Abstract
Neuronal ELAV-like proteins (HuB, HuC, and HuD) are highly conserved RNA-binding proteins able to selectively associate with the 3′ UTR of a subset of target mRNAs and increase their cytoplasmic stability and rate of translation. We previously demonstrated the involvement of these proteins in learning, reporting that they undergo a sustained up-regulation in the hippocampus of mice trained in a spatial discrimination task. Here, we extend this finding, showing that a similar up-regulation occurs in the hippocampus of rats trained in another spatial learning paradigm, the Morris water maze. HuD, a strictly neuron-specific ELAV-like protein, is shown to increase after learning, with a preferential binding to the cytoskeletal fraction. HuD up-regulation is associated with an enhancement of GAP-43 mRNA and protein levels, with an apparently increased HuD colocalization with the GAP-43 mRNA and an increased association of neuronal ELAV-like proteins with the GAP-43 mRNA. These learning-dependent biochemical events appear to be spatiotemporally controlled, because they do not occur in another brain region involved in learning, the retrosplenial cortex, and at the level of protein expression they show extinction 1 month after training despite memory retention. By contrast, HuD mRNA levels still remain increased after 1 month in the CA1 region. This persistence may have implications for long-term memory recall.
ELAV-like or Hu proteins are a small family of RNA-binding proteins representing vertebrate orthologues of the elav gene of Drosophila, whose mutation produces embryonic lethality and abnormal development of neural tissue in the fly (1). Four ELAV-like proteins have been identified in the genomes of vertebrates, named HuB, HuC, HuD, and HuR (2). The first three members of the family are characterized by a selective neuronal expression, whereas HuR is present in all tissues examined so far (3); all these proteins are highly homologous in sequence and highly evolutionarily conserved. The ELAV-like proteins share a common domain architecture, containing three RNA-binding domains of the RNA-recognition-motif consensus type (4); the first two, in tandem and very close to each other, are separated from the third by a hinge region (5). ELAV-like proteins are able to selectively bind to a subset of posttranscriptionally regulated mRNAs by recognizing, with the first two domains, AU-rich elements present in their 3′ UTRs (6, 7), whereas the third domain seems to have affinity, at least in vitro, for the poly(A) tail (8, 9). The biological activities of ELAV proteins have probably undergone a phylogenetic shift, because whereas Drosophila elav is a nuclear protein involved in the regulation of the alternative splicing of some target mRNAs (10–12), its vertebrate orthologues shuttle between the nucleus and the cytoplasm (13) and are able to promote gene expression up-regulation by cytoplasmic stabilization and/or enhancement of translation of their mRNA targets (14, 15). The “neural” ELAV-like proteins have been implicated from developmental analysis of expression and from in vitro data on model cell cultures (14, 16, 17) as key determinants of neuronal differentiation, in close analogy with the fly mutant phenotype (18). We recently provided the first demonstration of a physiological role for neuronal ELAV-like proteins by implicating their involvement in posttranscriptional regulation of gene expression after spatial learning in rodents (19). A paradigm for the activity of ELAV-like proteins on downstream gene targets is supported by data detailing the biochemical mechanisms and cellular consequences of the stabilization of GAP-43 mRNA mediated by HuD (20). The growth-associated protein GAP-43 is expressed in neurons principally during the process of establishment of connections, being an intrinsic determinant of neuronal development and plasticity (21). HuD is also strictly neuron-specific, developmentally regulated (3, 22, 23), and appears to act as a major determinant of GAP-43 expression, because ectopic modulation of HuD levels in rat PC12 cells results not only in coherent changes in GAP-43 mRNA stability and expression but also in the neurite outgrowth phenotype (16, 24, 25). Moreover, HuD overexpression in PC12 cells is able per se to induce differentiation in a GAP-43-dependent way in the absence of the nerve growth factor (16, 26, 27). In addition, HuD binds to the GAP-43 3′ UTR mRNA in a site that, if deleted, prevents HuD-induced stabilization of this mRNA (28–30).
After previously implicating all three neuronal ELAV-like proteins in spatial learning, in the present study, we have further implicated a mechanistic relationship among HuD protein, GAP-43 mRNA, and protein in neuronal plasticity established in vitro and in mammalian learning. Here, we have selected the Morris water maze (WM) as a well known spatial learning paradigm to explore learning-induced changes of the distribution and levels of expression of these genes in the rat hippocampus, where initial biochemical events related to learning are known to take place (31) 24 h or 1 month after the last training session. The emerging picture is that of a major up-regulation of the HuD protein specifically induced in the hippocampus by learning. This up-regulation of HuD is accompanied by an increase in the levels of GAP-43 mRNA and protein 24 h after training. The HuD up-regulation seems also to be accompanied by an increased colocalization of HuD protein and GAP-43 mRNA. One month in memory retention, only the HuD mRNA was still increased in the hippocampus.
Materials and Methods
Training Procedure. Eight-week-old male Wistar rats (250 g) were subjected to a WM task, as described (19). Spatial memory of rats was assessed by the time required to find the platform (escape latency). To evaluate retention of recent and remote spatial memories and to avoid a possible effect of probe tests (without the platform in the pool) on rats' behavior, we performed probe tests on a separate group of animals. That group (n = 5) demonstrated a clear spatial bias in rats' swim pathways 24 h and 1 mo after the last training session. Spatial bias was assessed by dwell time and the swim path length in the pool's quadrants. Animals were killed, and the tissues were quickly removed and kept at –80°C.
Subcellular Fractionation and Western Blotting. Subcellular fractioning (nuclei, cytosol, membrane, and cytoskeleton) from rat hippocampi and Western blots were performed as described (19). The anti-HuD mouse antibody (kind gift of Henry Furneaux, University of Connecticut Health Center, Farmington, CT) was used at 1:1,000; the biotin-conjugated 16A11 anti-Hu (Molecular Probes) at 6 μg/ml; the monoclonal anti-Gap43 (Sigma) at 1:3,000; and the anti-α-tubulin (Chemicon) at 1:1,000. The nitrocellulose membranes were developed through chemiluminescence. Experiments were performed at least three times on different cell preparations. Statistical analysis of Western blot data was performed on the densitometric values obtained with nih image (http://rsb.info.nih.gov/nih-image) software.
Immunohistochemistry. Twenty-micrometer-thick cryostat sections were mounted on silanated glass slides and processed as described (19). Nonspecific sites were blocked, and then the slices were exposed to the anti-HuD primary antibody, diluted at 1:100, at 4°C overnight, followed by incubation with an anti-mouse FITC-conjugated secondary antibody (Sigma) diluted at 1:400.
In Situ Hybridization. RNA probes were synthesized in vitro by using T7 or T3 polymerases in the presence of 1 μg of DNA template, 0.5 mM nucleotides, and 2 mM [α-35S]UTP (>1,000 Ci/mmol), and in situ hybridization was performed as described (32). In the fluorescence in situ hybridization protocol, for the preparation of the GAP-43 probe, the [α-35S]UTP was substituted with a digoxigenin-labeled UTP (Roche Molecular Biochemicals), and the signal was revealed with an antidigoxigenin horseradish peroxidase (Roche) at 1:3,000 dilution by using the Tyramide Signal Amplification system (Cyanine 3 TSA, Perkin–Elmer); the HuD protein signal was revealed by using the same conditions as described in Immunohistochemistry.
Hu Immunoprecipitation. Each fraction was precleared by incubation with 30 μl of protein A/G plus agarose (Santa Cruz Biotechnology) for 2 h at 4°C. The samples were centrifuged at 14,000 × g for 10 min, and the protein content was measured. Immunoprecipitation was performed overnight at 4°C with 1 μg of the mouse biotin-conjugated 16A11 anti-Hu antibody per 50 μg of protein diluted with an equal volume of 2× immunoprecipitation buffer (2% Triton X-100/300 mM NaCl/20 mM, pH 7.4/Tris-HCl/2 mM EDTA.2 mM EGTA.0.4 mM sodium vanadate and a protease inhibitor mixture; Boehringer–Mannheim). Fifty microliters of protein A/G plus agarose was then added, and the samples were incubated at 4°C for 2 h. Finally, the samples were centrifuged at 14,000 × g for 10 min, and the supernatant was discarded. The pellet was then subjected to RT-PCR to determine the GAP-43 mRNA content.
Real-Time Quantitative RT-PCR. Total RNA from dissected whole-rat hippocampi, from hippocampal subregions, or from hippocampal fractions was extracted, treated with DNAse, and subjected to reverse transcription following standard procedures. PCR amplifications were carried out by using the Lightcycler instrument (Roche), as described (19), with primers designed on the 3′ UTRs of the murine HuB, HuC, HuD, GAP-43, and GAPDH mRNA sequences by using the primer3 software (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Primer sequences were as follows. HuB, 5′-CCCACCTCCTTAGCTTTCCT-3′ (upstream) and 5′-CGTTTATAATGTCGCCCCC-3′ (downstream); HuC, 5′-GGTTCATGGTGATGGCTTTT-3′ (upstream) and 5′-TTTAAACCTFTTCCCTCCCC-3′ (downstream); HuD, 5′-GCGCACACACATACACAAAA-3′ (upstream) and 5′-AAAATCCTTTCCTGGTACACCTC-3′ (downstream); GAP-43, 5′-TCCTCTCCTGTCCTGCTCAC-3′ (upstream) and 5′-TCGCCATAACAACACCAAGA-3′ (downstream); GAPDH, 5′-CAGCAAGGATACTGAGAGCAAG-3′ (upstream) and 5′-GGATGGAATTGTGAGGGAGA-3′ (downstream).
Data Analysis. Analysis of the data was performed with ANOVA followed by the appropriate post hoc test as indicated in the figure legends, by using the statview (SAS Institute, Cary, NC) or origin 6.1 (OriginLab Corporation, Northampton, MA) statistical packages. Differences were considered statistically significant when P values were ≤0.05.
Results
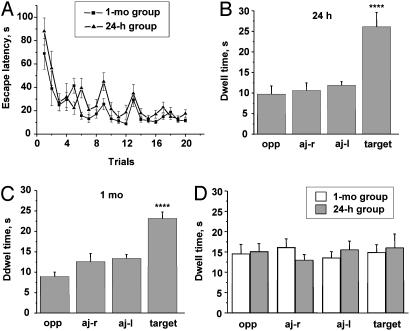
Wistar rats were trained (TR) for 5 days (four trials per day) to locate a submerged platform in a swimming pool and matched with control rats [swimming (SW)], which were left swimming in the same pool without the platform for 2 min. Both TR groups (killed 24 h and 1 mo after WM) displayed a similar time course of WM learning with a characteristic significant improvement in the escape latency over the first trials (Fig. 1A). A2(groups) × 5 (days of trials) ANOVA with repeated measures on the second factor revealed significant main effects of days [F(4, 95) = 27.34, P < 0.0001] and trials [F(3, 76) = 32.37, P < 0.0001] without a significant interaction between all factors. Rats reached an asymptotic performance simultaneously. There were no statistically significant differences between groups during the last two days of training [F(1, 18) = 2.98, P > 0.1]. Transfer tests performed on a separate group of rats both 24 h and 1 mo after the last training session indicated a clear spatial preference for the TR quadrant, as measured by dwell time [F(3, 16) = 19.7, P < 0.0001, Fisher's post hoc test; Fig. 1 B and C] and by distance swum (data not shown) in the target and control quadrants. There was no memory decline after a 1-mo delay for the second probe test, as compared to 24-h delay [F(1, 8) = 0.083, P > 0.78; compare Fig. 1 B and C]. On the contrary, SW rats for both 24-h and 1-mo groups did not demonstrate spatial bias in the dwell time [F(3, 76) = 0.058, P > 0.98] and swim track lengths [F(3, 76) = 0.3, P > 0.8, not shown] at the end of the behavioral procedures, and there were no differences between groups in the dwell time [F(1, 18) = 3.68, P > 0.07, Fig. 1D] and distance of swimming [F(1, 18) = 0.114; P > 0.74, not shown].
Fig. 1.
Acquisition and long-term retention of spatial-memory WM task. (A) Similar dynamics of escape latency in the groups of rats trained for biochemical studies of the recent and 1-mo-old spatial memories (n = 10; ref. 10). (B and C) Results of probe tests performed in the same animals 24 h (B) and 1 mo (C) after the last WM training session indicate no decline in spatial memory (n = 5). (D) Absence of spatial preference in both 24-h and 1-mo swimming controls as indicated by rats' dwell time in the pool quadrants (n = 10; ref. 10). Oppquadrant, opposite to training or target quadrant; Aj-r, adjacent right quadrant; Aj-l, adjacent left quadrant. Data represent means ± SE; n = number of tested animals. ****, P < 0.0001, repeated-measures ANOVA with Fisher's post hoc test.
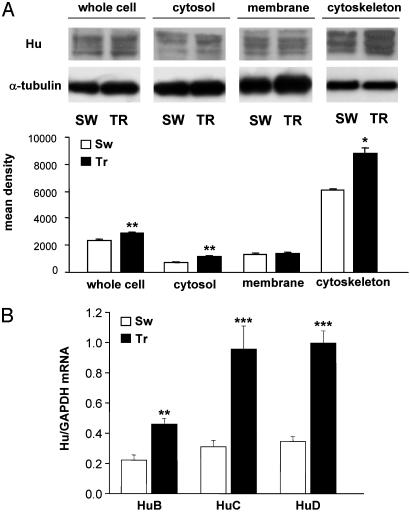
Hippocampal lysates of animals killed 24 h after the last training session were fractionated and subjected to Western blot analysis with a commercial monoclonal antibody, 16A11, which recognizes the three neuronal members of the ELAV-like protein family (6), namely HuB, HuC, and HuD. The results, reported in Fig. 2A, indicate a significant increase in the whole-cell ELAV-like content in the TR rats, due to the cytosolic and, most of all, cytoskeletal fractions and without any contribution of the cell membranes. This learning-related increase in the neuronal ELAV-like immunoreactivity is paralleled by an increase in the corresponding three single mRNA species, as assessed by real-time quantitative RT-PCR. The HuC and HuD mRNAs increased almost 3-fold in the whole hippocampi (Fig. 2B). This result confirms our previous in situ hybridization experiments performed on the HuC gene in the context of the same spatial learning paradigm (19) and indicates a marked increase also in the expression of the HuD gene.
Fig. 2.
Increased expression of the ELAV-like RNA-binding proteins in rats learning a Morris WM spatial task. (A) (Upper) Representative Western blots obtained by using the 16A11 anti-Hu mouse monoclonal 16A11 antibody from the whole-cell lysate or after subcellular fractionation of the hippocampus. (Lower) Mean gray level ratios (means ± SEM) of the determinations of the ELAV-like immunoreactivity measured by Western blots (white bar, SW animals; black bar, TR animals) in the tested hippocampal fractions (n = 6 for each group; *, P < 0.05, **, P < 0.005 Student's t test). Data were normalized to α-tubulin signal. (B) Steady-state levels of HuB, HuC, and HuD mRNAs evaluated by external-standard based real-time quantitative RT-PCR. Measurements obtained from hippocampal total RNA preparations of SW and TR rats were normalized to the signal of GAPDH mRNA and expressed as means ± SEM (n = 6 for each group; **, P < 0.01; ***, P < 0.001 Student's t test).
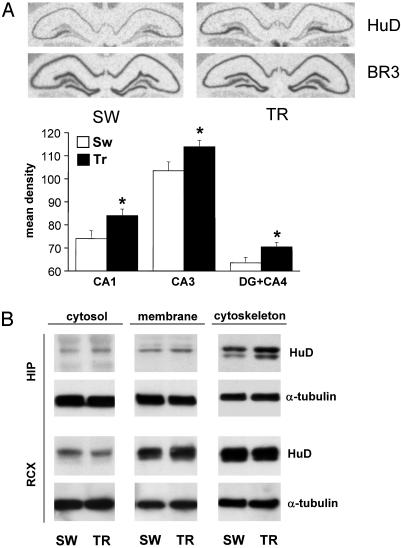
We therefore decided to assess specifically the profile of hippocampal HuD after rat WM learning. An in situ hybridization performed with the HuD probe (Fig. 3A) gave an independent indication of the increase in the steady-state levels of the mRNA of this gene in the three major hippocampal anatomical subregions (n = 6, Student's t test, P < 0.05 for all three determinations). Hybridization of adjacent brain slices from the same animals with a probe for the BRUNOL3 gene, belonging to the Bruno-like RNA-binding protein family (the closest to the ELAV-like family), showed no change in expression (see Fig. 3A; also confirmed by statistical analysis, CA1 P = 0.42; CA3 P = 0.95; DG+CA4 P = 0.10; n = 6). The availability of a HuD-specific monoclonal antibody allowed us to test HuD protein expression after spatial learning also at the protein level. The results are reported in Fig. 3B. Relative to α-tubulin immunoreactivity used for normalization, the cytosolic and the cytoskeletal fractions gave a significant increase of the HuD signal in the hippocampi of TR rats (relative signal units ± SEM; TR, 753.5 ± 62.3 vs. SW 558.5 ± 33.9, P < 0.05; and TR, 1,345.3 ± 32.1 vs. SW 968.3 ± 67.8, P < 0.005, in the cytosol and cytoskeletal fractions, respectively; n = 6, Student's t test). Therefore, the subcellular profile of the learning-induced increase in HuD immunoreactivity is analogous to that of the three neuronal ELAV-like proteins together. The signal obtained with purified nuclei was very faint; however, a significant increase of HuD protein was observed after training (relative to α-tubulin signal ± SEM; TR, 310 ± 4.0 vs. SW, 138 ± 22.5, P = 0.017, n = 3, not shown).
Fig. 3.
Specific up-regulation of the ELAV-like protein HuD, after Morris WM training, detected at the mRNA and protein levels. (A)(Upper) Representative in situ hybridization results reported for HuD and for a control phylogenetically related gene, BRUNOL3 (BR3), in the hippocampus of SW and TR rats. (Lower) Mean gray levels (±SEM) from autoradiograms of brain sections in the CA1, CA3, and DG+CA4 subfields for six animals in each group (white bar, SW; black bar, TR; *, P < 0.05, Student's t test). (B) Representative Western blots obtained by biochemical fractionation of protein lysates of the hippocampal (HIP) and retrosplenial cortex (RCX) brain regions. α-Tubulin levels for both regions are shown below and were used to normalize the data reported in the text; the statistical analysis is reported in the text.
To verify the regional specificity of learning-induced HuD upregulation for the hippocampal formation, we tested the retrosplenial cortex, a brain region suggested to be involved in path integration during spatial learning (33, 34). The results gave no statistically significant difference for the HuD protein in this area for any of the tested cellular fractions (see Fig. 3B; soluble P = 0.20, particulate P = 0.16, cytoskeletal fraction P = 0.34, n = 6).
To investigate the pattern of neuronal HuD, we performed fluorescent immunohistochemical staining for the HuD protein in the hippocampal slices. Fig. 4 shows the immunostaining pattern in the hippocampi of TR and SW animals in the CA1 (Fig. 4 A and B), CA3 (Fig. 4 C and D), and CA4 (Fig. 4 E and F) subregions. Fig. 4B Inset shows HuD staining in the proximal dendrites. In addition, HuD-positive cells are present in the DG granular cell layer after learning (Fig. 4F Inset).
Fig. 4.
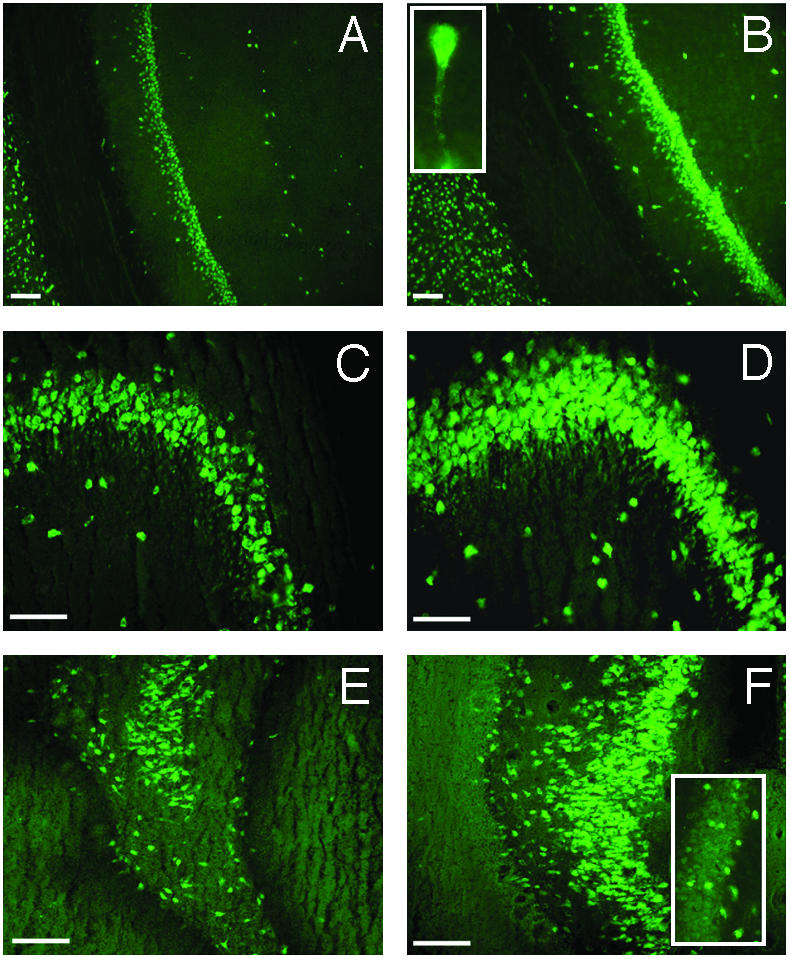
Expression of HuD protein in hippocampal pyramidal, polymorphic (hilar), and granular neurons after Morris WM training in rats. Fluorescence microscopy images showing HuD immunostaining in the CA1 (A and B), CA3 (C and D), and CA4 plus DG (E and F) hippocampal subregions from SW (A, C, and E) and TR (B, D, and F) rats. (Bars = 100 μm.) (B Inset) A pyramidal cell, at higher magnification, in the CA1 area from a TR rat; the staining in the proximal dendritic region is clearly evident. (F Inset) A detail of the granular cell layer in the DG region, at higher magnification, from a TR animal where it is possible to distinguish HuD-positive cells.
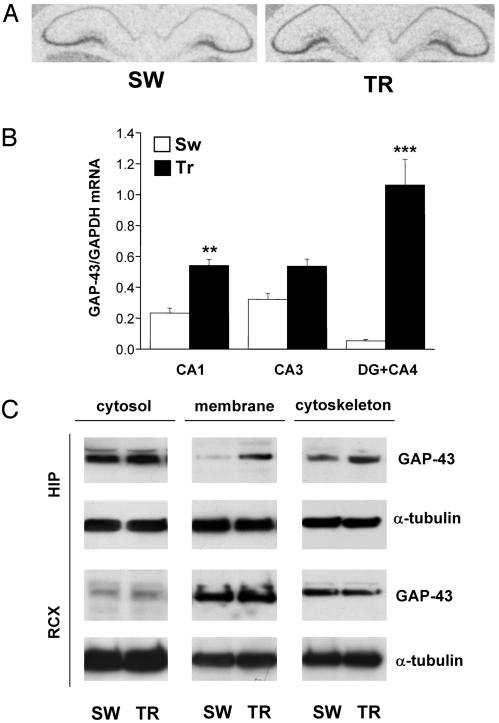
We next examined the effects of WM learning after 5 days of training on GAP-43 expression in the hippocampus at both mRNA and protein levels. In situ hybridization of rat hippocampi (Fig. 5A) showed an enhancement in the CA1 and CA3 subregions and a marked rise in the CA4+DG subregion (mean gray level ± SEM; CA1, TR 71.19 ± 2.76 vs. SW 60.83 ± 1.18, P < 0.005; CA3, TR 107.03 ± 2.69 vs. SW 99.01 ± 2.93, P = 0.06; CA4+DG, TR 71.99 ± 1.09 vs. SW 59.67 ± 0.88, P < 0.00001, n = 6) due essentially to the appearance of GAP-43 transcripts in the DG, a brain anatomical region where GAP-43 is normally not expressed (20). The quantitative RT-PCR results (Fig. 5B) confirm the in situ pattern, with a dramatic increase for GAP-43 expression in the TR animals in the CA4+DG area (≈20-fold). Looking at the GAP-43 protein levels in different cell compartments by Western blotting, we found a significant increase in the membrane-bound (TR, 1,810.3 ± 379.8 vs. SW, 690.5 ± 85.3; P < 0.05, n = 6) and cytoskeleton-associated (TR, 699.0 ± 72.4 vs. SW, 443.0 ± 24.7; P < 0.05, n = 6) protein for the TR animals. Nevertheless, in agreement with the results reported for HuD, no significant change was found for GAP-43 in the retrosplenial cortex, confirming the brain regional specificity of the observed learning-induced changes in mRNA and protein levels (see Fig. 5C; soluble P = 0.74, particulate P = 0.78, cytoskeletal fraction P = 0.34, n = 6).
Fig. 5.
Induction of GAP-43 expression by Morris WM training in the hippocampus, documented as mRNA and protein increase. (A) Representative in situ hybridization showing GAP-43 mRNA distribution in the hippocampus of SW and TR rats. (B) Determination of the steady-state relative levels of GAP-43 mRNA by external-standard based real-time quantitative RT-PCR in the CA1, CA3, and DG+CA4 subfields. Measurements obtained from total hippocampal RNA preparations of SW and TR rats were normalized to the signal of GAPDH mRNA and expressed as means ± SEM (n = six for each group; **, P < 0.005; ***, P < 0.0001, Student's t test). (C) Representative Western blots performed after fractionation of protein lysates of the hippocampal (HIP) and retrosplenial cortex (RCX) brain regions. α-Tubulin levels for both regions were used to normalize the data; the statistical analysis is reported in the text.
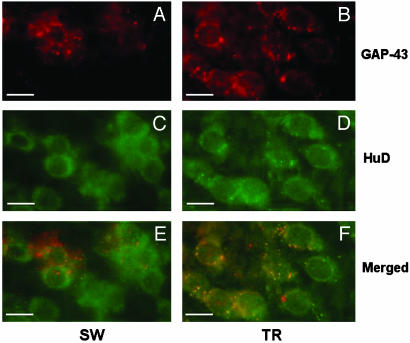
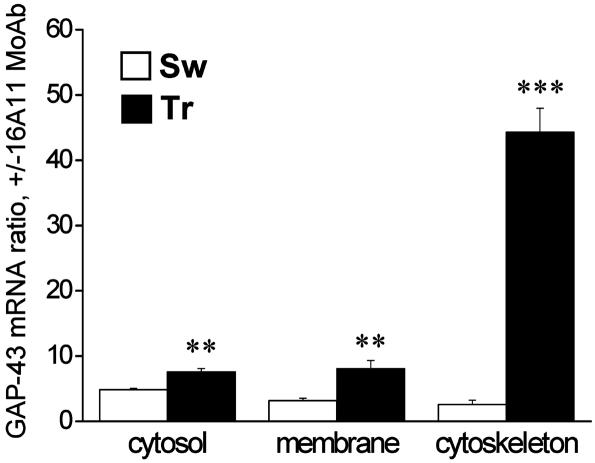
We then tested for cell distribution of HuD protein and GAP-43 mRNA in TR and SW animals to understand whether learning also coincided with an increased association between HuD protein and GAP-43 mRNA. Preliminary observations indicate this could be the case. Fig. 6, reporting a combined immunohistochemistry and in situ hybridization experiment on rat hippocampi (CA4 region), shows an increase in orange staining obtained by superimposition of the HuD and GAP-43 signals in the pyramidal cells of TR rats. To assess whether this increased overlapping of HuD protein and GAP-43 mRNA is at least partially the result of an increased physical association, we performed an immunoprecipitation with the 16A11 antibody on the same hippocampal fractions of SW and TR animals, and we quantified GAP-43 mRNA expression in the RNA extracted from the immunoprecipitated pellet. The results are reported in Fig. 7, expressed as the ratio between GAP-43 mRNA levels in the presence and in the absence (background signal) of MoAb 16A11, assumed to be an indirect measure of GAP-43 mRNA association to the Hu proteins. This association results in changes in the TR animals in all three fractions and remarkably in the cytoskeletal fraction, the interaction of GAP-43 mRNA with the Hu proteins appears to be enhanced >17-fold by learning. This result suggests that the reported learning-induced immunohistochemical increase in colocalization of HuD protein and GAP-43 mRNA (Fig. 6) is accompanied by increased binding and reaches its maximum in the cytoskeleton.
Fig. 6.
Hippocampal colocalization of GAP-43 mRNA and HuD protein after the Morris WM task. Fluorescence microscopy images taken in the CA4 region of the hippocampus of swimming SW (Left) and TR (Right) animals. (A and B) GAP-43 mRNA signal (red). (C and D) HuD protein signal (green). (E and F) Merged images (the orange color indicates overlapping of the two signals). (Bars = 50 μm.)
Fig. 7.
Increased coimmunoprecipitation of ELAV-like proteins and GAP-43 mRNA in the hippocampus after the Morris WM task. The levels of GAP-43 mRNA evaluated by external-standard based real-time quantitative RT-PCR were assessed in the pellet of hippocampal tissue fractions of SW and TR rats immunoprecipitated by the 16A11 MoAb. Values obtained in the presence of the antibody were normalized with the corresponding background values measured in the immunoprecipitation pellet in the absence of any antibody. Data are expressed as means ± SEM (n = 4 for each group; **, P < 0.005; ***, P < 0.001 Student's t test).
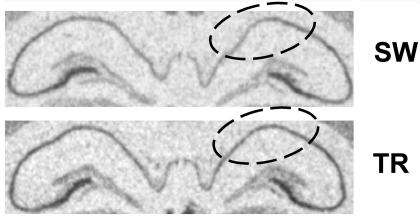
We finally assessed the involvement of HuD and GAP-43 in long-term memory retention by the analyses of a group of TR and SW animals 1 month after the last WM training session, with the evaluation of the mRNA (by in situ hybridization) and protein (by Western blotting) levels of HuD and GAP-43. Our results indicate that increased protein levels of HuD and GAP-43 in TR animals disappear in the hippocampus 1 month after the initial learning (see Table 1). Nevertheless, there was a statistically significant persistence (even after 1 month) of the increase in the HuD mRNA in the CA1 region (see Table 1 and Fig. 8).
Table 1. Expression of HuD and GAP-43 messenger and protein levels 1 mo after water maze spatial learning in rat hippocampus.
| Type of experiment | HuD | GAP-43 |
|---|---|---|
| In situ data | ||
| Hippocampal area | ||
| CA1 | Sw 74.07 ± 2.35 | Sw 70.78 ± 1.44 |
| Tr 81.67 ± 2.69* | Tr 72.33 ± 1.42 | |
| CA3 | Sw 103.80 ± 3.62 | Sw 119.56 ± 2.20 |
| Tr 112.53 ± 3.20 | Tr 120.53 ± 1.69 | |
| CA4+DG | Sw 64.78 ± 1.49 | Sw 64.58 ± 2.29 |
| Tr 64.14 ± 1.36 | Tr 64.92 ± 1.25 | |
| Western blot data | ||
| Cellular fraction | ||
| Soluble | Sw 600.24 ± 101.06 | Sw 680.07 ± 152.48 |
| Tr 590.67 ± 40.95 | Tr 776.43 ± 198 | |
| Membrane | Sw 803.75 ± 156.04 | Sw 996.00 ± 227.19 |
| Tr 905.75 ± 104.30 | Tr 1,149.21 ± 339.29 | |
| Cytoskeleton | Sw 1,011.12 ± 123.40 | Sw 648.75 ± 72.62 |
| Tr 1,055.03 ± 156.90 | Tr 817.18 ± 221.48 |
In situ data show mean gray levels (±SEM) from autoradiograms of the CA1, CA3, and DG+CA4 subfields in Sw and Tr animals. Western blot data show normalized levels of HuD and GAP-43 proteins. Western blots were performed on different fractions from hippocampi of SW and TR rats. Measurements were normalized to α-tubulin levels. n = six animals for each group. *, P < 0.05, Student's t test.
Fig. 8.
Increased expression of HuD mRNA in rat hippocampus 1 mo after WM spatial learning. Shown are representative in situ hybridization images reported for HuD in the hippocampus of swimming SW and TR rats. The circled area corresponds to the CA1 regions.
Discussion
Despite a large body of evidence pointing to the importance of posttranscriptional control of gene expression in the differentiation and physiology of mature neurons, the mechanisms underlying this control are almost entirely unknown. By in vitro experiments in differentiated neurons, dendrites have been shown to be targeted by a subset of cellular mRNAs and have been formally demonstrated to be the site of protein synthesis (for review, see ref. 35). Translational competence points to a possible role of confined pathways controlling local translation as a result of synaptic coincidence, thereby providing a possible biochemical basis for the learning-induced changes in synaptic strength that could underlie memory representation (36). Nevertheless, whereas candidate target mRNAs for this activity have been proposed (37), up to now neither related proteins nor biochemical pathways have been identified.
Recently we provided the first evidence for such a pathway involving posttranscriptional regulation of gene expression and the ELAV-like proteins in memory acquisition. We demonstrated a relationship between hippocampal up-regulation of the expression of the three neuronal HuB, HuC, and HuD genes and learning, after a radial arm maze task in the mouse (19). Here we confirmed this ELAV-like up-regulation in another mammalian spatial learning paradigm, the rat WM, showing similarly to the previous results that the ELAV-like proteins increase is associated with the cytoskeletal and to a lesser extent with the cytosolic cell component (Fig. 2). These results extend the previous findings to another species and another learning paradigm, strongly suggesting the generality of the involvement of ELAV-like proteins in spatial memory formation.
Of the three ELAV-like proteins, HuD is strictly expressed in neurons and is developmentally regulated, appearing in coincidence with the early stages of neuronal differentiation in chicken, mouse, and rat brain (3, 22, 23). A causal role for HuD in neuronal development has been also clearly established by inducing ectopic changes in HuD expression levels in PC12 cells and observing corresponding changes in neurite sprouting activity (16, 24, 25). With maturity, high levels of HuD are found to persist especially in pyramidal-like neurons of the hippocampus and of the cortex (ref. 23 and the present findings), suggesting a postdevelopmental specialized role in these neurons. Based on our previous results, we hypothesized that in the hippocampus, this role could be related to the memory-encoding phase. We therefore focused here on HuD and showed that the learning-induced increase in the HuD mRNA level by in situ hybridization involved the whole hippocampal pyramidal cell layer, from the CA1 to the CA4 regions. The increase in protein content was much more prominent in the cytoskeletal fraction as assessed by Western blotting. The association of ELAV-like proteins with the cytoskeleton has been already documented in vitro for HuB, suggesting a model in which HuB binds to microtubules and to the target mRNAs in messenger ribonucleoprotein particles which, in turn, associate with polysomes to form a translationally competent complex connected to the cytoskeletal network (38). Following this model, the increased cytoskeletal association we observed for HuD after learning could be interpreted as an engagement of the complex containing HuD and the bound mRNAs for translation. In addition, in mammals, HuD is also able to self-aggregate in dimers and trimers that associate with bound mRNAs or with other ELAV-like proteins to form multimeric complexes (39).
Among the HuD-bound mRNAs, GAP-43 mRNA is by far the most well characterized with respect to HuD activity. Several lines of evidence collected from in vitro experiments indicate the primary role of HuD in the posttranscriptional regulation of GAP-43 (20). HuD binds to the 3′ UTR of GAP-43 mRNA in a specific mapped site (28–30) and increases its half-life by a mechanism dependent on its affinity for the GAP-43 mRNA probably regulated by the length of the poly(A) tail (30). Consistent with this role, GAP-43 and HuD expression overlap in many sites of the mature brain (ref. 20 and this report), including the hippocampus. A learning-dependent increase in GAP-43 expression has been found previously by us (19) after the demonstration of an enhancement of memory performance in mice overexpressing GAP-43 in the brain (40). Because GAP-43 is a well known neuron-specific key controller of neural development, neural regeneration, and synaptic plasticity (41, 42), it could likely represent a HuD downstream gene in a pathway of regulation of gene expression elicited by behavioral training. In our learning model, we found a GAP-43 increase, at mRNA and protein levels, in the hippocampus. GAP-43 is basically absent in the dentate gyrus (ref. 20 and the present work), whereas the HuD mRNA signal is detected. Interestingly, after learning, we observed both GAP-43 mRNA and HuD protein in the granular cells, where they could not be previously detected. Our observations support previous findings (20) showing that, in this hippocampal region, GAP-43 expression can be at least partially controlled by a posttranscriptional mechanism involving HuD, although a concomitant transcriptional contribution cannot be excluded. Moreover, preliminary combined experiments on HuD protein and GAP-43 mRNA suggest a learning-associated increase in their colocalization (Fig. 6), paralleled by a learning-associated increase in coimmunoprecipitation of ELAV-like proteins with GAP-43 mRNA, especially in the cytoskeletal fraction (Fig. 7). These findings are consistent with our previous observation (19) of the in vivo binding of ELAV-like proteins to GAP-43 mRNA and can be interpreted as an increase of this binding promoted by spatial learning. The specificity of the HuD/GAP-43 pathway in memory formation seems to have spatiotemporal dimensions. The selectivity for the hippocampal area is shown by the absence of HuD activation in the retrosplenial cortex, with the concomitant absence of change in GAP-43 expression. During long-term memory retention, 1 month after memory acquisition, the increase in HuD and GAP-43 protein is no longer observable. However, the persistence of elevated HuD messenger in the CA1 region 1 month after learning does suggest that HuD may play a role in long-term memory storage and in a more rapid activation of the system in response to a recalling stimulus and subsequent consolidation of reactivated memory.
Learning-induced up-regulation of HuD and GAP-43 mRNA in mature hippocampal neurons suggests that the HuD protein can exert posttranscriptional control of gene expression through upstream molecular signals that could be selectively activated in neuronal microdomains during the encoding of a specific memory event.
Acknowledgments
We are very grateful to Dr. Umberto Laforenza (Istituto di Fisiologia Umana, Pavia, Italy) for kind help with cell fractioning and Dr. Henry Furneaux (University of Connecticut Health Center, Farmington, CT) for his helpful discussion and kind gift of the HuD antibody.
Abbreviations: WM, water maze; TR, trained rats; SW, swimming rats.
References
- 1.Robinow, S. & White, K. (1991) J. Neurobiol. 22, 443–461. [DOI] [PubMed] [Google Scholar]
- 2.Good, P. J. (1995) Proc. Natl. Acad. Sci. USA 92, 4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakamatsu, Y. & Weston, J. A. (1997) Development (Cambridge, U.K.) 124, 3449–3460. [DOI] [PubMed] [Google Scholar]
- 4.Birney, E., Kumar, S. & Krainer, A. R. (1993) Nucleic Acids Res. 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinow, S., Campos, A. R., Yao, K. M. & White, K. (1988) Science 242, 1570–1572. [DOI] [PubMed] [Google Scholar]
- 6.Levine, T. D., Gao, F., King, P. H., Andrews, L. G. & Keene, J. D. (1993) Mol. Cell. Biol. 13, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu, J., Dalmau, J., Szabo, A., Rosenfeld, M., Huber, J. & Furneaux, H. (1995) Neurology 45, 544–550. [DOI] [PubMed] [Google Scholar]
- 8.Abe, R., Sakashita, E., Yamamoto, K. & Sakamoto, H. (1996) Nucleic Acids Res. 24, 4895–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma, W. J., Chung, S. & Furneaux, H. (1997) Nucleic Acids Res. 25, 3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lisbin, M. J., Qiu, J. & White, K. (2001) Genes Dev. 15, 2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koushika, S. P., Lisbin, M. J. & White, K. (1996) Curr. Biol. 6, 1634–1641. [DOI] [PubMed] [Google Scholar]
- 12.Koushika, S. P., Soller, M. & White, K. (2000) Mol. Cell. Biol. 20, 1836–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan, X. C. & Steitz, J. A. (1998) EMBO J. 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antic, D., Lu, N. & Keene, J. D. (1999) Genes Dev. 13, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manohar, C. F., Short, M. L., Nguyen, A., Nguyen, N. N., Chagnovich, D., Yang, Q. & Cohn, S. L. (2002) J. Biol. Chem. 277, 1967–1973. [DOI] [PubMed] [Google Scholar]
- 16.Anderson, K. D., Morin, M. A., Beckel-Mitchener, A., Mobarak, C. D., Neve, R. L., Furneaux, H. M., Burry, R. & Perrone-Bizzozero, N. I. (2000) J. Neurochem. 75, 1103–1114. [DOI] [PubMed] [Google Scholar]
- 17.Akamatsu, W., Okano, H. J., Osumi, N., Inoue, T., Nakamura, S., Sakakibara, S., Miura, M., Matsuo, N., Darnell, R. B. & Okano, H. (1999) Proc. Natl. Acad. Sci. USA 96, 9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao, K. M., Samson, M. L., Reeves, R. & White, K. (1993) J. Neurobiol. 24, 723–739. [DOI] [PubMed] [Google Scholar]
- 19.Quattrone, A., Pascale, A., Nogues, X., Zhao, W., Gusev, P., Pacini, A. & Alkon, D. L. (2001) Proc. Natl. Acad. Sci. USA 98, 11668–11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrone-Bizzozero, N. & Bolognani, F. (2002) J. Neurosci. Res. 68, 121–126. [DOI] [PubMed] [Google Scholar]
- 21.Benowitz, L. I. & Routtenberg, A. (1997) Trends Neurosci. 20, 84–91. [DOI] [PubMed] [Google Scholar]
- 22.Okano, H. J. & Darnell, R. B. (1997) J. Neurosci. 17, 3024–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clayton, G. H., Perez, G. M., Smith, R. L. & Owens, G. C. (1998) Brain Res. Dev. Brain Res. 109, 271–280. [DOI] [PubMed] [Google Scholar]
- 24.Mobarak, C. D., Anderson, K. D, Morin M., Beckel-Mitchener, A., Rogers, S. L., Furneaux, H., King, P. & Perrone-Bizzozero, N. I. (2000) Mol. Biol. Cell. 11, 3191–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson, K. D., Sengupta, J., Morin, M., Neve, R. L., Valenzuela, C. F. & Perrone-Bizzozero, N. I. (2001) Exp. Neurol. 168, 250–258. [DOI] [PubMed] [Google Scholar]
- 26.Kasashima, K., Terashima, K., Yamamoto, K., Sakashita, E. & Sakamoto, H. (1999) Genes Cells 4, 667–683. [DOI] [PubMed] [Google Scholar]
- 27.Aranda-Abreu, G. E., Behar, L., Chung, S., Furneaux, H. & Ginzburg, I. (1999) J. Neurosci. 19, 6907–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung, S., Eckrich, M., Perrone-Bizzozero, N., Kohn, D. T. & Furneaux, H. (1997) J. Biol. Chem. 272, 6593–6598. [DOI] [PubMed] [Google Scholar]
- 29.Tsai, K. C., Cansino, V. V., Kohn, D. T., Neve, R. L. & Perrone-Bizzozero, N. I. (1997) J. Neurosci. 17, 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckel-Mitchener, A. C., Miera, A., Keller, R. & Perrone-Bizzozero, N. I. (2002) J. Biol. Chem. 277, 27996–28002. [DOI] [PubMed] [Google Scholar]
- 31.Burgess, N., Maguire, E. A. & O'Keefe, J. (2002) Neuron 35, 625–641. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, W., Chen, H., Xu, H., Moore, E., Meiri, N., Quon, M. J. & Alkon, D. L. (1999) J. Biol. Chem. 274, 34893–34902. [DOI] [PubMed] [Google Scholar]
- 33.Cooper, B. G., Manka, T. F. & Mizumori, S. J. (2001) Behav. Neurosci. 115, 1012–1028. [DOI] [PubMed] [Google Scholar]
- 34.Cooper, B. G. & Mizumori, S. J. (2001) J. Neurosci. 21, 3986–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberwine, J., Miyashiro, K., Kacharmina, J. E. & Job, C. (2001) Proc. Natl. Acad. Sci. USA 98, 7080–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter, J. D. & Lorenz, L. J. (2002) Curr. Opin. Neurobiol. 12, 300–304. [DOI] [PubMed] [Google Scholar]
- 37.Wittenberg, G. M. & Tsien, J. Z. (2002) Trends Neurosci. 25, 501–505. [DOI] [PubMed] [Google Scholar]
- 38.Antic, D. & Keene, J. D. (1998) J. Cell Sci. 111, 183–197. [DOI] [PubMed] [Google Scholar]
- 39.Kasashima, K., Sakashita, E., Saito, K. & Sakamoto, H. (2002) Nucleic Acids Res. 30, 4519–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Routtenberg, A., Cantallops, I., Zaffuto, S., Serrano, P. & Namgung, U. (2000) Proc. Natl. Acad. Sci. USA 97, 7657–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aarts, L. H., Schotman, P., Verhaagen, J., Schrama, L. H. & Gispen, W. H. (1998) Adv. Exp. Med. Biol. 446, 85–106. [DOI] [PubMed] [Google Scholar]
- 42.Aarts, L. H., Verkade, P., van Dalen, J. J., van Rozen, A. J., Gispen, W. H., Schrama, L. H. & Schotman, P. (1999) Mol. Cell Neurosci. 14, 85–97. [DOI] [PubMed] [Google Scholar]