Abstract
We analyze the evolutionary dynamics of three of the best-studied plant nuclear multigene families. The data analyzed derive from the genes that encode the small subunit of ribulose-1,5-bisphosphate carboxylase (rbcS), the gene family that encodes the enzyme chalcone synthase (Chs), and the gene family that encodes alcohol dehydrogenases (Adh). In addition, we consider the limited evolutionary data available on plant transposable elements. New Chs and rbcS genes appear to be recruited at about 10 times the rate estimated for Adh genes, and this is correlated with a much smaller average gene family size for Adh genes. In addition, duplication and divergence in function appears to be relatively common for Chs genes in flowering plant evolution. Analyses of synonymous nucleotide substitution rates for Adh genes in monocots reject a linear relationship with clock time. Replacement substitution rates vary with time in a complex fashion, which suggests that adaptive evolution has played an important role in driving divergence following gene duplication events. Molecular population genetic studies of Adh and Chs genes reveal high levels of molecular diversity within species. These studies also reveal that inter- and intralocus recombination are important forces in the generation allelic novelties. Moreover, illegitimate recombination events appear to be an important factor in transposable element loss in plants. When we consider the recruitment and loss of new gene copies, the generation of allelic diversity within plant species, and ectopic exchange among transposable elements, we conclude that recombination is a pervasive force at all levels of plant evolution.
Keywords: multigene family, alcohol dehydrogenase, chalcone synthase, rbcS, gene duplication
Plant molecular evolution has been dominated by studies of the chloroplast genome (cpDNA). There are several reasons for this focus on a single organelle that by itself accounts for less than 0.1% of the genetic complement of plants. First, cpDNA is an abundant component of total cellular DNA, and this facilitated the early molecular characterization of the cpDNA genome. Second, cpDNA turned out to have a conservative rate of nucleotide substitution (1), and slow rates of molecular evolution are ideal for the study of plant phylogenetic relationships at or beyond the family level. Because plant relationships are most controversial at deeper levels of evolution, cpDNA data promised to provide an important new tool for the reconstruction of plant phylogenies (2, 3). This promise has been borne out by extensive use of cpDNA-encoded genes to study plant phylogeny (4, 5). As a consequence, the bulk of research effort in plant molecular evolution has focused on problems in molecular systematics.
Despite a primary focus on molecular systematics, other important topics in cpDNA evolution have also been explored (6). For example, it is well established that cpDNA genes vary in rates of evolution among major plant lineages, violating the molecular clock hypothesis (7, 8). Studies of codon bias in chloroplast genes have uncovered a substantial bias of cpDNA genes favoring codons ending in A or T based on comparisons of liverwort, tobacco, and rice, which span the evolution of land plants (9). Recent studies also show a strong dependence on adjacent nucleotide site composition in transition/transversion rates in noncoding regions of cpDNA (10, 11). In addition, insertion/deletion mutations appear to be context-dependent because their rate of occurrence appears to increase in specific liable regions of both coding and noncoding DNA (9, 12, 13). Taken in combination, these and other studies reveal a complex mutational process that does not accord with simple models of molecular evolutionary change.
What about nuclear genes? Nuclear genes determine the vast range of phenotypes that are responsible for plant adaptation in nature, and yet knowledge of the molecular evolution of these genes is still at rudimentary stages. Our goal in this article is to discuss present knowledge of plant nuclear gene evolution. We do not attempt to be comprehensive, rather we select examples of genes and gene families that seem to us to illustrate important issues in gene evolution. Accordingly, we will restrict our discussion to a gene family that encodes a chloroplast enzyme component (small subunit of the enzyme ribulose-1,5-bisphosphate carboxylase, rbcS), a gene family that encodes an important component of plant secondary metabolism (chalcone synthase, Chs), and a gene family that encodes a glycolytic enzyme (alcohol dehydrogenase, Adh), and we will discuss pertinent facts that relate to the evolution of plant transposable elements.
It will be clear from the discussion that we lack detailed knowledge of the molecular evolution of plant nuclear genes. A comprehensive systematic sampling of plant nuclear genes has yet to be attempted, which makes it difficult to address many of the questions cited above for cpDNA evolution. However, one fact that does emerge from our consideration of plant nuclear gene evolution is the pervasive importance of recombinational processes at all levels of plant gene evolution.
Evolution of the rbcS Multigene Family
One interesting class of plant nuclear genes includes those that originally were components of the chloroplast genome but have been transferred to the nuclear genome and subsequently lost from the chloroplast genome. Within this class of genes are those encoding proteins involved in basic cellular processes, such as ribosomal proteins, and genes encoding proteins involved in photosynthesis. The best-studied among these transferred genes is rbcS, which encodes the small subunit of the enzyme ribulose-1,5-bisphosphate carboxylase. The enzyme is responsible for fixation of carbon in photosynthesis by catalyzing the condensation of carbon dioxide with the five-carbon sugar ribulose-1,5-bisphosphate to form two molecules of the three-carbon sugar 3-phosphoglycerate. The functional holoenzyme consists of eight identical, large subunits encoded by the chloroplast gene rbcL, and eight identical, small subunits encoded by the nuclear gene rbcS. Within cyanobacteria, rbcS and rbcL are adjacent and cotranscribed (14). However, sometime prior to the evolution of land plants rbcS was transferred to the nucleus, where it occurs in all lineages examined, including the green alga Chlamydomonas (15, 16).
Within diploid Angiosperms characterized so far, the rbcS gene family consists of two to eight copies. These copies are distributed among one or more loci, often on several chromosomes, and individual gene copies are frequently arranged in a tandem array at a locus. The gene typically consists of coding sequence for up to 189 amino acids (aa) that is interrupted by one intron in monocots and two or three introns in dicots. The carboxyl region of the translation product comprises a transit peptide necessary for targeting the polypeptide to the chloroplast stroma. The transit peptide is cleaved upon, or shortly following, arrival of the polypeptide into the chloroplast yielding the mature protein, which typically is 120–123 aa in length.
The sequence similarity is much higher for the mature protein than for the more variable transit peptide. Both 5′ and 3′ flanking sequences show very little sequence similarity within and between species. Sequence similarity among rbcS genes is hierarchical, with physically adjacent genes within a species showing the highest similarity, often identical in coding sequence, followed by genes at different loci within a species, and genes between species, which are the most diverged. There are no known functional differences associated with the very small differences in mature proteins within a species, and all gene products are considered to be functionally equivalent.
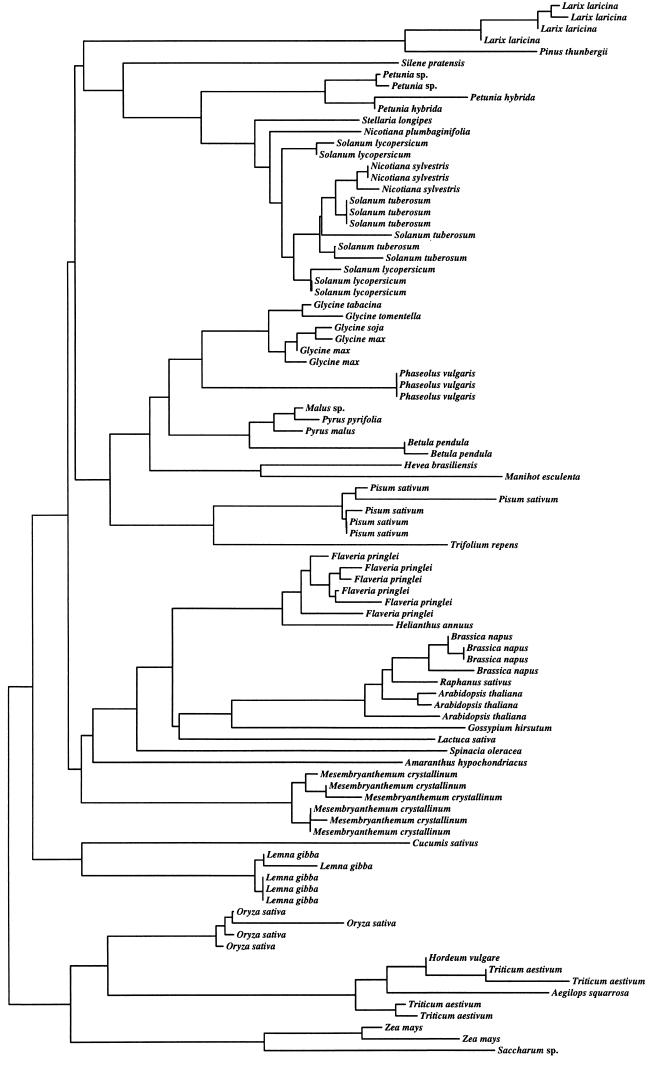
Although only a few species have been studied in detail, recombination appears to play an important role in the evolution of the rbcS gene family at several levels. The number of loci varies across Angiosperms in a pattern that implies both the loss and gain of loci (17). The data indicate expansions and contractions in number of gene copies, perhaps through slipped-strand mispairing, within tandem arrays. For example, locus 3 of petunia contains six copies of rbcS, whereas the homologous locus in tomato contains three (18). The overall view is that both loss and subsequent gain of gene copies, as well as homogenization of gene copies in situ via gene conversion, are important mechanisms that govern rbcS evolution within species. Interlocus gene-conversion events occur even for genes differing in number of introns (19). Thus, if we consider the evolutionary history of gene copies at different loci, we may observe first duplication followed by sequence divergence, and then recurrent returns to a state of complete identity owing to conversion events. This pattern of evolution causes all of the copies within a family or even a genus to be clustered together consistent with the rbcS phylogeny depicted in Fig. 1.
Figure 1.
A neighbor-joining tree depicting the relationships of the mature gene products of rbcS. The data were taken from GenBank subject to the following restrictions. (i) Only sequences that differed by 5% or more in primary nucleotide sequence were incorporated in the analysis to avoid the inclusion of allelic sequences. (ii) Only sequences that represented a minimum of 50% of the gene were included to avoid biases associated with very short sequences. Amino acid sequences were aligned and the neighbor-joining tree (20) was constructed based on corrected distances (21) using the program clustal w (22).
Evolution of Genes in the Flavonoid Biosynthesis Pathway
Another class of well studied nuclear genes that is important in plant adaptation and evolution is the genes of the flavonoid biosynthetic pathway. The pathway is shown in Fig. 2. The end product and side branches of the pathway lead to a general class of phenolic compounds known as flavonoids. Flavonoids have many functions in plants. The colored pigments localized in the vacuoles of flowers act as attractants to pollinators (23). Polymorphisms in flower color can influence pollinator behavior and, ultimately, genetic transmission (24). Flavonoids are also important in protection against UV light (25) and in defense against pathogens and insects (26, 27). Flavonoids are important in induction of nodulation (28), in auxin transport (29), and in pollen function (30). The products of this secondary metabolic pathway enable the plant to better adapt to a stressful environment. The study of the evolution of these genes thus is essential to our understanding of the processes that determine adaptive evolution.
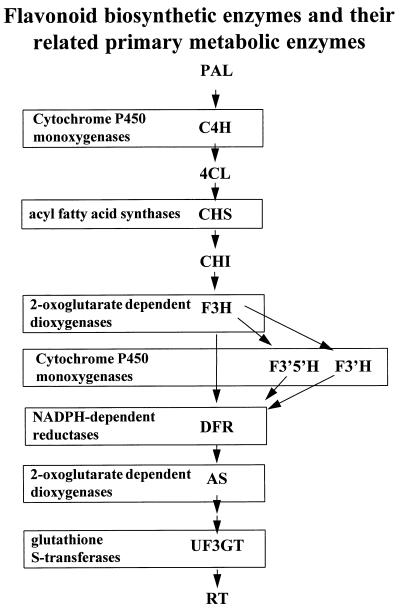
Figure 2.
Anthocyanin and flavonol biosynthetic pathway. The enzymes in bold represent the core genes of flavonoid biosynthesis. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-coenzyme A ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′5′-hydroxylase; DFR, dihydroflavonol 4-reductase; AS, anthocyanidin synthase; UF3GT, UDP-glucose flavonoid 3-oxy-glucosyl transferase; RT, rhamnosyl transferase. Also shown within the box associated with enzyme designations are the gene super families from which particular enzymes are thought to be derived.
Evolution of the Flavonoid Biosynthetic Pathway and Associated Regulatory Genes.
An important question is: “How do pathways composed of a series of sequential steps evolve to produce an end product?” Stafford (31) postulates that initially each step in plant secondary metabolism derived from an enzyme of primary metabolism and resulted in an intermediate product that was temporarily an end product that conferred some advantage to the plant. For example, chalcone synthase (CHS) is thought to share a common origin with fatty acid synthases of primary metabolism (32) (Fig. 2). The reaction catalyzed by CHS utilizes substrates from both the phenylpropanoid and malonyl CoA pathways of primary metabolism. Subsequent steps in the flavonoid pathway presumably “borrowed” hydroxylases, NADPH-reductases, and glutathione transferase from primary metabolism. As the flavonoid pathway expanded, each new intermediate resulted in some selective advantage such as defense from pathogens or herbivores and UV protection (31).
Another layer of complexity in the evolution of a pathway is the regulation of gene expression in terms of timing and tissue-specific expression. Fig. 3 shows the known regulatory genes for two dicot species in the genera Petunia and Antirrhinum (33, 34). The regulatory genes of the flavonoid pathway appear to regulate groups of genes as opposed to individual genes. This may act to organize the pathway into a biosynthetic complex or unit for more efficient functioning of the pathway. Whereas one might envision a single gene coming under new regulatory control by means of chromosomal rearrangement or a transposition event, it is hard to envision a mechanism by which several different genes might come under the control of the same regulatory gene. More evolutionary studies on the regulatory genes of flavonoid biosynthesis are needed to provide insight into the adaptive basis of this complex regulatory network.
Figure 3.
Regulation of the anthocyanin pathway in two dicot species. (Asterisks denote genes for which clones have been made from Ipomoea.) The regulatory genes that have been identified in Antirrhinum and Petunia are shown along with the genes that they regulate. Abbreviations are listed in the legend for Fig. 2.
Duplication and the Acquisition of New Functions.
The role of duplication and differentiation in evolution is well illustrated by the genes that encode the first committed step in flavonoid biosynthesis. This step is initiated by the enzyme chalcone synthase, which catalyzes the condensation of three molecules of malonyl CoA and one molecule of p-coumaroyl CoA to produce the 15-carbon naringenin chalcone molecule, which is then further modified in a series of enzymatic steps leading to the colored anthocyanin end product (33).
An example of such an event of duplication and differentiation is stilbene synthase (STS). Only a limited number of amino acid changes are required to convert CHS to STS (35). The resulting stilbene phytoalexins produced by the enzyme STS have antifungal properties that confer defense against plant pathogens (36). Phylogenetic analyses indicate that the recruitment of the stilbene synthase function from chalcone synthase has occurred independently several times in the course of land plant evolution (35). In addition, Durbin et al. (37) and Helariutta et al. (38) both report unusual CHS-like gene sequences that differ from both CHS and STS, suggesting that these enzymes are functionally divergent from both CHS and STS.
In the morning glory genus (Ipomoea), several Chs genes are more closely related to the unusual ChsB gene of Petunia than to other Petunia Chs genes (37). It has already been speculated that the ChsB Petunia gene is in the process of either acquiring a new function or being inactivated (39). Within Ipomoea these Chs genes appear to have diverged into at least two distinct groups based on amino acid substitutions. In addition, the ratio of synonymous-to-replacement polymorphism is low (about 5.6:1 in Ipomoea) compared with other taxa (e.g., 10:1 in grasses, 16:1 in legumes, and 42:1 in solaneaceous plants; ref. 40).
Molecular Population Genetics of Chs Genes.
Huttley et al. (41) sampled ChsA genes of the common morning glory (Ipomoea purpurea) from 18 lines that originated from broad geographic collections in Mexico and the southeastern United States. No nucleotide sequence diversity was detected at ChsA in the very limited sample of four lines from Georgia and North Carolina. The Mexican-derived materials had much higher levels of nucleotide sequence diversity with 11 distinct haplotypes. Further examination of the ChsA sequences reveals that the majority of variation resides in exons, whereas flanking sequences and introns show very little polymorphism. Finally, a comparison between the ChsA allele polymorphic sites indicates that at many of these sites one of the nucleotide states is present in all non-ChsA gene family members, and these observations strongly suggest that both the high level of nucleotide diversity present in the I. purpurea ChsA genealogy and the relatively low ratio of synonymous-to-replacement substitutions between ChsA alleles are probably derived from low to moderate rates of interlocus recombination/gene conversion among the different Chs gene family members.
To summarize, the Chs genes in Ipomoea have duplicated and diverged in function and they have diverged in specific expression patterns during Ipomoea evolution (ref. 37; unpublished data). In addition, the genes are quite variable both within and between species, but, unlike most cases analyzed, the variation is enhanced in coding regions rather than in noncoding regions (5′ and 3′ flanking regions and introns). The Ipomoea Chs genes also appear to be evolving at a very rapid rate (41). All the evidence indicates that some of the Chs genes have been subject to adaptive evolution, but the specific phenotypic effects associated with Chs evolution remain obscure.
The Alcohol Dehydrogenase Gene Family
Alcohol dehydrogenase (Adh) genes encode glycolytic enzymes that have been characterized at the molecular level in a wide range of flowering plant species and in one conifer species. Alcohol dehydrogenase (ADH) is an essential enzyme in anaerobic metabolism (42, 43). Transcription from Adh promoters increases under oxygen stress as well as in response to cold stress in both maize and Arabidopsis and to dehydration in Arabidopsis (43). Two or three isozymes are observed in all flowering plant species (44), with the exception of Arabidopsis, which appears to have a single Adh locus (45).
Duplication and Divergence Among Adh Gene Family Members.
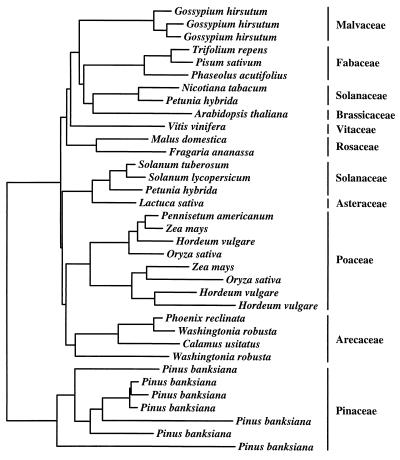
Unlike the previous examples of multigene families that we have considered, the Adh genes in flowering plants are encoded by small multigene families that generally appear to have approximately three duplicate members (46). Isozyme surveys covering an array of dicot and monocot species have revealed that most glycolytic enzymes have two forms in all species (44), probably reflecting a small, and stable, number of loci. The narrow range of gene family size for glycolytic enzymes suggests that additional constraints may also act to determine copy number for this important class of genes. Fig. 4 suggests a slow flux of gene duplication and loss that leads to an approximate dynamic equilibrium in copy number.
Figure 4.
Neighbor-joining tree depicting the relationships of ADH amino acid sequences. The data selection criteria and analytical methods are the same as those described for Fig. 1.
Molecular Clocks for Adh Genes.
The molecular clock hypothesis is one of the fundamental ideas of molecular evolution. The strict molecular clock hypothesis posits a linear rate of accumulation of nucleotide substitution over time (21). Most careful investigations have rejected the strict molecular clock, but a modification known as the generation–time–effect hypothesis, which posits constant rates of nucleotide substitution when time is measured in generation intervals rather than clock time, can also be examined (21, 47). The difficulty with a generation–time-based clock is that it has little practical utility because generation times vary widely both among major lineages and over evolutionary time within lineages.
Owing to the extensive database for the chloroplast gene rbcL, it has been possible to thoroughly investigate the molecular clock hypothesis among seed plants for this gene (7, 8). These investigations reject a strict molecular clock, and, within monocots, the data suggest a correlation of synonymous substitution rate with minimum generation time (7). The extent of rate variation within monocots is substantial where rate estimates are approximately 5-fold greater for grasses than for palms. This very large rate contrast presented the opportunity to ask whether synonymous rates showed similar variation for nuclear genes.
Because the Adh gene family is one of the most thoroughly studied plant gene families, it was natural to study both relative and absolute rates of nucleotide substitution for this gene family in grasses and palms. Two Adh loci, adh1 and adh2, have been sequenced for maize and rice as well as barley, from which a third locus, adh3, a recent duplication of adh2, has been isolated (citations in ref. 48). In addition, three Adh loci have been fully or partially sequenced from one or more of three palm genera (46, 48). Absolute synonymous rate estimates for the palm Adh loci are 2.6 × 10−9 in contrast to 7.0 × 10−9 for grass Adh loci. This difference is significant and indicates a deceleration of synonymous substitution rates in palms in parallel with the rbcL gene; however, the difference is closer to 3-fold rather than the 5-fold difference estimated for the rbcL gene. Replacement rates are much more complex. There is strong rate variation between gene family members within the grass lineage, suggesting that positive Darwinian selection played a role in determining rates. Thus, for example, the adh2 locus of grasses appears to be evolving at a rate about 3-fold higher than the adh1 locus of grasses, in the time interval since these two genes duplicated from a common ancestral gene. In addition, the palm adhA locus has a replacement rate that is approximately equal to the grass adh1 locus (48), suggesting a much more rapid replacement rate in palms when time is measured in generations.
Molecular Population Genetics of Adh Genes.
Gaut and Clegg (49, 50) have studied the evolution of adh1 within the genus Zea and within Pennisetum glaucum (pearl millet, a grass species in the same subfamily as Zea). Eight adh1 sequences were determined from a wide sample of Zea (including both inbred lines and three land race entries of Zea mays ssp. mays). The Zea sample also included the teosinte species Z. luxurians and Z. diploperennis. The sequence data spanned a 2.1-kb region of the gene between exon 3 and exon 10. Interestingly, the adh1 sequence data from Zea do not discriminate between the different Zea species in the sample. Indeed, the Z. luxurians and Z. diploperennis adh1 lineages were clustered within the range of Z. mays ssp. mays sequence variation.
The adh1 data were subjected to a series of statistical tests to detect selection or interallelic recombination within the Zea samples and within 21 pearl millet adh1 lineages. Tests for interallelic recombination revealed that a minimum of two adh1 alleles were derived from interallelic recombination, and it appears that at least one allele of the 21 pearl millet alleles also has a history of interallelic recombination (49, 50). These results establish that interallelic recombination in plants, as is also the case in Drosophila, can be an important source of allelic novelty. Tests for selection did not reject the null hypothesis of neutrality, although these tests have limited power and a failure to reject the null hypothesis for such small samples is to be expected. What is interesting is that both the coalescence times and the effective population sizes estimated from the data appear to be large. A large effective population size is somewhat surprising in view of the recent history of strong selection for domestication in both species (51).
Rates of Duplication and Loss Among Gene Families
What have we learned from a comparison of evolutionary patterns in three different gene families? One question of particular interest is to estimate the rate of recruitment of new genes within gene families and within major plant lineages. Before addressing this question, it is important to first discuss the limitations of the present data. To begin with, the great majority of present data was not collected with the goal of addressing evolutionary questions. Consequently, the data are very unevenly distributed across plant taxa. Second, gene family numbers are almost always underestimated. This follows because most investigators are not interested in an exhaustive description of all members of a gene family, and even when that is the goal the vagaries of cloning or PCR make a complete description difficult to accomplish. Finally, there are two processes that can lead to two identical gene copies at different loci (bifurcation of a gene lineage). The first is the origin of a new locus through duplication, and the second is the conversion of genes at two preexisting loci into identical copies. These two classes of events are indistinguishable when the data are solely based on evolutionary comparisons. Accordingly, we count the number of gene loci that have descended from a particular lineage within plant families, but this count confounds duplications and complete conversions. To ensure that our count is conservative, we assume that two gene sequences represent separate loci if and only if they differ by more than 5% in primary nucleotide sequence. With these strong caveats, we have calculated the average number of independent lineages for the Adh, Chs, and rbcS gene families within each of four plant families (Table 1). These data suggest a more rapid rate of duplication (and perhaps recruitment of new function) for the Chs and rbcS gene families than for the Adh gene family. The data also suggest that the appearance of new gene copies occurs infrequently at the family level. If we take the data at face value, and assume times of 70 million years (MY) for the origin of the Poaceae, 56 MY for the Fabaceae, 40 MY for the origin of the Asterceae, and 40 MY for the Solanaceae, we calculate new gene recruitment rates of 2.9 × 10−8 for Adh, 2.8 × 10−7 for Chs, and 2.7 × 10−7 for rbcS. This represents roughly a 10-fold range, with the highest rates observed for the Chs genes in the Fabaceae and the Solanaceae.
Table 1.
Numbers of new gene recruitments within plant families for the Adh, Chs, and rbcS gene families
| Family |
Adh
|
Chs
|
rbcS
|
|||
|---|---|---|---|---|---|---|
| Lineages | Copies | Lineages | Copies | Lineages | Copies | |
| Poaceae | 1 | 3 | 1 | 2 | 1 | 4 |
| Asteraceae | 1 | 1 | 2 | 4 | 2 | 5 |
| Fabaceae | 1 | 1 | 2 | 7 | 2 | 5 |
| Solanceae | 2 | 2 | 2 | 7 | 1 | 5 |
The actual range of gene family size is one to three copies for Adh, roughly two to seven for Chs, and approximately two to eight for rbcS; so why do we not see higher levels of recruitment for the rbcS genes, which appear to have slightly greater gene family sizes on average? One important consideration is the rate of homogenization among family members through recombination/conversion. The rbcS genes, for instance, are often organized in adjacent arrays where recombinational processes would be facilitated, and we have presented direct evidence for interlocus recombination/conversion in Chs genes in Ipomoea (41). The actual rate of duplication may be considerably above the recruitment rates calculated here, because most new duplicates are unlikely to escape and establish an independent lineage. Instead, the fate of many new duplicates may be conversion back to the sequence of a preexisting gene copy. Other factors such as cosuppression may also act as a barrier to the establishment of new duplicate genes (52).
There must be a strong pressure for divergence in function, or in expression patterns, before an escape is favored. It appears that the Chs genes satisfy these constraints, because divergence in function and expression pattern is frequently observed. Recurrent duplication of the rbcS genes may be favored, owing to a requirement for high rates of translation to match the synthesis of the chloroplast-encoded large subunit. Of course, these arguments beg the question of gene loss. We must assume a rough equilibrium between the recruitment of new gene copies and their loss because we observe a rough stability in copy number for each family. So what drives gene loss? Again, it seems likely that recombinational processes are a major factor. We will discuss evidence below that implicates ectopic exchange in transposable element loss, and the same processes are likely also to lead to the production of pseudogenes within gene families that quickly diverge (in evolutionary time) to unrecognizable sequences.
Evolution of Plant Transposable Elements
Transposable elements (TEs) are a heterogeneous assemblage of discrete DNA sequences that are capable of autonomous or semiautonomous movement from one genomic location to another. There have been very few systematic studies of TE evolution within plant species. Most current knowledge relates to element classification, modes of excision/replication, and genome abundance of various element classes.
Element Classification.
Transposable elements in plants, as in other organisms, fall into two broad categories, class I and class II. Class I transposable elements, often referred to as retrotransposons, are related to retroviruses but differ from them in that they do not form viron particles, which means they are not intrinsically transmissible between cells. As with all retroelements, the replication of class I elements involves reverse transcription, which is the synthesis of DNA from template RNA. Most of the DNA sequence of these elements encodes protein-coding sequences, including the enzyme reverse transcriptase, which catalyzes reverse transcription, and cis-acting sequences required for replication. Retrotransposons exhibit replicative transposition, i.e., transposition does not require excision of an element; rather, a new additional copy of the element is formed. These elements do not exhibit a precise excision process but rather appear to be lost through recombination between long terminal repeat (LTR) sequences or deletion events. Many retrotransposons exhibit a pattern of targeted integration whereby element insertions are much more frequent in chromosomal regions away from genes, thus decreasing the relative frequency of transposon-induced mutations (53, 54). Targeted integration may help explain both the very large population sizes and apparently low number of mutant phenotypes associated with class I elements.
Class I elements are divided into several broad categories based on arrangement of protein-coding domains and the presence or absence of LTRs. The non-LTR retrotransposons have been more extensively studied in mammals (the L1 or LINE elements) but do occur in plants, for example, Cin4 of maize (55) and del2 of lily (56). Retrotransposons with LTRs are usually referred to by the names given homologous elements first found in Saccharomyces cerevesiae and Drosophila melanogaster and are present in all plants examined (57, 58). Unrivaled in terms of numbers, LTR retrotransposons compose the largest class of transposable elements in plant genomes. The Ty1/copia elements constitute as much as 50% or more of the maize genome (53), and sizable fractions of the lily genome are composed of Ty3/gypsy elements (59). Within plants these elements have been most thoroughly studied in Arabidopsis thaliana (60–63). With low copy numbers in Arabidopsis and extremely high copy numbers in maize and lily, the abundance of retrotransposons singularly explains most of the observed variation in plant genome size.
There is a broad range of class II transposable elements (also referred to as short inverted repeat elements after a common characteristic of the group). Unlike class I elements, transposition of class II elements is directly from DNA to DNA (i.e., does not involve an RNA transposition intermediate), and replication is conservative or nonreplicative, meaning that transposition is coupled with excision. However, these elements may increase in copy number when an element-containing DNA strand is used as a template in double-strand gap repair of an empty target site (64). Several structural features typically characterize these elements, including a single open reading frame enclosed by inverted repeat sequences and sometimes containing introns, and often short, target-site duplications that are created upon integration. The protein product of the open reading frame is usually referred to as a transposase, although little is known about its functional attributes. Some members of this class consist of autonomous and nonautonomous elements within the same genome, for example, the Ac/Ds system of maize. The nonautonomous elements are capable of transposition in the presence of autonomous elements through trans activation.
Several groups of class II elements can be distinguished based on their amino acid sequences, for example, the Ac/hobo group, which includes the Ac/Ds system of maize and relatives (Zea spp.); the Tam3 of snapdragon (Antirrhinum majus); and hobo of Drosophila. Other examples of class II elements in plants include elements not yet recognized as members of widespread groups, such as Spm and Mu of maize.
Evolutionary Studies.
As noted above there are very few studies whose objective is to describe patterns of plant TE evolution. Among the rudimentary studies that do exist are several of Ac/Ds element evolution in the grass family (65). These data suggest that Ac elements have been in the genome grasses since the origin of the family, approximately 70 million years ago. They also suggest that ectopic exchange plays a major role in generating nonfunctional elements through illegitimate recombination. Owing to the complete absence of any comprehensive systematic study of plant TE distributions, there is no compelling evidence for or against horizontal transmission of plant transposable elements. However, factors such as large population sizes, high mutation rates, and frequent recombination make it very difficult to establish compelling evidence for horizontal transmission (66)
Unanswered Questions.
We can only speculate about the selective forces responsible for structuring transposable element populations within a genome. Clearly, factors such as negative selection associated with deleterious mutations and metabolic costs resulting from replication of increased genetic material have to be rethought given the very large portion of the genome that is composed of transposable elements. Possible positive selective forces that might influence transposable element dynamics also need to be considered, including the role of transposable elements in recombination and chromosome mechanics.
The dynamics of plant transposable element populations have yet to be clearly described. What proportion of the transposable element population is transpositionally active? For example, are most elements capable of transposition, or are there relatively few active elements that are the source for most transposition events, and what is the role, if any, of DNA methylation in regulating transposition? Following the previous points, how can models of transposable element evolution be improved? Most of the previous models are based on class II elements in Drosophila and Escherichia coli and are on the whole very inadequate for capturing principal features of class I elements, especially in plants. Given the evolutionarily and genetically heterogeneous nature of transposable elements, more specific models will have to be developed if congruence with empirical observation is to be improved. Current data do suggest that recombinational processes play a major role in TE replication and loss, so we may conclude that recombinational processes play a pervasive role in the fate of TEs just as recombination plays a major role in the evolution of plant gene families.
Conclusions
The elemental processes that govern plant gene evolution involve nucleotide substitution, the insertion or deletion of strings of nucleotides, and recombination/conversion between gene copies. As a consequence of these process, we observe increases and decreases in copy number, divergence in function, and divergence in expression patterns. Our study of plant nuclear gene evolution suggests that these processes are both necessary and sufficient to account for observed patterns of gene evolution. Nevertheless, many questions emerge from these data. In the case of the rbcS gene family, the original genes trace to a prokaryotic ancestor. We must assume that some kind of recombinational process facilitated the incorporation of the original plastid genes into the nuclear genome. Subsequent processes led to the duplication and elaboration of the rbcS gene family, but why are the rbcS genes constrained to an approximate upper bound of 10? Why not have many more copies to match the plastid-based synthesis of the large subunit polypeptide? There is good evidence that occasional gene conversion acts to homogenize rbcS gene family members. What other processes lead to the loss of gene copies? We can speculate that illegitimate recombination occasionally leads to pseudogenes that rapidly decay owing to nucleotide substitution. Is this speculation correct? Unfortunately, present data are too sparse to to allow us to measure gene-loss rates to ask whether an approximate equilibrium exists between the loss and gain of copy number.
The Chs gene family arose through the evolution of a novel function, which then precipitated the gradual elaboration of a new biosynthetic pathway. There is good evidence for repeated functional divergence of Chs genes based on patterns of amino acid substitution within flowering plants. Because the Chs gene copy number varies within reasonably narrow limits, we must assume that there are controls on copy number; so what determines the number of Chs gene copies in a typical plant genome? Why is this enzyme so plastic and so easily adapted to new uses? In contrast, alcohol dehydrogenase genes presumably have retained a unitary function and show less evolutionary elaboration through time, but why the relatively narrow limits on family size in flowering plants? Why is there a low rate of recruitment of new copies? Are low copy numbers characteristic of most or all glycolytic enzymes as speculated by Morton et al. (46), and, if so, what determines the optimum number of copies?
The data we have reviewed also shed new light on several important questions about plant gene evolution and about organismic evolution. For instance, we have learned that inter- and intrallelic recombination are important processes in generating allelic novelties. We have rejected the strict molecular clock hypothesis, and we have obtained crude estimates for the rates of recruitment of new gene copies for several important gene families. Finally, we have learned that historical species’ effective populations sizes are large for crop plants like maize and pearl millet.
What else have we learned from our brief survey of the ecology of plant genomes that might have surprised Dobzhansky and caused him to modify his views of evolutionary processes? The elemental processes of genetic change, enumerated above, were appreciated by Dobzhansky and his contemporaries, and there appears to be no need to invoke new processes of genetic change. But, genes and genomes have been revealed to be much more complex in their organization than suspected during Dobzhansky’s life. Introns were discovered in 1975, the year of Dobzhansky’s death, and molecular proof of transposable elements was also obtained in the mid-1970s. Genetic change now appears to occur at several levels. One level is the nucleotide and its associated influence on protein structure or on DNA-binding signals. A second level is the gene, because new copies may be recruited and elaborated through time. A third level is associated with the activity of mobile elements that infect and mutate genomes. Recombinational processes act at each of these levels to convert sequence information among loci, to disrupt transposon and other duplicate gene sequence continuity, to generate allelic diversity, and to recruit new gene copies. Perhaps Dobzhansky would have accorded a greater significance to the role of recombination in evolution were he writing today.
Acknowledgments
This work was supported in part by a grant from the Alfred P. Sloan Foundation.
ABBREVIATIONS
- ADH
alcohol dehydrogenase
- CHS
chalcone synthase
- TE
transposable element
- STS
stilbene synthase
- LTR
long terminal repeat
References
- 1.Curtis S E, Clegg M T. Mol Biol Evol. 1984;1:291–301. doi: 10.1093/oxfordjournals.molbev.a040319. [DOI] [PubMed] [Google Scholar]
- 2.Ritland K, Clegg M T. Am Nat. 1987;130:S74–S100. [Google Scholar]
- 3.Clegg M T. Proc Natl Acad Sci USA. 1993;90:363–367. doi: 10.1073/pnas.90.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chase M W, Soltis D E, Olmsted R G, Morgan D, Les D H, et al. Ann Missouri Bot Gard. 1993;80:528–580. [Google Scholar]
- 5.Soltis D E, Soltis P S. Evol Biol. 1995;28:139–194. [Google Scholar]
- 6.Clegg M T, Gaut B S, Learn J H, Jr, Morton B R. Proc Natl Acad Sci USA. 1994;91:6795–6801. doi: 10.1073/pnas.91.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaut B S, Muse S V, Clark W D, Clegg M T. J Mol Evol. 1992;35:292–303. doi: 10.1007/BF00161167. [DOI] [PubMed] [Google Scholar]
- 8.Bousquet J, Strauss S H, Doerksen A H, Price R A. Proc Natl Acad Sci USA. 1992;89:7844–7848. doi: 10.1073/pnas.89.16.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morton B R, Clegg M T. Curr Genet. 1993;24:357–365. doi: 10.1007/BF00336789. [DOI] [PubMed] [Google Scholar]
- 10.Morton B R. Proc Natl Acad Sci USA. 1995;92:9717–9721. doi: 10.1073/pnas.92.21.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morton B R, Clegg M T. J Mol Evol. 1995;41:597–603. doi: 10.1007/BF00175818. [DOI] [PubMed] [Google Scholar]
- 12.Golenberg E M, Clegg M T, Durbin M L, Doebley J, Ma D P. Mol Phyl Evol. 1993;2:52–64. doi: 10.1006/mpev.1993.1006. [DOI] [PubMed] [Google Scholar]
- 13.Cummings M P, King L M, Kellogg E A. Mol Biol Evol. 1994;11:1–8. doi: 10.1093/oxfordjournals.molbev.a040084. [DOI] [PubMed] [Google Scholar]
- 14.Nierzwicki-Bauer S A, Curtis SE, Haselkorn R. Proc Natl Acad Sci USA. 1984;81:5961–5965. doi: 10.1073/pnas.81.19.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldschmidt-Clermont M, Rahire M. J Mol Biol. 1986;191:421–432. doi: 10.1016/0022-2836(86)90137-3. [DOI] [PubMed] [Google Scholar]
- 16.Simard C, Lemieux C, Bellmare G. Curr Genet. 1988;14:461–470. doi: 10.1007/BF00521270. [DOI] [PubMed] [Google Scholar]
- 17.Manzara T, Gruissem W. Photosynth Res. 1988;16:117–139. doi: 10.1007/BF00039489. [DOI] [PubMed] [Google Scholar]
- 18.Dean C, Pichersky E, Dunsmir P. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:415–439. [Google Scholar]
- 19.Meagher R B, Berry-Lowe S, Rice K. Genetics. 1989;123:845–863. doi: 10.1093/genetics/123.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, MA: Cambridge Univ. Press; 1983. [Google Scholar]
- 22.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–5680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brouillard R. In: The Flavonoids. Harborne J B, editor. London: Chapman & Hall; 1988. pp. 525–538. [Google Scholar]
- 24.Clegg M T, Epperson B K. In: The Plant Evolutionary Biology. Gottlieb L, Jain S K, editors. London: Chapman–Hall; 1988. pp. 255–273. [Google Scholar]
- 25.Schmelzer E, Jahnen W, Hahlbrock K. Proc Natl Acad Sci USA. 1988;85:2989–2993. doi: 10.1073/pnas.85.9.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixon R A, Dey P M, Lamb C J. Adv Enzymol. 1983;55:1–136. doi: 10.1002/9780470123010.ch1. [DOI] [PubMed] [Google Scholar]
- 27.Lamb C J, Lawton M A, Dron M, Dixon R A. Cell. 1989;56:215–224. doi: 10.1016/0092-8674(89)90894-5. [DOI] [PubMed] [Google Scholar]
- 28.Long S. Cell. 1989;56:203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs M, Rubery P H. Science. 1988;241:346–349. doi: 10.1126/science.241.4863.346. [DOI] [PubMed] [Google Scholar]
- 30.Taylor L P, Jorgensen R. J Hered. 1992;83:11–17. [Google Scholar]
- 31.Stafford H A. Plant Physiol. 1991;96:680–685. doi: 10.1104/pp.96.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verwoert I, Verbree E C, Vanderlinden K H, Nijkamp H J, Stuitje A R. J Bacteriol. 1992;174:2851–2857. doi: 10.1128/jb.174.9.2851-2857.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forkmann G. In: The Flavonoids: Advances in Research Since 1986. Harborne J B, editor. London: Chapman & Hall; 1993. pp. 537–564. [Google Scholar]
- 34.Moyano E, Martinezgarcia J F, Martin C. Plant Cell. 1996;8:1519–1532. doi: 10.1105/tpc.8.9.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tropf S, Lanz T, Rensing S A, Schroder J, Schroder F. J Mol Evol. 1994;38:610–618. doi: 10.1007/BF00175881. [DOI] [PubMed] [Google Scholar]
- 36.Schroder J, Schanz S, Tropf S, Karfcher B, Schroder G. In: Mechanisms of Plant Defense Responses. Fritig B, Legrand M, editors. Dordrecht, The Netherlands: Kluwer; 1993. pp. 257–267. [Google Scholar]
- 37.Durbin M L, Learn G H, Huttley G A, Clegg M T. Proc Natl Acad Sci USA. 1995;92:3338–3342. doi: 10.1073/pnas.92.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helariutta Y, Kotilainen M, Elomaa P, Kalkkinen N, Bremer K, Teeri T H, Albert V A. Proc Natl Acad Sci USA. 1996;93:9033–9038. doi: 10.1073/pnas.93.17.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koes R R, Spelt C E, van den Elzen P J M, Mol J N M. Gene. 1989;81:245–257. doi: 10.1016/0378-1119(89)90185-6. [DOI] [PubMed] [Google Scholar]
- 40.Glover D, Durbin M L, Huttley G, Clegg M T. Plant Species Biol. 1996;11:41–50. [Google Scholar]
- 41.Huttley, G. A., Durbin, M. L., Glover, D. E. & Clegg, M. T. (1997) Mol. Ecol., in press.
- 42.Freeling M, Bennett D C. Annu Rev Genet. 1985;19:297–323. doi: 10.1146/annurev.ge.19.120185.001501. [DOI] [PubMed] [Google Scholar]
- 43.Dolferus R, deBruxelles G, Dennis E S, Peacock W J. Ann Bot (London) 1994;74:301–308. [Google Scholar]
- 44.Gottlieb L D. Science. 1982;216:373–380. doi: 10.1126/science.216.4544.373. [DOI] [PubMed] [Google Scholar]
- 45.Chang C, Meyerowitz E M. Proc Natl Acad Sci USA. 1986;83:1408–1412. doi: 10.1073/pnas.83.5.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morton B R, Gaut B S, Clegg M T. Proc Natl Acad Sci USA. 1996;93:11735–11739. doi: 10.1073/pnas.93.21.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W-H. Curr Opin Genet Dev. 1994;3:896–901. doi: 10.1016/0959-437x(93)90011-d. [DOI] [PubMed] [Google Scholar]
- 48.Gaut B S, Morton B R, McCaig B, Clegg M T. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaut B S, Clegg M T. Proc Natl Acad Sci USA. 1993;90:5095–5099. doi: 10.1073/pnas.90.11.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaut B S, Clegg M T. Genetics. 1993;135:1091–1097. doi: 10.1093/genetics/135.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clegg M T. J Hered. 1997;88:1–7. doi: 10.1093/oxfordjournals.jhered.a023048. [DOI] [PubMed] [Google Scholar]
- 52.Jorgensen R A. Science. 1995;268:686–691. doi: 10.1126/science.268.5211.686. [DOI] [PubMed] [Google Scholar]
- 53.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 54.Voytas D F. Science. 1996;274:737–738. doi: 10.1126/science.274.5288.737. [DOI] [PubMed] [Google Scholar]
- 55.Schwarz-Sommer Z, Leclerq L, Gobel E, Saedler H. EMBO J. 1987;6:3873–3880. doi: 10.1002/j.1460-2075.1987.tb02727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leeton P R, Smyth D R. Mol Gen Genet. 1993;237:97–104. doi: 10.1007/BF00282789. [DOI] [PubMed] [Google Scholar]
- 57.Voytas D F, Cummings M P, Konieczny A, Ausbel F M, Rodermel S R. Proc Natl Acad Sci USA. 1992;89:7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flavell A J, Dunnbar E, Anderson R, Pearce S R, Hartley R, Kumar A. Nucleic Acids Res. 1992;20:3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joseph J L, Sentry J W, Smyth D R. J Mol Evol. 1990;30:146–151. [Google Scholar]
- 60.Voytas D F, Ausbel F M. Nature (London) 1989;336:242–244. doi: 10.1038/336242a0. [DOI] [PubMed] [Google Scholar]
- 61.Voytas D F, Konieczny A, Cummings M P, Ausbel F M. Genetics. 1990;126:713–721. doi: 10.1093/genetics/126.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konieczny A, Voytas D F, Cummings M P, Ausbel F M. Genetics. 1991;30:801–809. doi: 10.1093/genetics/127.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wright D A, Ke N, Smalle J, Hauge B M, Goodman H M, Voytas D F. Genetics. 1996;126:569–578. doi: 10.1093/genetics/142.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engels W R, Johnson-Schlitz D M, Eggleston W B, Sved J. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 65.Huttley G, MacRae A F, Clegg M T. Genetics. 1995;139:1411–1419. doi: 10.1093/genetics/139.3.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cummings M P. Trends Ecol. 1994;9:141–145. doi: 10.1016/0169-5347(94)90179-1. [DOI] [PubMed] [Google Scholar]