Background: microRNAs 204/211 regulate retinal pigment epithelial cell phenotype.
Results: In RPE, MITF regulates miR-204/211 expression and down-regulation of MITF results in loss of RPE phenotype, which can be prevented by overexpressing miR-204/211.
Conclusion: MITF-mediated expression of miR-204/211 directs RPE differentiation.
Significance: miR-204/211-based therapeutics may be effective treatments for diseases that involve loss of RPE phenotype.
Keywords: Cell Differentiation, Epithelial to Mesenchymal Transition, Gene Regulation, MicroRNA, Retinal Degeneration, Transcription Factors, Microphthalmia-associated Transcription Factor, Proliferative Vitreoretinopathy
Abstract
The retinal pigment epithelium (RPE) plays a fundamental role in maintaining visual function and dedifferentiation of RPE contributes to the pathophysiology of several ocular diseases. To identify microRNAs (miRNAs) that may be involved in RPE differentiation, we compared the miRNA expression profiles of differentiated primary human fetal RPE (hfRPE) cells to dedifferentiated hfRPE cells. We found that miR-204/211, the two most highly expressed miRNAs in the RPE, were significantly down-regulated in dedifferentiated hfRPE cells. Importantly, transfection of pre-miR-204/211 into hfRPE cells promoted differentiation whereas adding miR-204/211 inhibitors led to their dedifferentiation. Microphthalmia-associated transcription factor (MITF) is a key regulator of RPE differentiation that was also down-regulated in dedifferentiated hfRPE cells. MITF knockdown decreased miR-204/211 expression and caused hfRPE dedifferentiation. Significantly, co-transfection of MITF siRNA with pre-miR-204/211 rescued RPE phenotype. Collectively, our data show that miR-204/211 promote RPE differentiation, suggesting that miR-204/211-based therapeutics may be effective treatments for diseases that involve RPE dedifferentiation such as proliferative vitreoretinopathy.
Introduction
The retinal pigment epithelium (RPE)2 is a monolayer of cells that forms the outer blood retinal barrier and performs a number of specialized functions that are critical for photoreceptor health and excitability (1). The RPE possesses long apical microvilli that ensheath the photoreceptor outer segments. These two structures are separated by a small volume of space (subretinal space ≈ 10 μl) that serves as a conduit for the transfer of nutrients and metabolic wastes between photoreceptors and the choroidal blood vessels (2). The RPE hosts a unique set of enzymes such as lecithin retinol acyltransferase (LRAT) and RPE65 that participates in the visual cycle by catalyzing the conversion of all-trans-retinol to 11-cis-retinal, the latter of which is critical for photoreceptor excitability (3). In addition, RPE cells phagocytose shed photoreceptor outer segments (4) and secrete growth factors to nourish the retina (5) and the choroidal blood vessels (6). Thus, loss of RPE functions, as occurs in age-related and proliferative diseases, invariably leads to photoreceptor degeneration and visual impairment (7).
Damage to the RPE or retinal detachment caused by trauma or intraocular diseases can trigger a repair process, in which RPE cells lose cell-cell contact and epithelial phenotype to become proliferative and motile fibroblast-like cells. In proliferative vitreoretinopathy (PVR), for example, unchecked proliferation of RPE cells and their migration into retinal layers and the vitreous result in formation of epiretinal membranes that can contract and cause retinal detachment and visual impairment (8). This switch from an epithelial to mesenchymal-like phenotype involves complex cellular reprogramming with significant alterations in core cellular functions (e.g. metabolism, cell-cell junctions, cell-cycle progression, cytoskeletal rearrangement) as well as gene and protein expression (9). In the search for potential regulators of this process, microRNAs (miRNAs) appeared to be excellent candidates because each miRNA potentially regulates expression of a large array of genes (∼300) that may be involved in a variety of cellular functions such as proliferation and metabolism (10). Recent studies in other biological systems have also established a role of miRNAs in cellular differentiation as positive and negative regulators of epithelial-to-mesenchymal transition (EMT) (11, 12).
MiRNAs are small (∼23 nucleotides) regulatory RNAs that suppress gene expression by binding to specific sequences in the 3′-untranslated region of their target mRNA. Studies in various organ systems revealed that certain miRNAs are highly enriched in a tissue-specific pattern (13–16). Furthermore, transfection of such miRNAs into stem cells (17, 18) or even fibroblasts (19) can induce differentiation into the cell type that normally expresses the miRNA at high levels. These findings support the notion that specific miRNAs may direct cell specification and differentiation of cells that normally express them at high levels (reviewed in Ref. 20). Because down-regulation of tissue-specific miRNAs is commonly associated with disease, their restoration may slow or inhibit disease progression. For example, miR-145 directs smooth muscle differentiation, and its expression was down-regulated in vascular walls with neointimal lesions induced by arterial injury (15). Formation of these lesions was inhibited when injured arteries were transfected with miR-145.
In the RPE, miR-204 and 211 are the two most highly enriched miRNAs, and their expression is critical for maintaining barrier properties and function (16). miR-211 resides in the sixth intron of TRPM1 (melastatin, transient receptor potential cation channel subfamily M member 1), the transcription of which is regulated by microphthalmia-associated transcription factor (MITF) (21), a master regulator of melanocyte and RPE differentiation (22, 23). Mice with homozygous null mutation in the MITF gene have white coats and microphthalmia (24). Furthermore, histological analysis of microphthalmia mouse eyes demonstrated that the absence of MITF prevented RPE differentiation (25). Because MITF and miR-204/211 are important regulators of RPE development and function, it was of interest to determine whether MITF regulates miR-204/211 expression in the RPE and whether expressing high levels of miR-204/211 alone is sufficient to direct RPE differentiation.
In this study, we used primary cultures of human fetal RPE cells (hfRPE) developed by Maminishkis et al. as a model system (26, 27). These cells exhibit properties (morphology, physiology, protein and mRNA profiles) characteristic of native fetal or adult human RPE. In our experiments, we mimicked RPE detachment and dedifferentiation (as occurs in PVR) by subculturing cells at low cell density and found that this process resulted in significant down-regulation of MITF and miR-204/211. Using this in vitro model of RPE dedifferentiation, we found that introduction of pre-miR-204/211 promoted RPE differentiation and protected them from dedifferentiation. Our findings may help facilitate development of miR-204/211-based therapies for human ocular diseases that involve RPE dedifferentiation such as age-related macular degeneration and PVR.
EXPERIMENTAL PROCEDURES
hfRPE Culture Model
hfRPE monolayers were cultured on T25 flasks (P0 hfRPE) as described previously (26). Briefly, hfRPE cells were trypsinized from a T25 flask and seeded onto 12-well Transwells at ≈1.25 × 105 cells/well. P1 hfRPE cells were cultured for 3–4 weeks to reach maturity (transepithelial resistance (TER) >500 ohms·cm2) prior to experimentation. TER was measured with an epithelial volt-ohm meter (EVOM) (WPI, Sarasota, FL) at room temperature. Media and Transwell resistances were taken into account by subtracting 122 ohms·cm2 from the EVOM readout. To test for choroidal fibroblast contamination, hfRPE cells were stained with collagen type I/procollagen antibody (Cell Sciences; Canton, MA). Human fetal choroidal fibroblast cells were used as positive controls and were cultured in the same medium as hfRPE cells.
Total mRNA Extraction
Total mRNA of samples was extracted using mirVana miRNA extraction kit (Ambion, Austin, TX) according to the manufacturer's protocol. RNA bound in the column matrix was treated with RQ1 DNase (5 units/sample; Promega) at 37 °C for 30 min followed by multiple wash steps according to the manufacturer's protocol. RNA was eluted with diethylpyrocarbonate-treated water preheated to 85 °C. Total RNA concentration was measuring using Qubit® fluorometer (Invitrogen).
miRNA Microarray and Data Analysis
Total mRNA of differentiated and dedifferentiated hfRPE samples were prepared using TRIzol (Invitrogen) as described previously (28), and 100 ng of total mRNA from each sample was labeled and hybridized to a human miRNA microarray (V2) from Agilent Technologies (Santa Clara, CA) according to the manufacturer's protocol. The microarray was scanned with an Agilent Microarray Scanner, and the data were processed using Feature Extraction software v10.7.3.1 (Agilent). The microarray was normalized to miR-24 and miR-130a, whose expression levels were the least different between the two RPE cell phenotypes. The normalized array was analyzed using Significance Analysis of Microarrays (SAM 4.0 with R2.14.1) (29) for two-class unpaired statistical analysis with Δ = 5.0 and -fold change >2. miRNAs with fluorescence <50 in both RPE sample types were eliminated. LOG2 fluorescence intensities of miRNAs were represented with a heatmap generated in MultiExperiment Viewer (MeV v4.8). The normalized version of the microarray data can be downloaded from NCBI GEO database (accession number GSE36137).
Reverse Transcription and Real-time Quantitative PCR (qPCR)
RNA (1 μg/sample) was reverse transcribed using oligo(dT)20 primers and SuperScript III (Invitrogen). qPCRs for gene expression studies were performed using ITaq SYBR Green Supermix with ROX (Bio-Rad) in 20-μl reactions (10 ng of cDNA/RxN). qPCR was performed using Eppendorf Mastercycler® ep realplex2. Primers were designed according to guidelines set by Dieffenbach et al. (30). Custom oligonucleotides were purchased from Eurofins MGW Operon (Huntsville, AL). Sequences for all primers used in this study are listed in supplemental Table 1.
qRT-PCR Using TaqMan miRNA Assays
miR-204, miR-211, miR-125b, let-7g, miR-21, and miR-31 TaqMan primers and probes were purchased from Applied Biosystems. 10 ng of total RNA was used in reverse transcription, and the PCRs were performed according to the manufacturer's protocol. qPCR data were analyzed using the comparative 2-ΔΔCt method (31).
qPCR Data Analysis
For SYBR Green qRT-PCR, ribosomal protein S18 (RPS18) gene was used as reference gene because the 2-Ct values of RPS18 from differentiated versus dedifferentiated hfRPE samples were statistically insignificant. For TaqMan assays, U18 snoRNA was used as reference gene because U18 lies within the intron of RPL4 and the mean Ct values (2-Ct) of RPL4 of dedifferentiated versus differentiated hfRPE samples were statistically insignificant. 2-ΔCt of treated versus control samples was analyzed for statistical significance using Student's t test (two-tailed; unpaired samples, unequal variances). p values of < 0.05 were considered statistically significant.
siRNA, Pre-miRNA, and Anti-miRNA Transfection
Ambion pre-miR miRNA precursors and anti-miRNA were purchased from Applied Biosystems. All siRNAs were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). In all experiments, pre-miRNA were transfected upon seeding and on the 3rd day after seeding. Dharmafect 4 was used as transfection reagent (0.2%) in antibiotic-free complete MEM containing 5% serum.
Western Blotting
hfRPE cells on Transwell filters were lysed and homogenized as described previously (28). 15 μg of total protein lysates was loaded onto a NuPAGE® 4–12% Tris-acetate gel (Invitrogen) for electrophoresis. Proteins were subsequently transferred onto PVDF membranes using XCell IITM Blot Module (Invitrogen). Nonspecific binding sites were blocked with TBS (+0.1% Tween 20) containing 5% w/v powdered milk. Antibodies used in this study are listed in supplemental Table 2.
Immunofluorescence and Imaging
hfRPE cells on Transwell filters were fixed with 4% formaldehyde in 1× PBS for 5 min at room temperature followed by 20 min at 4 °C. Samples were permeabilized for 5 min with 0.3% Triton X-100 and blocked with PBS + 0.1% Tween 20 (PBST) containing BSA (5% w/v). Samples were incubated overnight with antibodies against MCT3 and ZO-1 (clone ZO1-1A12; Invitrogen). Samples were washed with PBST and incubated for 1 h in secondary antibodies (Invitrogen). After washing with PBST, samples were stained with phalloidin (1 h at 1:100; Invitrogen) and DAPI (5 min at 1:1000) prior to mounting with gelvatol onto microscope slides. Confocal images on Figs. 3G and 5G were taken with Zeiss LSM 510 confocal microscope at ×40 (Plan-Neofluar ×40/1.3 oil differential interference contrast) with ×2 scanner zoom (×80 final) and 0.5-μm Z-stack intervals. All other images were taken using Nikon A1R confocal microscope at ×60 (Plan Apo VC ×60 WI differential interference contrast N2) with ×1.33 scanner zoom (×80 final) and 0.5-μm Z-stack intervals. Images were extracted from NIS-Elements and analyzed using Adobe Photoshop 7.0.
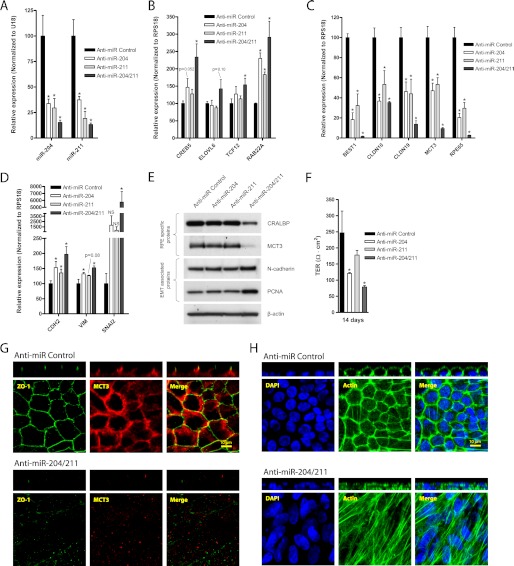
FIGURE 3.
Inhibition of miR-204/211 results in loss of RPE morphology and phenotype. A–D, hfRPE cells were seeded at 30% cell density on Transwell filters and transfected (twice; days 0 and 3) with control anti-miR (50 nm), anti-miR-204 (25 nm + 25 nm control anti-miR), anti-miR-211 (25 nm + 25 nm control anti-miR), or both anti-miR-204/211 (25 nm each) and cultured for 7 days. In these samples, qRT-PCR was performed to compare relative expression of mature miR-204/211 (A), miR-204/211 targets (B), RPE-specific genes (C), or EMT-associated genes (D). E, in a parallel experiment with identical treatment but grown over 10 days, Western blotting was performed to analyze protein expression RPE-specific and EMT-associated proteins. F, in a separate experiment with the same treatment (two transfections at days 0 and 3) but grown over 14 days, TER was measured with EVOM to evaluate RPE barrier function. G and H, from the experiment in which RPE cells were treated with anti-miRs and grown over 10 days, a set of samples was fixed and immunostained with ZO-1 and MCT3 (G) and DAPI and phalloidin (actin filaments) (H). The confocal vertical (Z-X) sections of the samples are shown in panels above their corresponding en-face (X-Y) representations. Statistically significant changes (p < 0.05) are marked with asterisks. Error bars, S.D.
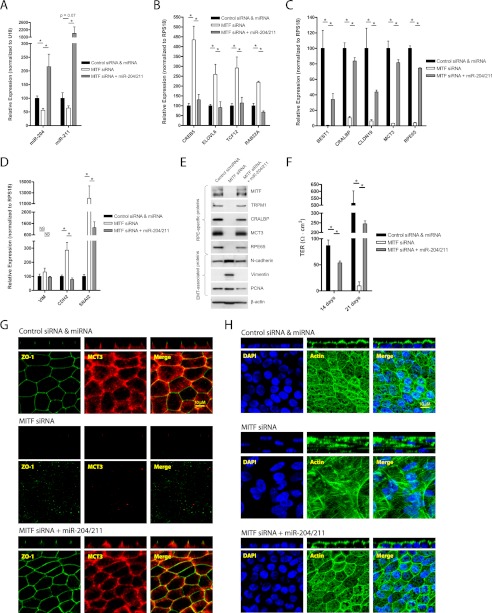
FIGURE 5.
MITF knockdown causes loss of miR-204/211 and RPE phenotype that can be prevented by transfection with pre-miR-204/211. A–D, hfRPE cells were seeded at 30% cell density and transfected (twice; days 0 and 3) with control siRNA (30 nm) + control pre-miRNA (30 nm), MITF siRNA (30 nm) + control pre-miRNA (30 nm), or MITF siRNA (30 nm) + pre-miR-204/211 (15 nm each) and cultured for 7 days. In these samples, qRT-PCR was performed to compare relative expression of mature miR-204/211 (A), miR-204/211 targets (B), RPE-specific genes (C), and EMT-associated genes (D). E, in a parallel experiment with identical treatment but grown over 21 days, Western blotting was performed to analyze protein expression RPE-specific and EMT-associated proteins. F–H, in these samples TER was measured with EVOM (14 and 21 days) to evaluate RPE barrier function (F), and a set of these samples was fixed and stained with ZO-1 and MCT3 antibodies (G) and DAPI and phalloidin (actin filaments) (H). The confocal vertical (Z-X) sections of the samples are shown in panels above their corresponding en-face (X-Y) representations. Statistically significant changes (p < 0.05) are marked with asterisks. Error bars, S.D.
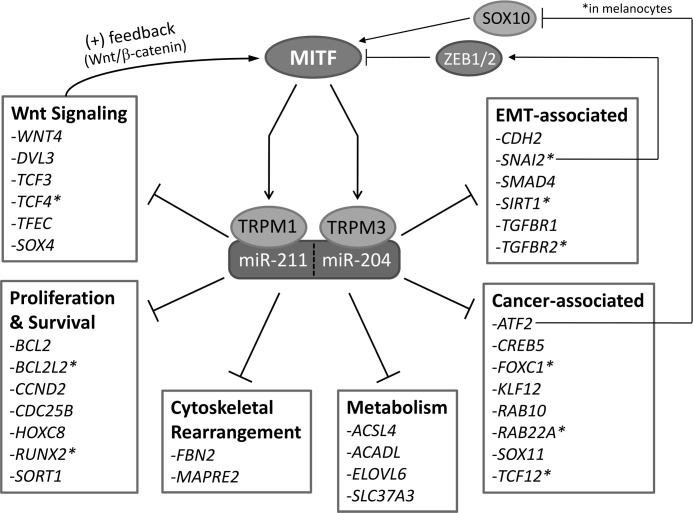
miRNA Target Prediction Analysis
Putative miR-204/211 targets were obtained from TargetScan, miRanda, PicTar, and miRDB. Because predictions made by TargetScan (and PicTar), among other algorithms, have been shown to be among the most accurate (as analyzed by proteomics) (32), miRNA target genes ranked in the “top 100” list of TargetScan were scored higher. Each gene was annotated with their respective ontology profiles and pathway profile (GenMAPP and KEGG data bases) that were extracted from the annotation files of Affymetrix human microarray chipset (HG-U133 Plus 2). This list of miR-204/211 targets is available in supplemental Table 3, and a selection of these targets was categorized and is listed in Fig. 6.
FIGURE 6.
miR-204/211 target genes are involved in various cellular functions. miR-204/211 targets were obtained from TargetScan, miRanda, PicTar, and miRDB. These targets were classified into genes that are known to be involved in Wnt signaling, proliferation and survival, cytoskeletal rearrangement, metabolism, cancer, and EMT. Experimentally confirmed miR-204/211 targets are marked with an asterisk.
RESULTS
RPE Dedifferentiation Involves Loss of RPE-specific Genes and miR-204/211
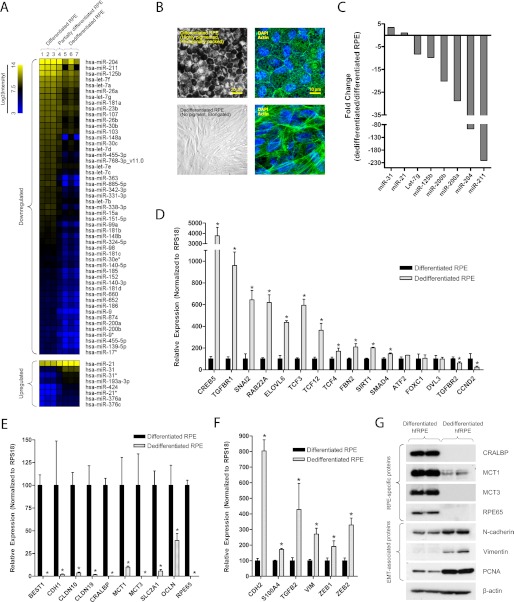
The RPE is normally quiescent and nonmigratory, but in disease conditions such as PVR, it can undergo dedifferentiation into fibroblast-like cells that are proliferative and motile. This phenomenon was observed in vitro as cells at the free edge of differentiated hfRPE monolayer dedifferentiate, migrate, and establish a new population of sparsely pigmented fibroblast-like cells (28). To study the role of miRNAs in RPE differentiation, we compared the miRNA expression profile of dedifferentiated versus differentiated hfRPE cells using miRNA microarray analysis (Fig. 1A). In this experiment, we also included a sample of partially differentiated hfRPE cells (pigmented but lost epithelial morphology) to represent RPE cells in an earlier stage of dedifferentiation. From this array, we found that the three most highly expressed miRNAs (miR-204, miR-211, and miR-125b) in the RPE (16) were significantly down-regulated in dedifferentiated hfRPE cells. miR-200a and miR-200b, which suppress EMT by targeting Zeb1 and Zeb2 transcription factors (33), were also down-regulated in dedifferentiated RPE cells. In addition, expression of miRNAs that are commonly down-regulated (let-7 family) or up-regulated (miR-21 and miR-31) in cancer was also altered. This model of RPE dedifferentiation, however, was difficult to manipulate because dedifferentiation and migration of cells from the edge of the RPE monolayer occur sporadically and therefore cannot be experimentally induced and controlled. Thus, we developed an alternative in vitro model of RPE dedifferentiation in which we passaged P1 hfRPE cells at low density (P2 at 1%, P3 at 30%) twice to produce a homogeneous population of dedifferentiated hfRPE cells (Fig. 1B). However, such seeding conditions also promote growth of choroidal fibroblast contaminants that may be present in the hfRPE culture. To address this concern, we examined the purity of our hfRPE cultures and showed that our dedifferentiated hfRPE cells did not express collagen type I (gene and protein), which was highly expressed in choroidal fibroblast cells (supplemental Fig. 2). Using this model of RPE dedifferentiation, we validated our microarray data with TaqMan qRT-PCR miRNA assay (Fig. 1C). In agreement with the microarray data, we found that miR-204/211 were among the most significantly down-regulated miRNAs in dedifferentiated RPE cells.
FIGURE 1.
RPE dedifferentiation is characterized by loss of epithelial phenotype, changes in miRNA profile, and significant alterations in mRNA and protein expression. Differentiated hfRPE cells (passage 1 (P1)) were grown on Transwell filters over 4 weeks to obtain a differentiated RPE monolayer. A, dedifferentiated RPE cells that migrated from the free edge of confluent hfRPE monolayer were isolated, and microRNA microarray was performed to compare their miRNA expression levels with that of differentiated RPE cells. B, in a different model, dedifferentiated RPE cells were obtained by passaging P1 hfRPE cells at low cell density twice (P1 to P2 at 1%, P2 to P3 at 30%) on 100-mm culture dishes. C, the expression of several miRNAs that were significantly altered in the microarray analysis was verified using TaqMan microRNA assay. D–F, to compare mRNA expression of differentiated versus dedifferentiated RPE cells, qRT-PCR was performed to evaluate the expression of miR-204/211 targets (D), genes involved in barrier, nutrient/ion transport, and RPE-specific functions (E), and genes that are commonly up-regulated in EMT (F). G, Western blot shows that RPE dedifferentiation involves a loss of RPE-specific proteins and an increase in EMT-associated proteins. Statistically significant changes (p < 0.05) are marked with asterisks. Error bars, S.D.
To further understand the biological role of miR-204/211 in the RPE, putative miR-204/211 target genes were identified using in silico miRNA target prediction tools (see Fig. 6). Next, we compared the expression of these genes in differentiated versus dedifferentiated RPE cells by qRT-PCR analysis. We found that several of the predicted miR-204/211 target genes (CDH2, CREB5, TCF3 (TCF7L1), TGFBR1, RAB22A, ELOVL6, TCF12, TCF4 (TCF7L2), SMAD4, and SIRT1) were up-regulated in dedifferentiated hfRPE cells, consistent with the loss of miR-204/211 (Fig. 1C). In addition, dedifferentiated hfRPE cells express low levels of genes that are known to be important for RPE function: blood-retinal barrier (CDH1, CLDN10, CLDN19, OCLN), ion and nutrient transport (BEST1, SLC16A1 (MCT1), SLC16A8 (MCT3), SLC2A1 (GLUT1)), and retinal cycle (RPE65, CRALBP) (Fig. 1E). Accompanying these changes was significant up-regulation of genes that are commonly associated with EMT (i.e. CDH2, CCND1, VIM, ZEB1, ZEB2, TGFBR1, TGFB2, and SNAI2) (Fig. 1F). The changes in gene expression were mirrored by changes in protein expression; RPE-specific proteins (CRALBP, MCT3, and RPE65) were down-regulated in dedifferentiated hfRPE cells, whereas vimentin and N-cadherin were up-regulated (Fig. 1G).
The study of miR-204/211 in RPE differentiation necessitates the use of a model system of RPE differentiation and dedifferentiation. We chose to vary seeding density (P1 to P2 at 7.5, 15, 30, or 60%) and found that hfRPE cells seeded at 30 and 60% densities developed characteristic RPE morphology (supplemental Fig. 1A) and expressed high levels of RPE-specific proteins such as MCT3 and CRALBP (supplemental Fig. 1B). On the other hand, hfRPE cells seeded at 7.5 and 15% densities exhibited fibroblast-like morphology and expressed low levels of MCT3 and CRALBP. In addition, qRT-PCR results show that expression of miR-204/211 (supplemental Fig. 1D) and RPE-specific genes (BEST1, CLDN19, CRALBP, MCT3, and RPE65) (supplemental Fig. 1E) were generally higher in RPE cells seeded at 60 versus 15% density. This trend was also observed at all three time points tested (3, 7, and 15 days after seeding). By 15 days, the expression of several RPE-specific genes (BEST1, CLDN19, and MCT3) in hfRPE cells seeded at 60% density reached levels comparable with that of P1 hfRPE cells, whereas RPE65 took significantly longer to achieve high levels of expression. These data showed that seeding hfRPE cells at 15% or lower led to RPE dedifferentiation whereas seeding at 30% or higher resulted in RPE differentiation.
miR-204/211 Promote Epithelial Phenotype
To determine whether miR-204/211 play a key regulatory role in directing RPE differentiation, we seeded P2 hfRPE cells at 15% density to induce RPE dedifferentiation and transfected them with pre-miR-control (50 nm), pre-miR-204 (25 nm), pre-miR-211 (25 nm), or both pre-miR-204/211 (25 nm each) and cultured for 7 days. RNA was extracted, and TaqMan qRT-PCR was performed to verify increased expression of mature miR-204/211 in samples transfected with corresponding miRNAs. In samples treated with pre-miR-204, -211, or both, the mRNA levels of several putative miR-204/211 target genes (CREB5, RAB22A, ELOVL6, and TCF12) were also down-regulated (Fig. 2B). Furthermore, cells transfected with miR-204/211 expressed significantly higher levels of RPE-specific genes (CLDN10, CLDN19, BEST1, MCT3, RPE65, and CRALBP) (Fig. 2C), and lower levels of genes associated with EMT (CDH2, VIM, and SNAI2) (Fig. 2D), suggesting that miR-204/211 promote RPE epithelial phenotype by suppressing genes that promote EMT. Western blot analysis also demonstrated that miR-204/211-transfected cells expressed higher levels of CRALBP and MCT3 and lower levels of N-cadherin (Fig. 2E).
FIGURE 2.
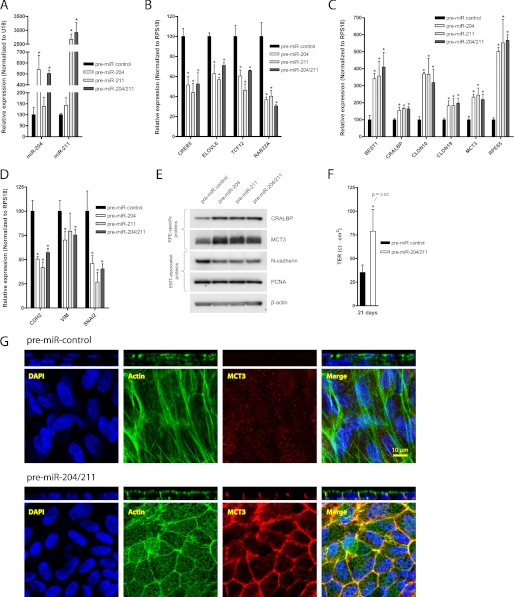
miR-204/211 promote RPE function and integrity. A–D, hfRPE cells were seeded at 15% density on Transwell filters and transfected (twice; days 0 and 3) with control pre-miRNA (50 nm), pre-miR-204 (25 nm + 25 nm control pre-miRNA), pre-miR-211 (25 nm + 25 nm control pre-miRNA), or pre-miR-204/211 (25 nm each) and cultured for 7 days. In these samples, qRT-PCR was performed to compare relative expression of mature miR-204/211 (A), miR-204/211 targets (B), RPE-specific genes (C), and EMT-associated genes (D). E, in a parallel experiment with identical treatment but grown over 21 days, Western blotting was performed to analyze relative expression of RPE-specific and EMT-associated proteins. F, TER of these samples were measured on the 21st day to evaluate barrier function. G, a set of these samples was fixed and immunostained with DAPI, phalloidin (actin filaments), and MCT3. The confocal vertical (Z-X) sections of the samples are shown in panels above their corresponding en-face (X-Y) representations. Statistically significant changes (p < 0.05) are marked with asterisks. Error bars, S.D.
To evaluate the role of miR-204/211 in establishing barrier functions of the RPE, we transfected hfRPE cells (15% density) with pre-miR-204/211 (25 nm each) or control miRNA (50 nm) on Transwell filters and measured TER after 21 days in culture. In this experiment, pre-miRNAs were transfected only twice (once upon seeding and another on the 3rd day). Fig. 2F shows that at the end of the 3rd week, hfRPE cells transfected with miR-204/211 had higher TER (79 ± 23 ohms·cm2) compared with control (35 ± 8 ohms·cm2; n = 4; p = 0.03). Immunostaining of these cells revealed that hfRPE cells transfected with control miRNA formed multiple layered fibroblast-like cells with stress fibers that did not express MCT3 (Fig. 2G, upper). In contrast, hfRPE cells transfected with pre-miR-204/211 formed a monolayer of hexagonally packed cells with circumferential bundles of actin filaments at the lateral junctions and apical microvilli as revealed by phalloidin staining. Furthermore, these cells reestablished proper epithelial polarity as MCT3 labeling was restricted to the basolateral membrane as observed in a mature and polarized RPE in situ. Taken together, our data indicate that miR-204/211 can prevent RPE dedifferentiation.
Inhibiting miR-204/211 Caused RPE Dedifferentiation
Because pre-miR-204/211 prevented RPE dedifferentiation, functional inhibition of miR-204/211 using miR-204/211 antagomers (anti-miRs) should block RPE differentiation. To test this hypothesis, we seeded hfRPE cells at 30% density and transfected them with anti-miR-204 (25 nm + 25 nm control anti-miR), 25 nm anti-miR-211 (25 nm + 25 nm control anti-miR), or both anti-miR-204/211 (25 nm each). qRT-PCR analysis showed that transfection of either anti-miR-204 or anti-miR-211 alone decreased expression of both miR-204 and miR-211 (Fig. 3A). However, the expression of miR-204/211 target genes (CREB5, ELOVL6, TCF12, and RAB22A) was significantly increased only in cells co-transfected with both anti-miR-204 and -211 (Fig. 3B). qRT-PCR results showed that transfection of anti-miR-204 or -211 resulted in down-regulation of RPE-specific genes (BEST1, CLDN10, CLDN19, MCT3, and RPE65) (Fig. 3C) and up-regulation of EMT-associated genes (CDH2, VIM, and SNAI2) (Fig. 3D). Our data are consistent with findings by Wang et al., who showed that inhibiting miR-204 or -211 in differentiated RPE cells resulted in loss of RPE-specific genes and up-regulation of EMT-associated genes (16). Importantly, we found that co-transfection of anti-miR-204 and -211 resulted in more significant down-regulation of RPE-specific genes and up-regulation of EMT-associated genes compared with anti-miR-204 or anti-miR-211 alone. Consistent with qRT-PCR data, Western blot analysis also demonstrated that inhibition of both miR-204 and -211 resulted in the most significant down-regulation of RPE-specific proteins (CRALBP and MCT3) and up-regulation of EMT-associated proteins (vimentin and N-cadherin) (Fig. 3E), indicating that miR-204 compensated for miR-211 activity and vice versa.
To test the effect of anti-miR-204/211 on RPE barrier function, we measured the TER of hfRPE cells cultured at 30% density transfected with anti-miR-204, -211, or both anti-miR-204/211 (two transfections over 14 days). After 14 days in culture, control hfRPE cells established a resistance of ≈250 ohms·cm2, whereas cells transfected with anti-miR-204, -211, or both 204/211 had significantly lower resistances (≈120, 180, and 80 ohms·cm2, respectively) (Fig. 3F). The morphology of cells transfected with either anti-miR-204 or -211 alone was not significantly different from control anti-miRNA-transfected cells (data not shown), but cells transfected with both anti-miR-204 and -211 exhibited dramatic loss of RPE phenotype as characterized by the complete loss of MCT3 and ZO-1 and the formation of multilayered cells with stress fibers (Fig. 3, G and H). Collectively, our data indicate that inhibition of both miR-204 and -211 is required to induce RPE dedifferentiation.
MITF Knockdown Decreased Expression of miR-204/211 and Their Host Genes, TRPM1 and TRPM3
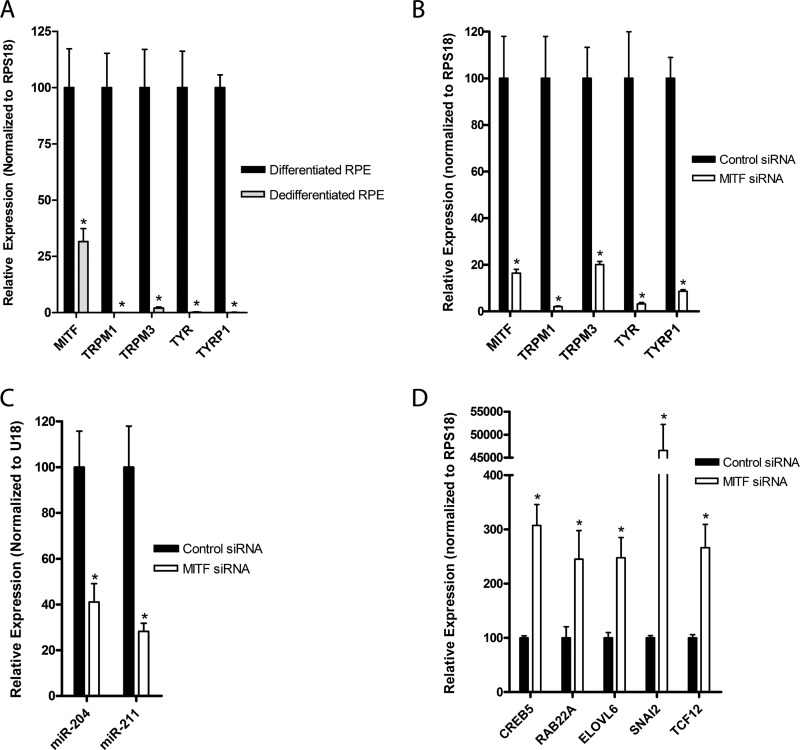
Because down-regulation of miR-204/211 caused RPE dedifferentiation, we examined upstream mechanisms that regulate miR-204/211 expression. miR-204 and miR-211 lie within the introns of TRPM3 and TRPM1, respectively, and early studies in melanocytes showed that transcription of miR-211 and its host gene, TRPM1, are coordinately regulated by MITF. Thus, we compared MITF gene expression in differentiated versus dedifferentiated RPE cells and found that MITF and its target genes (TRPM1, TRPM3, TYR, and TYRP1) were significantly down-regulated in dedifferentiated RPE cells (Fig. 4A). To further examine the role of MITF in miR-204/211 expression, we transfected hfRPE cells (30% density) with MITF siRNA (30 nm) versus control siRNA (30 nm) and found that MITF knockdown caused significant down-regulation of its target genes (TRPM1, TRPM3, TYR, and TYRP1) and miR-204/211 (Fig. 4, B and C). The MITF KD-induced decrease in miR-204/211 expression was accompanied by a concomitant up-regulation of miR-204/211 target genes (CREB5, RAB22A, ELOVL6, SNAI2, and TCF12) (Fig. 4D).
FIGURE 4.
MITF regulates miR-204/211 expression in RPE. A, qRT-PCR of was performed to compare expression of MITF and its target genes (TRPM1, TRPM3, TYR, TYRP1) in differentiated versus dedifferentiated RPE cells. B–D, hfRPE cells were seeded at 30% density on Transwell filters and transfected twice (days 0 and 3) with control versus MITF siRNA (30 nm each), and qRT-PCR was performed to determine relative expression of MITF and its target genes (B), mature miR-204/211 (C), and miR-204/211 target genes (D). Statistically significant changes (p < 0.05) are marked with asterisks. Error bars, S.D.
To determine whether miR-204/211 down-regulation was the primary cause for the loss of RPE phenotype in MITF KD cells, we examined whether addition of pre-miR-204/211 could prevent RPE dedifferentiation caused by MITF KD. hfRPE cells (30% density) were transfected with MITF siRNA (30 nm) + control miRNA (30 nm), MITF siRNA (30 nm) + pre-miR-204/211 (15 nm each), or control miRNA and siRNA (30 nm each) (once at time of seeding and again 3 days later) and cultured for 7 days. Consistent with miR-204/211 levels (Fig. 5A), expression of miR-204/211 targets (CREB5, ELOVL6, TCF12, and RAB22A) was up-regulated in MITF KD cells, and these genes were suppressed in hfRPE cells transfected with both MITF siRNA and pre-miR-204/211 (Fig. 5B). MITF siRNA also decreased expression of RPE-specific genes (BEST1, CRALBP, CLDN19, MCT3, and RPE65), and this effect was prevented by co-transfection with pre-miR-204/211 (Fig. 5C). Expression of EMT-associated genes that were up-regulated in MITF KD cells was also suppressed by pre-miR-204/211 (Fig. 5D). These effects were confirmed at the protein level by Western blot analysis (Fig. 5E; hfRPE cultured for 21 days).
Next, we examined whether hfRPE cells transfected with both MITF siRNA and miR-204/211 could reestablish barrier functions. TER was measured on the 14th and 21st day, and we observed that hfRPE cells with MITF KD had no detectable resistance at either time point (Fig. 5F). However, hfRPE cells transfected with both MITF siRNA and pre-miR-204/211 had resistances of ≈240 ohms·cm2 on the 21st day, demonstrating that miR-204/211 can prevent loss of RPE barrier function caused by MITF KD. Immunofluorescence staining of these samples showed that MITF KD resulted in loss of MCT3 and ZO-1 (Fig. 5G), whereas co-transfecting miR-204/211 with MITF siRNA maintained expression and polarized distribution of MCT3 and ZO-1 to the basolateral membrane and tight junction region, respectively. Phalloidin staining revealed that knockdown of MITF in hfRPE resulted in the formation of multilayered fibroblast-like cells with stress fibers. Co-transfection of hfRPE cells with MITF siRNA and pre-miR-204/211 rescued the RPE phenotype (Fig. 5H). Taken together, our data strongly suggest that loss of MITF led to miR-204/211 down-regulation and subsequent loss of RPE phenotype and function.
DISCUSSION
Dedifferentiation of RPE cells is a major contributing factor to the pathophysiology of proliferative ocular diseases such as PVR (8). Thus, we sought to understand the molecular mechanisms underlying RPE dedifferentiation and identify potential therapeutics that could inhibit this process. We focused our search to microRNAs because they are important regulators of gene expression and have well established roles in many biological processes including development and differentiation (17, 34–37). Previously, we and others demonstrated that RPE cells at the free edge of an intact monolayer can proliferate and migrate, giving rise to mesenchymal cells that express low levels of RPE-specific proteins and increased levels of EMT-associated proteins (28, 38). Microarray analysis comparing the miRNA profile of these samples with that of differentiated RPE cells revealed that miR-204 and miR-211 are among the most significantly down-regulated miRNAs in RPE dedifferentiation. Because different tissues have unique miRNA profiles that reflect their state of differentiation and functional activity, this finding is consistent with an early study by Wang et al., who demonstrated that miR-204/211 are the two most highly expressed miRNAs in the RPE and are also critical for maintaining its epithelial phenotype and function (16). Here, we extend upon the previously established role of miR-204/211 in maintaining RPE function by demonstrating that miR-204/211 could also direct RPE differentiation. Furthermore, we demonstrate that MITF regulates the transcription of miR-204/211 in the RPE and show for the first time that miR-204/211 act downstream of MITF to promote RPE differentiation.
In addition to miR-204/211, our microarray analysis revealed 49 additional miRNAs that were down-regulated by >2-fold in dedifferentiated RPE cells (Fig. 1A). Although any one of these miRNAs could potentially have a role in RPE differentiation, the let-7 family of miRNA was of particular interest as many of its members (isoforms a, b, c, d, e, f, and g) were significantly down-regulated in dedifferentiated RPE cells. Let-7 is a marker of cellular differentiation (39) that also has well established functions as a tumor suppressor (40). Earlier studies showed that let-7 inhibits tumor growth by suppressing the expression of high mobility group A2 (41–43), which induces transcription of two well established regulators of EMT, SNAIL, and TWIST (44, 45). Therefore, down-regulation of let-7 and the resultant increase in SNAIL and TWIST expression in RPE cells may contribute to the loss of RPE phenotype. In addition to let-7, miR-26a/b were also down-regulated in dedifferentiated RPE cells. Because miR-26a/b regulate cell cycle progression by targeting genes such as cyclin D2, D3, E1, and E2, and cyclin-dependent kinases (CDK4 and 6) (46, 47), down-regulation of miR-26a/b may also contribute to the increased proliferative potential that is characteristic of dedifferentiated RPE cells. MiR-204 and -211, the two most highly enriched miRNAs in the RPE, were most significantly down-regulated in dedifferentiated hfRPE cells. Because miR-204/211 target EMT-associated genes (SNAI2 and TGFBR2) and are necessary for maintaining RPE function (16), we asked whether they could also direct RPE differentiation. To test this idea, we developed a new model in which we can induce RPE dedifferentiation by subculturing hfRPE cells at low cell density and test whether overexpressing miR-204/211 in these cells could rescue the RPE phenotype.
This model system is based upon the finding that primary RPE cells have a limited number of divisions within which they can return to a differentiated state (48). Thus hfRPE cells seeded above a “threshold” density will differentiate whereas cells seeded below the threshold will dedifferentiate. By varying cell seeding density, we found that hfRPE cells seeded at 30% or higher achieved differentiation whereas hfRPE cells seeded at 15% density or lower resulted in dedifferentiation (supplemental Fig. 1). However, concerns arise when primary cells were seeded at low densities to induce dedifferentiation because these conditions favor the overgrowth of contaminating fibroblasts, which may be a confounding factor in our analysis. To address this issue, we first demonstrated that collagen type I/procollagen is a suitable fibroblast marker by showing that fibroblasts derived from human fetal choroid (the most likely source of contaminating cells) stained positive for collagen type I/procollagen whereas P1 RPE cells on Transwells do not (supplemental Fig. 2, A and B). However, we did find an average of 31 ± 7 randomly scattered collagen I-positive fibroblast cells embedded underneath the RPE monolayer (≈600,000 RPE cells/Transwell) (n = 9). RPE cells seeded at 15% density on Transwells (for 3 days; n = 3 each) had ∼2–3 collagen I-positive cells (supplemental Fig. 2C). Because fibroblast cells have a doubling time of ≈24 h, a small starting number of fibroblasts (1:20,000 RPE cells) could not have overtaken the RPE culture. Consistent with these observations, RT-PCR and Western blot analysis showed that choroid-derived fibroblasts express collagen type I, whereas dedifferentiated RPE cells (from RPE seeded at 1% density) do not (supplemental Fig. 2, D and E), thus confirming that dedifferentiated RPE cells were of RPE origin and that our model system is valid for the study of RPE dedifferentiation.
Using this model, we show that hfRPE dedifferentiation caused by seeding at 15% density can be prevented by transfecting with pre-miR-204, -211, or both -204/211, as evaluated by increases in RPE-specific gene and protein expressions, increase in TER, and formation of characteristic RPE morphology (Fig. 2). Of particular importance is the observation that transfecting pre-miR-204 or -211 individually had the same effect on RPE differentiation (mRNA, protein, morphology) as transfecting both pre-miR-204 and -211. In addition, anti-miR-induced loss of RPE phenotype (mRNA, proteins, and morphology) occurred only when cells were transfected simultaneously with both anti-miR-204 and -211, but not individually (Fig. 3). Collectively, these results suggest that miR-204 and -211 are functionally redundant in RPE cells, consistent with the fact that miR-204 and -211 possess an identical seed sequence and therefore have the same target genes. Using in silico computational programs, we obtained a list of potential miR-204/211 target genes. Among them are genes that are commonly associated with EMT, cancer, cytoskeletal rearrangement, proliferation, and survival (Fig. 6). However, the role of these genes in RPE differentiation is largely unknown. Future work will include using high throughput miRNA target validation methods such as RIP-ChIP (49) to verify these miR-204/211 targets and using a customized siRNA library to determine the functional role of these genes in RPE differentiation.
To understand how RPE dedifferentiation can lead to down-regulation of miR-204/211, we investigated upstream mechanisms that regulate miR-204/211 expression. MITF plays a key role in the differentiation of melanocytes and pigmented epithelial cells by regulating transcription of genes involved in melanogenesis such as tyrosinase (TYR) and tyrosinase-related protein 1 (TYRP1) (50, 51). Mice homozygous for mutant MITF are completely white and have underdeveloped eyes (microphthalmia) in which the RPE transdifferentiates into neural retina (24, 25). In addition to TYR and TYRP1, MITF also regulates expression of TRPM1, which hosts primary miR-211 (pri-miR) within its sixth intron. Coincidentally, the primary miR-204 hairpin sequence lies within the sixth intron of TRPM3. Earlier studies showed that miR-204 is co-expressed with TRPM3 mRNA in the choroid plexus (52), a cerebral spinal fluid secreting tissue in the brain that, like the RPE, is derived from the neural ectoderm. In hfRPE cells, we showed that MITF KD resulted in significant decreases in TRPM1, TRPM3, and miR-204/211. This effect was accompanied by a significant down-regulation of RPE-specific genes and a dramatic change in morphology from a polarized epithelial monolayer to multilayered mesenchymal cells. Importantly, co-transfection of pre-miR-204/211 into MITF KD cells prevented RPE dedifferentiation, indicating that MITF-mediated regulation of miR-204/211 expression is critical for RPE differentiation.
It is interesting to note that co-transfecting MITF siRNA with pre-miR-204/211 into hfRPE cells partially rescued expression of MITF (and protein) and its target genes (TRPM1, TRPM3, TYR, and TYRP1) (supplemental Fig. 3A). Consistent with these findings, inhibiting miR-204/211 decreased MITF, TRPM1, TRPM3, and miR-204/211 expression (supplemental Fig. 3B), suggesting that reduced miR-204/211 activity makes the RPE more susceptible to dedifferentiation. This helps explain why inhibition of miR-204 or miR-211 individually decreased the expression of both miR-204 and -211 (Fig. 3A). However, transfecting pre-miR-204/211 into dedifferentiating hfRPE cells (15% density) did not increase MITF, TRPM1, or TRPM3 expression (supplemental Fig. 3C). Collectively, our results show that miR-204/211 cannot increase MITF expression, but they can prevent loss of MITF. Thus it is not surprising to find that increasing miR-204/211 expressions in completely dedifferentiated hfRPE cells (P2 at 1%, P3 at 30%) was unable to restore the RPE phenotype (data not shown). The major implication of these findings is that maintaining high expression of miR-204/211 in RPE cells can confer resistance against dedifferentiation.
In conclusion, our results suggest targeted expression of miR-204/211 in RPE cells may be an effective preventive strategy for diseases that involve degeneration and dedifferentiation of RPE cells, such as age-related macular degeneration and proliferative vitreoretinopathy. With advances in adeno-associated virus (AAV) vector-based transgene delivery to specific tissues in the eye (for review, see Ref. 53) and the recent successes in AAV-mediated therapy for patients with Leber congenital amaurosis (54–57), developing an AAV-based miR-204/211 expression vector that specifically target RPE cells may be a viable strategy against ocular diseases that involve RPE dedifferentiation and loss of epithelial phenotype and function.
Acknowledgments
We thank Dr. Stephen Peiper for providing microRNA microarray chips; Alex Mingda Zhu, Vanessa Tercero, and Lea Hecht for technical assistance; George Purkins for help with microarray analysis; Dr. Ashiwel Undieh for permission to use the facilities of the Department of Pharmaceutical Sciences and Dr. Karen Pescatore for providing us with access to the equipment; and Dr. Kathy Boesze-Battaglia and Frank Stefano for training and use of the A1R Nikon confocal microscope at the University of Pennsylvania, School of Dental Medicine, live-cell imaging core.
This work was supported, in whole or in part, by National Institutes of Health Grant EY-012042 (to N. J. P.).

This article contains supplemental Figs. 1–3 and supplemental Tables 1–3.
- RPE
- retinal pigment epithelium
- AAV
- adeno-associated virus
- EMT
- epithelial-to-mesenchymal transition
- EVOM
- epithelial volt-ohm meter
- hfRPE
- human fetal RPE
- KD
- knockdown
- miRNA
- microRNA
- MITF
- microphthalmia-associated transcription factor
- PVR
- proliferative vitreoretinopathy
- qPCR
- quantitative PCR
- TER
- transepithelial resistance
- TRPM
- melastatin, transient receptor potential cation channel subfamily M member
- P0
- passage 0.
REFERENCES
- 1. Strauss O. (2005) The retinal pigment epithelium in visual function. Physiol. Rev. 85, 845–881 [DOI] [PubMed] [Google Scholar]
- 2. Adijanto J., Banzon T., Jalickee S., Wang N. S., Miller S. S. (2009) CO2-induced ion and fluid transport in human retinal pigment epithelium. J. Gen. Physiol. 133, 603–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lamb T. D., Pugh E. N., Jr. (2004) Dark adaptation and the retinoid cycle of vision. Prog. Retin. Eye Res. 23, 307–380 [DOI] [PubMed] [Google Scholar]
- 4. Kevany B. M., Palczewski K. (2010) Phagocytosis of retinal rod and cone photoreceptors. Physiology 25, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Steele F. R., Chader G. J., Johnson L. V., Tombran-Tink J. (1993) Pigment epithelium-derived factor: neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc. Natl. Acad. Sci. U.S.A. 90, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Witmer A. N., Vrensen G. F., Van Noorden C. J., Schlingemann R. O. (2003) Vascular endothelial growth factors and angiogenesis in eye disease. Prog. Retin. Eye Res. 22, 1–29 [DOI] [PubMed] [Google Scholar]
- 7. Sparrow J. R., Hicks D., Hamel C. P. (2010) The retinal pigment epithelium in health and disease. Curr. Mol. Med. 10, 802–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hiscott P., Sheridan C., Magee R. M., Grierson I. (1999) Matrix and the retinal pigment epithelium in proliferative retinal disease. Prog. Retin. Eye Res. 18, 167–190 [DOI] [PubMed] [Google Scholar]
- 9. Kalluri R., Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zavadil J., Narasimhan M., Blumenberg M., Schneider R. J. (2007) Transforming growth factor-β and microRNA: mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs 185, 157–161 [DOI] [PubMed] [Google Scholar]
- 12. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 13. Liang Y., Ridzon D., Wong L., Chen C. (2007) Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fineberg S. K., Kosik K. S., Davidson B. L. (2009) MicroRNAs potentiate neural development. Neuron 64, 303–309 [DOI] [PubMed] [Google Scholar]
- 15. Cheng Y., Liu X., Yang J., Lin Y., Xu D. Z., Lu Q., Deitch E. A., Huo Y., Delphin E. S., Zhang C. (2009) MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ. Res. 105, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang F. E., Zhang C., Maminishkis A., Dong L., Zhi C., Li R., Zhao J., Majerciak V., Gaur A. B., Chen S., Miller S. S. (2010) MicroRNA-204/211 alters epithelial physiology. FASEB J. 24, 1552–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cordes K. R., Sheehy N. T., White M. P., Berry E. C., Morton S. U., Muth A. N., Lee T. H., Miano J. M., Ivey K. N., Srivastava D. (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng L. C., Pastrana E., Tavazoie M., Doetsch F. (2009) miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee-Messer C., Dolmetsch R. E., Tsien R. W., Crabtree G. R. (2011) MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kosik K. S. (2010) MicroRNAs and cellular phenotypy. Cell 143, 21–26 [DOI] [PubMed] [Google Scholar]
- 21. Mazar J., DeYoung K., Khaitan D., Meister E., Almodovar A., Goydos J., Ray A., Perera R. J. (2010) The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One 5, e13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Steingrímsson E., Copeland N. G., Jenkins N. A. (2004) Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 38, 365–411 [DOI] [PubMed] [Google Scholar]
- 23. Tsukiji N., Nishihara D., Yajima I., Takeda K., Shibahara S., Yamamoto H. (2009) Mitf functions as an in ovo regulator for cell differentiation and proliferation during development of the chick RPE. Dev. Biol. 326, 335–346 [DOI] [PubMed] [Google Scholar]
- 24. Hodgkinson C. A., Moore K. J., Nakayama A., Steingrímsson E., Copeland N. G., Jenkins N. A., Arnheiter H. (1993) Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74, 395–404 [DOI] [PubMed] [Google Scholar]
- 25. Bumsted K. M., Barnstable C. J. (2000) Dorsal retinal pigment epithelium differentiates as neural retina in the microphthalmia (mi/mi) mouse. Invest. Ophthalmol. Vis. Sci. 41, 903–908 [PubMed] [Google Scholar]
- 26. Maminishkis A., Chen S., Jalickee S., Banzon T., Shi G., Wang F. E., Ehalt T., Hammer J. A., Miller S. S. (2006) Confluent monolayers of cultured human retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest. Ophthalmol. Vis. Sci. 47, 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Strunnikova N. V., Maminishkis A., Barb J. J., Wang F., Zhi C., Sergeev Y., Chen W., Edwards A. O., Stambolian D., Abecasis G., Swaroop A., Munson P. J., Miller S. S. (2010) Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 19, 2468–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gallagher-Colombo S., Maminishkis A., Tate S., Grunwald G. B., Philp N. J. (2010) Modulation of MCT3 expression during wound healing of the retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 51, 5343–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tusher V. G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dieffenbach C. W., Lowe T. M., Dveksler G. S. (1993) General concepts for PCR primer design. PCR Methods Appl. 3, S30–37 [DOI] [PubMed] [Google Scholar]
- 31. Schmittgen T. D., Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 32. Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008) The impact of microRNAs on protein output. Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Korpal M., Lee E. S., Hu G., Kang Y. (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen C. Z., Li L., Lodish H. F., Bartel D. P. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 35. Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007) The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu J., Guo S., Ebert B. L., Zhang H., Peng X., Bosco J., Pretz J., Schlanger R., Wang J. Y., Mak R. H., Dombkowski D. M., Preffer F. I., Scadden D. T., Golub T. R. (2008) MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev. Cell 14, 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. King I. N., Qian L., Liang J., Huang Y., Shieh J. T., Kwon C., Srivastava D. (2011) A genome-wide screen reveals a role for microRNA-1 in modulating cardiac cell polarity. Dev. Cell 20, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tamiya S., Liu L., Kaplan H. J. (2010) Epithelial-mesenchymal transition and proliferation of retinal pigment epithelial cells initiated upon loss of cell-cell contact. Invest. Ophthalmol. Vis. Sci. 51, 2755–2763 [DOI] [PubMed] [Google Scholar]
- 39. Thomson J. M., Parker J., Perou C. M., Hammond S. M. (2004) A custom microarray platform for analysis of microRNA gene expression. Nat. Methods 1, 47–53 [DOI] [PubMed] [Google Scholar]
- 40. Park S. M., Shell S., Radjabi A. R., Schickel R., Feig C., Boyerinas B., Dinulescu D. M., Lengyel E., Peter M. E. (2007) Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle 6, 2585–2590 [DOI] [PubMed] [Google Scholar]
- 41. Lee Y. S., Dutta A. (2007) The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 21, 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mayr C., Hemann M. T., Bartel D. P. (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315, 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shell S., Park S. M., Radjabi A. R., Schickel R., Kistner E. O., Jewell D. A., Feig C., Lengyel E., Peter M. E. (2007) Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 11400–11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan E. J., Thuault S., Caja L., Carletti T., Heldin C. H., Moustakas A. (2012) Regulation of transcription factor Twist expression by the DNA architectural protein high mobility group A2 during epithelial-to-mesenchymal transition. J. Biol. Chem. 287, 7134–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thuault S., Tan E. J., Peinado H., Cano A., Heldin C. H., Moustakas A. (2008) HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 283, 33437–33446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu J., He M. L., Wang L., Chen Y., Liu X., Dong Q., Chen Y. C., Peng Y., Yao K. T., Kung H. F., Li X. P. (2011) MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 71, 225–233 [DOI] [PubMed] [Google Scholar]
- 47. Zhu Y., Lu Y., Zhang Q., Liu J. J., Li T. J., Yang J. R., Zeng C., Zhuang S. M. (2011) MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grisanti S., Guidry C. (1995) Transdifferentiation of retinal pigment epithelial cells from epithelial to mesenchymal phenotype. Invest. Ophthalmol. Vis. Sci. 36, 391–405 [PubMed] [Google Scholar]
- 49. Keene J. D., Komisarow J. M., Friedersdorf M. B. (2006) RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1, 302–307 [DOI] [PubMed] [Google Scholar]
- 50. Yasumoto K., Yokoyama K., Takahashi K., Tomita Y., Shibahara S. (1997) Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J. Biol. Chem. 272, 503–509 [DOI] [PubMed] [Google Scholar]
- 51. Cheli Y., Ohanna M., Ballotti R., Bertolotto C. (2010) Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 23, 27–40 [DOI] [PubMed] [Google Scholar]
- 52. Deo M., Yu J. Y., Chung K. H., Tippens M., Turner D. L. (2006) Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 235, 2538–2548 [DOI] [PubMed] [Google Scholar]
- 53. Liu M. M., Tuo J., Chan C. C. (2011) Gene therapy for ocular diseases. Br. J. Ophthalmol. 95, 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bainbridge J. W., Smith A. J., Barker S. S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G. E., Stockman A., Tyler N., Petersen-Jones S., Bhattacharya S. S., Thrasher A. J., Fitzke F. W., Carter B. J., Rubin G. S., Moore A. T., Ali R. R. (2008) Effect of gene therapy on visual function in Leber's congenital amaurosis. N. Engl. J. Med. 358, 2231–2239 [DOI] [PubMed] [Google Scholar]
- 55. Cideciyan A. V., Hauswirth W. W., Aleman T. S., Kaushal S., Schwartz S. B., Boye S. L., Windsor E. A., Conlon T. J., Sumaroka A., Pang J. J., Roman A. J., Byrne B. J., Jacobson S. G. (2009) Human RPE65 gene therapy for Leber congenital amaurosis: persistence of early visual improvements and safety at 1 year. Hum. Gene Ther. 20, 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maguire A. M., Simonelli F., Pierce E. A., Pugh E. N., Jr., Mingozzi F., Bennicelli J., Banfi S., Marshall K. A., Testa F., Surace E. M., Rossi S., Lyubarsky A., Arruda V. R., Konkle B., Stone E., Sun J., Jacobs J., Dell'Osso L., Hertle R., Ma J. X., Redmond T. M., Zhu X., Hauck B., Zelenaia O., Shindler K. S., Maguire M. G., Wright J. F., Volpe N. J., McDonnell J. W., Auricchio A., High K. A., Bennett J. (2008) Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 358, 2240–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simonelli F., Maguire A. M., Testa F., Pierce E. A., Mingozzi F., Bennicelli J. L., Rossi S., Marshall K., Banfi S., Surace E. M., Sun J., Redmond T. M., Zhu X., Shindler K. S., Ying G. S., Ziviello C., Acerra C., Wright J. F., McDonnell J. W., High K. A., Bennett J., Auricchio A. (2010) Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 18, 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]