Abstract
Membrane proteins (MPs) play a critical role in many physiological processes such as pumping specific molecules across the otherwise impermeable membrane bilayer that surrounds all cells and organelles. Alterations in the function of MPs result in many human diseases and disorders; thus, an intricate understanding of their structures remains a critical objective for biological research. However, structure determination of MPs remains a significant challenge often stemming from their hydrophobicity.
MPs have substantial hydrophobic regions embedded within the bilayer. Detergents are frequently used to solubilize these proteins from the bilayer generating a protein-detergent micelle that can then be manipulated in a similar manner as soluble proteins. Traditionally, crystallization trials proceed using a protein-detergent mixture, but they often resist crystallization or produce crystals of poor quality. These problems arise due to the detergent′s inability to adequately mimic the bilayer resulting in poor stability and heterogeneity. In addition, the detergent shields the hydrophobic surface of the MP reducing the surface area available for crystal contacts. To circumvent these drawbacks MPs can be crystallized in lipidic media, which more closely simulates their endogenous environment, and has recently become a de novo technique for MP crystallization.
Lipidic cubic phase (LCP) is a three-dimensional lipid bilayer penetrated by an interconnected system of aqueous channels1. Although monoolein is the lipid of choice, related lipids such as monopalmitolein and monovaccenin have also been used to make LCP2. MPs are incorporated into the LCP where they diffuse in three dimensions and feed crystal nuclei. A great advantage of the LCP is that the protein remains in a more native environment, but the method has a number of technical disadvantages including high viscosity (requiring specialized apparatuses) and difficulties in crystal visualization and manipulation3,4. Because of these technical difficulties, we utilized another lipidic medium for crystallization-bicelles5,6 (Figure 1). Bicelles are lipid/amphiphile mixtures formed by blending a phosphatidylcholine lipid (DMPC) with an amphiphile (CHAPSO) or a short-chain lipid (DHPC). Within each bicelle disc, the lipid molecules generate a bilayer while the amphiphile molecules line the apolar edges providing beneficial properties of both bilayers and detergents. Importantly, below their transition temperature, protein-bicelle mixtures have a reduced viscosity and are manipulated in a similar manner as detergent-solubilized MPs, making bicelles compatible with crystallization robots.
Bicelles have been successfully used to crystallize several membrane proteins5,7-11 (Table 1). This growing collection of proteins demonstrates the versatility of bicelles for crystallizing both alpha helical and beta sheet MPs from prokaryotic and eukaryotic sources. Because of these successes and the simplicity of high-throughput implementation, bicelles should be part of every membrane protein crystallographer′s arsenal. In this video, we describe the bicelle methodology and provide a step-by-step protocol for setting up high-throughput crystallization trials of purified MPs using standard robotics.
Keywords: Molecular Biology, Issue 59, membrane proteins crystallization, bicelle, lipidic crystallization
Protocol
Bicelle based crystallization is comprised of four basic steps (Figure 2): i) preparation of a bicelle forming lipid:amphiphile mixture; ii) incorporation of purified protein into the bicelle medium; iii) crystallization trials (manually or robotically); and iv) visualization, crystal extraction and freezing. These steps are described in detail below
1. Preparation of bicelles
Bicelles can form in a variety of lipid:amphiphile combinations and over a wide range of concentrations. Therefore, an initial composition-based on previous successful conditions- is recommended (Table 1). The most successful mixture is the DMPC:CHAPSO bicelle formulation, which can either be purchased commercially as a premixed ready-to-use formulation (see Table of Reagents below) or prepared in the lab as described. For this exercise we will prepare 1 ml of 35% DMPC:CHAPSO mixture at a 2.8:1 molar ratio.
- Weigh out 0.26 g DMPC (Mr 677.9 g/mol), 0.09 g CHAPSO (Mr 630.9 g/mol) and add deionized water to a final volume of 1.0 ml.
- The bicelle percentage can vary between 10%-40% with a DMPC:CHAPSO molar ratio ranging from 2.6-3.0:1 (Table 1).
- Note: The higher the bicelle concentration the more difficult it is to dissolve the lipid resulting in a higher solution viscosity. However, a concentrated bicelle formulation can be advantageous when the protein concentration is low.
- Dissolving the lipid to obtain a homogenous solution requires considerable effort, making this step the most time consuming in the bicelle method. Cycle through the following steps until the DMPC is completely mixed:
- Warm the mixture to ~40°C using a water bath or an incubator and vortex for ~1 minute.
- Note: As more cycles are carried out, warming the mixture will result in a gel-like consistency making it difficult to vortex.
- Cool the mixture on ice and vortex for a few minutes. Cooling helps liquefy the solution making it easier to vortex.
- Note: As more cycles are performed, the mixture may become cloudy upon cooling.
- Repeat the steps listed above (1.2.1 and 1.2.2) until the lipid is completely dissolved.
- Note: This process may take several hours. Bicelle formation is indicated by the changes in phase behavior of the DMPC:CHAPSO formulation. Upon completion, the mixture will be a clear gel at or above room temperature and a viscous liquid on ice.
The bicelle mixture is now ready to use and can be kept at -20°C for long-term storage (up to 5 years). Due to the risk of hydrolysis of the phospholipid head group, it is not advisable to store bicelles at room temperature for extended periods.
2. Incorporation of protein into bicelles
Most MP structures obtained from bicelles were crystallized in DMPC:CHAPSO bicelle concentration ranging from 2 to 8% using a protein concentration of 8 to 12 mg/ml (Table 1). If possible, initial screens should use these guidelines and additional concentrations can be screened at the optimization stage. Compared to the LCP method, protein incorporation with bicelles is a simple process (Figure 3), which should be done the same day as crystallization trials.
- Thaw the DMPC:CHAPSO bicelle mixture at room temperature until the phase changes to a clear gel.
- Note: Multiple freeze-thaws will not affect bicelle behavior.
Place the mixture on ice to liquify and briefly vortex to reestablish a homogenous bicelle phase. When placed on ice the mixture may become cloudy.
From this point on, keep the bicelle mixture and purified protein on ice. This will keep the bicelle in a liquid phase making it amenable to pipetting.
- Add the bicelle mixture to the purified detergent-solubilized protein in a 1:4 (V/V) ratio.
- For example: 100 μl protein-bicelle mixture is obtained by mixing 80 μl protein with 20 μl bicelle. If the protein concentration is 15 mg/ml and the bicelle concentration is 35%, this will give a bicelle-incorporated protein mixture with a protein concentration of 12 mg/ml and a bicelle concentration of 7%.
- Mix by gently pipetting the contents up and down until the solution becomes clear and homogenous.
- Note: If bubbles appear, a quick spin (30-60 seconds, 13000 rpm, 4°C) with a tabletop centrifuge can help remove them.
Incubate the mixture on ice for at least 30 min to promote complete incorporation of protein into bicelles. The protein-bicelle mixture is now ready for crystallization trials.
3. Setting up crystallization trials
Other lipid crystallization techniques like the LCP require specialized equipment due to the medium′s high viscosity; but the unique phase behavior of bicelles permits implementation in virtually any standard crystallization format including robotics (Figure 3). Crystallization trials can be carried out in either hanging or sitting drop formats using standard commercially available screens.
Whether setting up trays manually or using a crystallization robot, maintain the protein-bicelle mixture on ice. This will keep the protein-bicelle mixture cold and ensure that the viscosity of the solution is minimal.
- Manual Crystallization Trials - Using a standard pipette, the protein-bicelle mixture can be mixed with reservoir solution in the same manner as normally performed for soluble or detergent-solubilized membrane proteins.
- Note: Keep the protein-bicelle mixture on ice during the trials.
- Robotic Crystallization Trials - We have tailored these trials for the Mosquito crystallization robot, but in principle (following the same precautions) the technique should be compatible with all crystallization robots. The following tips will ensure that the protein-bicelle mixture remains cool and accurately pipetted by the robot:
- Precool the plate holding the protein-bicelle mixture by placing it on ice.
- Pipette the protein-bicelle mixture into the plate and continue to keep the plate on ice. This plate should be the last item to go on the robot before commencing the run.
- Set-up the reservoir tray and crystallization lids on platform 3 and 5 respectively, of the mosquito robot.
- Place the plate containing protein-bicelle mixture on platform 4 of the mosquito robot. This assures the protein-bicelle mixture is the last to be picked up by the robot and it is immediately released.
- To prevent heating and increased viscosity, do not mix the reservoir with the protein-bicelle mixture.
- When performing multiple screens, immediately return the bicelle-protein plate to ice for cooling as soon as a run is completed.
Drop volume and ratio (protein:reservoir) can be chosen as for conventional crystallization trials. For example, initial crystal trials using the Mosquito robot can be set up using 0.25 μl protein:bicelle mixture plus 0.25 μl reservoir.
- Incubate the crystal trials in a chamber at 20°C. Temperature is a good screening and optimization parameter because the phase behavior of bicelles is temperature dependent.
- Higher temperatures induce the lamellar phase12 (Figure 1), which has the advantage of pre-organizing the protein in layers. Temperatures below 20°C can be screened, but you should not go below 4°C since this may cause the lipids to precipitate over extended periods of time.
In the same manner as traditional crystallization trials, bicelle trials should be monitored on a regular basis for crystal appearance and growth. We recommend to check trays on the 1st and 3rd day post-setup followed by weekly inspection.
Crystal optimization can be carried out using methods routinely used for detergent based crystals including grid screening, additive screening, changing temperatures etc. Additionally, bicelle percentage and protein:bicelle ratio can be varied. Furthermore, bicelles can be doped with specific lipids that may be necessary for protein stability or function.
4. Visualization, crystal extraction and freezing
Since crystal trials with the protein-bicelle mixture have a viscosity similar to protein-detergent drops, visualization and crystal extraction is routine and is carried out like traditional set-ups.
Visualization: In contrast to LCP media that often requires high-quality illumination under normal and polarized light for crystal detection, visualization is not hindered by bicelles. Colored as well as colorless protein crystal drops can be easily analyzed using standard microscopes and no special equipment is required.
Bicelles, like other lipidic media, tend to produce a high percentage of false positives. A UV microscope greatly helps in differentiating protein from salt crystals (Figure 4).
Extraction and Freezing: Crystal extraction and freezing is relatively straightforward and does not require dissolution of the surrounding bicelle media. Additionally, the bicelle phase itself provides some moderate cryo-protection.
5. Representative Results:
It generally takes 2-3 days for crystals to appear and approximately a week or more for them to grow to their maximum size. This was the case for bacteriorhodopsin and mouse voltage-dependent anion channel 1 (mVDAC1) crystals4,8. For other membrane proteins it can take several weeks for crystal growth, so it is important to continue monitoring the crystal trials well beyond the first weeks.
As with other lipidic media, bicelles tend to form shapes that may appear to be crystalline. It has also been observed that they lead to a higher percentage of salt and detergent crystals. A UV-microscope that detects tryptophan fluorescence can significantly help eliminate such non-proteinaceous false positives. Figure 4 shows lipid shapes, salt and protein crystals as viewed in visible and UV light to help distinguish the different results that may be observed.
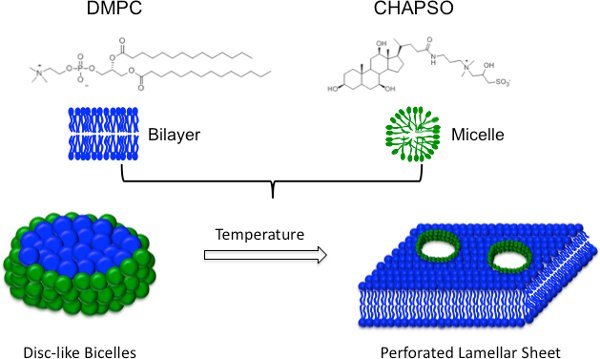 Figure 1.Bicelle Schematic. Bicelles are composed of a bilayer forming lipid molecule such as DMPC (blue) and an amphiphile such as CHAPSO (green), which protects the hydrophobic edges of the bilayer. As the temperature is increased, disc-like bicelles undergo a phase transformation into a perforated lamellar sheet12.
Figure 1.Bicelle Schematic. Bicelles are composed of a bilayer forming lipid molecule such as DMPC (blue) and an amphiphile such as CHAPSO (green), which protects the hydrophobic edges of the bilayer. As the temperature is increased, disc-like bicelles undergo a phase transformation into a perforated lamellar sheet12.
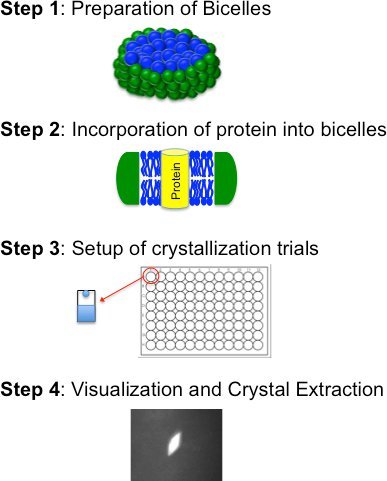 Figure 2.
Flowchart for the bicelle crystallization method outlining the four basic steps.
Figure 2.
Flowchart for the bicelle crystallization method outlining the four basic steps.
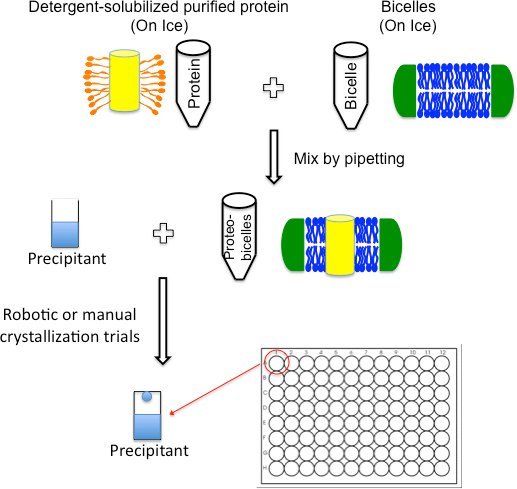 Figure 3. Crystal trials set-up schematic. Purified detergent-solubilized membrane proteins can be directly mixed with bicelles on ice by simply pipetting the contents together. After incubating the protein/bicelle mixture on ice for ~30 minutes, crystallization trials can be set up using any standard format including robotics.
Figure 3. Crystal trials set-up schematic. Purified detergent-solubilized membrane proteins can be directly mixed with bicelles on ice by simply pipetting the contents together. After incubating the protein/bicelle mixture on ice for ~30 minutes, crystallization trials can be set up using any standard format including robotics.
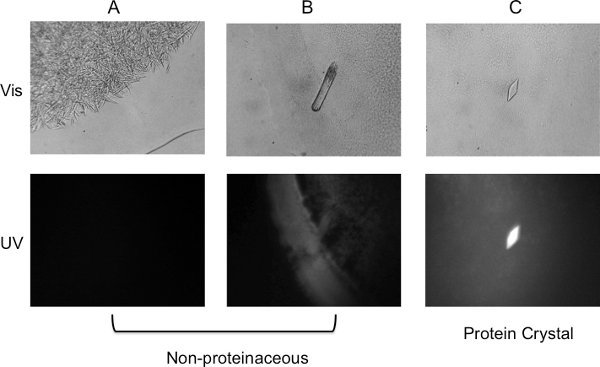 Figure 4. Visualization of crystal trials. Visible image (top panel) and UV image (bottom panel) of (A) Needle-shaped crystals observed in a salt-only condition. No fluorescence can be detected from the crystals, an indication of false positive. (B) Rod-shaped crystal formed in an MPD condition. The crystal fluoresced weakly but was found to be non-proteinaceous using X-ray diffraction. (C) Crystal observed about four weeks after setting up trials. The strong fluorescence under UV light confirms it is a protein crystal.
Figure 4. Visualization of crystal trials. Visible image (top panel) and UV image (bottom panel) of (A) Needle-shaped crystals observed in a salt-only condition. No fluorescence can be detected from the crystals, an indication of false positive. (B) Rod-shaped crystal formed in an MPD condition. The crystal fluoresced weakly but was found to be non-proteinaceous using X-ray diffraction. (C) Crystal observed about four weeks after setting up trials. The strong fluorescence under UV light confirms it is a protein crystal.
| No. | Protein | Source | Bicelle Formulation | Protein Concentration | Detergent1 | Resolution (Å) | Reference |
| 1 | Bacteriorhodopsin2 | Halobacterium salinarum | 8% DMPC:CHAPSO (2.8:1) | 8 mg/ml | 2.0 | Faham and Bowie, 2002 | |
| 8% DTPC:CHAPSO (3:1) | 8 mg/ml | 1.8 | Faham et al., 2005 | ||||
| 2 | β2-Adrenergic receptor/Fab complex | Homo sapiens | 8.3% DMPC:CHAPSO (3:1) | 10 mg/ml | DDM | 3.4/3.7 | Rasmussen et al., 2007 |
| 3 | Voltage-dependent anion channel 1 | Mus musculus | 7% DMPC:CHAPSO (2.8:1) | 12 mg/ml | LDAO | 2.3 | Ujwal et al., 2008 |
| 4 | Xanthorhodopsin | Salinibacter ruber | 4.2% DMPC, 5% NM | 4 mg/ml | DDM | 1.9 | Luecke et al., 2009 |
| 5 | Rhomboid protease | Escherichia coli | 2% DMPC:CHAPSO (2.6:1) | 9 mg/ml | Nonyl glucoside | 1.7 | Vinothkumar, 2011 |
1 Detergent used for membrane protein purification 2 Native lipids from purple membranes may be carried along during purification
Table 1. Summary of crystallization conditions for membrane protein structures solved using bicelles.
Discussion
Bicelles are a unique lipidic media that offer a native bilayer-like environment while behaving as if solubilized by detergents. This property gives bicelles a distinct advantage over other lipid-based crystallization methods since there is no learning curve or specialized equipment required for this technique. Once bicelles are available, either commercial or prepared in the lab, they can be directly mixed with purified protein and from this point on crystallization trials proceed almost exactly as with standard detergent based protocols. Furthermore, bicelles offer several practical advantages compared to other techniques, including extended storage periods, simple incorporation of protein, high-throughput capability using standard robotics and routine visualization and crystal extraction. Another distinct advantage is the ability to dope bicelles with specific lipids for optimization or if shown beneficial for the protein of interest. Taken together, the lipidic bicelle method offers considerable versatility in combination with practical ease-of-use, making it easily adoptable for all membrane protein crystallization projects.
Disclosures
No conflicts of interest declared.
Acknowledgments
We would like to thank Drs. James Bowie and Salem Faham for providing technical expertise and guidance on the bicelle method and Dr. Aviv Paz for useful discussions. We acknowledge Le Du for experimental support. Rachna Ujwal has financial interest in MemX Biosciences LLC, which, however, did not support this work. This work was supported in part by grants from the NIH (RO1 GM078844).
References
- Landau EM, Rosenbusch JP. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proceedings of the National Academy of Sciences. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M, Lyons J, Smyth T, Hart DJ. Chapter 4 Monoacylglycerols: The Workhorse Lipids for Crystallizing Membrane Proteins in Mesophases. Current Topics in Membranes. 2009;63:83–108. [Google Scholar]
- Nollert P, Landau EM. Enzymic release of crystals from lipidic cubic phases. Biochem. Soc. Trans. 1998;26:709–713. doi: 10.1042/bst0260709. [DOI] [PubMed] [Google Scholar]
- Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chem. Phys. Lipids. 1998;95:11–21. doi: 10.1016/s0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- Faham S, Ujwal R, Abramson J, Bowie JU. Chapter 5 Practical Aspects of Membrane Proteins Crystallization in Bicelles. Current Topics in Membranes. 2009;63:109–125. [Google Scholar]
- Faham S. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Science. 2005;14:836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke H. Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proceedings of the National Academy of Sciences. 2008;105:16561–16565. doi: 10.1073/pnas.0807162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proceedings of the National Academy of Sciences. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar KR. Structure of rhomboid protease in a lipid environment. J. Mol. Biol. 2011;407:232–247. doi: 10.1016/j.jmb.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SGF. Crystal structure of the human [bgr]2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Prosser RS, Hwang JS, Vold RR. Magnetically aligned phospholipid bilayers with positive ordering: a new model membrane system. Biophys. J. 1998;74:2405–2418. doi: 10.1016/S0006-3495(98)77949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


