG (Ovogest®, Intervet, Germany) for 22 h in humidified air with 5% CO2 at 38.5°C (Petersen et al., 2008). Additional IVM (16–22 h) was performed by transferring them into fresh NCSU-37 without dbcAMP, eCG, and hCG. Subsequently, selected oocytes with typical morphology and visible first body were enucleated by removing the first polar body and the metaphase plate. The serum starved (48 h; DMEM 0.5% FCS), cell cycle arrested (G0/G1) donor porcine fibroblasts (PF) (Kues et al., 2002) were trypsinised and subsequently transferred in the perivitelline space after centrifugation (200×g/3 min×2) and resuspension in Ca2+ free TL-HEPES. The couplet was fused in Ca2 free SOR2 medium (0.25 mol/L Sorbitol, 0.5 mmol/L Mg-acetate, 0.1% BSA) by providing a single electrical pulse of 1.1 kV/cm for 100 sec (Eppendorf Multiporator®, Eppendorf, Germany). The reconstructed oocytes were activated in an electrical field of 1.0 kV/cm for 45 s in SOR2 activation medium (0.25 mol/L Sorbitol, 0.1 mmol/L Ca-acetate, 0.5 mmol/L Mg-acetate, 0.1% BSA) and incubated with 2 mmol/L 6-dimethylaminopurine (DMAP, Sigma, Germany) in NCSU23 medium. The embryos were collected at 4–6 h (early one-cell stages), 16–20 h (late one-cell stages), day 2 (two-cell stages), day 3 (four-cell stages), day 4 (eight-cell/morulae), days 5–6 (early blastocysts), and days 6–7 (late blastocysts). The blastocyst rate for SCNT embryos in this experiment was 9.45% (59/624).
Processing for light and transmission electron microscopy
The collected embryos of all types were fixed in 3% glutaraldehyde in 0.1 M Na-phosphate buffer (pH 7.2) at 4°C for 1 h. Subsequently, the embryos were washed in buffer, postfixed in 1% OsO4 in 0.1 M Na-phosphate buffer, embedded in Epon, and serially sectioned into semithin sections (2 μm). The sections were stained with basic toluidine blue and evaluated by bright-field light microscopy. All embryos were serially sectioned into semithin sections that were carefully examined in order to assure the desired stages of embryonic development. Selected semithin sections were reembedded as described earlier (Hyttel and Madsen, 1987) and processed for ultrathin sectioning (70 nm). The ultrathin sections were contrasted with uranyl acetate and lead citrate and examined on a Philips CM100 transmission electron microscope (Darmstadt, The Netherlands).
Results
The numbers of IV, IVF, PA, and SCNT zygotes and embryos analyzed by TEM are presented in Table 1. An overview of morphological characteristics of IV, IVF, PA, and SCNT zygotes and embryos is presented in Figure 1.
Table 1.
The Numbers of Zygotes and Embryos Analyzed at Different Developmental Stages
| Stage/type | Zygote/one-cell | Two-cell | Four-cell | Eight-cell/morula | Early blastocyst | Late blastocyst |
|---|---|---|---|---|---|---|
| IV | 9 | 5 | 5 | 2 | 3 | 3 |
| IVF | 10 | 5 | 5 | 5 | 3 | 2 |
| PA | 9 | 5 | 7 | 3 | 5 | 5 |
| SCNT | 7 | 5 | 4 | 3 | 5 | 5 |
IV, in vivo developed; IVF, in vitro fertilized; PA, parthenogenetically activated; SCNT, somatic cell nuclear transfer.
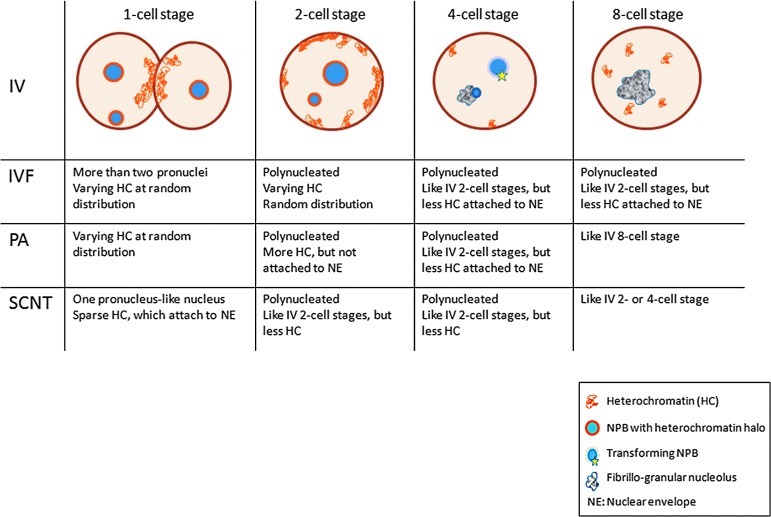
FIG. 1.
Schematic distribution of euchromatin and heterochromatin in porcine embryos of different origin. The drawings represent typical pronuclei (IV, one-cell stage) and nuclei. IV, in vivo developed; IVF, in vitro fertilized; PA, parthenogenetically activated; SCNT, somatic cell nuclear transfer; NE, nuclear envelope; NPB, nucleolar precursor body.
One-cell stage/zygote
The nine evaluated IV zygotes had either two individual peripheral pronuclei (1/9), two centrally apposed pronuclei (4/9) (Fig. 2; A1, arrowhead), or chromosomes at different phases of mitosis (2/9). The two remaining zygotes presented metaphase plate and condensed sperm. Both, the peripheral and the apposed pronuclei contained abundant euchromatin and sparse heterochromatin, located either as aggregates in the nucleoplasm and/or attached to the nuclear envelope (Fig. 2; A2, arrow). Heterochromatin density was highest at the sites of pronuclear apposition. On average, each zygote possessed two to five intact NPBs, equally distributed between the two pronuclei. The NPBs were surrounded by a halo of heterochromatin (Fig. 2; A2, arrowhead, detail in A3).
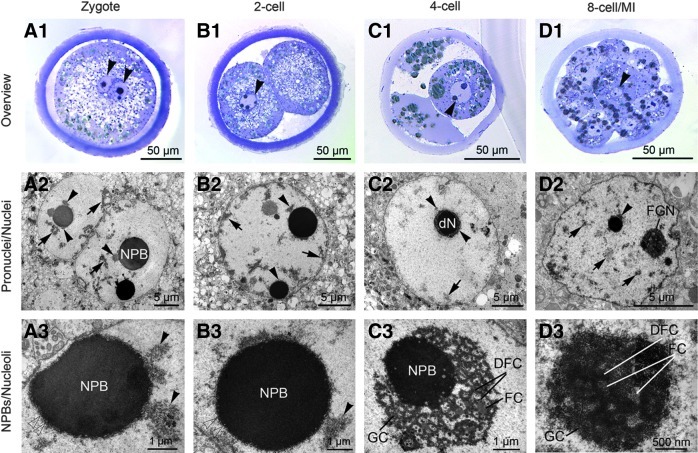
FIG. 2.
Representative light micrographic overviews of porcine IV zygote (A1), two-cell embryo (B1), four-cell embryo (C1), and eight-cell/morula stage (D1). Arrowheads indicate pronuclei and nuclei. (A2–D2) Transmission electron micrographs of pronuclei (zygote) and nuclei (embryos). Arrows indicate heterochromatin loose in the nucleoplasm or attached to the nuclear envelope. Arrowheads indicate heterochromatin clusters attached to the nucleolus precursor bodies (NPBs). dN, developing nucleolus; FGN, fibrillogranular nucleolus. (A3–D3) Transmission electron micrographs of NPBs/nucleoli in zygote (A3) and embryos (B3–D3). Arrowheads indicate heterochromatin clusters attached to the NPBs. Note the formation of fibrillar centers (FC), dense fibrillar component (DFC), and granular component (GC) around the NPB in the four-cell embryo (C3) and the presence of a fibrillogranular nucleolus with the same components in the eight-cell/Ml stage (D3).
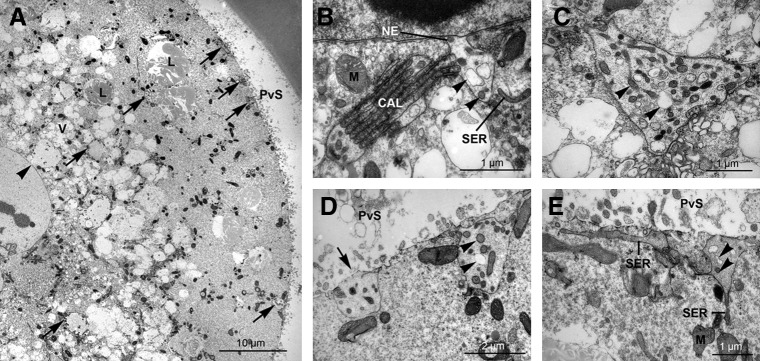
Large exvaginations of the outer membrane of the nuclear envelope were observed in all five IV zygotes with pronuclei and persisted throughout the two-cell stage (Fig. 3A). The exvaginated membrane was often connected to stacks of cytoplasmic annulate lamellae (CAL) or the smooth endoplasmic reticulum (Fig. 3B; arrowhead). At the exvaginations, the dilated perinuclear cisternae contained small vesicles with a content of varying electron-density (Fig. 3C; arrowhead). Numerous loose structures resembling these exvaginations were found in the deep and cortical cytoplasm detached from the nuclear envelope (Fig. 3D and E).
FIG. 3.
Electron micrographs of nuclear envelope exvaginations, their relationship to annulate lamellae and smooth endoplasmic reticulum, and their potential peripheral migration in IV zygotes and two-cell embryos. (A) Low-power micrograph from a porcine zygote showing a site of nuclear envelope exvagination (arrowhead) and apparently detached exvaginations (arrows) in the cytoplasm. (B) Detail from porcine two-cell embryo showing exvagination of the outer membrane of the nuclear envelope. The outer membrane of the nuclear envelope (NE) is continuous with cytoplasmic annulate lamellae (CAL). The dilated perinuclear cisterna contains vesicular structures containing material of varying electron-density (arrowheads). M, mitochondria; SER, smooth endoplasmic reticulum. (C) Apparently detached nuclear envelope exvagination found in the deep cytoplasm in the two-cell embryo. Note the content of vesicular structures containing material of varying electron-density (arrowheads). (D) Apparently detached nuclear envelope exvagination found in the cortical cytoplasm of zygote. Note the intimate spatial relationship between the detached exvagination and the plasma membrane (arrow). For arrowheads, see C. PvS, perivitelline space. (E) Apparently detached nuclear envelope exvagination found in the cortical cytoplasm of the two-cell embryo. Note the continuity between the membrane of the detached exvagination and the smooth endoplasmic reticulum (SER). For arrowheads, see C. M, mitochondria; PvS, perivitelline space.
Nine out of 10 IVF zygotes presented more than two pronuclei, which were located eccentrically, either solitarily or in apposition with each other (Suppl. Fig. 1A; see online supplementary data at www.liebertonline.com/cell). The pronuclei presented varying amounts of heterochromatin. On average, each zygote presented four to seven NPBs, which showed a complete lack of or less developed halo of heterochromatin (Fig. 4; A2, arrowhead).
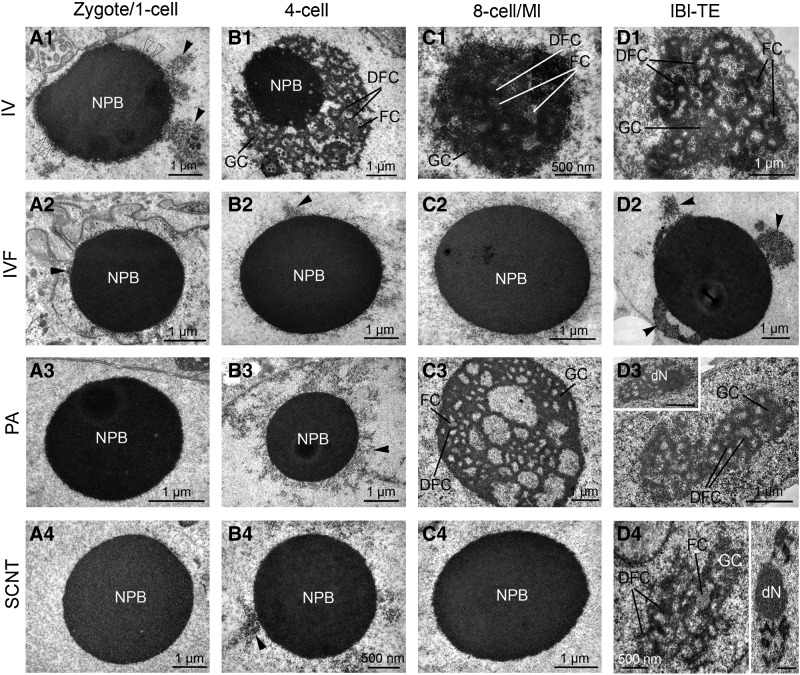
FIG. 4.
Electron micrographs of the nucleolar ultrastructure in in vivo developed (IV; A1–D1), in vitro fertilized (IVF; A2–D2), parthenogenetically activated (PA; A3–D3), and somatic cell nuclear transfer (SCNT; A4–D4) porcine embryos at the zygote/one-cell (A), four-cell (B), and eight-cell/morula (eight-cell/Ml; C) and TE of late blastocyst (lBl-TE; D) stages. Arrowheads indicate heterochromatin clusters attached to the nucleolous precursor body (NPB). Open arrowheads indicate heterochromatin halo around the NPB (A1). FC, fibrillar center; DFC, dense fibrillar component; GC, granular component; dN, developing nucleolus.
One-cell PA embryos had either one (2/9) or two (2/9) pronuclei (Suppl. Fig. 1B) or had progressed into mitosis (5/9). The nuclei possessed varying amounts of heterochromatin, which was distributed in the nucleoplasm. On average, each embryo presented two to four NPBs, which lacked the halo of heterochromatin (Fig. 4; A3).
One-cell SCNT embryos typically presented a single pronucleus-like nucleus (6/7), which showed decreased abundance of heterochromatin, exclusively attached to the nuclear envelope, as seen in the IV zygotes (Suppl. Fig. 1C). In general, one to three NPBs were found per nucleus, and the heterochromatin halo was lacking (Fig. 4; A4).
Two- to four-cell stage/embryonic genome activation period
In all IV embryos, each cell displayed a single nucleus (Fig. 2; B1, arrowhead). The two-cell stage nuclei displayed higher amounts of heterochromatin firmly attached to the nuclear envelope compared to the zygotes (Fig. 2; B2, arrow). Each nucleus had two to five NPBs with a halo of heterochromatin (Fig. 2; B3, arrowhead). Nuclear envelope exvaginations similar to the zygote were observed.
The nuclei of the four-cell embryos displayed the least amount of heterochromatin of all stages (Fig 2; C1, arrowhead). The rare heterochromatin clusters were attached to the nuclear envelope and/or to NPBs as a halo (Fig. 2; C2, arrow and arrowhead). All five evaluated four-cell embryos presented transformation of NPBs into fibrillogranular nucleoli: FCs surrounded by DFC and GC coated the NPBs (Fig. 2; C3). Developing fibrillogranular nucleoli were found together with intact nontransformed NPBs in the same nucleus. Nuclear envelope exvaginations were no longer observed.
All IVF embryos at two- and four-cell stages had bi- or polynucleated cells. Micronuclei were often closely associated with the primary nuclei (Suppl. Fig. 1D). In two-cell embryos, the nuclei presented varying amounts of heterochromatin, whereas in four-cell embryos the nuclei resembled the IV two-cell nuclear morphology, except for less attachment of the heterochromatin to the nuclear envelope. Both two- and four-cell embryos presented two to five NPBs per nucleus, and the heterochromatin halo around the NPBs was absent (4/5 at the two-cell, and 3/5 at the four-cell stage) or was less developed (1/5 at two-cell, and 2/5 at four-cell stage) (Fig. 4; B2, arrowhead).
Half of the PA embryos at the two-cell and one-third at the four-cell stage possessed bi- or polynucleated cells (Suppl. Fig. 1E). In two-cell embryos, the nuclei presented even more heterochromatin dispersed in the nucleoplasm than the IV two-cell nuclei, but heterochromatin was not attached to the nuclear envelope. Moreover, the heterochromatin halo around the NPBs was lacking. In the four-cell embryos, the nuclei resembled the IV two-cell nuclei. The heterochromatin halo around the NPBs, of which each cell in general presented two to four, was inconsistent and development of fibrillogranular nucleoli was not observed (Fig. 4; B3, arrowhead).
Most of the SCNT embryos presented bi- or polynucleated cells at the two-cell stage (4/5) and four-cell stage (2/4). Extra micronuclei were often located in close association with the primary nuclei (Suppl. Fig. 1F). Some embryos at the two- (1/5) and four-cell (3/4) displayed nuclei similar to the IV two-cell nuclei except for having slightly less heterochromatin. The remaining embryos presented chromatin similar to their IVF counterparts. In general, one to six NPBs were found per nucleus, and they lacked the consistent heterochromatin halo (Fig. 4; B4). Development of fibrillogranular nucleoli was not observed.
Eight-cell stage to morulae
The nuclei of IV embryos at the eight-cell/morula stage presented increased amounts of heterochromatin that was homogenously distributed throughout the nucleoplasm (Fig. 2; D2, arrow), and the fibrillogranular nucleoli (Fig. 2; D3). Occasionally, developing nucleoli and NPBs were observed together with fibrillogranular nucleoli.
The nuclear morphology in IVF embryos at the eight-cell stage was identical with that of four-cell stage embryos. A total of 50% of cells were polynucleated. Development of fibrillogranular nucleoli was not observed (Fig. 4; C2).
All PA embryos at the eight-cell/morula stage presented a single nucleus in all cells. Nuclear morphology was similar to the IV eight-cell/morula nuclei. Fibrillogranular nucleoli were observed in all blastomeres (Fig. 4; C3).
The eight-cell/morula SCNT embryos presented a single nucleus in all cell. The nuclei resembled the IV four-cell nuclear morphology without formation of functional nucleoli and attachment of heterochromatin to NPBs (Fig. 4; C4).
Blastocyst stage
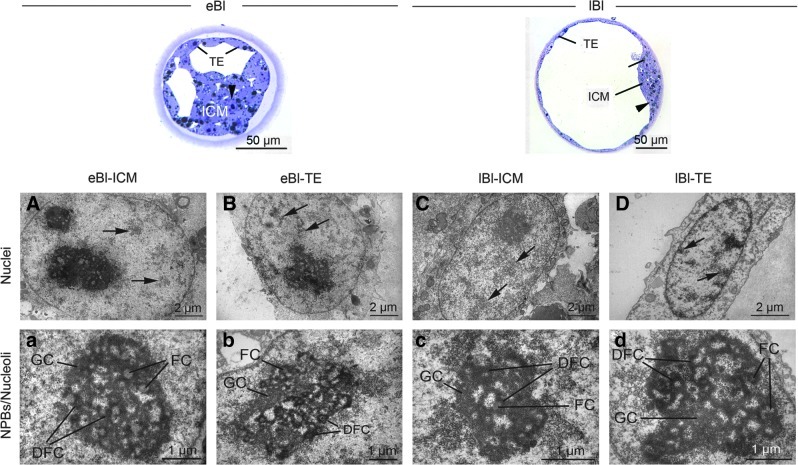
Early IV blastocysts presented segregation of the ICM and TE resulting in a well-defined ICM at the late blastocyst stage (Fig. 5, upper overview). In early blastocysts, both ICM and TE nuclei displayed the same chromatin distribution as at the eight-cell/morula stage, except for a slightly higher abundance of heterochromatin (Fig. 5A and B, arrows). In late blastocysts, the ICM nuclei were larger and round, but showed the same chromatin pattern as in early blastocysts (Fig. 5C, arrow). On the contrary, the TE nuclei were smaller and elongated and displayed higher amounts of heterochromatin, mainly attached to the nuclear envelope (Fig. 5D, arrow). Both cell types presented fibrillogranular nucleoli, which typically were larger in the ICM cells (Fig. 5a–d).
FIG. 5.
Light micrographic overviews of porcine IV early (eBl) and late blastocysts (lBl). Note the inner cell mass (ICM) and trophectoderm (TE). Arrowheads indicate nuclei. (A–D) Transmission electron micrographs of nuclei. Arrows indicate heterochromatin loose in the nucleoplasm or attached to the nuclear envelope. (a–d) Transmission electron micrographs of NPBs/nucleoli. FC, fibrillar center; DFC, dense fibrillar component (DFC); GC, granular component.
Neither early nor late IVF blastocysts presented well-defined ICMs, and both ICM and TE cells were extraordinary few and large and binucleated in at least one blastomere (Suppl. Fig. 2A, enlarged in 2B). Both early and late blastocysts presented ICM nuclei similar to the IV blastocyst, but the TE nuclei presented increased amounts of heterochromatin. Moreover, the late blastocyst TE nuclei often presented NPBs instead of fibrillogranular nucleoli (Fig. 4; D2). Occasionally, ring-shaped NPB-like structures were found in the cytoplasm.
All early and late PA blastocysts had only very few presumptive ICM cells, whereas the TE was morphologically well-defined and developed. Both ICM and TE cells were large and in three out of five blastocysts, binucleated cells were observed. In both early and late blastocysts, the nuclei resembled those of early IV blastocysts, that is, with sparse heterochromatin. However, both NPBs and developing fibrillogranular nucleoli were observed (Fig. 4; D3).
Both early and late SCNT blastocysts lacked a well-defined ICM. Only a few large ICM and TE cells were found which were in two out of five blastocysts binucleated. The nuclei resembled those of early IV blastocysts, that is, with sparse heterochromatin. However, both NPBs and developing fibrillogranular nucleoli were observed (Fig. 4; D4). Both nucleoli and NPBs were smaller compared to all other types of blastocysts.
Discussion
Proper chromatin dynamics is essential for reprogramming of the parental genomes at the onset mammalian development. Chromatin distribution in differentiated cells follows a strict pattern, placing transcriptionally silenced heterochromatin at the nuclear periphery in close contact with the nuclear envelope (Ellis et al., 1997; Izumi et al., 2000; Spann et al., 1997). Notably, our study revealed strong attachment of heterochromatin domains to the nuclear envelope (NE) in IV two-cell stages. This conformation was dramatically reorganized and replaced by fully decondensed chromatin at the four-cell stage, when the embryonic genome is activated and evidenced by the establishment of functional nucleoli (Hyttel et al., 2000b; Svarcova et al., 2009; Tomanek et al., 1989). Chromatin attachment to the nuclear envelope was first reestablished in the TE cells in the late blastocysts, coinciding with the first lineage formation and restriction of developmental potency of TE cells. In the blastomeres after the EGA and in the ICM cells, heterochromatin was more or less randomly distributed throughout the nucleoplasm similar to other pluripotent cell populations (Ahmed et al., 2010; Martin et al., 2006b).
The transient establishment of heterochromatin domains at the two-cell stage was not observed in IVF, PA, and, to so some extent in SCNT embryos before the four- to eight-cell stage. Moreover, the persistent localization of heterochromatin along the nuclear envelope in SCNT embryos indicates maintenance of somatic chromatin pattern, which has not been remodeled after SCNT (Ostrup et al., 2009; Pichugin et al., 2010). Furthermore, the subsequent extensive chromatin decondensation at the four-cell stage, synchronized with the EGA, was absent in the IVP embryos. Nuclear envelope proteins form tight complexes with heterochromatin components, for example, HP1 and cores of H3 and H4 (Guarda et al., 2009; Holmer and Worman 2001; Polioudaki et al., 2001). Interestingly, the envelope represents not only passive anchoring sites for condensed chromatin, but it also recruits chromatin actively by triggering gene silencing (Gruenbaum et al., 2005; Guarda et al., 2009; Sui and Yang 2011; Wang et al., 2002). Thus, the ability of the nuclear envelope to mediate interactions between chromatin remodeling proteins and the DNA may be critical for functional changes in activity and spatial remodeling of chromatin in early embryos. All three types of IVP embryos have decreased developmental potentials. The localization of heterochromatin to the nuclear envelope preceding the EGA and subsequent global chromatin decondensation are essential mechanisms for proper embryonic development. Moreover, the early deviations in chromatin dynamics suggest aberrations occurring already during in vitro oocyte maturation and/or initial embryonic cleavage.
Adding to the complexity of nuclear envelope function during initial embryogenesis, the exvaginations of the nuclear envelope (blebbing), observed only in IV zygotes and two-cell stages, are considered an important feature for successful genomic reprogramming and embryonic development (Kanka et al., 1991; Lavoir et al., 1997; Szollosi et al., 1988; Szollosi and Szollosi, 1988). As this feature is observed exclusively prior to the EGA, blebbing, interaction with annulate lamellae and smooth endoplasmic reticulum, and subsequent formation of extra-nuclear vacuoles indicate a critical role of the envelope in degradation of nuclear factors that might otherwise negatively impact the EGA (Szollosi and Szollosi, 1988), as also indicated by our data on IVF, SCNT, and PA embryos.
NPBs also contribute to the regulation of chromatin remodeling in early embryos. The perinucleolar chromatin contains centromeric and pericentric silent heterochromatin (Fulka and Fulka, 2010; Gavrilova et al., 2009; Ogushi et al., 2008) and α satellite DNA repeats contributing to the formation of chromocentres (Ahmed et al., 2010; Gavrilova et al., 2009; Martin et al., 2006a, 2006b; Pichugin et al., 2010). Hence, in mammalian embryos, the NPBs are likely to serve as anchors of specific chromatin domains as well as anlagen for nucleolar formation. The lack of a halo of heterochromatin around the NPBs in the majority of IVP embryos signals to some extend aberrations in the remodeling processes. Moreover, the occurrence of cytoplasmic NPB-like material further supports our observations on detrimental effect of culture condition on early porcine embryos (Hyttel and Niemann, 1990) and further substantiates our present observations of aberrant nucleolar functions under in vitro conditions.
The retained immaturity in nucleolar morphology, visible in IVP embryos from the four-cell stage by delayed nucleolar formation, and continuing toward the blastocyst stage with presence of extranuclear nucleolar-like material, may also indicate aberrant regulation of pluripotency. Nucleoli host proteins critical for establishment of pluripotency, for example, NUCLEOSTEMIN (Beekman et al., 2006), NANOG (He et al., 2006), and MLL2 (Andreu-Vieyra et al., 2010). Regular nucleolar function may therefore contribute to the first lineage segregation, which under normal circumstances correlates with formation of the pluripotent cell population in ICM. Thus, the absence of well-defined ICM in IV-produced blastocysts may also be a consequence of nucleolar dysfunctions.
In conclusion, porcine IV zygotes and embryos display a well-synchronized pattern of chromatin dynamics compatible with EGA and nucleolar formation at the four-cell stage. Production of porcine embryos under in vitro conditions by IVF, PA, or SCNT is associated with altered chromatin remodeling, delayed nucleolar formation, and poorly defined lineage segregation at the blastocyst stage, which in turn may impair their developmental capacity.
Supplementary Material
Acknowledgments
The project was supported by EU grants CLONET (MRTN-CT-2006-035468), PluriSys (FP7-HEALTH-2007-223485) and PartnErS (PIAP-GA-2008-218205); Carlsberg Foundation (2010-01-0452); DFG (NI256/31-1), and VEGA (1/0012/2010, 1/0077/11). We are grateful to Erika Lemme, Petra Hassel, and Anne Dorte Roed for expert technical assistance in embryo production and to Dr. Kirsten Schauser for organizing sows for the in vivo embryo collection. We also thank Hanne Holm for an excellent technical assistance in the processing of the samples for electron microscopy and preparing micrographs.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Ahmed K. Dehghani H. Rugg-Gunn P., et al. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu-Vieyra C.V. Chen R. Agno J.E., et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000453. , [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman C. Nichane M. De C.S., et al. Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell Biol. 2006;26:9291–9301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D. Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010;330:617–622. doi: 10.1126/science.1194776. [DOI] [PubMed] [Google Scholar]
- Corry G.N. Tanasijevic B. Barry E.R., et al. Epigenetic regulatory mechanisms during preimplantation development. Birth Defects Res. C Embryo Today. 2009;87:297–313. doi: 10.1002/bdrc.20165. [DOI] [PubMed] [Google Scholar]
- Cozzi E. Tallacchini M. Flanagan E.B., et al. The International Xenotransplantation Association consensus statement on conditions for undertaking clinical trials of porcine islet products in type 1 diabetes—chapter 1: key ethical requirements and progress toward the definition of an international regulatory framework. Xenotransplantation. 2009;16:203–214. doi: 10.1111/j.1399-3089.2009.00540.x. [DOI] [PubMed] [Google Scholar]
- Cremer T. Cremer M. Dietzel S., et al. Chromosome territories—a functional nuclear landscape. Curr. Opin. Cell. Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Efroni S. Duttagupta R. Cheng J., et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D.J. Jenkins H. Whitfield W.G., et al. GST-lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J. Cell Sci. 1997;110(Pt. 20):2507–2518. doi: 10.1242/jcs.110.20.2507. [DOI] [PubMed] [Google Scholar]
- Fulka H. Fulka J., Jr. Nucleolar transplantation in oocytes and zygotes: challenges for further research. Mol. Hum. Reprod. 2010;16:63–67. doi: 10.1093/molehr/gap088. [DOI] [PubMed] [Google Scholar]
- Gavrilova E.V. Kuznetsova I.S. Enukashvili N.I., et al. [Localization of satellite DNA and associated protein in respect to nucleolar precursor bodies in one- and two-cell mouse embryos] Tsitologiia. 2009;51:455–464. [PubMed] [Google Scholar]
- Gil M.A. Cuello C. Parrilla I., et al. Advances in swine in vitro embryo production technologies. Reprod. Domest. Anim. 2010;45(Suppl. 2):40–48. doi: 10.1111/j.1439-0531.2010.01623.x. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y. Margalit A. Goldman R.D., et al. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Guarda A. Bolognese F. Bonapace I.M., et al. Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp. Cell Res. 2009;315:1895–1903. doi: 10.1016/j.yexcr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- He S. Pant D. Schiffmacher A., et al. Developmental expression of pluripotency determining factors in caprine embryos: novel pattern of NANOG protein localization in the nucleolus. Mol. Reprod. Dev. 2006;73:1512–1522. doi: 10.1002/mrd.20525. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Roussel P. Thiry M., et al. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip. Rev. RNA. 2010;1:415–431. doi: 10.1002/wrna.39. [DOI] [PubMed] [Google Scholar]
- Holmer L. Worman H.J. Inner nuclear membrane proteins: functions and targeting. Cell. Mol. Life Sci. 2001;58:1741–1747. doi: 10.1007/PL00000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttel P. Madsen I. Rapid method to prepare mammalian oocytes and embryos for transmission electron microscopy. Acta Anat. (Basel) 1987;129:12–14. doi: 10.1159/000146370. [DOI] [PubMed] [Google Scholar]
- Hyttel P. Niemann H. Ultrastructure of porcine embryos following development in vitro versus in vivo. Mol. Reprod. Dev. 1990;27:136–144. doi: 10.1002/mrd.1080270208. [DOI] [PubMed] [Google Scholar]
- Hyttel P. Laurincik J. Rosenkranz C., et al. Nucleolar proteins and ultrastructure in preimplantation porcine embryos developed in vivo. Biol. Reprod. 2000a;63:1848–1856. doi: 10.1095/biolreprod63.6.1848. [DOI] [PubMed] [Google Scholar]
- Hyttel P. Laurincik J. Rosenkranz C., et al. Nucleolar proteins and ultrastructure in preimplantation porcine embryos developed in vivo. Biol. Reprod. 2000b;63:1848–1856. doi: 10.1095/biolreprod63.6.1848. [DOI] [PubMed] [Google Scholar]
- Hyttel P. Laurincik J. Zakhartchenko V., et al. Nucleolar protein allocation and ultrastructure in bovine embryos produced by nuclear transfer from embryonic cells. Cloning. 2001;3:69–82. doi: 10.1089/15204550152475572. [DOI] [PubMed] [Google Scholar]
- Izumi M. Vaughan O.A. Hutchison C.J., et al. Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A and C but not lamin B structure in mammalian cells. Mol. Biol. Cell. 2000;11:4323–4337. doi: 10.1091/mbc.11.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S. Rui R. Lu Q., et al. Analysis of apoptosis and methyltransferase mRNA expression in porcine cloned embryos cultured in vitro. J. Assist. Reprod. Genet. 2010;27:49–59. doi: 10.1007/s10815-009-9378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanka J. Fulka J., Jr. Fulka J., et al. Nuclear transplantation in bovine embryo: fine structural and autoradiographic studies. Mol. Reprod. Dev. 1991;29:110–116. doi: 10.1002/mrd.1080290204. [DOI] [PubMed] [Google Scholar]
- Koehler D. Zakhartchenko V. Froenicke L., et al. Changes of higher order chromatin arrangements during major genome activation in bovine preimplantation embryos. Exp. Cell. Res. 2009;315:2053–2063. doi: 10.1016/j.yexcr.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Kragh P.M. Nielsen A.L. Li J., et al. Hemizygous minipigs produced by random gene insertion and handmade cloning express the Alzheimer's disease-causing dominant mutation APPsw. Transgenic Res. 2009;18:545–558. doi: 10.1007/s11248-009-9245-4. [DOI] [PubMed] [Google Scholar]
- Kues W.A. Carnwath J.W. Paul D., et al. Cell cycle synchronization of porcine fetal fibroblasts by serum deprivation initiates a nonconventional form of apoptosis. Cloning Stem Cells. 2002;4:231–243. doi: 10.1089/15362300260339511. [DOI] [PubMed] [Google Scholar]
- Laurincik J. Hyttel P. Kopecny V. DNA synthesis and pronucleus development in pig zygotes obtained in vivo: an autoradiographic and ultrastructural study. Mol. Reprod. Dev. 1995;40:325–332. doi: 10.1002/mrd.1080400308. [DOI] [PubMed] [Google Scholar]
- Laurincik J. Kopecny V. Hyttel P. Detailed analysis of pronucleus development in bovine zygotes in vivo: ultrastructure and cell cycle chronology. Mol. Reprod. Dev. 1996;43:62–69. doi: 10.1002/(SICI)1098-2795(199601)43:1<62::AID-MRD8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Lavoir M.C. Kelk D. Rumph N., et al. Transcription and translation in bovine nuclear transfer embryos. Biol. Reprod. 1997;57:204–213. doi: 10.1095/biolreprod57.1.204. [DOI] [PubMed] [Google Scholar]
- Lorthongpanich C. Solter D. Lim C.Y. Nuclear reprogramming in zygotes. Int. J. Dev. Biol. 2010;54:1631–1640. doi: 10.1387/ijdb.103201cl. [DOI] [PubMed] [Google Scholar]
- Martin C. Beaujean N. Brochard V., et al. Genome restructuring in mouse embryos during reprogramming and early development. Dev. Biol. 2006a;292:317–332. doi: 10.1016/j.ydbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Martin C. Brochard V. Migne C., et al. Architectural reorganization of the nuclei upon transfer into oocytes accompanies genome reprogramming. Mol. Reprod. Dev. 2006b;73:1102–1111. doi: 10.1002/mrd.20506. [DOI] [PubMed] [Google Scholar]
- Mattout A. Meshorer E. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr. Opin. Cell Biol. 2010;22:334–341. doi: 10.1016/j.ceb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Miyamoto K. Nagai K. Kitamura N., et al. Identification and characterization of an oocyte factor required for development of porcine nuclear transfer embryos. Proc. Natl. Acad. Sci. USA. 2011;108:7040–7045. doi: 10.1073/pnas.1013634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogushi S. Palmieri C. Fulka H., et al. The maternal nucleolus is essential for early embryonic development in mammals. Science. 2008;319:613–616. doi: 10.1126/science.1151276. [DOI] [PubMed] [Google Scholar]
- Orkin S.H. Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrup O. Petrovicova I. Strejcek F., et al. Nuclear and nucleolar reprogramming during the first cell cycle in bovine nuclear transfer embryos. Cloning Stem Cells. 2009;11:367–375. doi: 10.1089/clo.2008.0076. [DOI] [PubMed] [Google Scholar]
- Petersen B. Lucas-Hahn A. Oropeza M., et al. Development and validation of a highly efficient protocol of porcine somatic cloning using preovulatory embryo transfer in peripubertal gilts. Cloning Stem Cells. 2008;10:355–362. doi: 10.1089/clo.2008.0026. [DOI] [PubMed] [Google Scholar]
- Pichugin A. Le B.D. Adenot P., et al. Dynamics of constitutive heterochromatin: two contrasted kinetics of genome restructuring in early cloned bovine embryos. Reproduction. 2010;139:129–137. doi: 10.1530/REP-08-0435. [DOI] [PubMed] [Google Scholar]
- Polioudaki H. Kourmouli N. Drosou V., et al. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2001;2:920–925. doi: 10.1093/embo-reports/kve199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. Kragh P.M. Li J., et al. Pregnancies and piglets from large white sow recipients after two transfer methods of cloned and transgenic embryos of different pig breeds. Theriogenology. 2010;74:1233–1240. doi: 10.1016/j.theriogenology.2010.05.026. [DOI] [PubMed] [Google Scholar]
- Spann T.P. Moir R.D. Goldman A.E., et al. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell. Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui L. Yang Y. Distinct effects of nuclear membrane localization on gene transcription silencing in Drosophila S2 cells and germ cells. J. Genet. Genomics. 2011;38:55–61. doi: 10.1016/j.jcg.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Suzuki M. Misumi K. Ozawa M., et al. Successful piglet production by IVF of oocytes matured in vitro using NCSU-37 supplemented with fetal bovine serum. Theriogenology. 2006;65:374–386. doi: 10.1016/j.theriogenology.2005.05.039. [DOI] [PubMed] [Google Scholar]
- Svarcova O. Laurincik J. Avery B., et al. Nucleolar development and allocation of key nucleolar proteins require de novo transcription in bovine embryos. Mol. Reprod. Dev. 2007;74:1428–1435. doi: 10.1002/mrd.20727. [DOI] [PubMed] [Google Scholar]
- Svarcova O. Strejcek F. Petrovicova I., et al. The role of RNA polymerase I transcription and embryonic genome activation in nucleolar development in bovine preimplantation embryos. Mol. Reprod. Dev. 2008;75:1095–1103. doi: 10.1002/mrd.20865. [DOI] [PubMed] [Google Scholar]
- Svarcova O. Dinnyes A. Polgar Z., et al. Nucleolar re-activation is delayed in mouse embryos cloned from two different cell lines. Mol. Reprod. Dev. 2009;76:132–141. doi: 10.1002/mrd.20936. [DOI] [PubMed] [Google Scholar]
- Szollosi D. Czolowska R. Szollosi M.S., et al. Remodeling of mouse thymocyte nuclei depends on the time of their transfer into activated, homologous oocytes. J. Cell Sci. 1988;91(Pt. 4):603–613. doi: 10.1242/jcs.91.4.603. [DOI] [PubMed] [Google Scholar]
- Szollosi M.S. Szollosi D. “Blebbing: of the nuclear envelope of mouse zygotes, early embryos and hybrid cells. J. Cell Sci. 1988;91(Pt. 2):257–267. doi: 10.1242/jcs.91.2.257. [DOI] [PubMed] [Google Scholar]
- Thomas S. Li X.Y. Sabo P.J., et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanek M. Kopecny V. Kanka J. Genome reactivation in developing early pig embryos: an ultrastructural and autoradiographic analysis. Anat. Embryol. (Berl.) 1989;180:309–316. doi: 10.1007/BF00315889. [DOI] [PubMed] [Google Scholar]
- Vajta G. Bartels P. Joubert J., et al. Production of a healthy calf by somatic cell nuclear transfer without micromanipulators and carbon dioxide incubators using the Handmade Cloning (HMC) and the Submarine Incubation System (SIS) Theriogenology. 2004;62:1465–1472. doi: 10.1016/j.theriogenology.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Wang X. Xu S. Rivolta C., et al. Barrier to autointegration factor interacts with the cone–rod homeobox and represses its transactivation function. J. Biol. Chem. 2002;277:43288–43300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
- Wilmut I. Sullivan G. Taylor J. A decade of progress since the birth of Dolly. Reprod. Fertil. Dev. 2009;21:95–100. doi: 10.1071/rd08216. [DOI] [PubMed] [Google Scholar]
- Yamanaka K. Sugimura S. Wakai T., et al. Acetylation level of histone H3 in early embryonic stages affects subsequent development of miniature pig somatic cell nuclear transfer embryos. J. Reprod. Dev. 2009;55:638–644. doi: 10.1262/jrd.20245. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Blau H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K. Suzuki C. Tanaka A., et al. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol. Reprod. 2002;66:112–119. doi: 10.1095/biolreprod66.1.112. [DOI] [PubMed] [Google Scholar]
- Zhao J. Hao Y. Ross J.W., et al. Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell Reprogram. 2010a;12:75–83. doi: 10.1089/cell.2009.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. Whyte J. Prather R.S. Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res. 2010b;341:13–21. doi: 10.1007/s00441-010-1000-x. [DOI] [PubMed] [Google Scholar]
- Zhou S. Ding C. Zhao X., et al. Successful generation of cloned mice using nuclear transfer from induced pluripotent stem cells. Cell Res. 2010;20:850–853. doi: 10.1038/cr.2010.78. [DOI] [PubMed] [Google Scholar]
- Zhu J. King T. Dobrinsky J., et al. In vitro and in vivo developmental competence of ovulated and in vitro matured porcine oocytes activated by electrical activation. Cloning Stem Cells. 2003;5:355–365. doi: 10.1089/153623003772032853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.