Abstract
Background and Objectives
Recurrence in lymph node-negative (pN0) colorectal cancer suggests the presence of undetected occult metastases. Occult tumor burden in nodes estimated by GUCY2C RT-qPCR predicts risk of disease recurrence. This study explored the impact of the number of nodes analyzed by RT-qPCR (analytic) on the prognostic utility of occult tumor burden.
Methods
Lymph nodes (range: 2–159) from 282 prospectively enrolled pN0 colorectal cancer patients, followed for a median of 24 months (range: 2–63), were analyzed by GUCY2C RT-qPCR. Prognostic risk categorization defined using occult tumor burden was the primary outcome measure. Association of prognostic variables and risk category were defined by multivariable polytomous and semi-parametric polytomous logistic regression.
Results
Occult tumor burden stratified this pN0 cohort into categories of low (60%; recurrence rate (RR)=2.3% [95% CI 0.1–4.5%]), intermediate (31%; RR=33.3% [23.7%–44.1%]), and high (9%; RR=68.0% [46.5%–85.1%], p<0.001) risk of recurrence. Beyond race and T stage, the number of analytic nodes was an independent marker of risk category (p<0.001). When >12 nodes were analyzed, occult tumor burden almost completely resolved prognostic risk classification of pN0 patients.
Conclusions
The prognostic utility of occult tumor burden assessed by GUCY2C RT-qPCR is dependent on the number of analytic lymph nodes.
Keywords: lymph node collection, colorectal cancer, molecular staging, occult tumor burden, guanylyl cyclase C
Introduction
Lymph node metastases are one of the most important prognostic factors determining disease recurrence and disease-related mortality in colorectal cancer [1,2]. Although theoretically rendered cancer free by surgery, patients with nodes devoid of histopathologic evidence of cancer (pN0) suffer recurrence rates of ~30%, while those rates exceed 50% in patients with ≥4 lymph nodes harboring macroscopic metastases (pN2) [1–4]. Adjuvant chemotherapy improves disease-free and overall survival in patients with histopathologically evident lymph node metastases, but its role in pN0 patients remains unclear [1–6].
Disease recurrence in a substantial fraction of node-negative colorectal cancer patients suggests the presence of occult metastases that escape conventional detection methods [7–11]. Conversely, patients who are free of lymph node metastases by any detection method may have a better prognosis [7–11]. More accurate assessment of occult metastases would improve clinical risk stratification. In addition, some patients with occult metastases might benefit from the increasingly effective adjuvant chemotherapy available for colorectal cancer.
The tumor suppressor GUCY2C (guanylyl cyclase C) is the receptor for the paracrine hormones guanylin and uroguanylin, gene products universally lost early in intestinal neoplasia [12,13]. Loss of hormone expression silences GUCY2C signaling which contributes to malignant transformation by promoting proliferation, crypt hypertrophy, metabolic remodeling and genomic instability [13]. Highly selective expression by normal intestinal epithelial cells, and universal over-expression by intestinal tumor cells [14–16], suggested that GUCY2C might be a specific molecular marker for metastatic colorectal cancer [11,17,18]. A recent prospective analysis revealed that pN0 colorectal cancer patients whose nodes expressed GUCY2C mRNA suffered recurrence more frequently than those with GUCY2C-negative nodes [11], an observation that has been independently validated [19–21]. Moreover, quantifying molecular tumor burden across the regional lymph node network dramatically improved the classification of recurrence risk in pN0 patients [18].
There is an established relationship between the number of nodes analyzed by histopathology and the accuracy of staging in colorectal cancer [1,2,22,23]. While molecular approaches to identifying occult tumor cells are emerging, the relationship between the number of nodes analyzed and the accuracy of molecular staging has not yet been explored. In that context, the accuracy of estimating occult tumor burden across the regional lymph node network theoretically depends on the number of nodes assessed [18,24]. The present analysis identifies the relationship between the number of nodes analyzed by GUCY2C RT-qPCR (analytic nodes) and the accuracy of risk stratification by occult tumor burden in pN0 colorectal cancer patients.
Methods and Materials
Study Design [11,18]
This prospective observational trial at nine centers in the U.S. and Canada explored the prognostic utility of GUCY2C RT-qPCR in lymph nodes of pN0 colorectal cancer patients. Investigators and clinical personnel were blinded to results of molecular analyses while laboratory personnel and analysts were blinded to patient and clinical information. To have at least 80% power to detect a hazard ratio of 1.6 (P≤0.05, 2-sided) employing categorical assessment of occult tumor metastases in the original study, 225 pN0 patients were required. The protocol was approved by the Institutional Review Board of each participating hospital. The 299 pN0 patients who met eligibility criteria provided 7,310 lymph nodes (range 2–159, median 21 lymph nodes/patient) for histopathologic examination, of which 2,774 nodes (range 1–87, median 8 lymph nodes/patient) were obtained by fresh dissection and eligible for analysis by RT-qPCR. Disease status obtained in routine follow-up by treating physicians, including local and distant recurrences, was provided for all patients through December 31, 2009.
Patients and Tissues [11,18]
Between March 2002 and June 2007, we enrolled 299 stage 0-II pN0 colorectal cancer patients who provided written informed consent before surgery at one of 7 academic medical centers and 2 community hospitals in the U.S. and Canada. Patients were ineligible if they had a previous history of cancer, metachronous extra-intestinal cancer, or perioperative mortality associated with primary resection. For all eligible patients, preoperative and perioperative examinations revealed no evidence of metastatic disease. Lymph nodes, and when available tumor specimens (51%), were dissected from colon and rectum resections and frozen at −80°C within one hour to minimize warm ischemia. Half of each resected lymph node was fixed with formalin and embedded in paraffin for histopathological examination. Lymph node specimens were subjected to molecular analysis if (1) tumor samples, where available, expressed GUCY2C mRNA above background levels in disease-free lymph nodes (>30 copies) and (2) at least one lymph node was provided which yielded RNA of sufficient integrity for analysis. Thus, analysis of the 2,774 lymph nodes available for molecular analysis from 299 patients revealed that 236 nodes from 76 patients yielded RNA of insufficient integrity by β-actin RT-qPCR. Elimination of these nodes from analysis resulted in the exclusion of two patients. Moreover, GUCY2C expression in tumors was below background levels in 6 patients who also were excluded from further analysis. Of the resulting 291 eligible patients, 23 were identified by their medical record as black, 259 as white (Table I), and 9 were of another race or their race could not be identified. These analyses focus on the 282 patients with known race.
Table I.
Patient Characteristics by Risk Group.
| Overall | Low Risk (n=170) |
Moderate Risk (n=88) |
High Risk (n=24) |
P† | |
|---|---|---|---|---|---|
| Characteristic | n | %++ | %++ | %++ | |
| Race | 0.007 | ||||
| Black | 23 | 47.8 | 26.1 | 26.1 | |
| White | 259 | 61.4 | 31.7 | 6.9 | |
| Age at Diagnosis | 0.83 | ||||
| <65 | 106 | 62.3 | 29.3 | 8.5 | |
| ≥65 | 175 | 58.9 | 32.6 | 8.5 | |
| Sex | 0.83 | ||||
| Male | 157 | 60.5 | 31.9 | 7.6 | |
| Female | 125 | 60.0 | 30.4 | 9.6 | |
| Location | 0.59 | ||||
| Colon | 243 | 59.2 | 31.7 | 9.1 | |
| Rectal | 39 | 66.7 | 28.2 | 5.1 | |
| Differentiation | 0.69 | ||||
| Poor/unknown | 45 | 60.0 | 33.3 | 6.7 | |
| Moderate | 217 | 59.0 | 31.8 | 9.2 | |
| Well | 20 | 75.0 | 20.0 | 5.0 | |
| T Stage | 0.008 | ||||
| T1/T2 | 117 | 66.7 | 30.8 | 2.5 | |
| T3/T4 | 165 | 55.8 | 31.5 | 12.7 | |
| Lymphovascular Invasion | 0.34 | ||||
| No | 224 | 61.2 | 29.4 | 9.4 | |
| Yes | 58 | 56.9 | 37.9 | 5.2 | |
| Treatment | 0.40 | ||||
| Surgery alone | 218 | 60.6 | 2.1 | 7.3 | |
| Surgery + chemotherapy | 64 | 59.4 | 28.1 | 12.5 | |
| Nodes Harvested for Histopathology | 0.003 | ||||
| ≤12 | 59 | 55.0 | 45.0 | 0.0 | |
| >12 | 223 | 62.2 | 27.0 | 10.8 | |
Row percentage.
P value from chi-square test of association.
RNA Isolation
RNA was extracted from tissues by a modification of the acid guanidinium thiocyanate-phenol-chloroform extraction method [15,17]. Briefly, individual tissues were pulverized in 1.0 mL Tri-Reagent (Molecular Research Center, Cincinnati, OH) with 12–14 sterile 2.5 mm zirconium beads in a bead mill (Biospec, Bartlesville, OK) for 1–2 min. Phase separation was performed with 0.1 mL bichloropropane, and the aqueous phase re-extracted with 0.5 mL chloroform. RNA was precipitated with 50% isopropanol and washed with 70% ethanol. Air-dried RNA was dissolved in water, concentration determined by spectrophotometry, and stored at −80°C.
RT-qPCR
GUCY2C mRNA was quantified by RT-PCR employing an established analytically validated assay [16]. The EZ RT-PCR kit (Applied Biosystems, Foster City, CA) was employed to amplify GUCY2C mRNA from total RNA in a 50 μL reaction. Optical strip-tubes were used for all reactions, which were conducted in an ABI 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). In addition to the kit components [50 mM Bicine (pH 8.2), 115 mM KOAc, 10 μM EDTA, 60 nM ROX, 8% glycerol, 3 mM Mg(OAc)2, 300 μM each dATP, dCTP, and dGTP, 600 μM dUTP, 0.5 U uracil N-glycosylase, and 5 U rTth DNA polymerase], the reaction master mix contained 900 nM each of forward (ATTCTAGTGGATCTTTTCAATGACCA) and reverse primers (CGTCAGAACAAGGACATTTTTCAT), 200 nM Taqman probe (FAM-TACTTGGAGGACAATGTCACAGCCCCTG-TAMRA), and 1 μg RNA template. The reference gene β-actin was amplified employing similar conditions except that forward (CCACACTGTGCCCATCTACG) and reverse (AGGATCTTCATGAGGTAGTCAGTCAG) primers were 300 nM each, while the Taqman probe (FAM-ATGCCC-X(TAMRA)-CCCCCATGCCATCCTGCGTp) was 200 nM. The thermocycler program employed for RT included: 50° × 2 min, 60° × 30 min, 95° × 5 min; and for PCR: 45 cycles of 94° × 20 sec, 62° × 1 min. Reactions were performed at least in duplicate and results averaged.
Statistical Methods
Statistical methods for estimating GUCY2C and β-actin mRNA by logistic regression analysis were described previously [11,18]. The primary endpoint was molecular risk category (low, intermediate, high) which was previously defined based on time to recurrence and recursive partitioning analysis [18]. Time to recurrence for those analyses was defined as the time from diagnosis to time of last follow-up, local or distant recurrence event, or death [11,25]. Analyses of molecular risk category by polytomous logistic regression included an established standard cut-off for the number of lymph nodes harvested for histopathology and the number of analytic lymph nodes available for molecular (RT-qPCR) evaluation [18,24]. Models were compared based on the Akaike Information Criteria (AIC=2k-2ln(L)), where k is the number of parameters and L is the maximized value of the Likelihood of the estimated model), an established metric for the comparison of non-nested models [26]. Multivariable model analyses were then completed using semi-parametric polytomous logistic regression [27,28] to define the relationship between risk level and number of analytic lymph nodes, using a smooth function for the relationship to analytic nodes [18], while controlling for other established factors [24]. Inference for this modeling approach is not incorporated in the software and properties are as yet undetermined. Thus, 5,000 bootstrap samples were utilized to compute confidence intervals and empirical p values based on the nonparametric bootstrap. The purpose of these multivariable analyses was to determine the potential impact of the number of analytic nodes available on patient risk stratification. The association of harvested lymph nodes to analytic lymph nodes was completed using Spearman's correlation, and visually plotted with a loess smoother. Confidence intervals for raw survival rates were computed by the exact method of Clopper-Pearson [29]. All tests were two-sided, and p<0.05 was considered statistically significant. All analyses were performed with R v 2.11.2, SAS v 9.2.
Results
Patient Characteristics
The 282 pN0 patients had a mean age of 68 years (26–90 years) at diagnosis and 56% were male (Table I). Clinicopathologic features, including depth of tumor penetration (T1/2, T3, T4), and tumor anatomical location (right, left, rectal) were similar to national experience [1–4]. Patients with colon cancer represented 86%, while those with rectal tumors comprised 14%. Black patients comprised 8.2% of the total population enrolled, nearly identical to the national average for disease-specific racial distribution [30–32]. In this cohort, 59 (21%) patients provided ≤12 harvested lymph nodes and 223 (79%) patients >12 harvested lymph nodes for histopathology (Table I), a staging quality standard [1–3]. There were no significant differences in clinicopathologic characteristics between patients providing different numbers of lymph nodes for histopathology (Table II).
Table II.
Patient Characteristics by Number of Lymph Nodes Harvested for Histopathology.
| Overall | ≤12 (n=59) | >12 (n=223) | p† | |
|---|---|---|---|---|
| Characteristic | n | %‡ | %‡ | |
| Age at Diagnosis | 0.73 | |||
| <65 | 106 | 21.1 | 78.3 | |
| ≥65 | 176 | 20.0 | 80.0 | |
| Sex | 0.22 | |||
| Male | 157 | 23.6 | 76.4 | |
| Female | 125 | 17.6 | 82.4 | |
| Location | 0.23 | |||
| Colon | 243 | 19.8 | 80.3 | |
| Rectal | 39 | 28.2 | 71.8 | |
| Differentiation | 0.31 |
|||
| Poor/unknown | 45 | 28.9 | 71.1 | |
| Moderate | 217 | 19.8 | 80.2 | |
| Well | 20 | 15.0 | 85.0 | |
| T Stage | 0.45 | |||
| T1/T2 | 117 | 23.1 | 76.9 | |
| T3/T4 | 165 | 19.4 | 80.6 | |
| Lymphovascular Invasion | 0.26 | |||
| No | 224 | 22.3 | 77.7 | |
| Yes | 58 | 15.5 | 84.5 | |
| Treatment | 0.57 | |||
| Surgery alone | 218 | 20.2 | 79.8 | |
| Surgery + chemotherapy | 64 | 23.4 | 76.6 | |
| Race | 0.66 | |||
| Black | 23 | 17.4 | 82.6 | |
| White | 259 | 21.2 | 78.8 | |
P value from chi-square test of association.
% of total for each category of characteristic.
Occult Tumor Burden and Risk Stratification
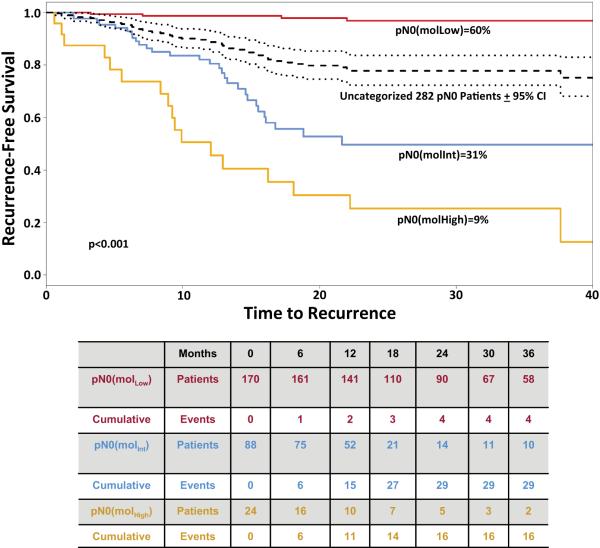
Clinical outcomes in pN0 colorectal cancer patients were analyzed by recursive partitioning using metrics of occult tumor burden estimated by GUCY2C RT-qPCR [18]. Based on time to recurrence, GUCY2C RT-qPCR stratified pN0 patients into categories in which 170 (60%) patients exhibited low (MolLow), 88 (31%) exhibited intermediate (MolInt), and 24 (9%) exhibited high (MolHigh) (p<0.001) risk of disease recurrence (Fig. 1). All but 4 of the MolLow patients remained free of disease during follow-up (recurrence rate (RR)=2.3% [95% CI 0.1–4.5%]); 29 MolInt patients developed recurrent disease (RR=33.3% [23.7%–44.1%]); and 16 (RR=68.0% [46.5%–85.1%]) MolHigh patients developed recurrent disease (p<0.001; Fig. 1). Univariate analysis revealed the expected relationship between advanced T stage and occult tumor burden (p=0.008; Table I) [18,24]. Similarly, black patients harbored a greater burden of occult metastatic tumor across their lymph node network, associated with greater risk, compared to white patients (p=0.007; Table I) [24].
Figure 1. Time to Recurrence in Patients with pN0 Colorectal Cancer Stratified by Occult Tumor Burden Analysis.
Time to recurrence in low (red line), intermediate (blue line) and high (yellow line) risk categories established by occult tumor burden (p<0.001). Time to recurrence in the uncategorized 282 pN0 patient cohort (black dashed line) ±95% confidence intervals (black dotted line) is provided for comparison. Censored values reflect death from another cancer, a non-cancer related death, death due to the cancer treatment, or loss of follow-up of individual patients [25]. Table below Kaplan Meier plot summarize the number of patients at risk as well as cumulative events for each outcome.
Occult Tumor Burden and Lymph Node Collections
The accuracy of molecular staging depended on the number of lymph nodes harvested for histopathologic analysis (Table I). Patients with ≤12 harvested lymph nodes exhibited occult tumor burden that stratified patients in low and intermediate risk categories, with no patients in the highest risk category (Table I). Conversely, analysis of >12 harvested lymph nodes reduced the number of patients with intermediate risk while revealing patients with the highest risk (p=0.002; Table I). This association of staging accuracy by RT-qPCR with increased lymph nodes harvested for histopathology is reminiscent of established improvements in histopathologic staging by increased nodal collections [1,2,22,23].
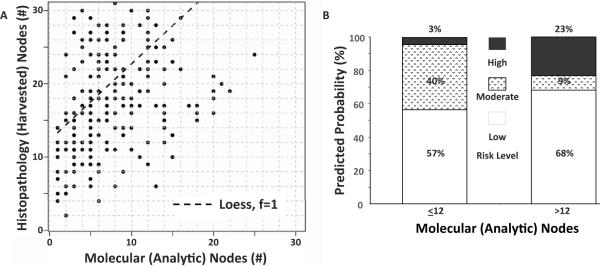
The 282 eligible pN0 patients provided 6,699 lymph nodes (range 2–159, median 21 lymph nodes/patient) for histopathologic examination, of which 2,570 (range 1–33, median 8 lymph nodes/patient) were analyzed by RT-qPCR. The greater number of lymph nodes available for histopathology (harvested), compared to molecular analysis (analytic), from pN0 patients includes those collected after formalin fixation or nodes <5 mm in diameter, smaller than the limit of bisection [11]. The relationship between accuracy of staging by occult tumor burden and number of nodes collected for histopathology suggested a direct association between total nodes harvested (6,699 lymph nodes) and nodes analyzed by RT-qPCR (2,570 lymph nodes; R=0.49, p<0.001; Fig. 2A). Moreover, molecular risk level depended on the number of lymph nodes analyzed per patient by RT-qPCR (p<0.001). Thus, patients providing ≤12 analytic nodes assessed by RT-qPCR exhibited occult tumor burden that stratified patients in low and intermediate risk categories, with only 3% of patients in the highest risk category (Fig. 2B). Conversely, >12 analytic nodes assessed by RT-qPCR improved prognostic resolution, minimizing the number of patients in the intermediate risk category while maximizing the identification of patients with the lowest and highest risk (Fig. 2B). Indeed, the polytomous model based on the number of analytic nodes (AIC=428.45) was preferred in predicting association with risk category compared to the polytomous model based on the number of nodes harvested for histopathology (AIC=480.37).
Figure 2. Occult tumor burden and the number of analytic lymph nodes assessed by RT-qPCR.
(A) Relationship between the number of lymph nodes harvested for histopathology and the number analyzed to quantify occult tumor burden (R=0.49, p<0.001). (B) Differential stratification of risk in patients with ≤12 or >12 lymph nodes evaluated by GUCY2C RT-qPCR to quantify occult tumor burden (p<0.001).
Occult Tumor Burden is an Independent Prognostic Variable Defined by Number of Analytic Lymph Nodes
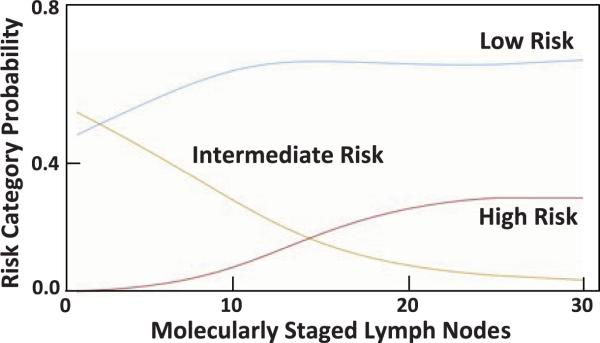
Multivariable analyses employing semi-parametric polytomous logistic regression confirmed that race, T stage, and number of analytic nodes assessed by RT-qPCR are independently associated with quantification of occult tumor burden and stratification into risk categories (Table III). This model used analytic nodes as a continuous variable with a smooth function of unspecified form. Black patients were more likely to be categorized as high risk on the basis of occult tumor burden compared to white patients (adjusted odds ratio 4.05 [1.01–16.67] p=0.03). Similarly, patients with T3 tumors were more likely to be categorized as high risk (adjusted odds ratio 5.51 [2.15–31.10]; p<0.001) compared to patients with T1/T2 tumors. Importantly, the number of analytic nodes analyzed by RT-qPCR was essential to accurately stratify risk by occult tumor burden (p<0.001; Table III). Quantification of occult tumor burden using >12 analytic nodes categorized ~70% of pN0 patients with low risk and ~30% of patients with intermediate and high risk (Fig. 3). Moreover, using ≤25 analytic nodes, which maximizes quantitative information for occult tumor burden analysis, completely resolved these latter categories, stratifying all patients who were not low risk as high risk, eliminating the intermediate risk category (Fig. 3). In that context, number of analytic lymph nodes assessed by RT-qPCR was an independent predictor of risk classification by occult tumor burden analysis (p<0.001, Table III).
Table III.
Multivariable Polytomous Logistic Regression Model.
| Overall | Adjusted Odds Ratio | 95% CI | p† | |
|---|---|---|---|---|
| Characteristic | n | (AOR) | ||
| Race | ||||
| White | 259 | Referent | --- | |
| Black (Moderate vs Low) | 23 | 1.34 | (0.21, 2.31) | 0.30† |
| Black (High vs Low) | 4.05 | (1.01, 16.67) | 0.03† | |
| T Stage | ||||
| T1/T2 | 117 | Referent | --- | |
| T3 (Moderate vs Low) | 145 | 1.38 | (0.86, 2.27) | 0.09† |
| T3 (High vs Low) | 5.51 | (2.15, 31.1) | <0.001† | |
| T4 (Moderate vs Low) | 20 | 2.12 | ||
| T4 (High vs Low) | 3.41 | (0.10, 37.13) | 0.34 | |
| Analytic Nodes (continuous) | 282 | |||
| Moderate vs Low | −0.05‡ | (−0.22, −0.02)‡ | <0.001 | |
| High vs Low | 0.08‡ | (0.04, 0.12)‡ | <0.001 | |
P value from multivariable logistic regression model, based on 5,000 bootstrap samples.
Values are in original scale, and represent coefficient of smooth function, , not odds ratio.
Figure 3.
Probability of risk classification associated with number of analytic lymph nodes evaluated by RT-qPCR.
Discussion
Lymph node metastases are one of the most important prognostic factors in patients with colorectal cancer [1–3]. In contrast, pN0 patients with lymph nodes ostensibly free of metastatic disease by histopathology have a recurrence rate approaching ~30% [1–3]. Recurrence in these patients suggests the presence of occult metastases that escape detection at the time of resection [8,10]. Under-staging by histopathology likely reflects sampling error [33], and the relative insensitivity of histopathology, which reliably detects only 1 cancer cell in 200 normal cells [34].
Molecular staging overcomes limitations in conventional detection of lymph node metastases by incorporating all available tissue into analyses, and improving detection sensitivity through quantifiable, highly sensitive, disease-specific molecular markers [8,10,35]. Previously, we demonstrated that categorical (yes/no) detection of occult metastases in regional lymph nodes using GUCY2C RT-PCR was an independent prognostic marker of recurrence risk in pN0 colorectal cancer patients [11]. Surprisingly, these studies revealed that ~87% of pN0 colorectal cancer patients harbored occult metastases in lymph nodes [11]. Occult nodal metastases were associated with an increased prognostic risk of recurrent disease [11].
These observations reveal an unanticipated diagnostic challenge inherent in the application of sensitive molecular techniques to lymph node staging, in which clinically insignificant metastases are detected in most patients [35]. Occult metastases were associated with increased prognostic risk, but most patients with involved lymph nodes did not progress to disease recurrence [11]. These considerations suggest a quantitative threshold of occult tumor metastases above which the prognostic risk for developing recurrent disease increases [18]. In that context, the prognostic utility of this paradigm was enhanced by quantifying molecular tumor burden (how much) across the regional lymph node network [18]. Quantification of occult tumor burden by GUCY2C RT-qPCR stratified pN0 patients into a low risk cohort representing ~60% of the population, with a very low (<5%) incidence of disease recurrence; an intermediate risk cohort with an incidence of disease recurrence of ~33% and a high risk cohort with >60% incidence of recurrence. Multivariable analyses revealed that occult tumor burden was a powerful independent prognostic marker of time to recurrence and disease-free survival [18].
While quantification of occult tumor burden offers a previously unavailable opportunity to identify patients at risk in the prognostically heterogeneous pN0 population, this staging paradigm classifies ~30% of patients as having intermediate risk [18]. Intermediate risk could reflect variations in tumor biology which influence recurrence beyond the quantity of occult tumor burden in lymph nodes [36,37]. Alternatively, systematic misclassification of high or low risk patients into the intermediate risk category could result from inaccurate quantification of occult tumor burden in inadequate collections of analytic lymph nodes. There is a well-established relationship between the accuracy of conventional staging and the number of lymph nodes analyzed by histopathology [1,2,22,23]. Collection of greater numbers of lymph nodes improves the likelihood of identifying macroscopic tumor deposits by histopathology, which depends on limited tissue sampling techniques [33]. In the context of molecular paradigms employing GUCY2C RT-qPCR, increased analytic lymph node collections improve the accuracy of occult tumor burden quantification across the regional lymph node network. This is underscored by the observation that quantification of occult tumor burden employing very small numbers of analytic lymph nodes only identifies patients with low or intermediate risk, but fails to identify patients with high risk (Fig. 3). In contrast, analyzing ≥25 lymph nodes eliminates the intermediate risk category, classifying patients in low or high risk groups (Fig. 3).
Current practice guidelines recommend the collection of ≥12 lymph nodes to optimize staging of colorectal cancer patients by conventional approaches [1,2]. In contrast, the present results suggest that analyzing ≥25 lymph nodes for occult tumor burden provides complete resolution of risk classification in the pN0 population. This approach identified ~70% of patients with near-zero risk, while ~30% of patients were classified with high risk. This recapitulates the actual risk of this population, in which ~70% of pN0 patients remain disease-free while up to ~30% of patients ultimately develop recurrent disease (e.g., see Fig. 1) [1–3]. It is noteworthy that this level of accuracy, with complete resolution of risk stratification, has not been achieved previously for pN0 colorectal cancer patients by any technique.
While analysis of ≥25 lymph nodes provides the most accurate classification of risk, the data suggest that patient management can be optimized using >12 lymph nodes. Analysis of >12 lymph nodes provides optimum resolution of patients with low risk and those who do not have low risk (see Fig. 3). Adding more lymph nodes to the analysis only improves the accuracy of classifying patients with high risk who were otherwise misclassified as having intermediate risk, without further improving the classification of low risk patients (see Fig. 3). The utility of >12 lymph nodes to optimally classify patients with low risk and those who do not have low risk (consequently, high risk) by occult tumor burden analysis suggests that this emerging molecular paradigm is compatible with current recommendations guiding lymph node collection [1,2].
The present observations demonstrate that the accuracy of staging pN0 colorectal cancer patients by occult tumor burden analysis employing GUCY2C RT-qPCR is critically dependent on the number of analytic lymph nodes. They suggest that with >12 lymph nodes for the analysis, occult tumor burden can provide near complete resolution of prognostic risk stratification in the otherwise heterogeneous pN0 cohort. These studies suggest a very close relationship between the quantity of tumor deposits in regional lymph nodes and the risk of metastatic disease. They underscore the importance of lymphatic spread of colorectal cancer as an essential process in tumor dissemination and metastatic disease [38]. Conversely, these observations challenge the presumed significance of hematogenous spread and circulating tumor cells as key elements underlying disease progression [38]. Further, they call into question the utility of genomic signatures, expression profiling, and other multi-component “omic” analyses of primary tumors, which to date have been only modestly useful in stratifying pN0 risk [36,37]. Most importantly, the ability of occult tumor burden analysis to resolve prognostic risk offers an unprecedented opportunity to identify patients who could most benefit from adjuvant treatment in the otherwise therapeutically ambiguous pN0 population [1–3,5,6].
Conclusion
There is emerging recognition of the utility of occult tumor burden in regional lymph nodes, quantified by molecular techniques, for staging to assess the prognostic risk of colorectal cancer patients. The present studies reveal that the number of lymph nodes available for molecular evaluation is an essential parameter that maximizes the prognostic value of occult tumor burden analysis. They suggest that including >12 lymph nodes in occult tumor burden analysis provides nearly complete resolution of risk classification in pN0 colorectal cancer patients.
Acknowledgments
These studies were supported by grants from NIH (CA75123, CA95026, CA112147, CA146033), the Pennsylvania Department of Health, and Targeted Diagnostic & Therapeutics, Inc. SAW is the Samuel M.V. Hamilton Endowed Professor of Thomas Jefferson University.
Footnotes
Disclosures SAW is the Chair of the Data Safety Monitoring Board for the C-Cure Trial™ sponsored by Cardio Biosciences, and the Chair (uncompensated) of the Scientific Advisory Board of Targeted Diagnostics & Therapeutics, Inc. which provided research funding that, in part, supported this work and has a license to commercialize inventions related to this work. DSW is a shareholder in Targeted Diagnostics & Therapeutics, Inc.
Reference
- 1.Compton CC, Greene FL. The staging of colorectal cancer: 2004 and beyond. CA Cancer J Clin. 2004;54:295–308. doi: 10.3322/canjclin.54.6.295. [DOI] [PubMed] [Google Scholar]
- 2.Edge SB, Byrd DR, Compton CC, et al. AJCC Cancer Staging Manual. Springer; New York: 2009. [Google Scholar]
- 3.Cunningham D, Atkin W, Lenz HJ, et al. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011 doi: 10.3322/caac.20121. caac.20121. [DOI] [PubMed] [Google Scholar]
- 5.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 6.Quasar Collaborative G, Gray R, Barnwell J, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 7.Bilchik AJ, Hoon DS, Saha S, et al. Prognostic impact of micrometastases in colon cancer: interim results of a prospective multicenter trial. Ann Surg. 2007;246:568–575. doi: 10.1097/SLA.0b013e318155a9c7. discussion 575–567. [DOI] [PubMed] [Google Scholar]
- 8.Iddings D, Ahmad A, Elashoff D, Bilchik A. The prognostic effect of micrometastases in previously staged lymph node negative (N0) colorectal carcinoma: a meta-analysis. Ann Surg Oncol. 2006;13:1386–1392. doi: 10.1245/s10434-006-9120-y. [DOI] [PubMed] [Google Scholar]
- 9.Liefers GJ, Cleton-Jansen AM, van de Velde CJ, et al. Micrometastases and survival in stage II colorectal cancer. N Engl J Med. 1998;339:223–228. doi: 10.1056/NEJM199807233390403. [DOI] [PubMed] [Google Scholar]
- 10.Nicastri DG, Doucette JT, Godfrey TE, Hughes SJ. Is occult lymph node disease in colorectal cancer patients clinically significant? A review of the relevant literature. J Mol Diagn. 2007;9:563–571. doi: 10.2353/jmoldx.2007.070032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldman SA, Hyslop T, Schulz S, et al. Association of GUCY2C expression in lymph nodes with time to recurrence and disease-free survival in pN0 colorectal cancer. JAMA. 2009;301:745–752. doi: 10.1001/jama.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P, Schulz S, Bombonati A, et al. Guanylyl cyclase C suppresses intestinal tumorigenesis by restricting proliferation and maintaining genomic integrity. Gastroenterology. 2007;133:599–607. doi: 10.1053/j.gastro.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 13.Pitari GM, Li P, Lin JE, et al. The paracrine hormone hypothesis of colorectal cancer. Clin Pharmacol Ther. 2007;82:441–447. doi: 10.1038/sj.clpt.6100325. [DOI] [PubMed] [Google Scholar]
- 14.Birbe R, Palazzo JP, Walters R, et al. Guanylyl cyclase C is a marker of intestinal metaplasia, dysplasia, and adenocarcinoma of the gastrointestinal tract. Hum Pathol. 2005;36:170–179. doi: 10.1016/j.humpath.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Carrithers SL, Barber MT, Biswas S, et al. Guanylyl cyclase C is a selective marker for metastatic colorectal tumors in human extraintestinal tissues. Proc Natl Acad Sci U S A. 1996;93:14827–14832. doi: 10.1073/pnas.93.25.14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz S, Hyslop T, Haaf J, et al. A validated quantitative assay to detect occult micrometastases by reverse transcriptase-polymerase chain reaction of guanylyl cyclase C in patients with colorectal cancer. Clin Cancer Res. 2006;12:4545–4552. doi: 10.1158/1078-0432.CCR-06-0865. [DOI] [PubMed] [Google Scholar]
- 17.Cagir B, Gelmann A, Park J, et al. Guanylyl cyclase C messenger RNA is a biomarker for recurrent stage II colorectal cancer. Ann Intern Med. 1999;131:805–812. doi: 10.7326/0003-4819-131-11-199912070-00002. [DOI] [PubMed] [Google Scholar]
- 18.Hyslop T, Weinberg DS, Schulz S, et al. Occult tumor burden predicts disease recurrence in lymph node-negative colorectal cancer. Clin Cancer Res. 2011;17:3293–3303. doi: 10.1158/1078-0432.CCR-10-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaulieu M, Desaulniers M, Bertrand N, et al. Analytical performance of a qRT-PCR assay to detect guanylyl cyclase C in FFPE lymph nodes of patients with colon cancer. Diagn Mol Pathol. 2010;19:20–27. doi: 10.1097/PDM.0b013e3181ad5ac3. [DOI] [PubMed] [Google Scholar]
- 20.Haince JF, Houde M, Beaudry G, et al. Comparison of histopathology and RT-qPCR amplification of guanylyl cyclase C for detection of colon cancer metastases in lymph nodes. J Clin Pathol. 2010;63:530–537. doi: 10.1136/jcp.2009.072983. [DOI] [PubMed] [Google Scholar]
- 21.Sargent DJ, Resnick MB, Meyers MO, et al. Evaluation of guanylyl cyclase C lymph node status for colon cancer staging and prognosis. Ann Surg Oncol. 2011 doi: 10.1245/s10434-011-1731-2. [DOI] [PubMed] [Google Scholar]
- 22.Le Voyer TE, Sigurdson ER, Hanlon AL, et al. Colon cancer survival is associated with increasing number of lymph nodes analyzed: a secondary survey of intergroup trial INT-0089. J Clin Oncol. 2003;21:2912–2919. doi: 10.1200/JCO.2003.05.062. [DOI] [PubMed] [Google Scholar]
- 23.Swanson RS, Compton CC, Stewart AK, Bland KI. The prognosis of T3N0 colon cancer is dependent on the number of lymph nodes examined. Ann Surg Oncol. 2003;10:65–71. doi: 10.1245/aso.2003.03.058. [DOI] [PubMed] [Google Scholar]
- 24.Hyslop T, Weinberg DS, Schulz S, et al. Occult tumor burden contributes to racial disparities in stage-specific colorectal cancer outcomes. Cancer. 2011 doi: 10.1002/cncr.26516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Punt CJ, Buyse M, Kohne CH, et al. Endpoints in adjuvant treatment trials: a systematic review of the literature in colon cancer and proposed definitions for future trials. J Natl Cancer Inst. 2007;99:998–1003. doi: 10.1093/jnci/djm024. [DOI] [PubMed] [Google Scholar]
- 26.Bozdogan H. Model selection and Akaike's information criterion (AIC): The general theory and its analytical extensions. Psychometrika. 1987;52:345–370. [Google Scholar]
- 27.Biesheuvel CJ, Vergouwe Y, Steyerberg EW, et al. Polytomous logistic regression analysis could be applied more often in diagnostic research. J Clin Epidemiol. 2008;61:125–134. doi: 10.1016/j.jclinepi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Yee TW. The VGAM package for categorical data analysis. Journal of Statistical Software. 2010;32:1–34. [Google Scholar]
- 29.Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 30.Chien C, Morimoto LM, Tom J, Li CI. Differences in colorectal carcinoma stage and survival by race and ethnicity. Cancer. 2005;104:629–639. doi: 10.1002/cncr.21204. [DOI] [PubMed] [Google Scholar]
- 31.Le H, Ziogas A, Lipkin SM, Zell JA. Effects of socioeconomic status and treatment disparities in colorectal cancer survival. Cancer Epidemiol Biomarkers Prev. 2008;17:1950–1962. doi: 10.1158/1055-9965.EPI-07-2774. [DOI] [PubMed] [Google Scholar]
- 32.White A, Vernon SW, Franzini L, Du XL. Racial disparities in colorectal cancer survival: to what extent are racial disparities explained by differences in treatment, tumor characteristics, or hospital characteristics? Cancer. 2010;116:4622–4631. doi: 10.1002/cncr.25395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hitchcock CL, Sampsel J, Young DC, et al. Limitations with light microscopy in the detection of colorectal cancer cells. Dis Colon Rectum. 1999;42:1046–1052. doi: 10.1007/BF02236701. [DOI] [PubMed] [Google Scholar]
- 34.Ratto C, Sofo L, Ippoliti M, et al. Accurate lymph-node detection in colorectal specimens resected for cancer is of prognostic significance. Dis Colon Rectum. 1999;42:143–154. doi: 10.1007/BF02237119. discussion 154–148. [DOI] [PubMed] [Google Scholar]
- 35.Mejia A, Schulz S, Hyslop T, et al. Molecular staging estimates occult tumor burden in colorectal cancer. Adv Clin Chem. 2010;52:19–39. doi: 10.1016/s0065-2423(10)52007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connell MJ, Lavery I, Yothers G, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28:3937–3944. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 38.Grothey A. Does stage II colorectal cancer need to be redefined? Clin Cancer Res. 2011;17:3053–3055. doi: 10.1158/1078-0432.CCR-11-0574. [DOI] [PubMed] [Google Scholar]