Abstract
Alu and L1 are families of non-LTR retrotransposons representing ≈30% of the human genome. Genomic distributions of young Alu and L1 elements are quite similar, but over time, Alu densities in GC-rich DNA increase in comparison with L1 densities. Here we analyze two processes that may contribute to this phenomenon. First, DNA duplications in the human genome occur more frequently in Alu- and GC-rich than in AT-rich chromosomal regions. Second, most Alu elements tend to be coclustered with each other, but recently retroposed elements are likely to be inserted outside the existing clusters. These “stand-alone” elements appear to be rapidly eliminated from the genome. We also report that over time, the densities of recently retroposed Alu families on chromosome Y decline rapidly, whereas Alu densities on chromosome X increase relative to autosomal densities. We propose that these changes in the chromosomal proportions of Alu densities and the elimination of stand-alone Alus represent the same process of paternal Alu selection. We also propose that long-term Alu accumulation in GC-rich DNA is associated with DNA duplication initiated by elevated recombinogenic activities in Alu clusters.
Alu and L1 are families of non-LTR retrotransposons that have been actively retroposed throughout the evolutionary history of primates (1–3) and together contributed ≈30% of the DNA in the human genome (4). Both Alu and L1 elements are transcribed from a limited number of active source genes, reverse transcribed, and integrated to host DNA. The retroposed elements form subfamilies that share characteristic features with their source genes. The Alu source genes are ≈300 bp long and GC-rich, whereas L1 source genes are 6–7 kb long and AT-rich. Alu retrotransposition depends on reverse transcriptase encoded by active L1 retroelements (5–9), but the overall chromosomal distributions of Alu and L1 elements are quite different (10–12). L1s tend to be preserved in AT-rich DNA, whereas Alu are more abundant in GC-rich DNA. No standard biological mechanism has so far been able to explain this difference in the retroelement distribution (13, 14).
It has been shown recently that the chromosomal distributions of young Alu and L1 elements initially resemble each other but, unlike L1, the Alu distribution shifts toward GC-rich DNA over time (4). Based on this GC bias, it has been proposed that originally Alu elements are inserted relatively randomly throughout the genome but over time, they accumulate in GC-rich DNA (4, 15). The accumulation is particularly active for younger Alu, <5 million years (Myrs) old, but the mechanism of this process involving positive Alu selection in gene-rich regions appears to be controversial (4, 16, 17).
In this paper, we discuss Alu-mediated DNA duplication and selection against young Alus as two basic processes that might have contributed to the postinsertional evolution of Alu distribution. DNA duplications, also known as segmental duplications or low copy repeats, represent ≈5% of the human genome and have been studied extensively due to their association with genetic diseases (18–24). As demonstrated in this paper, segmental duplications frequently occur in GC-rich and Alu-dense DNA and, therefore, they can affect Alu distribution. However, they are unlikely to drive the initial accumulation of young Alus in GC-rich DNA, which appears to be caused by selection against Alu elements inserted outside the existing Alu clusters. This selection is probably related to paternal elimination of young Alu elements.
Materials and Methods
Nonredundant segmental duplications >1 kb long were downloaded directly from the publicly available database (http://humanparalogy.gene.cwru.edu/SDD) (19) and were analyzed in conjunction with the June 2002 version of the human genome sequence downloaded from the University of California, Santa Cruz, genome web site (www.genome.ucsc.edu) (25). Some analyses were based on the July 2003 version of the human genome, as indicated in the text or figure captions.
The human genome sequence was annotated by using both censor (26) (Version 4.1; www.girinst.org) and repeatmasker (A. F. A. Smit and P. Green, unpublished work; http://repeatmasker.genome.washington.edu). The annotation of Alu and L1 repeats was then crosschecked between the two programs. In most analyses, average Alu densities were calculated for 50-kb nonoverlapping windows. In the analysis of segmental duplications, we chose a range of intervals 20–50 kb long, because many duplications are relatively short. We also analyzed densities of transposable elements in overlapping windows to calculate Alu densities around reference sequences. The reference sequences were chosen from the relatively young AluYa5 and Yb8 subfamilies as well as from much older AluS and J subfamilies (27). Alu content was computed in 25-kb windows on both sides of each reference sequence (28). The pairs of windows were combined to give 50-kb intervals and sorted by base composition. The average Alu content was calculated for each group falling within the same 2% range of GC content. An analogous analysis was performed for primate-specific L1 elements.
The annotation of young Alu elements was based on direct alignment to recently published consensus sequences (29, 30). Classification of major subfamilies was verified by analyzing diagnostic positions. Repeat annotations, detailed Alu classification, and other large-scale data were downloaded to a mysql database and analyzed by using the database tools.
The tools for finding exact duplicates without alignment (adup and vdup), classifying Alu elements based on diagnostic positions (valu), and Perl scripts for analyzing Alu densities around other Alus are available on request. All supporting information quoted in this paper (Tables 1–3 and Figs. 6–8, which are published as supporting information on the PNAS web site) is also available at www.girinst.org/server/publ/PNAS.2004.
Results
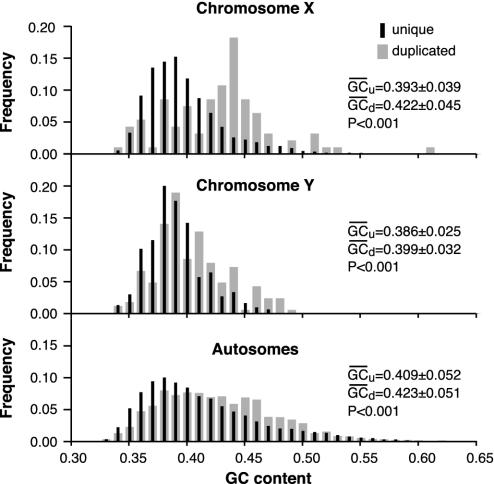
DNA Duplications Occur Primarily in GC-Rich DNA. We analyzed the base composition of long intrachromosomal duplications (>1 kb), also known as low copy repeats or segmental duplications (18, 19). The mean GC content of the segmental duplications is significantly higher than the GC content of the remaining genomic DNA (Fig. 1). The bias toward GC-rich DNA appears to be significant on all chromosomes and is highest on chromosome X and lowest on chromosome Y (Fig. 1).
Fig. 1.
Relative frequency distribution of GC content in duplicated and nonduplicated (unique) DNA. Base composition of segmental duplications (24) and of the remaining genomic DNA for segments >20 kb long. The corresponding mean GC content and the standard deviation are indicated. The percentage of GC content for nonredundant segmental duplications without any length limitations is: for chromosome X, 42.47 (39.13); Y, 39.98 (38.47); and autosomes, 42.57 (40.52). Numbers in parentheses indicate the corresponding genomic base composition after the duplicated segments were masked. Based on the two-tailed t test, all differences between base compositions are significant (P < 0.001).
A similar GC bias was detected between 50-kb segments (nonoverlapping windows) containing duplicated and nonduplicated Alu elements (Fig. 6). In this case, the pool of duplicated Alus included only identical elements with 50-bp flanks on each side. DNA segments containing such duplicated Alus are more GC-rich than segments with the remaining “unique” Alu repeats.
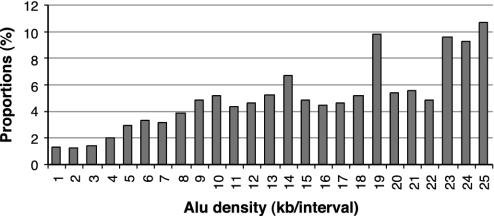
Alu duplications are more likely to occur in Alu-rich than in Alu-poor chromosomal regions. This is shown in Fig. 2, which illustrates the proportions of nonoverlapping windows that contain Alus also containing duplicated Alus, to all Alu-containing 50-kb windows for different ranges of Alu content per 50-kb window. Despite substantial variations, Alu duplications appear to be generally enhanced in Alu-dense regions in comparison to Alu-poor regions. The observed variations may be attributed to outliers such as sex chromosomes or chromosome 7. The former contain a large number of segmental duplications, whereas chromosome 7 includes a large Alu-rich segmental duplication. Nevertheless, even after removing the main outliers from the data analyzed, the basic pattern remains essentially the same (Tables 1 and 2 and Figs. 7 and 8). The observed variations in the proportions presented in Fig. 2 can also be caused by random loss of Alu-rich DNA segments mediated by Alu–Alu recombinations or other nonallelic homologous recombinations.
Fig. 2.
Proportions of nonoverlapping 50-kb segments harboring duplicated Alu to all Alu-containing segments, for different content of Alu per 50 kb. The duplicated Alu include at least one 50-bp flanking region (either 5′ or 3′).
Alu-mediated DNA recombination can lead to both duplications and deletions. Typically, Alu elements directly involved in recombination should retain the similarity of either the 5′ or the 3′ flanking sequence, but not both, to their duplicated copies, whereas Alu elements with both flanks duplicated are considered to be passively duplicated. We analyzed all categories of duplicated Alu present in the July 2003 version of the human genome. Of 17,362 Alu elements with either their 5′ or 3′ 50-bp flank exactly duplicated, as many as 15,205 (87.6%) had both flanks identical. This shows that ≈90% of duplicated Alus are not involved in Alu-mediated recombinations process (i.e., they are passively duplicated).
The remaining question is about the relative frequency of Alu duplications and their potential impact on overall Alu density. We analyzed proportions of duplicated Alu elements within and between different chromosomes by using the July 2003 sequence version of the human genome. The proportion of identical AluYs, each flanked by a 50-bp sequence on the 5′ side only, did not exceed 1% of all AluYs extracted from the July 2003 version of the human genome (by count or length). The relative proportions of duplicated AluYa5 and Yb8 elements determined during the same analysis were even smaller (≈0.5% and ≈0.1%, respectively). The figure was somewhat higher (≈1.4%) for duplicated genomic Alus from all subfamilies pooled together and higher in GC- than AT-rich DNA. In GC-rich DNA (GC >41%), the proportion of duplicated Alus from all subfamilies was ≈1.8%, with less than half of this (≈0.8%) in the remaining AT-rich DNA. In conclusion, the overall fraction of Alu elements duplicated in GC-rich DNA is relatively small. More importantly, it is even smaller for the young AluYa5 and Yb8 families, which are known to accumulate most rapidly in GC-rich DNA (4). A separate analysis of AluY repeats that are <10% diverged from each other, including their 50-bp-long 5′ flanks, yielded only 2.6% duplicated elements. This number, even if doubled due to possible underestimates, is still very small in comparison with the previously reported several-fold increase of AluY densities in GC-rich DNA (4). Therefore, DNA duplications are unlikely to be responsible for rapid early Alu accumulation in GC-rich DNA. However, even a small rate of Alu duplications in Alu-rich regions can substantially affect their densities over the long term, as discussed below.
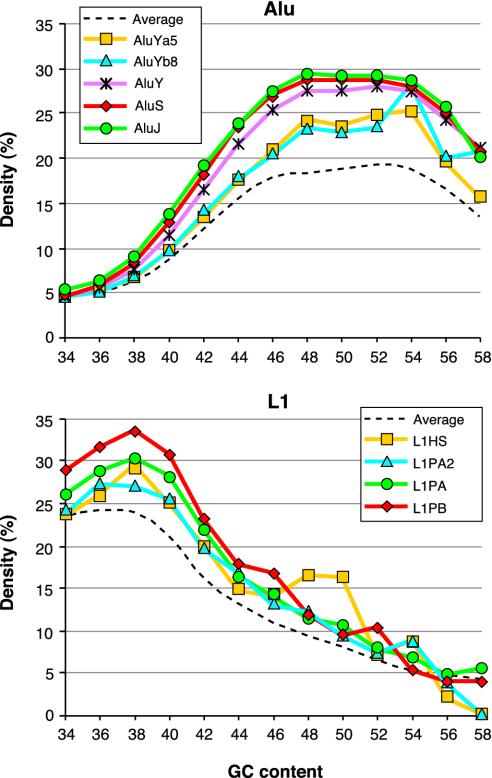
Clustering of Alu and L1 Elements. On average, there is about one Alu element in every 3 kb of the human genomic sequence. However, Alu elements are not uniformly distributed and tend to cocluster with each other (28, 29, 31). An interesting exception is young Alu elements, which are more often found outside the existing Alu clusters than are old ones. This is shown in Fig. 3, which compares Alu densities around elements from relatively young AluYa5 and Yb8 subfamilies with those around elements from older Alu subfamilies. The AluYa5 and Yb8 subfamilies are ≈<5 Myrs old and contain many recently retroposed elements. The AluY subfamily is ≈20 Myrs older, and the remaining two groups of subfamilies, AluS (excluding AluSc) and AluJ, are ≈20 and 40 Myrs older than AluY, respectively (32).
Fig. 3.
Alu–Alu and L1–L1 coclustering in the human genome sequence (July 2003 version). Densities of all Alu near the reference Alu sequences (Upper) and all L1 near the reference L1 sequences (Lower) were determined as described in Materials and Methods. They are plotted against average base composition of the analyzed 50-kb intervals. The subfamily classification of the reference sequences is indicated. Broken lines show average Alu and L1 densities in nonoverlapping 50-kb windows.
The figure shows that the AluYa5 and Yb8 subfamilies are present in less Alu-dense regions than are the remaining AluY elements, in perfect agreement with previous observations (28). The differences in Alu densities around the elements from AluS and J subfamilies are much smaller despite their substantial age difference. Therefore, the major shift toward Alu clusters can be observed in young subfamilies. The shift occurs in a wide range of base compositions but is smaller in AT- than in GC-rich DNA.
The trend toward coclustering can also be seen for primate-specific L1 subfamilies (28). As is shown in Fig. 3 Lower, the youngest subfamilies, L1HS and L1PA2, are in regions with a lower L1 density compared with older L1PA and still older L1PB elements (3). Unlike in the case of Alu, the largest shift toward L1-dense regions occurs in AT-rich DNA.
The data suggest that young Alu and L1 elements tend to be eliminated unless they are inserted in Alu- and L1-rich regions, respectively. This process may not be limited to Alu and L1 families, but its general significance remains to be substantiated. The elimination appears to be particularly active in young actively retroposing subfamilies, probably due to their random insertion patterns. However, it appears to continue in older AluY elements, albeit at a slower rate. Elimination of older elements may be affected by the ongoing insertions of younger ones (33).
Chromosomal Proportions of Young Alu Elements. The density of recently retroposed human AluY retroelements is approximately three times higher on chromosome Y than on chromosome X and about two times higher than on autosomes. The analogous ratio of Alu densities on chromosome X relative to autosomes is ≈2/3. These proportions suggest that Alu elements are retroposed primarily in paternal germlines (29).
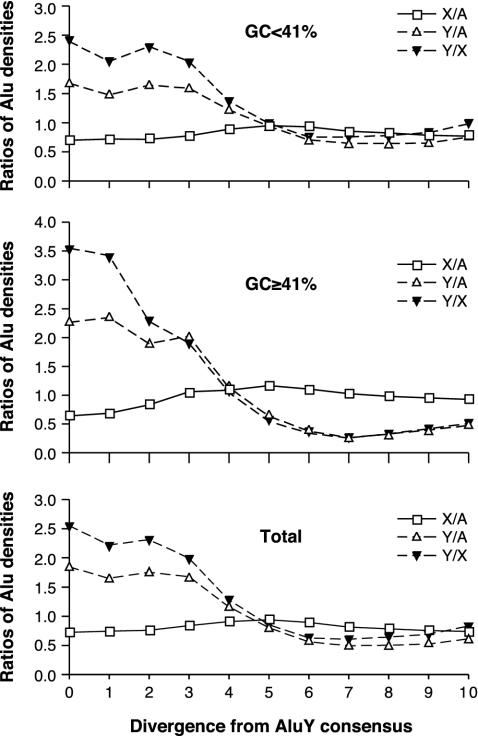
The proportions of young Alu elements appear to follow the paternal model of Alu insertions in both GC-poor and -rich DNA (29). This is illustrated in Fig. 4, where the chromosomal proportions of very young Alu elements are comparable between regions with above- and below-average genomic GC content (41%), suggesting similar Alu insertion patterns in DNA with different base compositions. However, recently inserted elements appear to be unstable and, with increasing Alu diversity, their density on chromosome Y declines rapidly relative to autosomal density. This decline is accompanied by a slight increase in relative Alu density on chromosome X.
Fig. 4.
Changes of Alu chromosomal proportions with their increasing average divergence from consensus. The younger the Alu elements, the more closely they follow the model of paternal retroposition in both AT- and GC-rich regions. Each point represents a ratio of cumulative Alu counts per 50 kb, from chromosomes indicated. For example, X/A indicates a ratio of Alu densities on chromosome X and autosomes. Intervals from left to right include fractions of Alu elements <1% divergent, 2% divergent, etc., from their respective subfamily consensus sequences, and the point corresponding to 10% divergence represents all Alu elements analyzed. No corrections were made for CpG or chromosome-specific mutation rates. All duplicated Alu elements were eliminated from the original set based on 5′ flanking regions. The dataset is based on the June 2002 version of the human genome. Supporting information is in Table 3.
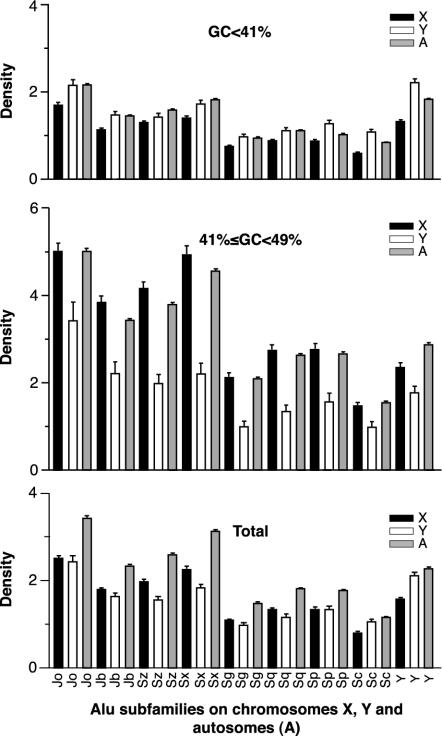
In Fig. 5, we compare the chromosomal densities of all major Alu subfamilies ordered from the oldest, AluJo, to the youngest, AluY. Because there are no multiple 50-kb segments on chromosome Y with GC content >49%, we chose 49% of GC as an upper limit for these comparisons. Fig. 5 Bottom shows Alu densities for all base compositions. As expected from the data in Fig. 4, Alu densities on sex chromosomes continue to change in opposite directions in both AT- and GC-rich regions. In AT-rich regions (GC <41%), the densities of Alu elements are still higher on chromosome Y than on chromosome X, although the original Y/X density ratios are reduced from the expected three to less than two. In GC-rich regions (41% ≤ GC < 49%), the Y/X ratios are less than one for all subfamilies. Thus changes of the original Alu proportions appear to be much more dramatic in GC- than in AT-rich regions. Overall, the most dynamic changes in chromosomal densities are between the youngest of the major subfamilies, particularly AluY and AluSc, and the rest (see Fig. 5 Bottom). Older subfamilies also tend to be more abundant in GC-rich regions than the younger ones both on sex chromosomes and autosomes. For example, the ratio of autosomal AluJo density in GC-rich DNA compared to the autosomal density in AT-rich DNA is 2.34. An analogous ratio for AluY elements is 1.57 and for AluSc, 1.86.
Fig. 5.
Densities of Alu elements in AT- and GC-rich regions and all regions combined on chromosomes X, Y, and autosomes. Each bar represents the average number of Alu per 50 kb of genomic DNA. Segments with GC content >49% are not included in these comparisons, because chromosome Y does not contain any significant DNA fragments with GC content over this limit. Higher error bars in GC-rich DNA reflect higher variations in Alu densities. The dataset is based on the July 2003 version of the human genome, with duplicated regions included.
Discussion
Segmental duplications occur most frequently in GC-rich chromosomal regions that are also Alu rich. In principle, such duplications can produce a systematic shift of Alu distribution toward GC-rich DNA. However, Alu repeats from the most rapidly accumulating young subfamilies such as AluYa5 and Yb8 appear to be underrepresented among duplicated Alu elements. By most estimates, the entire duplicated DNA represents 5–10% of the human genome, indicating that DNA duplications are unlikely to be responsible for the initial rapid accumulation of young Alu in GC-rich DNA.
The initial Alu accumulation can be explained by a selection process operating on young Alus. As is shown in Fig. 3, young AluYa5 and Yb8 elements tend to occupy less Alu-dense environment than their older relatives from the AluY subfamily, which in turn are in less-dense Alu regions than still-older AluS and AluJ elements (27). This systematic shift toward higher Alu densities appears to be larger in GC- than in AT-rich regions. It parallels the dynamics of changes in Alu densities on sex chromosomes and autosomes, as discussed below. The shift can be caused by the elimination of Alu repeats inserted outside preexisting Alu clusters. It can lead to highly nonuniform distribution of Alu densities, which is particularly visible in GC-rich chromosomal regions.
As is shown in Fig. 4, the density of recently retroposed Alu elements declines rapidly on chromosome Y relative to autosomes and slightly increases on chromosome X. A higher Alu deletion rate on chromosome Y relative with chromosome X may be a result of chromosome Y-specific processes, such as the recently described phenomenon of intrachromosomal gene conversion (34). Elimination of Alu elements may also be facilitated by the relatively low gene density on chromosome Y in comparison to autosomes and chromosome X (35). Although we cannot exclude chromosome Y-specific processes at this point, the correlated changes in Alu densities on sex chromosomes can also be attributed to paternal elimination of young Alus. The paternal model of Alu elimination predicts that the observed loss of Alus in the offspring genome should be the fastest on chromosome Y and half as fast on autosomes. The rate of Alu loss on chromosome X should be one-third of that on chromosome Y, because chromosome X is passed through the paternal germline about one-third of the time. Thus, higher rates of Alu removal in male germlines should cause decline in Alu densities on chromosome Y accompanied by a parallel increase in Alu densities on chromosome X relative to autosomal densities. This is the general trend that can be observed from the data in Fig. 5. Like the previously discussed shift of young Alus toward Alu clusters (Fig. 3), changes of the chromosomal Alu densities are also more striking in GC- than in AT-rich DNA. Therefore, we propose that fixation of Alus in clusters and changes in the Alu densities on different chromosomes represent the same process of Alu elimination in paternal germlines. The process may be driven by Alu fixation at neutral possibly duplicated chromosomal sites. A similar fixation of repetitive DNA in insects was proposed to occur in regions of restricted crossing over (36).
The detailed mechanism of paternally biased elimination of young Alus remains to be determined. One distinct possibility is that young CpG-rich Alus inserted outside the existing clusters can affect Alu methylation patterns on paternal chromosomes (37), which may lead to their elimination. A promising way to approach the problem may be analysis of Alu underrepresentation in imprinted regions (38).
Because Alu coclustering increases over time, particularly in GC-rich chromosomal regions, so does their instability. This is indicated by more frequent duplications in Alu-rich than in Alu-poor regions (Fig. 2). The instability can be caused by nonallelic homologous recombinations among Alu elements. Such recombinations can lead to deletions, duplications, and complex rearrangements not only of Alu elements but also of the adjacent regions (39). The longer the duplicated region that participates in the recombination, the less likely it is to be deleted from gene-rich DNA. This may be the basic mechanism generating long segmental duplications rather than deletions in GC- and gene-rich chromosomal regions (40). DNA deletions, particularly the long ones, are more likely to be nonlethal in AT-rich and gene-poor DNA. The same recombination initiated in Alu-dense regions can produce a small net Alu accumulation in GC-rich DNA and net elimination in AT-rich DNA.
Overall Alu density in GC-rich (>41% GC) regions is ≈15.5%. GC-rich DNA represents ≈36% of the human genome and ≈55% of all genomic Alu. The density of Alu elements in the remaining genomic regions is ≈7%. The difference in Alu densities can be attributed to both duplications and deletions in GC- and AT-rich regions, respectively. Assuming that half of the difference is due to Alu duplications in GC-rich DNA, one can calculate that ≈15% of all genomic Alu could be added by DNA duplication over ≈65 Myrs, in good qualitative agreement with 5–6% of segmental duplications accumulated during the last 30–35 Myrs (24).
Conclusion
The extensively debated accumulation of Alu elements in GC-rich DNA (4, 13, 14, 41) can reflect paternally driven selection against newly inserted Alus at nonneutral chromosomal sites. The process is likely to be responsible for Alu clustering, which in turn can stimulate nonallelic homologous recombination between Alu elements that can produce DNA duplications and deletions of different lengths. Large DNA deletions are more likely to be lethal in gene- and GC-rich regions than in gene-poor and AT-rich chromosomal regions. This may explain the more frequent occurrence of DNA duplications in GC-rich than in AT-rich regions, as reported in this paper. DNA duplications in Alu-dense regions can also lead to a limited accumulation of Alu elements in GC-rich DNA, consistent with the observed data.
While this manuscript was in review, we became aware of another article reporting Alu enrichment near or within junctions of segmental duplications (42). This report is consistent with our hypothesis that formation of segmental duplications may be triggered by intrachromosomal homologous recombinations in Alu clusters.
Supplementary Material
Acknowledgments
We thank Alison McCormack and Jolanta Walichiewicz for help with editing of the manuscript. We also thank Bernice Morrow for constructive criticism and advice, reviewer no. 2 for substantial improvements of the text, and all anonymous reviewers for excellent suggestions. This work was supported in part by National Institutes of Health Grant 2 P41 LM 06252-04A1.
Abbreviation: Myrs, million years.
References
- 1.Jurka, J. (1995) in The Impact of Short Interspersed Elements (SINEs) on the Host Genome, ed. Maraia, R. J. (Landes, Austin, TX), pp. 25–41.
- 2.Ostertag, E. M. & Kazazian, H. H., Jr. (2001) Annu. Rev. Genet. 35, 501–538. [DOI] [PubMed] [Google Scholar]
- 3.Smit, A. F. A., Toth, G., Riggs, A. D. & Jurka, J. (1995) J. Mol. Biol. 246, 401–417. [DOI] [PubMed] [Google Scholar]
- 4.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 5.Dombroski, B. A., Mathias, S. L., Nanthakumar, E., Scott, A. F. & Kazazian, H. H., Jr. (1991) Science 254, 1805–1808. [DOI] [PubMed] [Google Scholar]
- 6.Mathias, S. L., Scott, A. F., Kazazian, H. H., Jr., Boeke, J. D. & Gabriel, A. (1991) Science 254, 1808–1810. [DOI] [PubMed] [Google Scholar]
- 7.Moran, J. V., Holmes, S. E., Naas, T. P., DeBerardinis, R. J., Boeke, J. D. & Kazazian, H. H., Jr. (1996) Cell 87, 917–927. [DOI] [PubMed] [Google Scholar]
- 8.Jurka, J. (1997) Proc. Natl. Acad. Sci. USA 94, 1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewannieux, M., Esnault, C. & Heidmann, T. (2003) Nat. Genet. 35, 41–48. [DOI] [PubMed] [Google Scholar]
- 10.Soriano, P., Meunier-Rotival, M. & Bernardi, G. (1983) Proc. Natl. Acad. Sci. USA 80, 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, T. L. & Manuelidis, L. (1989) Chromosoma 98, 309–316. [DOI] [PubMed] [Google Scholar]
- 12.Korenberg, J. R. & Rykowski, M. C. (1988) Cell 53, 391–400. [DOI] [PubMed] [Google Scholar]
- 13.Wichman, H. A., Van den Bussche, R. A., Hamilton, M. J. & Baker, R. J. (1992) Genetica 86, 287–293. [DOI] [PubMed] [Google Scholar]
- 14.Medstrand, P., van de Lagemaat, L. N. & Mager, D. L. (2002) Genome Res. 12, 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazzarella, R. & Schlessinger, D. (1997) Gene 205, 29–38. [DOI] [PubMed] [Google Scholar]
- 16.Brookfield, J. F. (2001) Curr. Biol. 11, R900–R901. [DOI] [PubMed] [Google Scholar]
- 17.Bernardi, G. (2001) Gene 276, 3–13. [DOI] [PubMed] [Google Scholar]
- 18.Mazzarella, R. & Schlessinger, D. (1998) Genome Res. 8, 1007–1021. [DOI] [PubMed] [Google Scholar]
- 19.Bailey, J. A., Yavor, A. M., Massa, H. F., Trask, B. J. & Eichler, E. E. (2001) Genome Res. 11, 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichler, E. E. (2001) Trends Genet. 17, 661–669. [DOI] [PubMed] [Google Scholar]
- 21.Eichler, E. E. (2001) Genome Res. 11, 653–656. [DOI] [PubMed] [Google Scholar]
- 22.Ji, Y., Eichler, E. E., Schwartz, S. & Nicholls, R. D. (2000) Genome Res. 10, 597–610. [DOI] [PubMed] [Google Scholar]
- 23.Lupski, J. R. (1998) Trends Genet. 14, 417–422. [DOI] [PubMed] [Google Scholar]
- 24.Bailey, J. A., Gu, Z., Clark, R. A., Reinert, K., Samonte, R. V., Schwartz, S., Adams, M. D., Myers, E. W., Li, P. W. & Eichler, E. E. (2002) Science 297, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 25.Karolchik, D., Baertsch, R., Diekhans, M., Furey, T. S., Hinrichs, A., Lu, Y. T., Roskin, K. M., Schwartz, M., Sugnet, C. W., Thomas, D. J., et al. (2003) Nucleic Acids Res. 23, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jurka, J., Klonowski, P., Dagman, V. & Pelton, P. (1996) Comput. Chem. 20, 119–121. [DOI] [PubMed] [Google Scholar]
- 27.Jurka, J. & Smith, T. (1988) Proc. Natl. Acad. Sci. USA 85, 4775–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlicek, A., Jabbari, K., Paces, J., Paces, V., Hejnar, J. V. & Bernardi, G. (2001) Gene 276, 39–45. [DOI] [PubMed] [Google Scholar]
- 29.Jurka, J., Krnjajic, M., Kapitonov, V. V., Stenger, J. E. & Kohkanyy, O. (2002) Theor. Popul. Biol. 61, 519–530. [DOI] [PubMed] [Google Scholar]
- 30.Jurka, J. (2000) Trends Genet. 16, 418–420. [DOI] [PubMed] [Google Scholar]
- 31.Shakhmuradov, I. A., Kolchanov, N. A. & Kapitonov, V. V. (1989) Mol. Biol. 23, 526–536. [PubMed] [Google Scholar]
- 32.Kapitonov, V. V. & Jurka, J. (1996) J. Mol. Evol. 42, 59–65. [DOI] [PubMed] [Google Scholar]
- 33.Hayakawa, T., Satta, Y., Gagneux, P., Varki, A. & Takahata, N. (2001) Proc. Natl. Acad. Sci. USA 98, 11399–11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozen, S., Skaletsky, H., Marszalek, J. D., Minx, P. J., Cordum, H. S., Waterston, R. H., Wilson, R. K., Page, D. C., Kuroda-Kawaguchi, T., Hillier, L., et al. (2003) Nature 423, 873–876. [DOI] [PubMed] [Google Scholar]
- 35.Quintana-Murci, L. & Fellous, M. (2001) J. Biomed. Biotechnol. 1, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charlesworth, B., Langley, C. H. & Stephan, W. (1986) Genetics 112, 947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid, C. W. (1998) Nucleic Acids Res. 26, 4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greally, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babcock, M., Pavlicek, A., Spiteri, E., Kashork, C. D., Ioshikes, I., Shaffer, L. G., Jurka, J. & Morrow, B. E. (2003) Genome Res. 13, 2519–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoubak, S., Clay, O. & Bernardi, G. (1996) Gene 174, 95–102. [DOI] [PubMed] [Google Scholar]
- 41.Jurka, J. & Kapitonov, V. V. (1999) Genetica 107, 239–248. [PubMed] [Google Scholar]
- 42.Bailey, J. A., Liu, G. & Eichler, E. E. (2003) Am. J. Hum. Genet. 73, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.