Abstract
Functional characterization of protein interactions in mammalian systems has been hindered by the inability to perform complementation analyses in vivo. Here, we use functional replacement of the vesicle docking protein p115 to separate its essential from its nonessential interactions. p115 is required for biogenesis of the Golgi apparatus, but it is unclear whether its mechanism of action requires its golgin and/or SNARE interactions. Short interfering RNA-mediated knockdown of p115 induced extensive Golgi fragmentation and impaired secretory traffic. Reassembly of a structurally and functionally normal Golgi occurred on expression of a p115 homologue not recognized by the short interfering RNA. Strikingly, versions of p115 lacking its phosphorylation site and the golgin-binding domains also restored the Golgi apparatus in cells lacking endogenous p115. In contrast, the p115 SNARE-interacting domain was required for Golgi biogenesis. This suggests that p115 acts directly, rather than via a tether, to catalyze trans-SNARE complex formation preceding membrane fusion.
Keywords: Golgi apparatus, RNA interference, giantin, GM130
Genome-wide protein interaction studies and other large scale approaches are rapidly expanding the number of interactions attributed to particular gene products. This places a premium on establishing in vivo procedures that validate and determine the function of putative interactions. Gene replacement, or complementation, can be invaluable in testing the importance of a given interaction by determining the consequence of replacing the endogenous gene with a version mutated such that it is specifically defective for the interaction in question. In mammalian cells, this approach has had limited use because of the difficulty of inhibiting endogenous genes. However, the advent of RNA interference (1), which effectively knocks down expression of any gene of choice, suggests that mammalian gene replacement is poised to become an important approach.
The vesicle docking protein p115 provides a significant example of a protein exhibiting multiple interactions of uncertain functional importance. p115 is essential for biogenesis of the Golgi apparatus (2–4). Part of its mechanism of action may be to tether transport vesicles to Golgi stacks at considerable distances by simultaneous binding of two highly elongated golgins: vesicle-localized giantin and Golgi-localized GM130 (5, 6). Phosphorylation of p115 enhances this binding (6), whereas phosphorylation of GM130 inhibits it (7). During mitosis, p115 is dephosphorylated by an unknown phosphatase (8), whereas GM130 is phosphorylated by the mitotic kinase Cdc2 (9). This suggests that the vesiculation of the Golgi apparatus that occurs during cell division may be caused by inhibition of p115 tethering such that Golgi vesicles form but do not dock and fuse (7). In addition to golgins, p115 directly interacts with several endoplasmic reticulum (ER)- and Golgi-localized SNARE proteins including syntaxin5, gos28, membrin, sly1, and rbet1 (10, 11) and can enhance SNARE complex formation in vitro (11). Thus, p115 may mediate vesicle fusion during Golgi biogenesis in two sequential reactions, initially forming a mitotically regulated giantin-p115-GM130 tether, and later mediating pairing of SNAREs (11).
Surprisingly, however, in vivo tests of the p115/tether interaction have generated controversy concerning its role in Golgi biogenesis and mitotic Golgi disassembly, and in vivo tests of the role of p115/SNARE interactions have not been reported. Several observations suggest that interactions of p115 with giantin and GM130 are not required. Microinjection of peptide and antibody inhibitors directed against GM130 or giantin neither cause Golgi disassembly nor prevent reassembly (3, 12). Also, even in cells lacking detectable GM130 because of an unknown genetic lesion, the Golgi apparatus is present (13). Because Golgi biogenesis depends on interactions of p115 in vivo, this raises two critical questions. First, is the role of p115 in vesicle tethering truly dispensable in vivo, or, as many believe, was previous work misleading because of insufficient inhibition? Second, if the role of p115 in vesicle tethering is dispensable, what is its essential role? It may be p115 binding to SNAREs that is required, but given that general models of SNARE-mediated fusion do not include an activity similar to that proposed for p115, it remains critical to test whether p115/SNARE interactions are required in vivo for ER-to-Golgi transport and Golgi biogenesis.
Materials and Methods
Constructs and Reagents. Bovine p115 cDNA and the phosphorylation mutant S941A were provided by V. Malhotra (University of California at San Diego, La Jolla). Mutant versions of p115 were generated either by restriction digests or by using the QuikChange site-directed mutagenesis kit (Stratagene). Constructs were confirmed by restriction analyses and sequencing. For RNA interference, p115 was targeted by using the sequence AAGACCGGCAATTGTAGTACT. Both p115 and control short interfering RNAs (siRNAs) for transfection were either generated by using the Silencer siRNA construction kit (Ambion, Austin, TX) or purchased from Xeragon (Valencia, CA).
Cell Culture and RNA Interference Treatment. HeLa cells were cultured on glass coverslips in MEM (Invitrogen) supplemented with 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin at 37°C in a 5% CO2 incubator. Culture medium for HeLa stably expressing GalNacT2-GFP (kindly provided by J. White, Cancer Research Center, Massachusetts General Hospital, Charlestown) contained 0.5 mg/ml G418 (Invitrogen). siRNA at a final concentration of 30 nM was transfected into cells with Oligofectamine (Invitrogen) by using published guidelines (14). For immunoblotting, cells were plated in 24-well dishes, treated with siRNA for various times, and lysed in a buffer containing 1% SDS, 50 mM Tris (pH 6.8), and protease inhibitors.
Microinjection and Immunofluorescence Analysis. Cells treated with siRNA for 90 h were microinjected by using the conditions described (3). For nuclear injections, corresponding plasmids at 100 ng/μl in water were used. After desired incubation times, cells were fixed in methanol at –20°C for 10 min and blocked in PBS containing 2.5% calf serum and 0.1% Tween 20 (Fisher Scientific). Rabbit polyclonal anti-p115 and anti-GM130, and mouse monoclonal anti-GPP130, anti-giantin, and anti-p115 (8A6) were used as described (3, 15). Rhodamine- and Cy5-conjugated goat secondary antibodies were purchased (Zymed). Cells were analyzed by using a Deltavision restoration microscopy system (Applied Precision, Issaquah, WA) and a ×60 [numerical aperture (n.a.) 1.4] PlanApo objective (Zeiss) unless noted.
Vesicular Stomatitis Virus Glycoprotein (VSVG) Trafficking Assay. HeLa cells treated with siRNA were microinjected with a plasmid encoding ts045 VSVG-GFP cDNA, either alone or with constructs encoding p115, and incubated at 40°C for 5 h. The cells were then shifted to 32°C for 0, 20, or 60 min. For determination of surface VSVG, live cells were stained with an antibody (8G5) directed against the luminal domain of VSVG (16) for 10 min at 4°C, washed extensively, fixed, and stained by using Cy5-conjugated secondary antibodies. Cells were analyzed by using a standard fluorescence microscope (Nikon) equipped with a Hamamatsu black-and-white cooled charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan). Digital images in Cy5 and GFP channel were acquired in photoshop (Adobe Systems, Mountain View, CA) by using a ×40 objective (n.a. 1.3). The images were merged, and the mean fluorescence value in each channel was measured by manually selecting the whole cell. Average pixel values in areas of the coverslip lacking expressing cells were taken as background fluorescence and subtracted. The ratio of surface-to-total fluorescence was used to calculate the extent of VSVG transport.
Results and Discussion
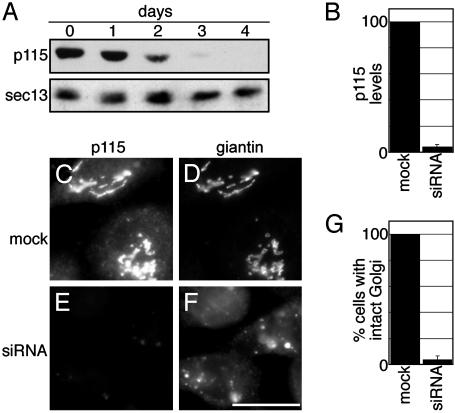
We reasoned that the questions raised above could be answered by functional replacement of p115 in living cells, using RNA interference inhibition followed by expression of mutant versions of p115 specifically defective for binding to either tether or SNARE components. Therefore, the role of p115 was first confirmed by using siRNA-mediated knockdown. Cultured HeLa cells were transfected with siRNA oligonucleotides designed to be specific against human p115. Immunoblotting showed time-dependent loss of p115 in cells treated with siRNA for 24, 48, 72, or 96 h (Fig. 1A). At 96 h posttransfection, expression was reduced by 96% compared to mock transfectants (Fig. 1B). p115 is normally present on Golgi and ER-Golgi intermediate compartment (ERGIC) membranes (17). A parallel analysis at the 96-h time point by immunofluorescence indicated that loss of detectable p115 expression was observed in 95% of the cells. This implies that the cells lacking staining by immunofluorescence were essentially depleted of p115. In contrast to mock-transfected cells (Fig. 1 C and D) or control siRNA-transfected cells (not shown), all of the cells that showed depletion of p115 showed a fragmented Golgi apparatus (Fig. 1 E–G). Golgi fragmentation was confirmed by using a variety of Golgi markers including GPP130, giantin, GM130, and GalNAcT2. Golgi proteins were present in both dispersed membranes and membranes that costained with the ERGIC marker ERGIC53, suggesting redistribution and accumulation of the Golgi in the ERGIC (not shown). Microtubule organization was normal, arguing against microtubule destabilization as the cause of Golgi fragmentation (not shown). Thus, as expected, a requirement for p115 in maintenance of Golgi structure was revealed by using RNA interference.
Fig. 1.
Depletion of p115 by siRNA induces Golgi fragmentation. HeLa cells stably expressing GalNacT2-GFP were treated with p115 siRNA for the indicated times. p115 and sec13 (as a control) were detected by immunoblotting (A). Average levels of p115 (±SD, n = 4) after 96 h of siRNA treatment were determined by quantitative immunoblot and normalized to mock-transfected cells (B). After 96 h, mock or siRNA-treated cells were stained for p115 to detect knockdown, and the Golgi was visualized in the same cells by using giantin staining (C–F, as labeled). Cells were imaged by using a standard fluorescence microscope and a ×40 objective. Average percentages (±SD, n = 2, >90 cells each) of mock-transfected and siRNA-treated cells exhibiting a normal Golgi apparatus were calculated (G). (Bar = 10 μm.)
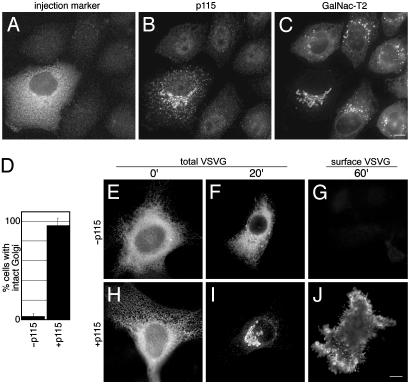
Next, we attempted to functionally complement the Golgi fragmentation phenotype by expressing exogenous bovine p115 in cells depleted of endogenous p115. Bovine p115 was chosen, as it is 97% identical to human p115, yet it contained silent sequence mismatches in three base-pair positions corresponding to the selected siRNA. Cells treated with siRNA for 90 h were microinjected with bovine p115 cDNA and further incubated. By 5 h postinjection, cells labeled with the injection marker (Fig. 2A) exhibited a restored p115 localization on Golgi and ERGIC membranes (Fig. 2B) and a reassembled Golgi apparatus (Fig. 2C). Indeed, 96% of the cells expressing the transgene reassembled a normal Golgi apparatus (Fig. 2D). Noninjected or control injected cells showed no detectable p115 and no rescue of the fragmentation phenotype.
Fig. 2.
Replacement of human p115 with bovine p115 restores a functional Golgi apparatus. After 90 h, siRNA-treated HeLa-T2GFP cells were microinjected with bovine p115 cDNA with a fluorescent injection marker. After a 5-h incubation, the cells were stained for p115. Injected and noninjected cells were distinguished in the rhodamine channel (A), the levels of endogenous and exogenous p115 were determined in the Cy5 channel (B), and Golgi integrity was assessed by visualizing GalNAcT2-GFP (C). Average percentages of control-injected (n = 2, 120 cells) or p115-expressing siRNA-treated cells (n = 6, 139 cells) exhibiting an intact Golgi ribbon are presented (D). siRNA-treated HeLa cells were injected with either VSVG-GFP cDNA alone (–p115) or VSVG-GFP in conjunction with p115 cDNA (+p115), kept at 40°C for 5 h, and transferred to 32°C for 0, 20, or 60 min (E–J, as labeled). As indicated, VSVG-GFP or surface VSVG staining is shown; the cells were also stained to confirm exogenous and endogenous p115 levels (not shown). All images were acquired by using an API Deltavision system with a ×60 objective and deconvolved. (Bar = 10 μm.)
To assess the functionality of the restored Golgi apparatus, we measured trafficking of a temperature-sensitive version of VSVG tagged with GFP (VSVG-GFP). At 40°C, VSVG-GFP is misfolded and retained in the ER. Shifting cells to 32°C leads to correct folding and concerted trafficking of the protein through the Golgi apparatus to the plasma membrane (18, 19). Cells depleted of endogenous p115 were microinjected with VSVG-GFP cDNA, either alone or in combination with bovine p115 cDNA. Cells were then kept at 40°C for 5 h and shifted to 32°C for various times. In the absence of exogenous p115, cells depleted of endogenous p115 exhibited severely impaired VSVG trafficking. Whereas control cells yielded VSVG-GFP in the ER at 0 min, in the Golgi at 20 min, and on the surface at 60 min (not shown), p115-depleted cells yielded only a minor fraction of VSVG-GFP trafficking out of the ER at 20 min and no detectable surface staining at 60 min (Fig. 2 E–G). Not surprisingly, the VSVG-GFP appeared to accumulate in punctate structures that, based on costaining with giantin (not shown), corresponded to the Golgi fragments identified above as ERGIC. This suggests that cargo traffic is blocked at an early step in the absence of p115, which confirms in vivo the role of p115 in ER-to-Golgi traffic (2, 10). Although profound, the block appeared to be kinetic, rather than absolute, as some VSVG was detected on the surface after longer incubations (not shown). This may explain why previous work carried out in cultured Drosophila cells by using longer incubations failed to reveal a role for p115 in trafficking (4). Nevertheless, our findings are consistent with the previous work, in that unlike Golgi biogenesis, trafficking may persist, albeit at an impaired level, in the absence of p115.
Exogenous full-length p115 completely reversed the traffic block induced by p115 depletion. In cells coinjected with VSVG-GFP and bovine p115 cDNA, VSVG-GFP was efficiently transported at 32°C from the ER at 0 min (Fig. 2H) to the Golgi apparatus by 20 min (Fig. 2I) and to the cell surface by 60 min (Fig. 2 J). Restored transport was further confirmed by quantitative analysis of surface to total VSVG-GFP ratios (see Fig. 4). Thus, bovine p115 functionally complemented human p115 causing reassembly of a normal-appearing and functionally competent Golgi apparatus from dispersed membrane fragments.
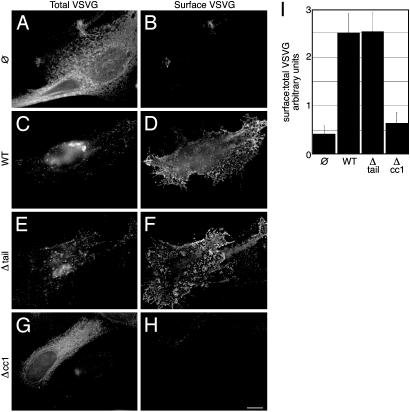
Fig. 4.
p115 requires its SNARE-binding domain, but not its golgin-binding domain, to support secretory traffic. siRNA-treated cells (90 h) were injected with cDNA for VSVG-GFP alone (A and B) or VSVG-GFP in conjunction with wild-type p115 (C and D), Δtail (E and F), or Δcc1 (G and H). Cells were incubated at 40°C for 5 h and transferred to 32°C for 60 min. Total and surface VSVG-GFP were visualized in the same cells by using deconvolution microscopy (A–H, as labeled). Surface-staining was carried out by using anti-VSVG antibodies at 4°C before fixation. The ratio of surface to total fluorescence was estimated as a measure of forward traffic (I). (Bar = 10 μm.)
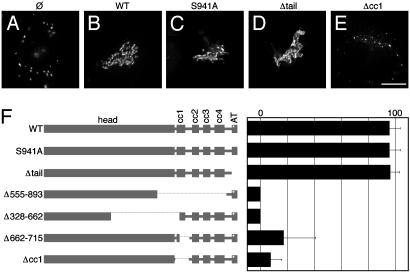
Homodimeric p115 contains two N-terminal globular head domains linked by a central coiled-coil domain (20). The extreme C termini form an acidic tail domain that mediates p115 binding to giantin and GM130 (21, 22). The serine residue that is phosphorylated is also located at the acidic tail. To test whether the phosphorylation site and/or the tether-forming acidic tail domain of exogenous p115 was required to rescue the Golgi apparatus in p115-depleted cells, we used two mutant versions of p115: one that cannot be phosphorylated because of a serine-to-alanine conversion at its phosphorylation site (S941A) and another that was truncated just before the start of the acidic tail (Δtail). Strikingly, neither abolition of phosphorylation nor deletion of the acidic tail impaired the ability of the exogenous version to replace endogenous p115. Unlike the disassembled Golgi complex in noninjected cells (Fig. 3A), and in common with intact Golgi ribbon in wild-type injected cells (Fig. 3B), cells expressing either S941A (Fig. 3C) or Δtail (Fig. 3D) showed complete reassembly of the Golgi apparatus. Similar to wild type, rescue was robust for both mutated p115 versions, with ≈96% of cells showing restored Golgi complexes (Fig. 3F). In addition, both mutated constructs localized to the Golgi and the ERGIC in a pattern indistinguishable from endogenous or exogenous full-length p115 (not shown). These results indicate that p115 binding to giantin and GM130 is not required for Golgi assembly in vivo and that inhibition of giantin-p115-GM130 interactions cannot explain mitotic Golgi disassembly.
Fig. 3.
p115-mediated Golgi reassembly requires its SNARE-binding domain but not its phosphorylation site or its golgin-binding domain. siRNA-treated cells (90 h) were injected with cDNA for wild-type p115, S941A, Δtail, or Δcc1, and the Golgi was imaged after5hby using GalNAcT2-GFP (A–E, as labeled). The cells were also stained to confirm exogenous and endogenous p115 levels (not shown). The p115 constructs indicated by name (numbers refer to amino acids) and in the schematic (F) were expressed by microinjection into siRNA-treated cells. The Golgi was imaged by using deconvolution microscopy after 5 h with GalNAcT2-GFP. Exogenous and endogenous p115 levels were confirmed by p115 staining. Average percentages of cells showing intact Golgi are shown (±SD, n > 3). (Bar = 10 μm.)
The SNARE-binding domain on p115 has been mapped to coiled-coil domain 1 (cc1), an N-terminal subdomain of the entire p115 coiled-coil domain (11). Hence, constructs lacking the entire coiled-coil domain, the entire cc1 subdomain (Δcc1), the distal part of cc1, or the proximal part of cc1 were generated and tested for their capacity to replace endogenous p115 in vivo. Expression of Δcc1 failed to restore the Golgi apparatus in cells depleted of endogenous p115 (Fig. 3E). Indeed, deletion of any part of the cc1 domain significantly diminished the capacity of the exogenous version to replace endogenous p115 (Fig. 3F). Thus, the p115 SNARE-binding domain is essential for Golgi biogenesis. Not surprisingly, the presence of the acidic tail did not confer any capacity to rescue the phenotype in the absence of the SNARE-binding domain. Importantly, as evidence against dominant negative effects and in support of stable folding, all of the constructs tested colocalized with Golgi markers when expressed in both untreated and siRNA-treated cells. In the case of untreated cells, the constructs were identified by using antibodies specific for bovine p115 (except Δ555–893, which was not recognized by this antibody). These results are consistent with previous work demonstrating multiple p115 domains sufficient to mediate its Golgi localization (17).
The VSVG-GFP trafficking assay described above was used to confirm that Δtail, but not Δcc1, restored trafficking-competent Golgi complexes. For each coinjected cell after 60 min at 32°C, the total VSVG-GFP fluorescence pattern was compared to the VSVG-GFP cell-surface pattern as determined by staining live cells with an antibody against the extracellular domain of VSVG. In p115-depleted cells, only minimal amounts of VSVG were detected on the surface (Fig. 4 A and B). In contrast, siRNA-treated cells expressing wild-type p115 showed strong surface expression (Fig. 4 C and D). Significantly, deletion of the acidic tail did not affect the rescue of surface traffic by exogenous p115, as cells expressing Δtail efficiently transported VSVG to the surface (Fig. 4 E and F). In contrast, deletion of the SNARE-interacting domain significantly diminished the capacity of exogenous p115 to rescue surface traffic (Fig. 4 G and H). To quantify these results, the ratio of surface to total fluorescence was determined. Cells expressing Δtail transported VSVG to the surface at a rate indistinguishable from those expressing wild-type p115, whereas surface traffic in cells expressing Δcc1 was comparable to that of p115-depleted cells (Fig. 4I). Thus, p115 interactions with ER/Golgi SNAREs are vital for biogenesis of a Golgi apparatus that supports forward trafficking of cargo, whereas its interactions with giantin and GM130 are expendable.
Catalysis of SNARE complex formation by p115 is somewhat surprising, as trans-SNARE pairs have high affinities for each other and are able to drive membrane fusion on their own (23, 24). Therefore, confirmation of the physiological role of p115/SNARE interactions was critical. Interposition of p115 may provide a level of regulation to SNARE function. Possibly, it is p115 activity toward the SNARE proteins that is cell cycle regulated. One model is that p115 is recruited to membranes by Rab1 before binding SNAREs (10). As Rab1 is mitotically phosphorylated, this recruitment might be inhibited, thus preventing subsequent SNARE pairing and contributing to mitotic Golgi vesiculation.
The siRNA-based approach described here for complementation analysis is a powerful approach to extend in vitro analyses of structure–function relationships of any protein to living cells. It also provides a much-needed strategy to confirm the specificity of any given phenotype observed by using siRNA-mediated knockdown. Although in this report we used microinjection for expression of the replacement gene, other modes also work and allow biochemical, as well as morphological, assays of function (unpublished observations). In the case of p115, further gene replacements can be used not only to dissect the role of p115/SNARE interactions but also to test the role of p115 interactions with other important proteins such as the ARF1 exchange factor GBF1 (25) and phospholipase Cγ1 (26). We anticipate that the combined use of high resolution assays, which are readily applied to cultured cells, and sophisticated replacement constructs will significantly further the elucidation of cellular function.
Acknowledgments
We thank Drs. V. Malhotra, G. Waters, J. White, and O. A. Weisz for contributing essential reagents, and T. H. Lee for critical reading of the manuscript. Funding was provided by American Cancer Society and National Institutes of Health Grants RSG-03-148-01-CSM and GM-56779-02 (to A.D.L.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; siRNA, short interfering RNA; VSVG, vesicular stomatitis virus glycoprotein.
References
- 1.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, C., Fujita, H., Hubbard, A. & Sztul, E. (1999) J. Cell Biol. 147, 1205–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puthenveedu, M. A. & Linstedt, A. D. (2001) J. Cell Biol. 155, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondylis, V. & Rabouille, C. (2003) J. Cell Biol. 162, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnichsen, B., Lowe, M., Levine, T., Jamsa, E., Dirac-Svejstrup, B. & Warren, G. (1998) J. Cell Biol. 140, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dirac-Svejstrup, A. B., Shorter, J., Waters, M. G. & Warren, G. (2000) J. Cell Biol. 150, 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura, N., Lowe, M., Levine, T. P., Rabouille, C. & Warren, G. (1997) Cell 89, 445–455. [DOI] [PubMed] [Google Scholar]
- 8.Sohda, M., Misumi, Y., Yano, A., Takami, N. & Ikehara, Y. (1998) J. Biol. Chem. 273, 5385–5388. [DOI] [PubMed] [Google Scholar]
- 9.Lowe, M., Rabouille, C., Nakamura, N., Watson, R., Jackman, M., Jamsa, E., Rahman, D., Pappin, D. J. & Warren, G. (1998) Cell 94, 783–793. [DOI] [PubMed] [Google Scholar]
- 10.Allan, B. B., Moyer, B. D. & Balch, W. E. (2000) Science 289, 444–448. [DOI] [PubMed] [Google Scholar]
- 11.Shorter, J., Beard, M. B., Seemann, J., Dirac-Svejstrup, A. B. & Warren, G. (2002) J. Cell Biol. 157, 45–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann, J., Jokitalo, E. J. & Warren, G. (2000) Mol. Biol. Cell 11, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasile, E., Perez, T., Nakamura, N. & Krieger, M. (2003) Traffic 4, 254–272. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., Harborth, J., Weber, K. & Tuschl, T. (2002) Methods 26, 199–213. [DOI] [PubMed] [Google Scholar]
- 15.Waters, M. G., Clary, D. O. & Rothman, J. E. (1992) J. Cell Biol. 118, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevier, C. S., Weisz, O. A., Davis, M. & Machamer, C. E. (2000) Mol. Biol. Cell 11, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson, D. S., Alvarez, C., Gao, Y. S., Garcia-Mata, R., Fialkowski, E. & Sztul, E. (1998) J. Cell Biol. 143, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Silva, A. M., Balch, W. E. & Helenius, A. (1990) J. Cell Biol. 111, 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Presley, J. F., Cole, N. B., Schroer, T. A., Hirschberg, K., Zaal, K. J. & Lippincott-Schwartz, J. (1997) Nature 389, 81–85. [DOI] [PubMed] [Google Scholar]
- 20.Sapperstein, S. K., Walter, D. M., Grosvenor, A. R., Heuser, J. E. & Waters, M. G. (1995) Proc. Natl. Acad. Sci. USA 92, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesa, G. M., Seemann, J., Shorter, J., Vandekerckhove, J. & Warren, G. (2000) J. Biol. Chem. 275, 2831–2836. [DOI] [PubMed] [Google Scholar]
- 22.Linstedt, A. D., Jesch, S. A., Mehta, A., Lee, T. H., Garcia-Mata, R., Nelson, D. S. & Sztul, E. (2000) J. Biol. Chem. 275, 10196–10201. [DOI] [PubMed] [Google Scholar]
- 23.Weber, T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., Sollner, T. H. & Rothman, J. E. (1998) Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- 24.Hu, C., Ahmed, M., Melia, T. J., Sollner, T. H., Mayer, T. & Rothman, J. E. (2003) Science 300, 1745–1749. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Mata, R. & Sztul, E. (2003) EMBO Rep. 4, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, S. J., Lee, J. H., Kim, C. G. & Hong, S. H. (2003) Biochem. Biophys. Res. Commun. 300, 649–655. [DOI] [PubMed] [Google Scholar]