Abstract
Bacillus thuringiensis Cry8Ea toxin is specifically toxic to larvae of the Asian cockchafer, Holotrichia parallela. Here we investigated the mechanism of transcriptional regulation of the cry8Ea1 gene. Reverse transcription-PCR (RT-PCR) results indicated that cry8Ea1 and an upstream gene (orf1) were cotranscribed. Transcriptional fusions with the lacZ gene demonstrated that transcription of the cry8Ea1 gene started from two promoters: Porf1, which is located upstream of the orf1 gene, and Pcry8E, located in the intergenic region mapping between orf1 and cry8Ea1. Of the known, similar orf1-cry operons, this is the first report of the existence of a promoter in the intergenic region between the orf1 and cry genes. The transcriptional activity of Porf1 was found during sporulation in B. thuringiensis subsp. kurstaki HD-73 and was almost abolished in the sigE mutant, while the transcriptional activity of Pcry8E was detected after the end of the exponential phase in HD-73 and was considerably lower in the sigH mutant. The transcription start sites generated by the two cry8Ea1 promoters were determined by the 5′ -SMARTer rapid amplification of cDNA ends (RACE) method. The −35 and −10 regions of Porf1 and Pcry8E showed high sequence similarity with the σE and σH promoters, respectively. These results indicated that Porf1 is controlled by the σE factor and Pcry8E by the σH factor.
INTRODUCTION
Bacillus thuringiensis is a Gram-positive, spore-forming bacterium that produces parasporal crystal proteins. These parasporal crystal proteins are encoded by cry genes and possess a highly specific insecticidal activity against a great number of insect species. To date, more than 500 cry genes have been discovered (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/). Most B. thuringiensis cry genes have a monocistronic structure, with the exceptions of cry2Aa (25), cry2Ac (27), cry9Ca (15), cry9Ec (23), cry11A (11), and cry15A (8), which form operons with at least one upstream open reading frame (ORF), named orf1 or p19. However, no promoter activity in the intergenic region between the orf1 and cry genes was reported in the previous studies.
Transcription of cry1A depends on the BtI promoter in the early stage of the stationary phase and on the BtII promoter in the late stage of the stationary phase (26). They are regulated by sporulation-specific sigma factors, σE and σK, respectively (6, 9, 10). Most of the known crystal protein genes (3, 5) contain either BtI alone or both BtI and BtII. Among these, some cry genes (i.e., cry1Ac and cry4A) with two or more promoters are weakly controlled by σH during the transition phase and strongly controlled by sporulation-specific sigma factors (18, 28). In contrast, cry3A expression is not dependent on sporulation-specific sigma factors. The cry3A promoter is similar to promoters recognized by the main sigma factor of vegetative cells, σA (1, 2).
B. thuringiensis strain BT185 produces spheric inclusions that exhibit specific toxicity against larvae of the Asian cockchafer, Holotrichia parallela (29). Shu et al. cloned two homologous 130-kDa insecticidal crystal protein genes, designated cry8Ea1 and cry8Fa1. Cry8Ea1 toxin is toxic to H. parallela, whereas no target insect was found to be susceptible to the Cry8Fa1 protein (21). The three-dimensional structure of activated Cry8Ea1 toxin was determined by X-ray crystallographic methods (14). However, the regulation of cry8Ea1 gene expression still remains unknown. For this study, we report that cry8Ea1 is transcribed from two promoters: one is located upstream of the orf1 gene, and the second maps in the intergenic region between orf1 and cry8Ea1. Transcription of the cry8Ea1 gene is controlled by σE and σH.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Escherichia coli JM110 was used for the cloning experiments, while SCS110 was used to produce nonmethylated plasmid DNA for B. thuringiensis transformations (22). B. thuringiensis subsp. kurstaki HD-73 was used as a recipient strain to monitor promoter activity (12).
E. coli was cultured at 37°C in Luria-Bertani medium (1% NaCl, 1% tryptone, 0.5% yeast extract). B. thuringiensis strains were grown at 30°C in Schaeffer's sporulation medium (SSM) (20). Ampicillin (100 μg/ml) or erythromycin (50 μg/ml) was added to the media, when appropriate, for selection of antibiotic-resistant strains of E. coli and B. thuringiensis.
DNA manipulation and transformation.
Chromosomal DNA was extracted from B. thuringiensis as described previously (16). Plasmid DNA was extracted from E. coli by the standard alkaline lysis procedure. Restriction enzymes and T4 DNA ligase were purchased from TaKaRa Biotechnology Corporation (Dalian, China). PCR product purification kits were purchased from Axygen Inc. (Hangzhou, China). Taq DNA polymerase and KOD DNA polymerase were purchased from New England BioLabs Ltd. (Beijing, China). All oligonucleotide primers used in this study are listed in Table S2 in the supplemental material. Standard procedures were used for E. coli transformation. B. thuringiensis cells were transformed by electroporation as previously described by Lereclus et al. (16).
Construction of sigE and sigH deletion mutant.
The DNA fragments corresponding to the downstream and upstream regions of the sigH gene were amplified by PCR, using chromosomal DNA from HD-73 as the template and sigH1/sigH2 and sigH3/sigH4 as primers. The corresponding DNA fragments were fused by overlapping PCR, using sigH1 and sigH4 as the primers. The PCR products were then digested with BamHI-SalI restriction enzymes. The digested fragments were purified and ligated with the temperature-sensitive suicide plasmid pMAD (4), which was also treated with BamHI-SalI, to generate the recombinant plasmid pMADΩsigH. The recombinant plasmid was electroporated into the HD-73 strain. In-frame deletion of the sigH gene in B. thuringiensis was conducted following a modification of a previously described procedure (4). One verified transformant of this deletion mutant was cultured at 40°C. Colonies with no erythromycin resistance were selected, and one mutant strain, HDΔsigH, was verified by PCR identification using the primers YZsigH5 and YZsigH3, mapping outside the deletion frame, and DNA sequencing was performed.
The primers sigE1/sigE2 and sigE3/sigE4 were used to construct sigE deletion mutation cassettes. This fragment was integrated into pMAD to give pMADΩsigE. The deletion mutant, HDΔsigE, was selected and confirmed by PCR and Southern blotting (24).
RNA extraction and reverse transcription-PCR (RT-PCR) analyses.
Total RNA was extracted from B. thuringiensis BT185 cells grown in SSM at stages T0, T3, and T7 (T0 is the end of the exponential phase; Tn is n hours after the end of the exponential phase). The bacterial cells were harvested from 1 ml of culture by centrifugation in 2-ml tubes (13,000 × g, 30 s, 4°C), and the cell pellets were immediately resuspended in 1 ml cold TRI Reagent (Invitrogen, San Diego, CA). The RNA was extracted with the Qiagen Easy RNA kit according to the manufacturer's instructions. The residual DNA was removed using RNase-free DNase I(New England BioLabs), and the resulting RNA samples were stored at −70°C. cDNA was synthesized from 0.5-μg aliquots of total RNA using the PrimeScript II 1st-strand cDNA synthesis kit (TaKaRa, Dalian, China) with the Random 6mer primers according to the manufacturer's instructions. The following primers were used to detect expression of the orf1 gene and cry8Ea1 gene: orf1, RTorf1-5/RTorf1-3; cry8Ea1, RT8E5/RT8E3; and 16S rRNA genes, 16SrDNA5/16SrDNA3. Conditions for amplification were as follows: one incubation for 5 min at 94°C, followed by 30 cycles for 1 min at 94°C for denaturation, 1 min at 54°C for annealing, and 2 min at 72°C for extension, with an extra extension at 72°C for 10 min. The amplification products were separated on 1.5% agarose gels. Negative control samples were subjected to amplification.
Determination of transcriptional start sites.
To determine the transcriptional start sites, we employed the technique switching mechanism at the 5′ end of the RNA transcript-rapid amplification of cDNA ends (SMARTer RACE) cDNA amplification kit (Clontech, Mountain View, CA), following the manufacturer's instruction. Gene-specific primers, 8ERace and NestRace, were used to amplify the 5′ end of cry8Ea mRNA.
Construction of Porf1 and Pcry8E promoter fusions with the lacZ gene.
The first putative promoter fragment of Porf1 (559 bp), which is located upstream of the orf1 gene, was cloned from B. thuringiensis BT185 genomic DNA using the specific primers Porf1-5 (with a PstI restriction site) and Porf1-3 (with a BamHI restriction site). The PstI-BamHI fragment of the Porf1 promoter was then integrated into the vector pHT304-18Z, harboring a promoterless lacZ gene. The resulting plasmid, pHTPorf1, was introduced into HD-73, the sigE mutant, and the sigK mutant (13). The corresponding strains, HD(Porf1-lacZ), HDΔsigE(Porf1-lacZ), and HDΔsigK(Porf1-lacZ), were selected by erythromycin resistance and PCR identification. Similarly, the primers Pcry8E5 and Pcry8E3 were used to construct the second putative promoter fragment, Pcry8E (280 bp), which was inserted into the pHT304-18Z vector to generate pHTPcry8E. The recombinant plasmid pHTPcry8E was electroporated into HD-73 and the sigH mutant to give HD(Pcry8E-lacZ) and HDΔsigH(Pcry8E-lacZ), respectively.
Primers Porf1-5 and Pcry8E3 were used for PCR amplification of the 1,352-bp fragment containing the upstream region of orf1 gene and the region between the orf1 and cry8Ea1 genes. After digestion with PstI-BamHI, the PCR product was integrated into pHT304-18Z to generate pHTPorf1-cry8E. The resulting plasmid, pHTPorf1-cry8E, was introduced into HD-73 to generate HD(Porf1-cry8E-lacZ).
β-Galactosidase assay.
B. thuringiensis strains containing lacZ fusions were grown in SSM at 220 rpm and 30°C, supplemented with appropriate antibiotics. Samples of 2 ml were taken at T−2 and at 1-h intervals until T9. The cells were harvested, and the specific β-galactosidase activity of the samples was measured as previously described and expressed as Miller units per milligram of protein (17). The values reported are the means for at least three independent experiments.
RESULTS AND DISCUSSION
Sequence analysis and RT-PCR.
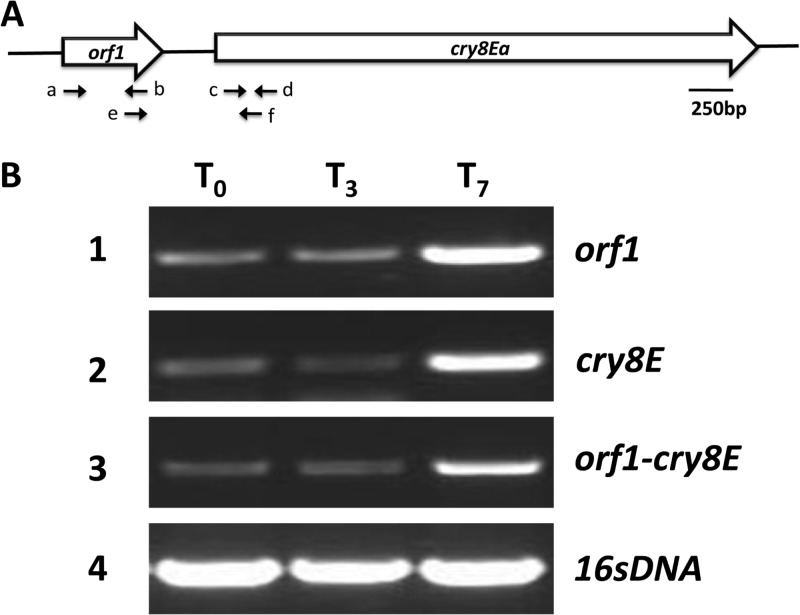
The sequence analysis showed that orf1 was located 286 bp immediately upstream of cry8Ea1 (GenBank accession number AY329081) and that no potential transcriptional terminator was present between the two genes. This finding suggested that the orf1 and cry8Ea1 genes may form one operon. To test this hypothesis, we analyzed the transcription of the two genes. Three pairs of primers were designed according to the orf1 and cry8Ea1 gene sequence to detect their transcription (Fig. 1A; see also Table S2 in the supplemental material). RT-PCR was carried out with total RNA extracted from B. thuringiensis strain 185 at T0, T3, and T7 from cultures grown in SSM (Fig. 1B). The data suggest that the orf1 and cry8Ea1 genes are transcribed together as one transcription unit. This coincided with the cry2A, cry11A, cry15A, and cry18A operons (8, 11, 25, 30).
Fig 1.
Analysis of cry8Ea operon in B. thuringiensis BT185. (A) cry8E gene locus. The letters below summarize the main primers used for RT-PCR. Primers a (RTorf1-5), b (RTorf1-3), c (RTcry8E5), d (RTcry8E3), e (RTCO), and f (RTCO) were used for RT-PCR analysis of the orf1 and cry8E genes (see Table S2 in the supplemental material). (B) RT-PCR for the transcription linkages of the orf1 and cry8E genes. The RNA samples were prepared at different time points of bacterial growth in SSM, as indicated. Rows 1 to 3 denote the RT-PCR products of orf1, cry8E, and orf1-cry8E, and the 16S rRNA gene (row 4) was used as a positive control.
Transcriptional activity of promoters from cry8Ea operon.
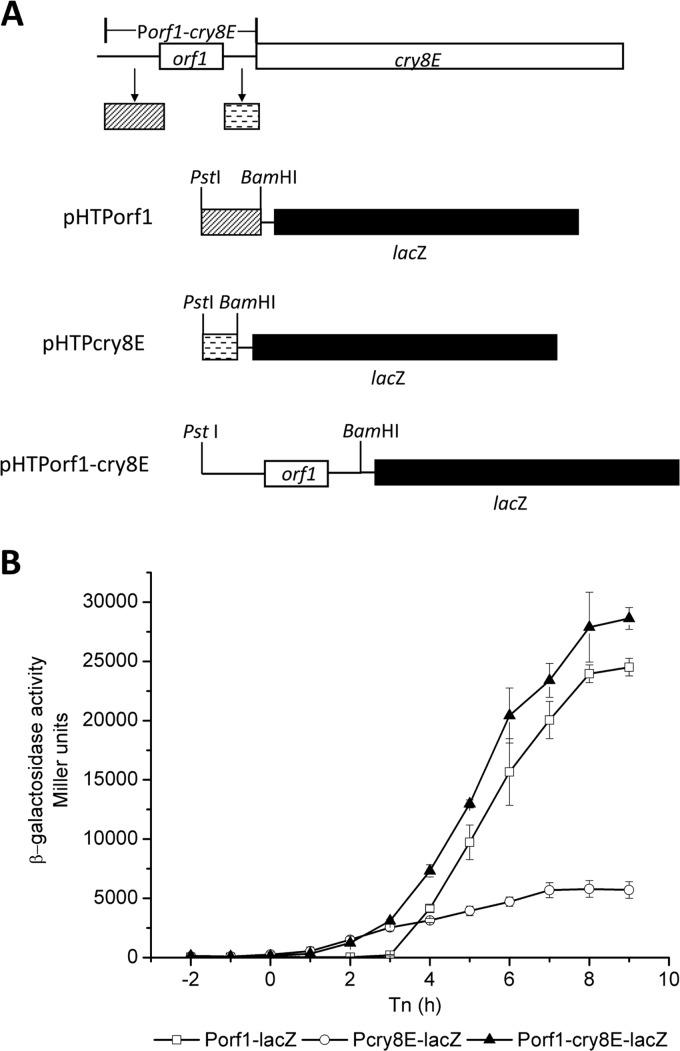
The promoter region(s) of the cry8Ea operon was analyzed by constructing transcriptional fusions with the lacZ gene (Fig. 2A). The HD-73 strains containing pHTPorf1, pHTPcry8E, and pHTPorf1-cry8E were cultured in SSM, and β-galactosidase activities of the strains were assayed (Fig. 2B). The β-galactosidase activities of HD(Pcry8E-lacZ) and HD(Porf1-cry8E-lacZ) were detected at the onset of sporulation (T0), while that of HD(Porf1-lacZ) was detected only 3 h later (T3). The activity of the Pcry8E promoter was approximately 5-fold lower than that of the Porf1 promoter after T3. The higher transcriptional activity obtained with the complete cry8Ea1 gene promoter, Porf1-cry8E, is likely due to the combined activities of Porf1 and Pcry8E. These results suggest that cry8Ea1 has two promoters, Porf1, which has a high level of activity during sporulation, and Pcry8E, which is active at the onset of sporulation. The first promoter, Porf1, is located upstream of the orf1 gene. The second promoter, Pcry8E, is located in the intergenic region between the orf1 and cry8Ea1 genes. Additionally, the cry2A, cry11A, cry15A, and cry18A operons have a single promoter, located upstream of the first orf operon. No promoter activity was found in the intergenic region between the orf1 and cry genes (cry2A, cry11A, cry15A, and cry18A) (8, 11, 25, 30), suggesting that the cry8Ea1 operon promoter is a novel cry gene promoter. To our knowledge, this is the first report of the existence of a promoter in the intergenic region between the orf1 and cry genes.
Fig 2.
Characterization of the promoter region of the cry8Ea operon. (A) Construction of promoter fusions with the lacZ gene. The upstream regions of the orf1 gene were amplified by PCR using the primers Porf1-5 and Porf1-3. The intergenic region between orf1 and cry8Ea was amplified using the primers Pcry8E5 and Pcry8E3. The complete upstream region, including the upstream region of the orf1 gene, the orf1gene, and the region between the orf1 and cry8E genes, was amplified using the primers Porf1-5 and Pcry8E3. The three amplified fragments were inserted into pHT304-18Z to generate the transcriptional fusion plasmids pHTPorf1, pHTPcry8E, and pHTPorf1-cry8E. (B) Transcriptional analysis of the promoter region of the cry8Ea operon. The promoter-directed β-galactosidase synthesis of three clones was determined at the indicated times after growing the cells in SSM at 30°C. Each value represents the mean for at least three independent replicates.
Porf1 is a σE-dependent promoter.
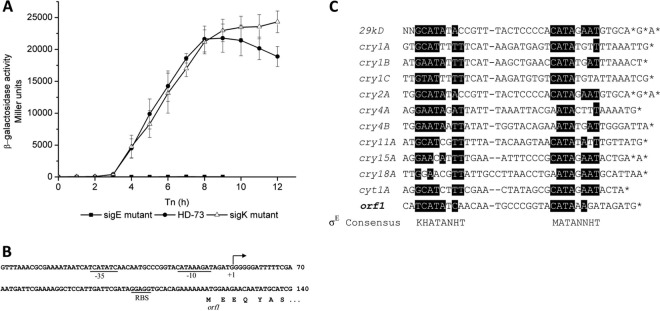
The time course of Porf1 activity increased sharply at T3, suggesting the possibility of a sporulation-specific sigma factor. Therefore, the σE and σK dependence of Porf1 was tested. The recombinant plasmid pHTPorf1 was introduced into the HDΔsigE and the HDΔsigK strains, respectively. The β-galactosidase assay showed that Porf1 activity was almost abolished in the HDΔsigE strain and was similar in the wild-type and HDΔsigK strains before T10 (Fig. 3A).
Fig 3.
(A) Analysis of Porf1 promoter activities in the HD-73 wild-type strain (●), the sigE mutant (■), and the sigK mutant (△). The promoter-directed synthesis of β-galactosidase in the strains was determined at the indicated times after growing the cells in SSM at 30°C. Each value represents the mean for at least three independent replicates. (B) The promoter region of the operon. Transcriptional initiation (+1) is marked. The putative promoter region of the operon (−35 and −10) and the putative ribosome binding site of orf1 (RBS) are underlined and marked. “orf1” below the arrow indicates the 5′-end of orf1 and the deduced amino acid sequence. (C) Sequence similarity among the cry gene promoters and the consensus sequences obtained by aligning the σE-dependent promoters of B. thuringiensis. “*” indicates the transcriptional initiation sites. Consensus sequences of σE-dependent promoters are presented above and below the alignment, respectively. D is A, G, or T; H is A, C, or T; K is G or T; M is A or C; N is A, C, G, or T; V is A, G, or C.
The transcriptional start site was determined by 5′-RACE–PCR to be a single 5′-end G residue located 64 nucleotides (nt) upstream of the orf1 translational start codon (Fig. 3B). The nucleotide sequences of the −35 and −10 regions of the orf1 gene are similar to those of some σE-dependent cry genes previously described (30) (Fig. 3C). Taken together, these results suggested that high-level transcription of the cry8Ea1 gene is controlled by the σE factor during sporulation, similar to regulation of the cry2A and cry15A genes (8, 25).
Pcry8E is a σH-dependent promoter.
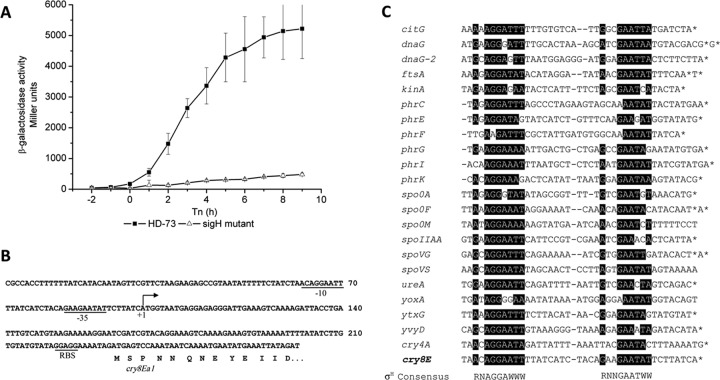
The intergenic region between orf1 and cry8Ea1 encodes a promoter with low-level activity at the onset of sporulation (Fig. 2B). The nucleotide sequences of the −35 and −10 regions of the cry8Ea1 gene are highly similar to those of several σH-dependent genes in Bacillus subtilis (6) (Fig. 4C). To test whether the cry8E gene promoter is σH dependent, the plasmid pHTPcry8E was introduced into HDΔsigH. β-Galactosidase activity in the HDΔsigH strain was dramatically lower than that in the wild-type strain (Fig. 4A). The results of 5′-RACE–PCR mapping performed on total RNA isolated from BT185 cells at T1 indicated that a transcription start site mapped 133 bases upstream of the cry8Ea1 gene start codon (Fig. 4B). The nucleotide sequence of the promoter region is similar to that of σH-dependent genes (7) (Fig. 4C). These results suggested that the Pcry8E promoter with low-level activity located in the intergenic region between orf1 and cry8Ea1 is controlled by the σH factor during the transition phase. The cry4A and cry11A genes are also weakly transcribed by the σH form of RNA polymerase, and the σH-dependent promoter overlaps the σE-dependent promoter (19, 28). This is not the case with the cry8Ea1 operon promoter, which is located upstream of two transcriptional start sites of the cry8Ea1 gene. This work describes the unique character of the cry8Ea1 operon promoter, which is composed of Porf1 and Pcry8E, controlled by the σE and σH factors, respectively.
Fig 4.
(A) Analysis of Pcry8E promoter activities in the HD-73 wild-type strain (■) and the sigH mutant (△). The promoter-directed synthesis of β-galactosidase in the strains was determined at the indicated times after growing the cells in SSM at 30°C. Each value represents the mean for at least three independent replicates. (B) The promoter region of the operon. Transcriptional initiation (+1) is marked. The putative promoter region of the operon (−35 and −10) and the putative ribosome binding site of orf1 (RBS) are underlined and marked. “cry8Ea” below the arrow indicates the 5′ end of orf1 and the deduced amino acid sequence. (C) Sequence similarity between the Pcry8E promoter and the consensus sequences was obtained by aligning the σH-dependent promoters of B. thuringiensis. “*” indicates the transcriptional initiation sites. Consensus sequences of σH-dependent promoters are presented above and below the alignment, respectively. R is A or G; W is A or T.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Key Project of Chinese National Programs for Fundamental Research and Development (973 Program) (2009CB118902) and State Key Laboratory Foundation for Biology of Plant Diseases and Insect Pests (SKL2011OP04).
Footnotes
Published ahead of print 13 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Agaisse H, Lereclus D. 1994. Expression in Bacillus subtilis of the Bacillus thuringiensis cryIIIA toxin gene is not dependent on a sporulation-specific sigma factor and is increased in a spo0A mutant. J. Bacteriol. 176:4734–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agaisse H, Lereclus D. 1994. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13:97–107 [DOI] [PubMed] [Google Scholar]
- 3. Agaisse H, Lereclus D. 1995. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J. Bacteriol. 177:6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baum JA, Malvar T. 1995. Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol. Microbiol. 18:1–12 [DOI] [PubMed] [Google Scholar]
- 6. Bravo A, Agaisse H, Salamitou S, Lereclus D. 1996. Analysis of cryIAa expression in sigE and sigK mutants of Bacillus thuringiensis. Mol. Gen. Genet. 250:734–741 [DOI] [PubMed] [Google Scholar]
- 7. Britton RA, et al. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown KL. 1993. Transcriptional regulation of the Bacillus thuringiensis subsp. thompsoni crystal protein gene operon. J. Bacteriol. 175:7951–7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown KL, Whiteley HR. 1988. Isolation of a Bacillus thuringiensis RNA polymerase capable of transcribing crystal protein genes. Proc. Natl. Acad. Sci. U. S. A. 85:4166–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown KL, Whiteley HR. 1990. Isolation of the second Bacillus thuringiensis RNA polymerase that transcribes from a crystal protein gene promoter. J. Bacteriol. 172:6682–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dervyn E, Poncet S, Klier A, Rapoport G. 1995. Transcriptional regulation of the cryIVD gene operon from Bacillus thuringiensis subsp. israelensis. J. Bacteriol. 177:2283–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Du C, Nickerson KW. 1996. Bacillus thuringiensis HD-73 spores have surface-localized Cry1Ac toxin: physiological and pathogenic consequences. Appl. Environ. Microbiol. 62:3722–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Du L, et al. 2011. Characterization of Bacillus thuringiensis sigK disruption mutant and its influence on activation of cry3A promoter. Acta Microbiol. Sin. 51:1177–1184 (In Chinese.) [PubMed] [Google Scholar]
- 14. Guo S, et al. 2009. Crystal structure of Bacillus thuringiensis Cry8Ea1: an insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela. J. Struct. Biol. 168:259–266 [DOI] [PubMed] [Google Scholar]
- 15. Lambert B, et al. 1996. A Bacillus thuringiensis insecticidal crystal protein with a high activity against members of the family Noctuidae. Appl. Environ. Microbiol. 62:80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lereclus D, Arantes O, Chaufaux J, Lecadet M. 1989. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 51:211–217 [DOI] [PubMed] [Google Scholar]
- 17. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 18. Perez-Garcia G, Basurto-Rios R, Ibarra JE. 2010. Potential effect of a putative sigma(H)-driven promoter on the over expression of the Cry1Ac toxin of Bacillus thuringiensis. J. Invertebr. Pathol. 104:140–146 [DOI] [PubMed] [Google Scholar]
- 19. Poncet S, Dervyn E, Klier A, Rapoport G. 1997. Spo0A represses transcription of the cry toxin genes in Bacillus thuringiensis. Microbiology 143(Pt 8):2743–2751 [DOI] [PubMed] [Google Scholar]
- 20. Schaeffer P, Millet J, Aubert JP. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shu C, et al. 2009. Characterization of two novel cry8 genes from Bacillus thuringiensis strain BT185. Curr. Microbiol. 58:389–392 [DOI] [PubMed] [Google Scholar]
- 22. Wang G, et al. 2006. Engineered Bacillus thuringiensis GO33A with broad insecticidal activity against lepidopteran and coleopteran pests. Appl. Microbiol. Biotechnol. 72:924–930 [DOI] [PubMed] [Google Scholar]
- 23. Wasano N, et al. 2005. Cloning and characterization of a novel gene cry9Ec1 encoding lepidopteran-specific parasporal inclusion protein from a Bacillus thuringiensis serovar galleriae strain. Can. J. Microbiol. 51:988–995 [DOI] [PubMed] [Google Scholar]
- 24. Wei J, et al. 2008. Construction and characteristics of the sigE mutation in Bacillus thuringiensisHD-73. Microbiology 35:1581–1586 (In Chinese.) [Google Scholar]
- 25. Widner WR, Whiteley HR. 1989. Two highly related insecticidal crystal proteins of Bacillus thuringiensis subsp. kurstaki possess different host range specificities. J. Bacteriol. 171:965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong HC, Schnepf HE, Whiteley HR. 1983. Transcriptional and translational start sites for the Bacillus thuringiensis crystal protein gene. J. Biol. Chem. 258:1960–1967 [PubMed] [Google Scholar]
- 27. Wu D, Cao XL, Bai YY, Aronson AI. 1991. Sequence of an operon containing a novel delta-endotoxin gene from Bacillus thuringiensis. FEMS Microbiol. Lett. 65:31–35 [DOI] [PubMed] [Google Scholar]
- 28. Yoshisue H, Ihara K, Nishimoto T, Sakai H, Komano T. 1995. Expression of the genes for insecticidal crystal proteins in Bacillus thuringiensis: cryIVA, not cryIVB, is transcribed by RNA polymerase containing sigma H and that containing sigma E. FEMS Microbiol. Lett. 127:65–72 [DOI] [PubMed] [Google Scholar]
- 29. Yu H, Zhang J, Huang D, Gao J, Song F. 2006. Characterization of Bacillus thuringiensis strain Bt185 toxic to the Asian cockchafer: Holotrichia parallela. Curr. Microbiol. 53:13–17 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J, Schairer HU, Schnetter W, Lereclus D, Agaisse H. 1998. Bacillus popilliae cry18Aa operon is transcribed by sigma E and sigma K forms of RNA polymerase from a single initiation site. Nucleic Acids Res. 26:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.