Abstract
MCF10A mammary epithelial cells form growth-arrested structures when cultured in three-dimensional basement membrane gels. Activation of the receptor tyrosine kinase ErbB2 induces formation of proliferative structures that share properties with noninvasive early stage lesions. We conducted a genetic screen to identify cDNAs that can cooperate with ErbB2 to induce migration in these cells, with the hypothesis that they would represent candidate “second hits” in the development of invasive breast carcinomas. We found that expression of transforming growth factor (TGF)β1 and TGFβ3 in cells expressing activated ErbB2 induces migration in transwell chambers and invasive behavior in both basement membrane cultures and invasion chambers. The ability of ErbB2 to cooperate with TGFβ correlated with sustained, elevated activation of extracellular signal-regulated kinase (Erk)-mitogen-activated protein kinase. Pharmacological reduction of Erk activity inhibited the cooperative effect of TGFβ and ErbB2 on migration and expression of activated Erk kinase was sufficient to cooperate with TGFβ to induce migration and invasion, suggesting that sustained Erk activation is critical for ErbB2/TGFβ cooperation. In addition, we show that costimulation of ErbB2 and TGFβ induces autocrine secretion of factors that are sufficient to induce migration, but not invasion, by means of both epidermal growth factor receptor-dependent and -independent processes. These results support the role of TGFβ as a pro-invasion factor in the progression of breast cancers with activated ErbB2 and suggest that activation of the Erk and epidermal growth factor receptor pathways are key in mediating these events.
ErbB2 (HER2/Neu) is overexpressed in 20% to 30% of invasive breast tumors and up to 85% of comedo type ductal carcinoma in situ (DCIS), an early stage in breast cancer (1, 2). The ErbB receptor family has four members: ErbB1 (EGFR/HER1), ErbB2, ErbB3, and ErbB4. When these receptors are stimulated with epidermal growth factor (EGF) family ligands, receptor activation occurs by means of both homo- and heterodimerization among the receptor family members (3). However, under conditions where ErbB2 is amplified or overexpressed, activation can occur by ligand-independent homodimerization (4, 5).
Previously, we investigated the consequences of activation of ErbB2 homodimers in the context of a nontransformed human mammary epithelial cell line, MCF10A (6, 7). When grown in basement membrane cultures, MCF10A cells form growth arrested three-dimensional structures, termed acini (8). These structures are comprised of a single layer of polarized epithelial cells surrounding a hollow lumen and resemble the mammary acini that form terminal ductal lobular units in the adult breast (7, 8).
To study the contributions of ErbB receptors in these cells, we generated chimeras of ErbB1 (p75.B1) and ErbB2 (p75.B2), which are inducibly activated by means of homodimerization by using the dimeric FKBP ligand AP1510 (ARIAD Pharmaceuticals, Cambridge, MA; refs. 9 and 10). The chimeras consist of the extracellular and transmembrane domains of the p75 low-affinity nerve growth factor receptor, the cytoplasmic domain of ErbB1 or ErbB2 and the FKBP ligand-binding domain.
The phenotypes of MCF10A cells expressing p75.B1 or p75.B2 (10A.B1 or 10A.B2 cells) have been extensively characterized (7). Dimerization of either p75.B1 or p75.B2 is sufficient to promote proliferation of MCF10A cells in monolayer cultures in the absence of EGF. Both chimeric receptors induce activation of extracellular signal-regulated kinase (Erk) and are competent to bind Shc and Grb2, whereas only ErbB1 dimers induce significant phosphorylation of Cbl. This pattern of activation of downstream signaling molecules is consistent with that of wild-type ErbB1 or ErbB2 receptors (3). Activation of p75.B2 in growth-arrested acini reinitiates proliferation and induces formation of structures consisting of multiple acini-like units with filled lumen. These cells are not transformed, because they are not anchorage-independent, do not invade basement membrane, and are not migratory. However, the high proliferative index, maintenance of E-cadherin adherens junctions, and lack of invasion of the ErbB2-induced structures are properties associated with DCIS lesions. MCF10A cells expressing p75.B1, in contrast, do not reinitiate proliferation after p75.B1 dimerization, and p75.B1 homodimers have no effect on preformed acini.
Given that DCIS tumors with amplified or overexpressed ErbB2 frequently progress to invasive tumors, it is of interest to find genes that confer a migratory or invasive phenotype on 10A.B2 cells to identify cellular pathways that may be involved in the invasive conversion of ErbB2 structures. Here, we describe a screen for genes that cooperate with ErbB2 activation to induce migration of MCF10A cells. By using this approach, two members of the transforming growth factor (TGF)β family, TGFβ1 and TGFβ3, were identified. Furthermore, coactivation of ErbB2 and TGFβ signaling pathways induced invasive activity of MCF10A cells. Erk activation was required for the observed synergy between ErbB2 and TGFβ, but was not sufficient to induce migration or invasion. Analogous to ErbB2, however, activated Erk collaborated with TGFβ to induce migration and invasion. Additionally, costimulated MCF10A cells secreted factors that were sufficient to induce migration but not invasion.
Materials and Methods
Cell Culture and Materials. MCF10A cells were cultured as described (11). The following reagents were used: recombinant human (rh)TGFβ1 (R & D Systems); purified porcine TGFβ2(a gift from L. Wakefield, National Cancer Institute, Bethesda); PD98059, U0126, and AG1478 (Calbiochem); AP1510 (ARIAD Pharmaceuticals; ref. 9); α-actin, α-Erk1, α-Erk2, and α-Mek1/2 (Santa Cruz Biotechnology); α-phospho Erk1/2 (BioSource); α-phospho Mek 1/2 (Cell Signaling); α-Vimentin, α-E-cadherin and α-N-cadherin (BD Biosciences); mAb 225 hybridoma cells (American Type Culture Collection); and purified mAb 225 [a gift of D. Lauffenburger (Massachusetts Institute of Technology, Boston; ref. 12)].
Expression Vectors and Cell Lines. The generation of MCF10A cells expressing the chimeric receptors p75.B2 or p75.B1 has been described (7).
The full-length experession library of cDNAs was assembled in the Gateway system (Invitrogen). mRNA was isolated from human brain and placenta and converted into first-strand cDNA according to the manufacturer's protocol; this library served as the template for PCR amplification. Specific PCR primers were designed by using the nearest-neighbor algorithm (http://flex.med.harvard.edu:8080/oligo). Recombination sites were added in a second PCR. DNA was captured in the Gateway vector according to the manufacturer's protocol. Clones were sequenced by primer walking. All cDNAs were transferred into the pBabe-puro vector according to Invitrogen's protocol. TGFβ1 was also subcloned into the MSCV-IRES-GFP vector (O. Witte, University of California, Los Angeles). pBabe-Mek2DD was provided by S. Meloche (University of Montreal). MCF10A cells expressing pBabe-HRasV12 were generated by means of stable transfection. Stable lines expressing TGFβ were not generated, because its expression inhibits cell proliferation. Fresh infections were performed for each experiment and the cells cultured for <96 h postinfection.
Transwell Migration Assay. Cells (10A.B2) expressing full-length expression library genes or TGFβ were starved overnight in assay media (MCF10A media containing no EGF and only 1% serum). Cells (1 × 105) were added to the top chambers of 24-well transwell plates (BD Biosciences; 8-μm pore size), and assay media, with or without 5 ng/ml EGF or 500 nM AP1510, was added to the bottom chambers. After overnight incubation, top (nonmigrated) cells were removed, and bottom (migrated) cells were fixed and stained with 4′,6-diamidino-2-phenylindole (5 μg/ml) to visualize nuclei. The number of migrating cells in five fields were counted under ×20 magnification, and the mean for each chamber determined. Experiments were repeated a minimum of three times.
For assays with chemical inhibitors, cells were incubated with inhibitor or vehicle 15 min before plating. For assays with inhibitory antibodies, cells were incubated with antibody 30 min before plating. For experiments with conditioned media, cells were seeded in assay media in the chambers and conditioned media placed in the bottom well.
Transwell Invasion Chambers. BD BioCoat invasion chambers coated with growth factor reduced Matrigel were purchased from BD Biosciences. Assays were conducted according to BD's protocol, by using 5% horse serum (GIBCO) as the chemoattractant. When required, 5 ng/ml EGF or 500 nM AP1510 was added. Invading cells were quantified as for migration assays. Assays using inhibitors were conducted as for the migration assays.
Three-Dimensional Culture. Cells were cultured in basement membrane gels by using the overlay method as described (11), except a mixture of bovine dermal collagen I (Vitrogen; Cohesion Technologies, Palo Alto, CA) and growth factor-reduced Matrigel, rather than 100% Matrigel, was used as the underlay. Final collagen concentration was kept constant at 1.6 mg/ml, and the remainder of the underlay was Matrigel. Before mixing, collagen I was neutralized by addition of 100 mM NaOH and 10× PBS to final concentrations of 10 mM and 1×, respectively, and the pH was brought to 7.5 by using 0.1 M HCl. Four days after seeding, EGF was withdrawn from cells to be stimulated with AP1510, and 500 nM AP1510 was added.
Conditioned Medium. Cells were starved overnight in assay medium. Starvation medium was removed, and fresh assay medium, containing 500 nM AP1510 when necessary, was added to the cells (4 ml of medium was used for a 10-cm culture dish). After 18 h, medium was passed through a 2-μm filter and was stored at –20°C.
Immunoblotting. Cells were lysed in Nonidet P-40 (NP-40; for phosphospecific antibody blots) or radioimmunoprecipitation assay (RIPA; for vimentin and cadherin blots) lysis buffer for 10 min and clarified at 16,000 × g for 10 min. NP-40 buffer contained 50 mM Tris·HCl (pH 7.6), 150 mM NaCl, 10 mM NaF, 10% glycerol, 1 mM Na3VO4, and 1% NP-40. RIPA buffer contained 10 mM Tris·HCl (pH 7.2), 158 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 0.1% SDS, 1% sodium deoxycholate, and 1% Triton X-100. Both buffers were supplemented with protease inhibitors (1 mM PMSF/2 μg/ml leupeptin/2 μg/ml aprotinin) before use. NP-40-insoluble fractions were recovered by solublizing in SDS/PAGE sample buffer.
Fifteen micrograms of protein was resolved on SDS/PAGE and was transferred to PVDF membranes (Bio-Rad). Membranes were blocked with 5% BSA in Tris-buffered saline/Tween 20 (TBST) and incubated with primary antibodies overnight at 4°C. Membranes were washed with TBST, incubated with secondary antibodies conjugated to horseradish peroxidase, and antibody binding detected by chemiluminescence.
Results
Screen to Identify cDNAs That Induce Migration of Cells Expressing Activated ErbB2. We have shown previously that activation of a chimeric ErbB2 receptor by using the dimerizing ligand AP1510 in MCF10A cells (10A.B2 cells) is sufficient to induce proliferation and multi-acini formation in the absence of EGF; however ErbB2 dimerization does not induce cell migration in the absence of EGF (7). To identify genes that could contribute to migratory behavior of breast epithelial cells, we took advantage of the sensitized genetic background of 10A.B2 cells to conduct a screen for genes that could cooperate with ErbB2 to induce cell migration, with the hypothesis that these genes might represent candidate “second hits” in the development of invasive breast carcinomas.
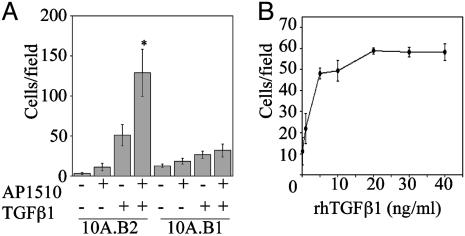
Cells (10A.B2) were infected with retroviral vectors encoding a set of 30 cDNAs from the breast cancer cDNA collection of the full-length expression library (Table 1, which is published as supporting information on the PNAS web site). Cells expressing these genes were assayed for migration in medium lacking EGF but containing AP1510 to activate ErbB2. Three cDNAs induced migration of 10A.B2 cells in the presence of AP1510: TGFβ1, TGFβ3, and hepatocyte growth factor (HGF) (Fig. 1A and data not shown). HGF induced migration of MCF10A cells in the absence of AP1510; however, TGFβ1 and β3 enhanced migration only in the presence of AP1510, suggesting that ErbB2 and TGFβ collaborate to induce migration.
Fig. 1.
ErbB2 and TGFβ cooperate to induce migration of MCF10A cells. (A) Cells (10A.B2 or 10A.B1) expressing TGFβ1 or control vector were seeded in transwell migration chambers –/+ AP1510, incubated for 18 h, and quantified as detailed in Materials and Methods. *, P < 0.0001 and P = 0.048 as compared with ErbB2 or TGFβ alone, respectively. (B) Cells (10A.B2) were seeded in transwell chambers in the presence of 500 nM AP1510 and indicated doses of rhTGFβ1 for 18 h and quantified as for A.
Expression of TGFβ1 in 10A.B2 cells enhanced AP1510-induced migration 24.7-fold relative to 10A.B2 cells lacking TGFβ1 (P < 0.0001; Fig. 1 A). TGFβ1 expression also enhanced migration of 10A.B2 cells in the absence of dimerizer (2.8-fold, P = 0.048), likely due to leaky activation of ErbB2 (7). In comparison, TGFβ1 enhanced migration of parental MCF10A cells by <2-fold (data not shown).
To determine whether ErbB1 could also collaborate with TGFβ to induce migration, we examined whether TGFβ1 and TGFβ3 induced migration in MCF10A cells expressing the chimeric ErbB1 receptor (10A.B1 cells). Although comparable levels of TGFβ were expressed in the 10A.B1 and 10A.B2 cells, neither TGFβ1 nor TGFβ3 induced migration of the 10A.B1 cells after stimulation with AP1510 (Fig. 1 A and data not shown). These results indicate that coactivation of ErbB1 and TGFβ receptors in MCF10A cells cannot promote cell migration and suggests that the ErbB2 and TGFβ collaboration involves ErbB2-specific activities.
To further examine the nature of the migration induced by TGFβ and ErbB2, we analyzed the kinetics of migration. Migration of 10A.B2 cells expressing TGFβ1 (10A.B2.TGFβ cells) commenced ≈8 h and reached a maximum after 18 h of AP1510 treatment (data not shown). We detected similar levels of migration whether AP1510 was present in the top or bottom chamber alone or in both during the transwell assay (data not shown). These data suggest that the observed migration is not chemotactic but chemokinetic.
To examine whether soluble, exogenous TGFβ could recapitulate the effects of autocrine TGFβ expression, we treated cells with rhTGFβ1 during the migration assay. rhTGFβ1 induced migration of AP1510-treated 10A.B2 cells in a dose-dependent manner, with the maximal response occurring between 10 and 20 ng/ml rhTGFβ1 (Fig. 1B). Purified porcine TGFβ2 also induced migration of MCF10A cells in the presence of activated ErbB2 (data not shown). Whereas soluble TGFβ recapitulated the migratory phenotype in cooperation with ErbB2, the total number of cells that migrated after acute stimulation with soluble TGFβ was lower than that seen in cells constitutively producing TGFβ. This finding suggests that autocrine production of TGFβ induces migration more effectively than soluble TGFβ.
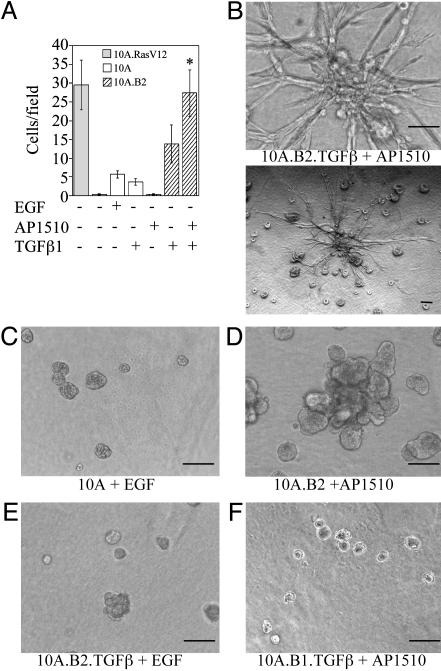
TGFβ1 Cooperates with ErbB2 to Induce Invasive Activity in Both Two- and Three-Dimensional Assays. Because the goal of our screen was to identify genes that might represent second hits in the development of invasive breast cancers, we examined whether 10A.B2.TGFβ cells displayed invasive activity in Matrigel-coated invasion chambers or in three-dimensional basement membrane cultures when treated with AP1510. Treatment with AP1510 significantly enhanced the invasive activity of 10A.B2.TGFβ cells in invasion chambers (Fig. 2A). As in the migration assay, untreated 10A.B2.TGFβ cells displayed greater activity than 10A.TGFβ cells, most likely because of baseline activation of the ErbB2 chimera.
Fig. 2.
ErbB2 and TGFβ cooperate to induce invasion. (A) Cells (10A.B2 and 10A.B2.TGFβ; hatched bars) were seeded in transwell invasion chambers with or without AP1510 and incubated for 24 h. MCF10A cells expressing RasV12 (10A.RasV12; gray bar) served as a positive control and MCF10A cells with or without EGF or TGFβ (white bars) as negative controls. *, P = 0.0023 and 0.0188 as compared with ErbB2 or TGFβ (in 10A.B2 cells) alone, respectively. (B–F) Representative three-dimensional structures of 10A.B2.TGFβ (B) cells treated with 500 nM AP1510; the same structure is shown at high (Upper) and low (Lower) magnification. MCF10A cells plus EGF (C), 10A.B2 cells plus AP1510 (D), 10A.B2.TGFβ cells plus EGF (E), and 10A.B1.TGFβ cells plus AP1510 (F). Structures were photographed at day 17. (Bars, 100 μm.)
The 10A.B2.TGFβ cells also exhibited invasive activity in Matrigel and collagen I basement membrane cultures after 8 days of treatment with AP1510 (Fig. 2B). Invasive projections were observed in 30–40% of the large structures in each well. Invasive activity depended on expression of TGFβ and activation of ErbB2, because cells with activated ErbB2 alone, activated ErbB1 and TGFβ, or neither ErbB2 nor TGFβ did not display invasive activity (Fig. 2 C–F). Treatment of 10A.B2 structures with AP1510 and recombinant TGFβ1 or 2 also induced invasive activity in this culture system (data not shown). We did not observe invasive activity in response to ErbB2 and TGFβ stimulation by using cultures containing 100% Matrigel, suggesting that the presence of collagen I is critical for the observed invasive activity.
Analysis of Markers of Epithelial-Mesenchyme Transition (EMT). Previous studies have demonstrated that certain epithelial cells undergo an EMT when exposed to TGFβ, thus increasing migratory and invasive activity (13, 14). Other cells undergo EMT only after TGFβ treatment combined with activation of the Ras pathway (15–20). Therefore, we examined whether MCF10A cells exposed to TGFβ or TGFβ combined with ErbB2 activation displayed phenotypic effects commonly observed with EMT, such as up-regulation of vimentin and N-cadherin, down-regulation of E-cadherin, and morphological EMT (21). Expression of TGFβ in MCF10A or 10A.B2 cells caused spreading of the cells; however, they did not assume a fibroblastoid morphology, and treatment of 10A.B2.TGFβ cells with AP1510 for 18 h did not alter this morphology (Fig. 5, which is published as supporting information on the PNAS web site). Analogously, TGFβ expression increased vimentin and N-cadherin expression in both MCF10A and 10A.B2.TGFβ cells, but no change in expression of these proteins was observed after stimulation of the 10A.B2.TGFβ cells with AP1510 for 18 h (Fig. 5B). Total E-cadherin levels remained constant in 10A.B2.TGFβ, cells even in the presence of AP1510 (Fig. 5B). However, analysis of E-cadherin levels in NP-40-soluble and -insoluble fractions revealed that TGFβ reduced insoluble E-cadherin (Fig. 5C). The partial reduction in insoluble E-cadherin or the increase in N-cadherin or vimentin may be required for migration induced by ErbB2 and TGFβ. These alterations, however, are not sufficient to induce migration, because they are induced by TGFβ expression alone and are not enhanced in ErbB2-stimulated cells.
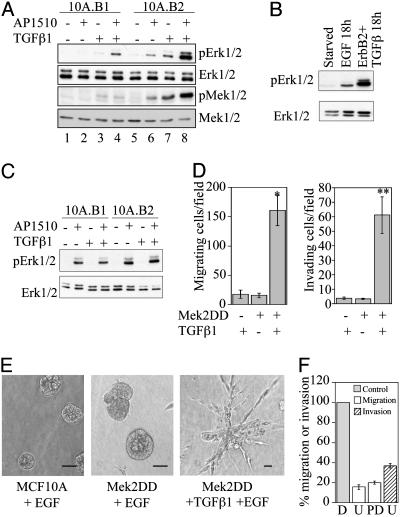
Erk Activation Is Critical for ErbB2- and TGFβ-Induced Migration and Invasion. The Erk-mitogen-activated protein kinase pathway has been implicated in cell migration and can be activated by both ErbB2 and TGFβ (3, 22, 23). Therefore, we compared the levels of Erk1/2 and Mek1/2 phosphorylation in 10A.B1.TGFβ and 10A.B2.TGFβ cells stimulated with AP1510 for 18 h, which is the time period of the migration assay. Both Erk and Erk kinase (Mek) displayed higher levels of phosphorylation when ErbB2 and TGFβ were activated for 18 h, as compared with activation of ErbB1 and TGFβ or ErbB2 or TGFβ alone (Fig. 3A, compare lane 8 with lanes 4, 6, and 7). This finding suggests that ErbB2 and TGFβ induce sustained, elevated Erk pathway activation. TGFβ coexpression with ErbB1 also increased the level of phosphorylated Erk at 18 h; however, the level of activation was significantly lower than that induced by ErbB2 and TGFβ.
Fig. 3.
Role of the Erk pathway in migration and invasion induced by ErbB2 and TGFβ. (A) Immunoblot analysis of phosphorylated Erk1/2 (pErk1/2) and Mek1/2 (pMek1/2) in 10A.B1 and 10A.B2 cells with or without TGFβ expression and AP1510 treatment for 18 h. (B) pErk1/2 levels in 10A.B2 cells treated with 20 ng/ml EGF or with TGFβ expression and AP1510 treatment for 18 h. (C) pErk1/2 levels in 10A.B1 and 10A.B2 cells with or without TGFβ expression and AP1510 treatment for 15 min. (D) MCF10A cells expressing Mek2DD or control vector were analyzed in transwell migration (Left) and invasion (Right) assays with or without TGFβ expression. *, P < 0.0002; **, P < 0.001 as compared with either Mek2DD or TGFβ alone. (E) Representative three-dimensional structures of MCF10A cells expressing control vector, Mek2DD, or Mek2DD plus TGFβ from day 15. (Bars, 40 μm.) (F) Transwell migration and invasion assays were performed with 10A.B2.TGFβ cells in the presence of AP1510 and Mek inhibitors U0126 (U; 5 μM) or PD98059 (PD; 50 μM), or DMSO control (D). Data are expressed as the percent of control cells (gray bar, normalized to 100%) that migrate (white bars) or invade (hatched bar) in the presence of inhibitor. PD98059 was not tested in invasion.
Because EGF also induces MCF10A cell migration, phosphorylated Erk levels were examined in MCF10A cells stimulated with EGF for 18 h and compared with 10A.B2.TGFβ cells stimulated with AP1510 for 18 h (Fig. 3B). Whereas long-term stimulation of MCF10As with EGF induced Erk activation, long-term stimulation with ErbB2 and TGFβ led to greater activation. Both 10A.B1 and 10A.B2 cells activated Erk after acute AP1510 stimulation, and Erk activation was not enhanced in 10A.B2.TGFβ cells (relative to 10A.B2 cells) by acute AP1510 stimulation (Fig. 3C).
To directly evaluate the role of sustained, elevated activation of the Erk pathway in ErbB2- and TGFβ1-induced migration and invasion, we expressed an activated variant of Mek2 (Mek2DD) in MCF10A cells or MCF10A cells expressing TGFβ1. Expression of Mek2DD was not sufficient to induce MCF10A cell migration or invasion to the levels induced by ErbB2 and TGFβ (Fig. 3 D and E); however, the combined expression of Mek2DD and TGFβ caused a significant increase in MCF10A cell migration and invasion (Fig. 3 D and E). The invasive structures formed in three-dimensional culture by cells expressing Mek2DD and TGFβ do not exactly phenocopy those formed with activation of ErbB2 and TGFβ, possibly due to ErbB2's ability to affect other cellular pathways. These data suggest that sustained Erk activation, while not sufficient to induce migration or invasion, can mimic the effects of ErbB2 activation in inducing migration and invasion in cooperation with TGFβ.
To examine the requirement for Erk activation in TGFβ- and ErbB2-induced migration and invasion, transwell assays were performed in the presence of Mek inhibitors. Inhibition of Mek caused an 85% reduction of migration and 65% inhibition of invasion through Matrigel induced by ErbB2 and TGFβ (Fig. 3F), suggesting that Erk activation is required for ErbB2- and TGFβ-collaborative effects.
Because Mek and Erk are required for intrinsic processes associated with cell motility, the inhibition of TGFβ1- and ErbB2-induced motility by Mek inhibitors could merely reflect the involvement of Erk in these fundamental migratory processes (22). Thus, we performed a dose–response analysis to determine whether the enhanced motility of the ErbB2- and TGFβ-costimulated cells depends on the elevated levels of activated Erk found in the costimulated cells (Fig. 6, which is published as supporting information on the PNAS web site). The levels of Erk phosphorylation directly correlated with migratory activity of the 10A.B2.TGFβ cells. Importantly, migration of AP1510-treated 10A.B2.TGFβ cells was inhibited at concentrations of UO126 that retained significant levels of Erk phosphorylation, which is higher than those found in EGF-treated cells that are fully competent for migration. Reduction of Erk phosphorylation in AP1510-treated 10A.B2.TGFβ cells to the levels observed in 10A.B2.TGFβ cells without AP1510 (compare boxed samples) eliminated the synergism observed between TGFβ and ErbB2. These results support the possibility that the elevated, sustained activity of Erk induced by TGFβ and ErbB2 is critical for enhanced migration.
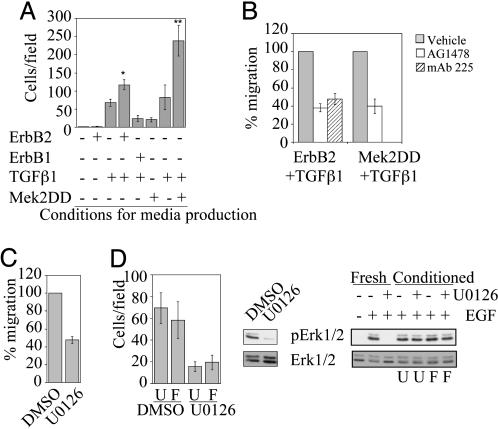
Conditioned Medium from Cells Stimulated with ErbB2 and TGFβ Contains Multiple Soluble Migratory Factors. Constitutive activation of Raf, an upstream regulator of Erk, induces secretion of EGF family ligands in MCF10A cells (24). To examine whether costimulation of ErbB2 and TGFβ receptors induces secretion of soluble migratory factors, we assayed the migration-stimulating activity of conditioned medium collected from 10A.B2.TGFβ cells stimulated with AP1510 for 18 h. This medium induced significantly more migration of parental MCF10A cells than media from MCF10A cells expressing activated ErbB2 without TGFβ or expressing TGFβ alone (Fig. 4A). Conditioned medium from MCF10A cells expressing Mek2DD and TGFβ also contained soluble migratory factors, whereas medium from cells expressing Mek2DD alone induced migration only slightly above background (Fig. 4A). To determine whether cooperation of ErbB2 and TGFβ is necessary to produce the entire complement of secreted factors, media from cells expressing either activated ErbB2 or TGFβ alone were mixed. The mixed media did not enhance migration of MCF10A cells, suggesting that cooperation between TGFβ and ErbB2 within the same cell environment is necessary (data not shown). Whereas media from cells expressing TGFβ together with either activated ErbB2 or Mek2DD was sufficient to induce migration, it was not sufficient to induce invasion (data not shown).
Fig. 4.
The combined activation of ErbB2 plus TGFβ or Mek2 plus TGFβ leads to secretion of both EGFR-dependent and -independent soluble migratory factors. (A) MCF10A cells were seeded in transwell migration chambers, and conditioned media produced from cells as indicated was added to the bottom chambers and incubated for 18 h. *, P = 0.0056 as compared with TGFβ alone; P = 0.004 as compared with ErbB1 plus TGFβ. **, P = 0.008 as compared with TGFβ alone; P = 0.002 as compared with MEK2DD alone. (B) Migration assays were performed as described in A, except EGFR inhibitors AG1478 (300 nM) or mAb 225 (10 μg/ml) were added. Data are expressed as the percent of control cells that migrate or invade with inhibitor. Migration with Mek 2DD plus TGFβ-conditioned medium was not tested with mAb 225. (C) Migration assays were performed by using 10A.B2.TGFβ plus AP1510-conditioned medium and 5 μM U0126. Data are expressed as the percent of control cells that migrate with inhibitor. (D) Conditioned media from 10A.B2.TGFβ cells treated with AP1510 was made in the presence of 5 μM U0126 or DMSO and inhibitor removed by filtration. Transwell migration assays were performed with this media and MCF10A cells (Left; U, unfiltered medium; F, filtered medium). To show inhibition of Erk phosphorylation by U0126 in the cells used to make the media, they were lysed after media collection and pErk1/2 levels were analyzed by immunoblot (Center). To show that migration inhibition was not due to carryover of U0126 after filtration, MCF10A cells were pretreated for 15 min with conditioned medium or fresh U0126 and were stimulated for 15 min with 20 ng/ml EGF. Lysates of these cells were analyzed by immunoblotting pErk1/2 (Right; U, unfiltered medium; F, filtered medium).
To determine whether the soluble migratory factors are epidermal growth factor receptor (EGFR) ligands, we examined whether inhibition of EGFR (ErbB1) affects migration induced by conditioned media from either 10A.B2.TGFβ cells treated with AP1510 or MCF10A cells coexpressing Mek2DD and TGFβ. Both AG1478, an ErbB1-specific pharmacological inhibitor, and mAb 225, an ErbB1-inhibitory antibody, reduced the migration stimulating activity of both conditioned media samples by 50–60%, suggesting that both EGFR-dependent and -independent motogenic factors are secreted (Fig. 4B). Inhibition of ErbB1 also partially inhibited AP1510-stimulated migration of 10A.B2.TGFβ cells (data not shown). In addition, immunoblots probed with an antibody to phosphotyrosine residues indicated that 10A.B2.TGFβ plus AP1510-conditioned medium induced phosphorylation of ErbB1 (data not shown). These results support the hypothesis that at least one EGF family ligand is secreted from cells expressing activated ErbB2 and TGFβ.
Because increased Erk pathway activation is required for enhanced migration of MCF10A cells expressing activated ErbB2 and TGFβ, we examined whether hyperstimulation of Erk is required for migratory factor secretion or for factor-induced motility. Mek inhibition blocked migration induced by conditioned medium from 10A.B2.TGFβ cells treated with AP1510, and conditioned medium from 10A.B2.TGFβ cells treated with AP1510 and U0126 did not induce migration (Fig. 4 C and D). To determine whether the lack of migration under the latter condition was due to residual U0126 in the conditioned media, MCF10A cells were stimulated in parallel with conditioned media that was first filtered to remove small molecules. The filtered media displayed no Erk-inhibitory activity on EGF-stimulated cells (Fig. 4D Right), yet was still unable to stimulate MCF10A migration, indicating that Erk activity is required for the production of the soluble migratory factors. These data suggest that Erk activity is required both up- and downstream of migratory factor production in MCF10A cells expressing activated ErbB2 and TGFβ.
Discussion
Here, we describe the identification of TGFβ as a promigratory factor by using a sensitized genetic screen to identify genes that enhance MCF10A cell migration in the context of ErbB2 activation. The costimulation of ErbB2 and TGFβ receptors is also sufficient to induce invasive activity in these cells. Several lines of evidence support a role for the Erk pathway in these phenotypic effects. Hyperactivation of Erk is required, but not sufficient, to mediate migration and invasion induced by costimulation of ErbB2 and TGFβ, and reduction of Erk activation inhibits the enhancement of migration and invasion. In addition, activation of ErbB2 and TGFβ signaling pathways induces secretion of both EGFR-dependent and -independent soluble motogenic factors, and Erk activation is required both for factor production and factor-induced motility.
TGFβ is involved in two opposing activities: suppression of cell proliferation and enhancement of tumor cell metastasis (25–28). Given these dual roles, dissecting TGFβ's link to cancer progression has been difficult. TGFβ usually inhibits proliferation of epithelial cells, and can function as a tumor suppressor. Paradoxically, TGFβ produced by tumor cells can enhance tumorigenesis by means of multiple mechanisms, involving either direct effects on tumor cells or paracrine effects on other cells (29, 30). During the course of our studies, several reports provided evidence that TGFβ can collaborate with ErbB2 in mice to promote metastasis. Inhibition of TGFβ by using mouse mammary tumor virus (MMTV) expression of a soluble antagonist in mice expressing oncogenic ErbB2 (Neu) under the control of the MMTV promoter causes a significant reduction in the formation of metastatic lesions (31). Mice derived from crossing MMTV-Neu transgenic mice with mice expressing inducible TGFβ1 display increased numbers of metastatic lesions (32). In addition, expression of an activated TGFβ receptor increases extravasation of Neu-induced tumor cells from pulmonary vessels (33). The metastasis and invasion promoting effects of TGFβ have also been observed in other mouse models of tumorigenesis (34–36).
In mouse models, it is difficult to dissect the precise mechanisms whereby TGFβ promotes metastasis because it could act directly on tumor epithelial cells or indirectly on the tumor microenvironment. Our experiments have specifically addressed the effects of TGFβ on epithelial cells that express activated ErbB2. The data presented here complement the studies of ErbB2 and TGFβ in mice and indicate that TGFβ can act directly on epithelial cells expressing ErbB2 to induce invasive behavior.
EMT conversion is typically accompanied by an increase in migratory activity, a change to spindle-like cell morphology, a loss of epithelial-specific proteins, and a concurrent gain of mesenchymal markers (37). Whereas we observed an increase in mesenchymal marker expression in TGFβ-treated MCF10A cells, this alone was not sufficient to induce migration, nor was it accompanied by a loss of expression of the epithelial marker E-cadherin or conversion to a spindle-like morphology. Also, activation of ErbB2 in TGFβ-expressing cells did not cause additional changes in mesenchyme marker expression or cell morphology. However, E-cadherin expression was reduced in the detergent-insoluble fraction of cells expressing TGFβ, indicating that TGFβ induces a reduction in E-cadherin linkage to the actin cytoskeleton. HGF induces a similar change in E-cadherin solubility in MDCK cells; this change is required for HGF-induced cell motility and depends on activation of Erk (38). Analogously, we hypothesize ErbB2 and TGFβ-induced migration may require a loss in E-cadherin linkage to the cytoskeleton.
Investigation into the mechanisms whereby the combination of signals from ErbB2 and TGFβ induce events not promoted by each alone led us to the observation that Erk activation is sustained at a high level in MCF10A cells exposed to both ErbB2 and TGFβ. This enhanced activation of Erk is required for cell migration and invasion in response to ErbB2 and TGFβ. This finding is interesting in light of the observation that Erk is both hyperactivated and overexpressed in breast carcinomas (39). In MCF10A cells, hyperactivation of Erk appears to influence two mechanisms that increase cell motility; it is required for the basal migration machinery of the cells and for secretion of motogenic factors.
TGFβ has been shown to collaborate with activated mutants of Ras, which induce constitutive Erk activation, to induce EMT in numerous epithelial cell types (15–20). In EpH4 mouse mammary epithelial cells, induction of EMT by activated Ras and TGFβ is coincident with induction of cell migration and invasion (20). In the MCF10A system described here, the enhanced migration in response to ErbB2 and TGFβ is not accompanied by a typical EMT conversion. However, it is possible that perturbations in E-cadherin linkage to the cytoskeleton in response to TGFβ may represent a partial EMT and may contribute to migratory activity.
In this report we demonstrate that MCF10A cells expressing activated ErbB2 and TGFβ secrete both EGFR-dependent and -independent factors that are sufficient to induce migration of parental MCF10As. The involvement of EGFR-dependent factors presents a conundrum. Homodimerization of either ErbB1 or ErbB2 alone cannot induce cell migration (7), yet induction of migration in response to ErbB2 and TGFβ requires secretion of at least one ErbB1 ligand. It is possible that endogenous ErbB1 ligands stimulate migration through induction of heterodimers of ErbB1 with other ErbB family members, whereas homodimers do not induce a migratory response. Differences in signaling and phenotypic responses have been described after treatment of cells with different ErbB1 ligands, some of which are attributable to the activation of distinct homo- or heterodimers of ErbB family members (3, 40). Thus, ErbB2 and TGFβ stimulation may induce production of a factor which stimulates heterodimerization of ErbB receptors, leading to cell migration.
These results support the role of TGFβ in the progression of breast cancers with activated ErbB2 and suggest that activation of the Erk and EGFR pathways are key in mediating these events. We have successfully used two- and three-dimensional cultures of MCF10A cells to model invasive processes and dissect the signaling pathways and cellular changes that are important for ErbB2 and TGFβ cooperation. Further analysis of the mechanisms underlying invasion induced by ErbB2 and TGFβ and other genes in such models may provide important insights into the processes of tumor cell migration and invasion.
Supplementary Material
Acknowledgments
We thank D. Lauffenburger, L. Wakefield, S. Meloche, O. Witte, and S. Gutkind for providing reagents and ARIAD Pharmaceuticals for providing AP1510 (www.ariad.com/regulationkits). This work was supported by the National Cancer Institute, the Ludwig Cancer Research Foundation, the National Institutes of Health (to J.S.B.), and a National Institutes of Health Program Project Grant (to J.S.B. and J.L.).
Abbreviations: TGF, transforming growth factor; rh, recombinant human; EGF, epidermal growth factor; EGFR, EGF receptor; EMT, epithelial-mesenchyme transition; NP-40, Nonidet P-40; Erk, extracellular signal-regulated kinase; Mek, Erk kinase.
References
- 1.Harari, D. & Yarden, Y. (2000) Oncogene 19, 6102–6114. [DOI] [PubMed] [Google Scholar]
- 2.Hynes, N. E. & Stern, D. F. (1994) Biochim. Biophys. Acta 1198, 165–184. [DOI] [PubMed] [Google Scholar]
- 3.Olayioye, M. A., Neve, R. M., Lane, H. A. & Hynes, N. E. (2000) EMBO J. 19, 3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Fiore, P. P., Pierce, J. H., Kraus, M. H., Segatto, O., King, C. R. & Aaronson, S. A. (1987) Science 237, 178–182. [DOI] [PubMed] [Google Scholar]
- 5.Samanta, A., LeVea, C. M., Dougall, W. C., Qian, X. & Greene, M. I. (1994) Proc. Natl. Acad. Sci. USA 91, 1711–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soule, H. D., Maloney, T. M., Wolman, S. R., Peterson, W. D., Jr., Brenz, R., McGrath, C. M., Russo, J., Pauley, R. J., Jones, R. F. & Brooks, S. C. (1990) Cancer Res. 50, 6075–6086. [PubMed] [Google Scholar]
- 7.Muthuswamy, S. K., Li, D., Lelievre, S., Bissell, M. J. & Brugge, J. S. (2001) Nat. Cell Biol. 3, 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen, O. W., Ronnov-Jessen, L., Howlett, A. R. & Bissell, M. J. (1992) Proc. Natl. Acad. Sci. USA 89, 9064–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amara, J. F., Clackson, T., Rivera, V. M., Guo, T., Keenan, T., Natesan, S., Pollock, R., Yang, W., Courage, N. L., Holt, D. A. & Gilman, M. (1997) Proc. Natl. Acad. Sci. USA 94, 10618–10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muthuswamy, S. K., Gilman, M. & Brugge, J. S. (1999) Mol. Cell. Biol. 19, 6845–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debnath, J., Muthuswamy, S. K. & Brugge, J. S. (2003) Methods 30, 256–268. [DOI] [PubMed] [Google Scholar]
- 12.Sato, J. D., Kawamoto, T., Le, A. D., Mendelsohn, J., Polikoff, J. & Sato, G. H. (1983) Mol. Biol. Med. 1, 511–529. [PubMed] [Google Scholar]
- 13.Miettinen, P. J., Ebner, R., Lopez, A. R. & Derynck, R. (1994) J. Cell Biol. 127, 2021–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Portella, G., Cumming, S. A., Liddell, J., Cui, W., Ireland, H., Akhurst, R. J. & Balmain, A. (1998) Cell Growth Differ. 9, 393–404. [PubMed] [Google Scholar]
- 15.Grunert, S., Jechlinger, M. & Beug, H. (2003) Nat. Rev. Mol. Cell Biol. 4, 657–665. [DOI] [PubMed] [Google Scholar]
- 16.Ellenrieder, V., Hendler, S. F., Boeck, W., Seufferlein, T., Menke, A., Ruhland, C., Adler, G. & Gress, T. M. (2001) Cancer Res. 61, 4222–4228. [PubMed] [Google Scholar]
- 17.Fujimoto, K., Sheng, H., Shao, J. & Beauchamp, R. D. (2001) Exp. Cell Res. 266, 239–249. [DOI] [PubMed] [Google Scholar]
- 18.Gotzmann, J., Huber, H., Thallinger, C., Wolschek, M., Jansen, B., Schulte-Hermann, R., Beug, H. & Mikulits, W. (2002) J. Cell Sci. 115, 1189–1202. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann, K., Janda, E., Pierreux, C. E., Rytomaa, M., Schulze, A., McMahon, M., Hill, C. S., Beug, H. & Downward, J. (2000) Genes Dev. 14, 2610–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oft, M., Peli, J., Rudaz, C., Schwarz, H., Beug, H. & Reichmann, E. (1996) Genes Dev. 10, 2462–2477. [DOI] [PubMed] [Google Scholar]
- 21.Boyer, B., Valles, A. M. & Edme, N. (2000) Biochem. Pharmacol. 60, 1091–1099. [DOI] [PubMed] [Google Scholar]
- 22.Stupack, D. G., Cho, S. Y. & Klemke, R. L. (2000) Immuol. Res. 21, 83–88. [DOI] [PubMed] [Google Scholar]
- 23.Hartsough, M. T. & Mulder, K. M. (1995) J. Biol. Chem. 270, 7117–7124. [DOI] [PubMed] [Google Scholar]
- 24.Schulze, A., Lehmann, K., Jefferies, H. B., McMahon, M. & Downward, J. (2001) Genes Dev. 15, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumont, N. & Arteaga, C. L. (2000) Breast Cancer Res. 2, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derynck, R., Akhurst, R. J. & Balmain, A. (2001) Nat. Genet. 29, 117–129. [DOI] [PubMed] [Google Scholar]
- 27.Massague, J., Blain, S. W. & Lo, R. S. (2000) Cell 103, 295–309. [DOI] [PubMed] [Google Scholar]
- 28.Wakefield, L. M. & Roberts, A. B. (2002) Curr. Opin. Genet. Dev. 12, 22–29. [DOI] [PubMed] [Google Scholar]
- 29.Roberts, A. B., Sporn, M. B., Assoian, R. K., Smith, J. M., Roche, N. S., Wakefield, L. M., Heine, U. I., Liotta, L. A., Falanga, V., Kehrl, J. H., et al. (1986) Proc. Natl. Acad. Sci. USA 83, 4167–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torre-Amione, G., Beauchamp, R. D., Koeppen, H., Park, B. H., Schreiber, H., Moses, H. L. & Rowley, D. A. (1990) Proc. Natl. Acad. Sci. USA 87, 1486–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, Y. A., Dukhanina, O., Tang, B., Mamura, M., Letterio, J. J., MacGregor, J., Patel, S. C., Khozin, S., Liu, Z. Y., Green, J., et al. (2002) J. Clin. Invest. 109, 1607–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muraoka, R. S., Koh, Y., Roebuck, L. R., Sanders, M. E., Brantley-Sieders, D., Gorska, A. E., Moses, H. L. & Arteaga, C. L. (2003) Mol. Cell. Biol. 23, 8691–8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel, P. M., Shu, W., Cardiff, R. D., Muller, W. J. & Massague, J. (2003) Proc. Natl. Acad. Sci. USA. 100, 8430–8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muraoka, R. S., Dumont, N., Ritter, C. A., Dugger, T. C., Brantley, D. M., Chen, J., Easterly, E., Roebuck, L. R., Ryan, S., Gotwals, P. J., Koteliansky, V. & Arteaga, C. L. (2002) J. Clin. Invest. 109, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui, W., Fowlis, D. J., Bryson, S., Duffie, E., Ireland, H., Balmain, A. & Akhurst, R. J. (1996) Cell 86, 531–542. [DOI] [PubMed] [Google Scholar]
- 36.Weeks, B. H., He, W., Olson, K. L. & Wang, X. J. (2001) Cancer Res. 61, 7435–7443. [PubMed] [Google Scholar]
- 37.Hay, E. D. (1995) Acta Anat. (Basel) 154, 8–20. [DOI] [PubMed] [Google Scholar]
- 38.Potempa, S. & Ridley, A. J. (1998) Mol. Biol Cell. 9, 2185–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivaraman, V. S., Wang, H., Nuovo, G. J. & Malbon, C. C. (1997) J. Clin. Invest. 99, 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riese, D. J., III & Stern, D. F. (1998) BioEssays 20, 41–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.