Abstract
The extreme osmotic conditions prevailing in hypersaline environments result in decreasing metabolic diversity with increasing salinity. Various microbial metabolisms have been shown to occur even at high salinity, including photosynthesis as well as sulfate and nitrate reduction. However, information about anaerobic microbial iron metabolism in hypersaline environments is scarce. We studied the phylogenetic diversity, distribution, and metabolic activity of iron(II)-oxidizing and iron(III)-reducing Bacteria and Archaea in pH-neutral, iron-rich salt lake sediments (Lake Kasin, southern Russia; salinity, 348.6 g liter−1) using a combination of culture-dependent and -independent techniques. 16S rRNA gene clone libraries for Bacteria and Archaea revealed a microbial community composition typical for hypersaline sediments. Most-probable-number counts confirmed the presence of 4.26 × 102 to 8.32 × 103 iron(II)-oxidizing Bacteria and 4.16 × 102 to 2.13 × 103 iron(III)-reducing microorganisms per gram dry sediment. Microbial iron(III) reduction was detected in the presence of 5 M NaCl, extending the natural habitat boundaries for this important microbial process. Quantitative real-time PCR showed that 16S rRNA gene copy numbers of total Bacteria, total Archaea, and species dominating the iron(III)-reducing enrichment cultures (relatives of Halobaculum gomorrense, Desulfosporosinus lacus, and members of the Bacilli) were highest in an iron oxide-rich sediment layer. Combined with the presented geochemical and mineralogical data, our findings suggest the presence of an active microbial iron cycle at salt concentrations close to the solubility limit of NaCl.
INTRODUCTION
Hypersaline aquatic environments are abundant worldwide and include inland salt lakes and marine coastal areas such as marshes and solar salterns. On a global scale, nearly as much water is stored in salt lakes as in freshwater lakes (24). Due to climate change, the area covered by salt lakes is expected to increase in the near future. Many freshwater lakes will turn into salt lakes and existing salt lakes will increase in salinity due to increased evaporation (99). With salinities ranging from below seawater salinity (i.e., 35 g/liter−1 dissolved salt) to NaCl saturation (i.e., 304 g/liter−1 or 5.2 M) (56), salt lakes represent extreme habitats for microbial life (72). However, high rates of primary production show that salt-adapted microorganisms are active regardless of the extreme environmental osmotic conditions (80).
A major challenge for microorganisms living in hypersaline environments is to prevent desiccation caused by osmotic stress. The various modes of adaptation to extreme osmotic conditions can pose a high energy burden on microorganisms in hypersaline environments (56, 57, 61). The high energy requirements to counteract the osmotic pressure have been used to explain the often-observed decrease in metabolic diversity with increasing salinity and led to the hypothesis of an upper limit of salinity for every metabolic process as determined by thermodynamic constraints (55, 56, 58). In other words, metabolic processes from which sufficient energy can be gained, such as nitrate reduction (2,716 kJ is generated per 4.8 mol of nitrate reduced with 1 mol of glucose under standard conditions), are expected to occur up to higher environmental salt concentrations than processes from which less metabolic energy can be derived, such as methanogenesis from H2 and HCO3− (−34 kJ/mol of HCO3− [56]). So far, the majority of observations made in hypersaline environments strengthen the aforementioned hypothesis (58). Denitrification, for example, has been observed in laboratory cultures in medium with more than 300 g NaCl liter−1, while sulfate reduction has been shown to occur up to 240 g NaCl liter−1, and methanogenesis from H2 and HCO3− could not be detected at salinities beyond 120 g NaCl liter−1 (58). While metabolic processes of microorganisms involved in the biogeochemical cycling of sulfur, including dissimilatory sulfate reduction in salt lake water and sediments (36, 78), have been studied extensively, knowledge of the use of oxidized metal ions, such as iron (Fe) in the form of Fe(III), as electron acceptors in anaerobic hypersaline environments is scarce. An upper salinity limit for microbial Fe(III) reduction has not been defined yet (58). The use of Fe(III) in the form of Fe(OH)3 as electron acceptor is more thermodynamically favorable than the use of sulfate [ΔG = −48 kJ per mole of electrons transferred from acetate to Fe(III) at pH 7 in comparison to −7 kJ per mole of electrons transferred from acetate to sulfate under the same conditions (44)]. Consequently, from a thermodynamic point of view, as long as bioavailable Fe(III) is not limiting, microbial Fe(III) reduction should be more energetically favorable than sulfate reduction in hypersaline environments. With typical concentrations of 1 to 5% of sediment dry matter, Fe(III) (hydr)oxides have indeed been shown to represent important electron acceptors in freshwater (74) and also in marine sediments, where Fe can constitute up to 20% of the sediment by weight (93). In hypersaline sediments, amounts of Fe(III) hydroxides similar to those in freshwater lake sediments have been found (12). In a previous study, these iron oxide minerals were identified as jarosite, goethite, and hydrous iron oxides (41). However, these studies consider only abiotic aspects of Fe geochemistry and neglect the possible role of microorganisms in Fe mineral transformations. To date, a few isolates from hypersaline environments have been reported to be capable of Fe(III) reduction. Often the isolates were shown to reduce dissolved Fe(III) but were not tested for the reduction of any solid Fe(III) phases (63, 87, 88), which represent the dominant form of Fe(III) at neutral or even alkaline pH values of most salt lake environments (85). In other cases, isolates were found to be halotolerant rather than halophilic. Bacteria such as Geoalkalibacter ferrihydriticus can only grow and reduce Fe(III) in up to 50 g/liter−1 NaCl (103). The same applies to several Bacillus strains (31, 88) as well as to an Fe(III)-reducing isolate from the sediment of the hypersaline Lake Chaka (China), because the strain originated from 880-cm depth, where salinity was only slightly elevated compared to freshwater (29).
While knowledge about microbial Fe(III) reduction in hypersaline environments is scarce, even less is known about Fe(II) oxidizers in these habitats. Previous reports indicated an inhibitory effect of Cl− at seawater concentration on microaerophilic (4) and phototrophic (54) Fe(II) oxidizers. McBeth et al. (46) recently presented the first study of a microaerophilic Fe(II)-oxidizing strain associated with the Zetaproteobacteria isolated from a microbial mat in the Great Salt Bay, where salinity ranges between 0% and 2.5% (46). However, our current knowledge on microbial Fe(II) oxidation, Fe(III) reduction, and the role of both processes in iron cycling in hypersaline sediments is very limited (8, 76, 84). Therefore, the goals of the present study were (i) to determine the diversity of Fe-metabolizing microorganisms in pH-neutral, NaCl-saturated Lake Kasin sediments, (ii) to analyze the abundance and distribution of Fe-metabolizing microorganisms within a geochemically heterogeneous sediment profile, and (iii) to determine the activity of microbial Fe(II) oxidizers and Fe(III) reducers in order to evaluate their ecological role in the cycling of iron at a salt lake in southern Russia, Lake Kasin.
MATERIALS AND METHODS
Field measurements and sampling.
Lake Kasin is a shallow hypersaline lake located approximately 250 km southeast of Volgograd (Russia) within the district of Astrachan. The region forms a part of the North- or Pre-Caspian Depression, a low-elevation flatland north of the Caspian Sea (Global Positioning System [GPS]: 47°36.165′N 047°27.129′E). A detailed description of the study site is given in the supplemental material. The sampling site was located about 50 m east of the water-covered area of Lake Kasin and can be described as exposed lakebed (see Fig. S1 in the supplemental material). In addition to samples from Lake Kasin, reference samples were taken from two other salt lakes in the same area (Lake Elton and Lake Baskunchak).
From all visually distinguishable horizons of the individual sampling sites (see Fig. S1 in the supplemental material), sediment was mixed in a 1:1 ratio with deionized water for determination of pH with color indicator strips (pH range 5.0 to 10; Merck) and electrical conductivity with a portable electrode (SenTix ORP as part of a MultiLine P4 universal meter). Groundwater samples were tested for NO3− and NO2− with color indicator strips (Merckoquant) and amended with 1 M HCl and ferrozine in a ratio of sample:HCl:ferrozine of 1:1:2 in order to do a qualitative test for the presence of Fe(II) (82).
Zero- to 10-cm composite samples of the salt pan and sediment sites were taken with a spatula and transported in UV-sterilized plastic bags. Lake water was sampled with a UV-sterilized plastic bottle. The water-covered sediment was found to release some gas upon tramping, which was sampled in 20-ml glass vials that were closed under water with polytetrafluoroethylene (PTFE)-layered butyl rubber septa and metal crimp caps. Sediment composite samples as well as water and gas samples were immediately placed into an insulated transport box, cooled on site, and kept at about 4°C during transportation to Tübingen, Germany. In Tübingen, composite samples were sieved (2-mm diameter) and stored in plastic bags at 4°C in the dark.
At site Kasin, two 15-cm sediment cores (named A and B) were drilled with UV-sterilized plastic tubes of 3-cm diameter and cut into 0.5-cm-thick (up to 5 cm of depth) or 1-cm-thick (from 5 to 15 cm of depth) segments. Samples were transferred into sterile 15-ml plastic tubes on site and immediately transferred to a battery-driven portable refrigerator box set to −20°C. The same was done with a 0- to 10-cm composite sample for the clone library construction. Samples were transported to Tübingen, Germany within a few days after sampling and remained frozen until arrival. In Tübingen, Germany, samples were stored at −20°C until further analysis.
Laboratory chemical analysis of sediment samples.
All geochemical analyses of sediment samples were performed in duplicate. Determination of the water content, pH, total organic carbon (TOC), and total inorganic carbon (TIC) as well as X-ray fluorescence analysis (XRF) of the sieved composite samples were performed as previously described by Porsch and Kappler (64).
One gram of each sieved, freeze-dried, and milled sample from the different horizons of Lake Kasin sediment was amended with 10 ml of double distilled water (ddH2O) and shaken horizontally for 24 h. After centrifugation and 1:20-to-1:40 (vol/vol) dilution with ddH2O, the eluates were used to quantify HCO3−, Cl−, Br−, IO3−, NO3−, PO43−, and SO42− by ion chromatography (Dionex DX 120 equipped with an AS9HC column and an AG9HC precolumn). From this analysis, the concentrations of water-leachable ions in the sediment samples were calculated. Lake water was filtered through a cellulose ester filter with a pore size of 0.45 μm and diluted 1:5,000 with Millipore water before determining concentrations of dissolved ions by ion chromatography with a Dionex DX 120 device equipped with an AS14 column, an ASRS 300 suppressor, and a conductivity detector (Dionex, Germering, Germany). From the sieved, freeze-dried, and milled samples of the different horizons, pH was measured according to DIN ISO 10390 (21) and electrical conductivity was determined following DIN ISO 11265 (20) in order to confirm the values measured on site.
In order to quantify bioavailable (0.5 M HCl extractable) versus crystalline (6 M HCl extractable) iron in the sediment profile, Fe extractions were performed using the sequential extraction protocol previously described by Porsch and Kappler (64). The sequential extractions were performed in an anoxic (N2) glove box in order to prevent samples from oxidizing. Fe concentrations were quantified with the ferrozine assay (82).
Since the sediment samples were too dry to obtain pore water by established methods, “artificial pore water” was created by leaching 1 g of the 0- to 10-cm composite samples from Lake Kasin three times with 30 ml of autoclaved Millipore water for 4 h. Analysis was performed in triplicate as described by Jiang et al. (30). Ionic composition of this “artificial pore water” was determined by ion chromatography (see above for analytical details) and organic acids (e.g., acetate, lactate, formate) were quantified by high-pressure liquid chromatography (HPLC) (instrument type “class vp” from Shimadzu, Duisburg, Germany, equipped with a Microguard cation H cartridge precolumn and an Aminex HPX-87H ion-exclusion column of 300 mm by 7.8 mm from Bio-Rad, Munich, Germany). The total amount of ions and organic acids leached from 1 g of sediment was back-calculated to the water content of the sample.
Analysis of gas samples from the water-covered part of the lake sediment for methane was performed with a Varian 3800 gas chromatograph equipped with an Alltech 13939 column (length: 30 m; inner diameter: 0.53 mm; AT-Q) and a flame ionization detector (FID). The injector had a temperature of 200°C, and the temperature program was as follows: 60°C, hold 3 min, 60°C to 200°C at 75°C/min, hold 3 min. The flow rate was 5 ml/min at a flow pressure of 3.2 lb/in2.
Mössbauer spectroscopy.
A Fe oxide-rich layer between 1 and 2.5 cm in depth was observed in the sediment profile of Lake Kasin. Because the sample contained less than 3% (wt/wt) of total Fe, unlikely to be detected by X-ray diffraction (XRD), Fe speciation was analyzed by 57Fe Mössbauer spectroscopy. For this purpose, about 1 g of sample was transferred into an anoxic glove box and preserved between two layers of O2-impermeable Kapton tape (Polyfluor Plastics BV, Oosterhout, the Netherlands). The measurement was performed with a 57Co source at room temperature with linear acceleration in transmission mode as described previously (39). The spectrometer was constructed by WissEL (Wissenschaftliche Elektronik GmbH, Starnberg, Germany). A Janis closed-cycle cryostat with a helium atmosphere was used to vary the temperature of the sample. Spectra were calibrated against spectra of α-Fe(0) foil. Recoil software (University of Ottawa, Canada) and Voigt-based models were used for spectrum interpretation.
Most-probable-number (MPN) counts and enrichments of Fe(II)-oxidizing and Fe(III)-reducing microorganisms.
Anoxic salt water medium (SWM) containing 5 M and 0.5 M NaCl (pH 7.2 to 7.4) was used to enumerate anaerobic nitrate-reducing Fe(II) oxidizers (anFeOx) as well as anaerobic Fe(III) reducers (FeRed) by MPN counts. Dilution series of sediment suspensions of the 0- to 10-cm composite sample were set up in 96-well plates. The anoxic salt water medium contained 100 μM MgSO4 · 7H2O, 3 mM KCl, 100 μM KBr, 5 mM NH4Cl, 1.9 mM MgCl2, 15 mM NaHCO3, 1 mM CaCl2, 1 mM NaHPO4, 0.2 μM NH4VO3, and 0.5 mM Na2S2O3 as well as 1× vitamin solution (98), trace element solution (92), and selenate-tungstate solution (97).
The following electron donors and acceptors were added to the medium from stock solutions. For anFeOx, the final medium contained 4 mM NaNO3 and 10 mM FeCl2 as well as 0.5 mM Na-acetate to allow growth of mixotrophic Fe(II) oxidizers. For FeRed, the medium contained 5 mM 2-line ferrihydrite prepared according to Straub et al. (83) and a mixture of 5 mM Na-acetate and 5 mM Na-lactate. Setup and analysis of the MPN counts are described in detail in the supplemental material.
Gradient tubes for enumeration and enrichment of microaerophilic Fe(II) oxidizers were prepared as described by Emerson and Floyd (17) with some modifications, which are also described in the supplemental material.
DNA extractions.
For denaturing gradient gel electrophoresis (DGGE) and quantitative PCR (qPCR) analyses, DNA from sediment samples was extracted as follows: in order to remove salts, 0.3 g of sediment was washed three times with 1.5 ml of TE buffer (10 mM Tris-EDTA, pH 7.0). Following centrifugation for 10 min at 7,200 × g, the supernatant was collected and filtered through a 0.22-μm polyethersulfone (PES) membrane filter. Washed sediments and membrane filters were separately extracted using the PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) according to the protocol of the manufacturer. At the end of the protocol, DNA extracted from sediment and filters was sequentially eluted in 2× 25-μl elution buffer (buffer C6).
For 16S rRNA gene clone library construction, DNA was also extracted following the protocol described by Zhou et al. (104). Each extraction was performed in duplicate. The two DNA extracts obtained using the PowerSoil kit and the protocol of Zhou et al. were pooled in order to minimize a methods-immanent extraction bias. Pooled DNA extracts were further purified using the QIAEX II kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA from cell pellets of the liquid enrichment cultures was extracted using the UltraClean DNA isolation kit (MoBio).
DGGE.
Partial 16S rRNA genes from the DNA extracts of the liquid enrichment cultures were amplified using the primers 341GCF (51) and 907R (14) for amplification of general bacterial 16S rRNA gene fragments. The primers 20F (45) and 1392R (38) were used for amplification of general archaeal 16S rRNA gene fragments. Amplified archaeal 16S rRNA gene fragments were used as the template for a nested PCR with primers 344GCF (81) and 519R (38).
Details about PCR and DGGE conditions can be found in the supplemental material. Prominent DGGE bands were cut from the gel, reamplified, cloned, and sequenced.
Clone library construction and phylogenetic analysis.
DNA from the 0-to 10-cm composite sample from Lake Kasin sediment was PCR amplified with general primers GM3F (52) and 1392R (38) for bacterial 16S rRNA gene fragments and 20F (45) and 958R (13) for archaeal 16S rRNA gene fragments. Reaction mixtures contained 1× PCR buffer with 1.5 mM MgCl2 final concentration (Promega), 200 μM deoxynucleoside triphosphate (dNTP) mix (New England BioLabs), 200 nM each primer, 1.25 U Taq DNA polymerase (Promega), and 10 ng of DNA in a total volume of 25 μl. The following thermocycler program was used for amplification of 16S rRNA gene sequences: hot start at 70°C; initial denaturation at 95°C for 5 min; 25 cycles of denaturing (95°C for 1 min Bacteria/2 min Archaea), annealing (44°C Bacteria/58°C Archaea for 1 min), and elongation (72°C for 3 min Bacteria/1.5 min Archaea); and a final elongation at 72°C for 10 min. PCR products were purified with the Wizard PCR Clean-Up System (Promega Laboratories). All PCRs were performed in duplicate and pooled after purification. The purified PCR products were cloned using the TOPO TA cloning kit and TOP10 competent cells (Invitrogen). Escherichia coli clones were sent for sequencing of the 16S rRNA gene insert to GATC Biotech (Konstanz, Germany). Forward and reverse reads were assembled and trimmed using the program “DNA Baser” (http://www.dnabaser.com/). The total length of the sequences after trimming was ∼900 bp for Archaea and ∼1,300 bp for Bacteria. All sequences were checked for chimeras by using Bellerophon (26) and Slayer (23). Chimeras were removed from the data sets. Sequences were aligned and analyzed using the SINA aligner of the SILVA rRNA database project (67) and the ARB software package (version 5.2) (42) with the corresponding SILVA SSURef 106 database (67) according to the standard operating procedure published by Peplies et al. (62). Tree construction was carried out with up to 200 sequences using the neighbor joining and maximum likelihood (ML; RAxML, AxML, and fastDNAml) methods in ARB. Tree topology was further tested by the application of positional variability filters for Archaea and Bacteria, respectively, as well as with 50% positional conservatory filters that were created for Archaea and Firmicutes, respectively (62). The archaeal tree was calculated with 152 sequences based on 7,123 valid columns (50% conservation filtering) with fastDNAml. The bacterial tree was calculated with 200 sequences based on 6,370 valid columns (50% conservation filtering) with RAxML (model: GTRCAT). Partial sequences were added to both trees using the ARB parsimony tool. A multifurcation was introduced manually into the bacterial tree in one case where the tree topology could not be unambiguously resolved. For clarity, only selected subsets of the sequences used for treeing are shown in the figures. Rarefaction curves were constructed using the software MOTHUR (75).
qPCR.
Copy numbers of 16S rRNA genes in the environmental DNA extracts were quantified on an iQ5 real-time PCR cycler (Bio-Rad Laboratories GmbH, Munich, Germany) using the SsoFast Eva Green detection kit (Bio-Rad). Table S1 in the supplemental material lists the primers used in the different qPCR assays. For quantification of general bacterial 16S rRNA gene copies, 75 nM primer 341F and 225 nM primer 797R were used per reaction mix. All other primers were added to final concentrations of 250 nM. In addition, reaction mixes contained 10 μl of SsoFast Eva Green master mix, 2 μl of template, and standard or DNase-free water in a final volume of 20 μl. All thermal cycler programs started with 2 min at 98°C followed by 40 cycles of the respective programs listed in Table S2 in the supplemental material. Table S2 also contains information on the standards used in the different qPCR assays. Data analysis was performed with the iQ5 optical system software, version 2.0 (Bio-Rad, 2006) as described by Behrens et al. (2).
Quantification of archaeal 16S rRNA gene copies was performed once in triplicate. All other qPCRs were performed in triplicate twice. Cell numbers per g dry sediment were calculated from the qPCR 16S rRNA gene copy numbers considering the average rRNA operon numbers of the respective taxa (Bacteria, Archaea, Bacillaceae, Peptococcaceae, Halobacteriaceae) as listed in the rRNA Operon Copy Number Database (http://rrndb.mmg.msu.edu/index.php).
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences determined in this study have been deposited in GenBank under accession numbers HE604643 to HE604939 (Bacteria) and HE604411 to HE604642 (Archaea). 16S rRNA gene sequences of the Fe(III)-reducing enrichments have the accession numbers HE604940 to HE604952.
RESULTS
Geochemistry of Lake Kasin, southern Russia.
The exact geographic location of the sampling site at Lake Kasin is given in Table 1 and Fig. S1 in the supplemental material. The ionic strength of the wet sediment was 6.09 M mainly comprised of sodium and chloride ions (about 5 M). The normalized electrical conductivity at 25°C of the 0- to 10-cm composite sediment sample was 438.5 mS cm−1. Based on the measured conductivity, the salinity of the sediment was 348.6 g liter−1 (100) (Table 1). A ferrozine test performed on site revealed the presence of 100 μM Fe(II) in the pore water at about a 20-cm depth. Neither NO2− nor NO3− could be detected in the groundwater with color indicator strips, which had a detection limit of 160 μM for NO3− and 45 μM for NO2−. However, as indicated in Table 1, 130 μM water-leachable NO3− was measured using ion chromatography. The leachate also contained 37.2 ± 4.6 mM acetate and 10.6 ± 1.6 mM formate, but no lactate. During sampling, a strong scent of H2S was noticeable at Lake Kasin (odor detection threshold, 4.7 ppb or 15 μM [66]). Together with the high concentration of water-leachable sulfate in the sediment (968 mM), this might be an indication for the presence of an effective redox cycling of sulfur at Lake Kasin. About 1.87 mmol/liter of methane was measured in gas samples from the water-covered part of the sediment. This corresponds to approximately 45,000 ppm of methane, which exceeds atmospheric concentrations by 3 orders of magnitude. These observations suggest that in addition to sulfate reduction, methanogenesis takes place in the sediments as well. In addition, based on thermodynamics, Fe(III) reduction is also expected to occur given the fact that bioavailable Fe(III) is present. Fe extractions of the composite sample gave a total Fe content of 0.85% (wt/wt) (dry weight) in the top 10 cm of Lake Kasin, of which 19% was Fe(II). The total Fe content determined by XRF was slightly higher (1.13% [wt/wt] [dry weight]) (Table 1). Upon visual inspection of the sediment profile, an Fe oxide-rich layer could be identified between 1 and 3 cm of depth (see Fig. S1). We quantified both 0.5 M HCl-extractable (“bioavailable”) and 6 M HCl-extractable (“crystalline”) Fe throughout the sediment profile and found clear maxima of both bioavailable and crystalline Fe at 1.5 cm of depth, where the two iron phases constituted 0.85% (wt/wt) and 1.30% (wt/wt) of the sediment dry weight, respectively (Fig. 1).
Table 1.
Location and geochemical properties of Lake Kasin sediment (0- to 10-cm depth composite sample)
| Location or geochemical property | Value |
|---|---|
| Geographic position | 47°36.165′N 047°27.129′E |
| pHa | 7.86 |
| Fe contentb | 1.13% |
| Ionic strengthc | 6.09 M |
| Conductivityd | 438.5 mS cm−1 |
| Salinitye | 348.6 g liter−1 |
| H2O content | 14.65% |
| Clb | 1.51% |
| SO42− | 968 mMc |
| NO3−c | 130 μM |
| Cinorgf | 1.85% |
| Corgg | 0.11% |
Determined with 0.01 M CaCl2.
Weight % of dry sediment quantified by XRF.
Quantified by IC from modified pore water after reference 30.
Average value horizon H1 to H4 (0- to 10-cm depth) at 25°C.
Calculated from conductivity after Williams and Sherwood (100).
Weight % determined by weight loss during titration with HCl.
Weight % as quantified by a C/N analyzer using a HCl-titrated sample.
Fig 1.
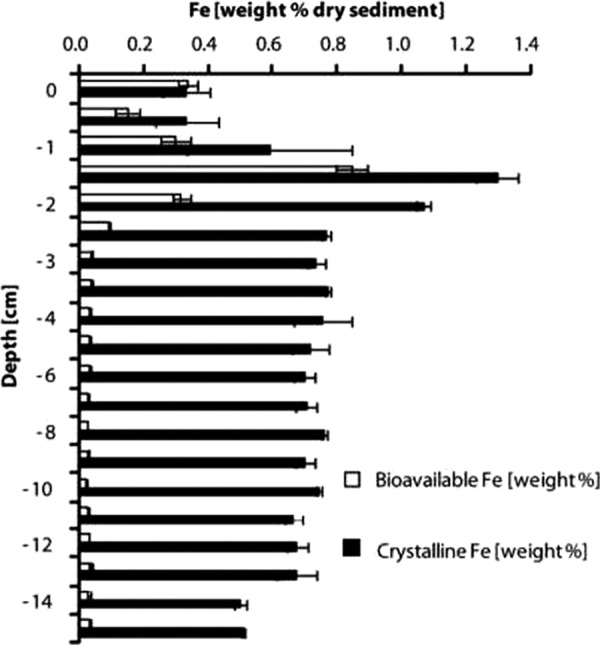
Concentration of different iron fractions in a sediment profile of Lake Kasin. White bars, 0.5 M HCl-extractable (bioavailable) iron. Black bars, crystalline iron, extracted during incubation in 6 M HCl at 70°C for 24 h. Iron concentrations in the extracts were determined with the ferrozine assay (82). Error bars represent standard deviations calculated from duplicate samples.
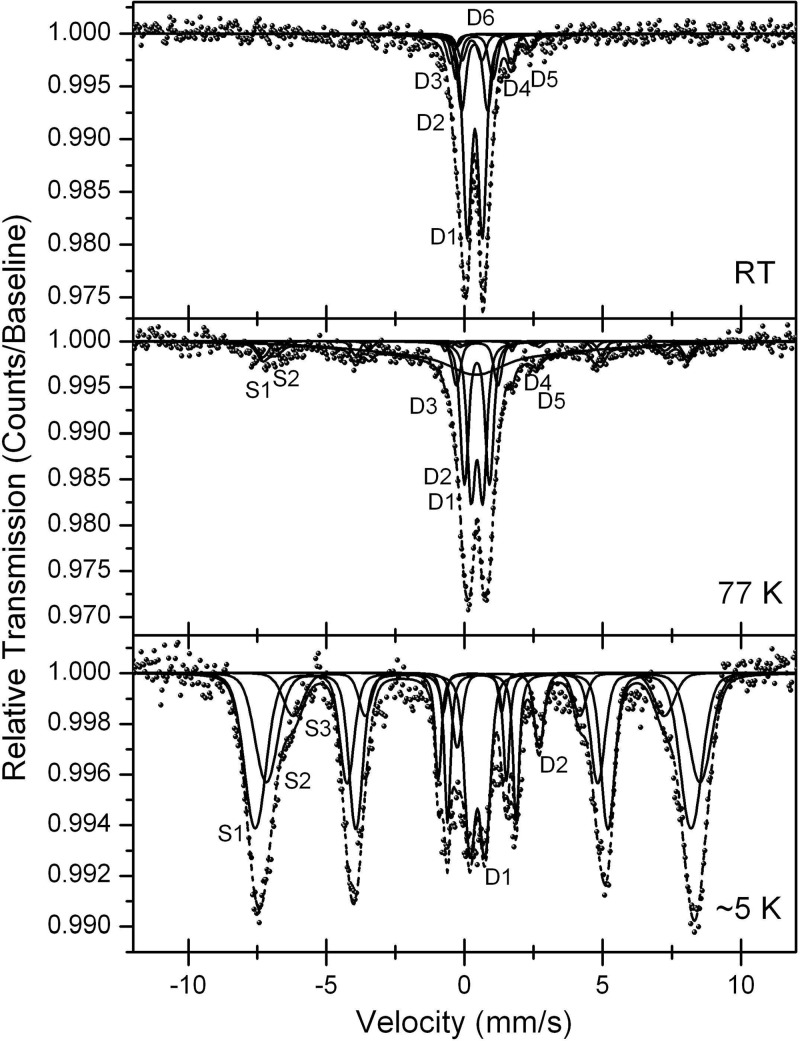
Figure 2 shows Mössbauer spectra of the Fe-rich layer at 1.5 cm of depth obtained at room temperature (RT), 77 K, and ∼5 K. The spectra show predominantly Fe(III) phases, but also small amounts of Fe(II). We modeled all three spectra of the Fe(III) phase using parameters for akaganéite reported by Murad and Johnston (50), Chambaere et al. (7), and Barrero et al. (1) and parameters reported by Feder et al. (18), Genin et al. (19), Refait et al. (70), and Rusch et al. (73) for the green rust Fe(II) phases (see Table S3 in the supplemental material). Akaganéite [β-FeO(OH)] can incorporate up to 7 mol% of Cl− ions as additional constituents that stabilize tunnels within the crystal structure (9). Since the presence of Cl− or F− ions is necessary for the formation of akaganéite (9), its natural occurrence is generally restricted to environments with high concentrations of Cl− or F− ions such as, e.g., hypersaline lakes. However, the Mössbauer parameters reported for lepidocrocite (e.g., reference 49) and for Fe(III) in green rust (18, 19, 70) are very similar to those of the akaganéite doublet D1 (see Table S3 in the supplemental material), the akaganéite sextets S3 (lepidocrocite), and S1 (green rust). A quantification of these different iron mineral phases based on Mössbauer spectra was therefore not possible. The Mössbauer spectra did not provide any evidence for the presence of iron sulfide minerals.
Fig 2.
Mössbauer spectra of the Fe-rich layer of Lake Kasin sediment (1.5 cm of depth) recorded at room temperature (RT) (top), 77 K (middle), and 5 K (bottom). Sextets (S) and doublets (D) are labeled as listed in Table S3 in the supplemental material.
Abundance and activity of culturable Fe(III) reducers and Fe(II) oxidizers.
The results of the most-probable-number (MPN) counts are shown in Fig. 3. The main finding of this experiment was that similar numbers of anaerobic Fe(II) oxidizers (anFeOx) and Fe(III) reducers (FeRed) (210 and 943 cells per g dry sediment, respectively) grew in medium with 0.5 M NaCl and in medium with 5 M NaCl. We also performed MPN counts to quantify microaerophilic Fe(II) oxidizers from Lake Kasin in gradient tubes, but we did not observe growth in any tube. From selective wells of the FeRed and anFeOx MPN plates, cultures were transferred into fresh medium in order to pursue further enrichment. Enrichment cultures were consecutively transferred into fresh medium as soon as Fe(II)-oxidizing or Fe(III)-reducing activity was observed, which was on average every 8 to 12 weeks. While the activity of anFeOx could not be maintained over repeated transfers, several FeRed enrichments performed well. After three transfers, the most dominant microorganisms in these enrichment cultures were identified by sequencing prominent DGGE bands.
Fig 3.
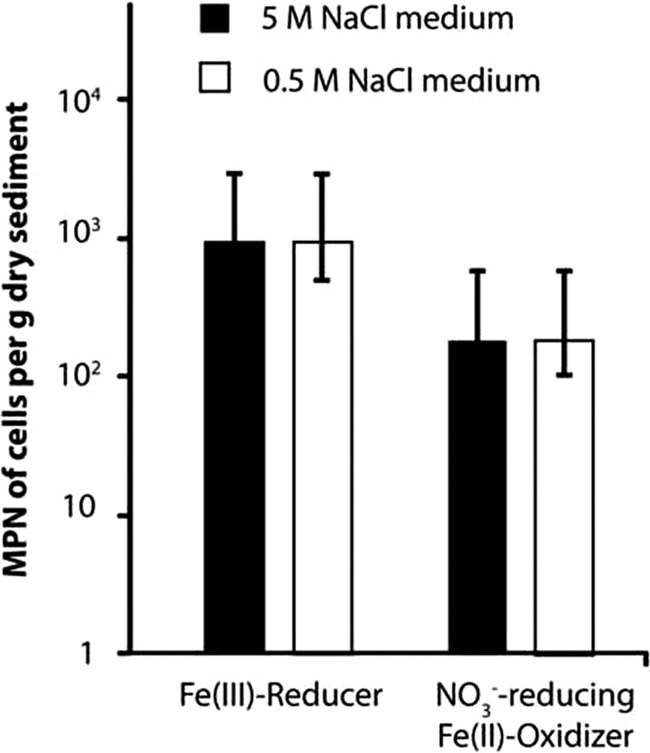
Most-probable-number (MPN) counts of Fe(III)-reducing (FeRed) and anaerobic Fe(II)-oxidizing (anFeOx) microorganisms from the top 10 cm of Lake Kasin in mineral medium with 5 M (black bars) and 0.5 M NaCl (white bars), respectively. Medium for FeRed was supplemented with 0.5 M ferrihydrite as electron acceptor and 0.5 M lactate and acetate each as electron donors. For anFeOx, 10 mM FeCl2 was added as the electron donor and 0.4 M NO3− as the electron acceptor. The Fe(II) oxidizer medium further contained 0.05 M acetate as a carbon source. Error bars denote 95% confidence intervals determined from seven replicate samples according to the work of Klee (34).
From DNA extracts of an Fe(III)-reducing enrichment culture in medium with 5 M NaCl, only archaeal 16S rRNA genes could be amplified by PCR. The two most prominent DGGE bands (see Fig. S3A in the supplemental material) were excised from the gel, reamplified, cloned and sequenced. However, since these 175-bp sequences were too short to allow accurate classification, full-length 16S rRNA gene amplicons were generated from the original DNA extract of the enrichment culture. Six out of the 10 sequenced clones had 97 to 98% 16S rRNA gene sequence identity to Halobaculum gomorrense strain JCM 9908. Halobaculum gomorrense was first isolated by Oren et al. in 1995 from the Dead Sea (59). The other four sequences grouped within the Halobacteriaceae family. The closest cultivated relatives to these sequences were Halogranum rubrum, Halomicrobium katesii, and Halobacterium noricense.
Only bacterial but no archaeal 16S rRNA gene amplicons could be amplified from DNA extracts of another Fe(III)-reducing enrichment in medium with 0.5 M NaCl. Only one dominant band was visible on a DGGE gel (see Fig. S3B in the supplemental material). The band was excised, reamplified, cloned, and sequenced. Even though they originated from one single DGGE band, the three clones that were sequenced were found to be phylogenetically distinct from each other: one of the 16S rRNA gene sequences was 97% identical to a Dehalobacter restrictus strain from an anaerobic coculture that had been enriched from a hexachlorocyclohexane-polluted soil (94). The second sequence was 99% identical to the 16S rRNA gene sequence of Lactobacillus fabifermentans (11). The closest cultivated relative (97% 16S rRNA gene sequence identity) to the third sequence from our enrichment was Desulfosporosinus lacus, a sulfate-reducing bacterium isolated from sediments of Lake Stechlin, Germany (68).
In the two Fe(III)-reducing enrichment cultures, between 0.46 and 2.49 mM Fe(II) was formed within 44 days. These values correspond to 0.01 to 0.06 μmol of Fe(II) produced per ml of culture daily (Table 2).
Table 2.
Comparison of rates of Fe(II) production in Fe(III)-reducing enrichment cultures with Fe(II) concentrations in Lake Kasin sediment
| Fe(III)-reducing enrichment cultures | Cell numbersb,c | Fe(II) production ratesd [μmoles Fe(II) ml−1 day−1] | μmoles Fe(II)b |
|---|---|---|---|
| Halobaculum gomorrense relatives | 1.86 × 105 | 0.01 | 28 |
| Desulfosporosinus spp. | 7.13 × 104 | 0.02 | |
| Bacilli (containing Bacillus alkalidiazotrophicusa) | 5.48 × 105 | 0.06 |
Culture originates from sediment of salt Lake Baskunchak, southern Russia.
Per g dry sediment of 0- to 10-cm composite sample from Kasin sediment.
Determined by qPCR with primers “Hgomorr,” “Desulfosp.,” and “Bacilli.”
In the enrichment cultures.
Archaeal and bacterial diversity as determined by 16S rRNA gene clone libraries.
We constructed 16S rRNA gene clone libraries for both Bacteria and Archaea. These libraries revealed a very high diversity among Bacteria (12 different phyla and five uncultured “candidate divisions”) compared to Archaea (2 phyla).
In the bacterial clone library, Gammaproteobacteria represented the most abundant group (169 out of 299 sequences) (Fig. 4B; also see Table S4 in the supplemental material). The majority of the gammaproteobacterial clones (144 sequences) belonged to the genus Halothiobacillus. Members of this genus are obligate chemolithoautotrophs. They tolerate high concentrations of solutes and obtain energy from oxidizing reduced sulfur species (32). The second major phylum within the Bacteria was the Firmicutes, comprising 35 clone sequences, which fell into two different classes, namely, Bacilli (30 sequences) and Clostridia (5 sequences). Firmicutes comprise many halophilic, thermophilic, anaerobic, and fermentative bacteria capable of forming spores and toxins (16).
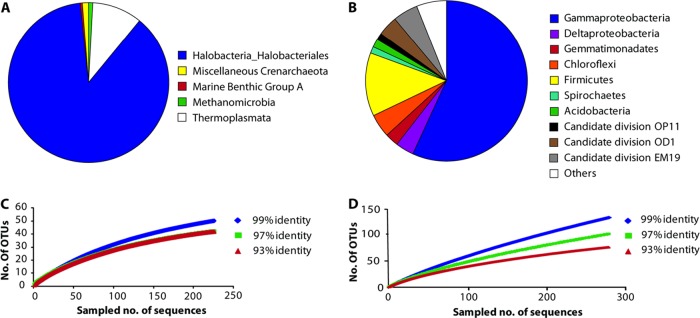
Fig 4.
(A and B) Classification of 231 archaeal (A) and 299 bacterial (B) full-length 16S rRNA gene sequences retrieved from a 0- to 10-cm composite sample of Lake Kasin. (C and D) Rarefaction curves for the archaeal and bacterial sequences from the respective clone libraries for three different sequence identity cutoff values (99%, 97%, and 93%). The archaeal rarefaction curve for the 97% cutoff value in C is not visible because it exactly resembles the 93% curve. Rarefaction curves were calculated with the program MOTHUR (75).
As for the archaeal clone library (Fig. 4A), 16S rRNA gene sequences of both Euryarchaeota (227 sequences) and Crenarchaeota (4 sequences) were found. Of the euryarchaeotal sequences, 209 belonged to the class of Halobacteria, extreme halophiles that grow even at saturated salt concentrations (10).
Rarefaction curves (Fig. 4C and D) and Chao indices indicated that the diversity of Archaea in Lake Kasin has been covered by the 16S rRNA gene clone library to a larger extent than the bacterial diversity. Interestingly, none of the bacterial sequences recovered were closely related to known and cultured dissimilatory Fe(III) reducers or Fe(II) oxidizers. The sequences obtained from the DGGEs of the bacterial Fe(III)-reducing enrichments were distinct from those in the clone library. However, 10.4% of the sequences in the clone library were affiliated with the class Bacilli, a representative of which, Bacillus infernus, has been shown to reduce Fe(III) (3). Furthermore, Fe(III)-reducing enrichment cultures that were inoculated with sediment from other Russian salt lakes (Lake Elton and Lake Baskunchak) were also dominated by strains which showed 98% 16S rRNA gene sequence identity to Bacillus alkalidiazotrophicus and Anaerobacillus alkalilacustris, respectively (data not shown). For these reasons and since most of the cultivated Bacilli have not been tested for their ability to reduce Fe(III), it is conceivable that Bacilli might be contributing to Fe(III) reduction at Lake Kasin.
With respect to the Archaea, the dominant strain in our Fe(III)-reducing enrichment culture with 5 M NaCl, Halobaculum gomorrense, was also the closest cultivated relative of six clones from our archaeal 16S rRNA gene clone library, with sequence identities exceeding 97%. Based on the results from our cultivation-dependent and -independent experiments, species affiliated with the Bacilli and Desulfosporosinus spp. as well as species affiliated with Halobaculum gomorrense were considered potential candidates contributing to Fe(III) reduction in Lake Kasin sediment.
Abundance and distribution of Fe(III)-reducing microorganisms along the sediment profile.
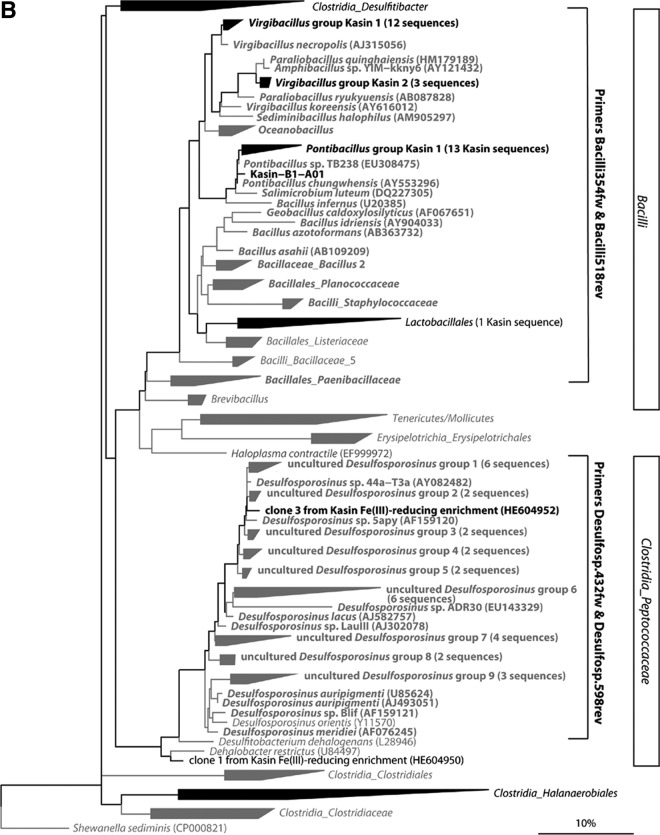
In order to analyze the abundance and vertical distribution of Desulfosporosinus sp. relatives, Bacilli, and Halobaculum gomorrense relatives in the sediment, we designed 16S rRNA gene primers specific to the obtained sequences, their cultivated relatives, and a few sequences from uncultivated strains that belonged to the same sequence clusters. While the “Bacilli” quantitative PCR (qPCR) primers targeted most sequences within this class, the numbers of targeted sequences of primers “Desulfosp.” and “Hgomorr” were much smaller and did not include all species of the genera Desulfosporosinus and Halobaculum, respectively (see Table S1 in the supplemental material). Figure 5 shows maximum likelihood trees of the archaeal (Fig. 5A) and bacterial (Fig. 5B) 16S rRNA gene sequences from Lake Kasin sediment and the Fe(III)-reducing enrichment cultures. Brackets indicate target sequences of the group-specific qPCR primers.
Fig 5.
Maximum likelihood trees of archaeal (A) and bacterial (B) 16S rRNA gene sequences directly amplified from Lake Kasin sediment or obtained from the Fe(III)-reducing enrichment cultures. (A) ML tree of the Halanaerobiaceae. (B) ML tree of the Bacilli and the Peptococcaceae family of the Clostridia. Groups for which at least one representative sequence was found in Lake Kasin sediment or enrichments are printed in black. Groups with no representatives from Lake Kasin sediment or enrichments are printed in gray. (Groups of) sequences that match both forward and reverse qPCR primers designed for Halobaculum gomorrense (A)- and Desulfosporosinus sp. or Bacilli (B)-related sequences are shown in boldface.
The total number of cells (16S rRNA gene copy numbers of Bacteria and Archaea corrected for average rRNA operon numbers) ranged from 1.1 × 106 to 6.7 × 107 cells per g dry sediment, with the highest numbers found at 1.5 to 2 cm of depth. Bacteria outnumbered Archaea by factors of two to three throughout the sediment profile.
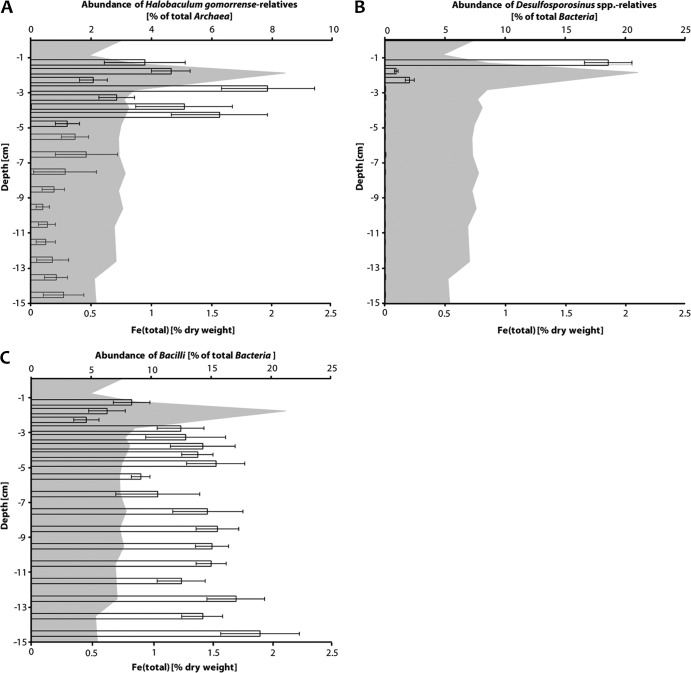
With respect to the distribution of putative Fe(III) reducers, Halobaculum gomorrense relatives comprised 2 to 6% of all Archaea in the top 4.5 cm of the sediment (Fig. 6A). However, there was no significant correlation between cell numbers of Halobaculum gomorrense relatives and the distribution of total Fe (% [wt/wt] dry weight) in the upper sediment layers. Desulfosporosinus sp. relatives, on the other hand, were detected only in the Fe-rich top 3 cm of the sediment. Their abundance was 3 orders of magnitude higher at 2.5 cm of depth than further below (Fig. 6B). Between 1 and 1.5 cm of depth, up to 20% of all Bacteria were Desulfosporosinus sp. relatives. Bacilli represented between 5 and 20% of all Bacteria throughout the sediment profile without an obvious cell number increase in the upper Fe-rich layers (Fig. 6C).
Fig 6.
Changes in relative abundance of potential Fe(III)-reducing taxa in a sediment profile of Lake Kasin. Bars indicate the abundance of Halobaculum gomorrense relatives (A), Desulfosporosinus sp. relatives (B), and Bacilli (C) relative to total Archaea (A) or total Bacteria (B and C) cell numbers for each sediment layer. Error bars refer to standard deviations of six individual qPCR measurements recorded in triplicate during two independent runs. The gray-shaded area in the background shows Fe (total) concentrations in % dry weight of the sediment.
DISCUSSION
Occurrence and speciation of Fe in Lake Kasin sediments.
A substantial fraction of the Fe in Lake Kasin occurred in the form of the mineral akaganéite (Fig. 2). Akaganéite has a Néel temperature of 299 K (49) and should appear as magnetically split sextets in the spectrum recorded at 77 K. Under these conditions, lepidocrocite, which has a Néel temperature of 77 K (49), may have contributed to the akaganéite spectrum.
Furthermore, green rust minerals, which can contain Cl−, SO42−, and CO32−, also have similar Mössbauer parameters. The chemistry of Lake Kasin sediment would allow the formation of all three mineral phases. Therefore, a mixture of the three mineral phases seems most likely. The fact that the Mössbauer spectra did not reveal any FeS precipitated in this system could be due to the precipitation of other Fe(II)-bearing minerals that have a lower solubility product than FeS and are therefore favored to precipitate under in situ conditions.
Only minor amounts of akaganéite can be extracted with 1 M hydroxylamine-HCl within 48 h (65). This implies that akaganéite is not expected to contribute to the “bioavailable” Fe fraction of Lake Kasin sediment extracted with 0.5 M HCl. However, the mineral can still be subject to microbial transformations: we observed that the Fe(III)-reducing strain Shewanella oneidensis MR1 can reduce akaganéite almost as efficiently as poorly crystalline ferrihydrite (unpublished data). This means that akaganéite could also serve as an electron acceptor for Fe(III)-reducing microorganisms in Lake Kasin. Therefore, it is most likely that we underestimated the amount of bioavailable Fe by extraction with 0.5 M HCl.
Apart from Lake Kasin, sediments of other salt lakes have also been found to contain high amounts of Fe minerals: the sediment of Lake Tyrrell (Australia) contains hydrous Fe oxides such as goethite (41), which can also be subject to microbial reduction (71). In addition to goethite, alunite, and jarosite were identified in Lake Tyrrell sediments (41). The formation of jarosite in low-temperature environments has been related to microbial oxidation of iron and sulfur (27). In other salt lake sediments, Fe(II) rather than Fe(III) minerals were found to dominate; e.g., in the sediment of Lake Qinghai (China), pyrite was the dominating Fe mineral (37). About 1% (dry weight) of the sediment of the SO42−-rich Salton Sea (United States) was found to consist of reduced Fe phases, which were dominated by greigite (Fe3S4; sulfur analogue to magnetite) and pyrite (FeS2) (12). Fe(II) released during oxidation of pyrite has been shown to also serve as an electron donor for microorganisms (89). The presence of bioavailable Fe(II) and Fe(III) mineral phases in various salt lake sediments could be indicative of both microaerophilic and anaerobic microbial Fe(II) oxidation and Fe(III) reduction in these environments.
Fe-metabolizing microorganisms in Lake Kasin.
The MPN counts revealed as many Fe(III) reducers as nitrate-reducing Fe(II) oxidizers in medium with 5 M NaCl as in medium with 0.5 M NaCl. This was the case for inocula from Lake Kasin sediment (Fig. 3) as well as for two other reference sediments we investigated (Lake Elton and Lake Baskunchak) (see Fig. S2 in the supplemental material). Since the microorganisms were exposed to considerable osmotic stress in medium with 5 M NaCl, these results showed that the Fe-metabolizing microorganisms in Lake Kasin sediments were well adapted to a broad range of NaCl concentrations. A comparison of results from MPN counts to qPCR measurements of total bacterial 16S rRNA gene copies revealed that culturable anaerobic Fe(II) oxidizers and Fe(III) reducers represent <0.1% of the total Bacteria present in Lake Kasin sediment.
The conditions in our Fe(III)-reducing enrichment cultures did not allow distinction between dissimilatory and fermentative Fe(III) reduction. As a consequence, part of the observed Fe(III) reduction in the sediment might be due to fermentation, also explaining the relatively high abundance of Bacilli and other microbial taxa capable of fermentation.
The absence of growth of microaerophilic Fe(II) oxidizers in gradient tubes might have been due to limited diffusion of Fe(II) to the upper layers of the gradient tube in the high-salinity medium. Since we measured 50 μM total Fe in the upper part of the agar tubes, it seems more likely that microaerophilic Fe(II) oxidizers either did not grow under the applied conditions or were not present in Lake Kasin sediments.
Total cell numbers and total Fe content showed a strong linear correlation (R2 = 0.91; P = 1.77 × 10−8; according to Spearman's test for nonparametric data). Both parameters were highest at the same sediment depth. The Fe content refers to both bioavailable and crystalline Fe, even though the difference in Fe concentration between the Fe-rich layer and the lower parts of the sediment was more pronounced for the bioavailable than for the crystalline Fe fraction (Fig. 1). Another strong linear correlation (R2 value of 0.76) was found between cell numbers and NaCl content. NaCl-dominated salinity continually decreases from 48.7 g Cl− per kg dry weight in the topmost horizon (0 to 2.5 cm) to 10.8 g Cl− per kg dry weight at 5 cm of depth. Correlations between other geochemical parameters (such as temperature, pH, sulfate or carbonate concentration) and cell numbers were not found.
The number of total cells (Bacteria and Archaea) in Lake Kasin sediment (1.1 × 106 to 6.7 × 107 cells per g dry sediment) was rather low in comparison to cell numbers determined in other hypersaline environments, such as sediments of saline Lakes Chaka and Qinghai (15, 29), a Californian salt marsh (5), and sediments of a hypersaline mud volcano (40). In Lake Qinghai, China, for example, which has a salinity of 12.5 g/liter, cell counts revealed 107 cells per g wet sediment at 50 cm of depth (15). Although these numbers refer to wet sediment, the cell numbers at 50 cm of depth in Lake Qinghai are still higher than the numbers quantified for the iron-rich layer (1 to 3 cm of depth) of Lake Kasin. This might be explained by the low amount of total organic carbon in Lake Kasin sediments (0.11% [wt/wt]) compared to, e.g., Lake Quinghai (1.8 to 2.4% [wt/wt]) sediment (15).
In Lake Kasin, Bacteria were more abundant than Archaea throughout the sediment profile. This finding is in contrast to results for sediments of Lake Chaka (29) and the Salton Sea (86) but similar to those for La Sal del Rey sediments, where more than 97% of all 16S rRNA gene copies belonged to the Bacteria (25). Only a few comprehensive data sets are available from hypersaline sediments, and currently it remains unclear what physicochemical factors determine whether Bacteria or Archaea dominate numerically.
The presence of anaerobic Fe oxidizers and Fe reducers in our MPNs and enrichment cultures as well as the strong correlation between Fe content and total cell numbers in Lake Kasin sediments suggests that next to sulfur Fe might also serve as electron donor and acceptor of microbial respiration at high salinities. However, a strong correlation between Fe content and total cell numbers could also reflect sedimentary deposition of iron and carbon over time.
Microbial diversity in Lake Kasin sediments.
The 16S rRNA gene clone library revealed a higher bacterial than archaeal diversity in the top 10 cm of Lake Kasin sediment. This is in agreement with clone library data from other saline sediments such as those from the salt Lakes Chaka (29, 30) and Qinghai (15, 28) as well as sediments from soda lakes of the Wadi An Natrun (47), the Kenyan-Tanzanian Rift Valley (69), and Lonar Lake (96). Based on 97% 16S rRNA gene sequence identity, higher numbers of bacterial than archaeal operational taxonomic units (OTUs) were defined in all clone libraries from the above-mentioned saline lakes.
The majority of the sequences in our bacterial clone library were affiliated with Gammaproteobacteria. Gammaproteobacteria also comprised up to 10% of all bacterial 16S rRNA gene sequences in clone libraries of other saline sediments (15, 25, 30, 35, 43, 47, 69, 96). Although high numbers of gammaproteobacterial 16S rRNA gene sequences are also common in clone libraries of other saline sediments, the gammaproteobacterial genus Halothiobacillus, which comprised 38% of all bacterial 16S rRNA gene sequences in our library (see Table S4 in the supplemental material), to date has not occurred in any other published clone library of saline sediments. The absence of Halothiobacillus sequences in all other clone libraries from saline sediments might be due to the fact that we constructed our clone library from exposed lakebed sediment samples, while most other published clone libraries (except for a transect of hypersaline La Sal del Rey, Texas, United States) were constructed from samples of waterlogged and anoxic saline sediments. Halothiobacillus is an obligatory aerobic genus which grows by oxidizing reduced sulfur species at up to 4 M NaCl (79). This means that the presence of H2S, a salinity close to saturation, and the availability of O2 in the top few cm of the sediment constitute nearly optimal growth conditions for Halothiobacillus in Lake Kasin. Halothiobacillus species have been described to outcompete other species on the basis of their high growth rate (79), which may further explain why so many sequences in our clone library are affiliated with this genus.
Firmicutes accounted for 12% of all bacterial 16S rRNA gene sequences and were the second-most abundant bacterial phylum in Lake Kasin sediments. Firmicutes were also abundant in other saline lake sediments. In sediments from the saline Lakes Chaka (29, 30) and Qinghai (15, 28) as well as the soda lakes of the Wadi An Natrun (47), the Kenyan-Tanzanian Rift Valley (69), and Lonar Lake (96), Firmicutes were the most abundant phylum (constituting 19 to 42% of all sequences in these libraries).
What is notable in our clone library is the absence of sequences representing the Cytophaga/Flexibacter/Bacteroidetes (CFB) group. This group is reported to dominate in many different saline environments including marine water (33), salt lakes (60), soda lake sediments (69), and hypersaline endovaporitic mats (77, 79). The main ecological role of the heterotrophic CFB group has been described to be the degradation of organic material due to the capability of many of its members to degrade biopolymers such as cellulose and lignin (33). Since the sediment of Lake Kasin is particularly poor in organic material, this lack of substrate might explain the low abundance of CFB representatives in this environment.
With respect to the Archaea, our clone library is dominated by the order Halobacteriales, with most representatives belonging to the Halobacteriaceae family. Eighty-six percent of the archaeal 16S rRNA gene sequences from Lake Kasin sediment clustered with this family. Halobacteriaceae showed a similar abundance in clone libraries of the Great Salt Plains in Oklahoma (100% of 166 sequences belong to the Halobacteriaceae [6]) as well as in sediments of the soda lakes of the Wadi An Natrun (91%) (47) and the Kenyan-Tanzanian Rift Valley (93%) (69). In a recent study, Youssef and coworkers describe that both the alpha and beta diversity of Halobacteriales populations of five different saline sediments were much higher than previously thought, suggesting a profound ecological role of this order in saline ecosystems (102).
In other archaeal clone libraries from saline sediments such as the Antrim Shale in Michigan (95), Lake Lonar in India (96), Lake Chaka in China (29, 30), and the Salton Sea in California (86), sequences from methanogenic orders constituted at least 10% of all archaeal 16S rRNA gene sequences. In our library, only one sequence clustered with the order Methanobacteriales and two sequences belonged to the Methanosarcinales. In the Antrim Shale 67% of all archaeal sequences belonged to methanogenic groups (95). The relatively high concentrations of SO42− and Fe(III) in the sediment of Lake Kasin seem to favor microbial sulfate and iron reduction over methanogenesis.
From the archaeal sequences retrieved from Lake Kasin sediment, 10% belonged to the order Thermoplasmatales, which has also been shown to be present in other saline sediments (15, 28–30, 86, 95). In summary, both the composition of the archaeal and bacterial 16S rRNA clone libraries from Lake Kasin sediment revealed the presence of microbial taxa that have been shown to exist also in other saline habitats. Given the fact that we did not find any sequences of known photoautotrophs together with the scarce vegetation observed at this site, we expect primary production by eukaryotic algae to be the main source of carbon input into the ecosystem.
Interestingly, none of the obtained 16S rRNA gene sequences of the Lake Kasin library were closely affiliated with any known taxa of Fe(II) oxidizers or Fe(III) reducers.
Abundance, distribution, and activity of putative Fe(III) reducers in the sediment.
Notably, 16S rRNA gene sequences from our bacterial Fe(III)-reducing enrichments did not occur in the clone library. Even though we tried to limit biases introduced by DNA extraction and PCR by combining DNA extracts from different extraction methods and pooling amplicons from several independent PCRs, we cannot completely rule out that this phenomenon is due to method-related limitation.
Unfortunately, little is known about the Fe(III)-reducing capability of species that are most closely related to the microorganisms we found in our Fe(III)-reducing enrichment cultures. Halobaculum gomorrense described by Oren et al. (59) has not been tested for its capability to reduce Fe(III), but it could not grow with nitrate as the electron acceptor or fermentatively with arginine. Dehalobacter restrictus has been described as an anaerobic bacterium that can grow by dehalogenating halogenated phthalides. However, its capability to reduce Fe(III) has not been tested either (101). Lactobacillus fabifermentans has been described as a facultative anaerobe, but its capability to reduce Fe(III) has also not been shown (11). Desulfosporosinus lacus, on the other hand, can use Fe(III) but also SO42− as the terminal electron acceptor (68). The electron acceptor preferences of Desulfosporosinus under in situ conditions are not known. As mentioned in the introduction, Fe(III) reduction is thermodynamically more favorable than sulfate reduction. Even though for exact calculations, activities would need to be considered, the relatively higher energy yield of Fe(III) reduction over sulfate reduction is not expected to change.
Overall, absolute cell numbers of the enriched putative Fe(III) reducers affiliated with Desulfosporosinus spp., Bacilli, and Halobaculum gomorrense increased in the Fe-rich sediment layer (1.5 to 2.5 cm of depth) of Lake Kasin. Also, total cell numbers of Bacteria and Archaea were highest at this particular depth. While the relative abundance of Bacilli is more or less constant throughout the sediment profile, Halobaculum gomorrense-related Archaea and Desulfosporosinus sp.-related Bacteria were more abundant in the upper sediment layers. Interestingly, the highest numbers of 16S rRNA gene copies of the later group were found between 1 and 1.5 cm just above the iron-rich layer. It could be that it is particularly the high content of bioavailable Fe(III) that supports the growth of Desulfosporosinus spp. in this sediment layer.
The cumulative rate of Fe(II) production in all three Fe(III)-reducing enrichment cultures reached 1 μmol ml−1 day−1 (Table 2). Based on this rate, it would take the three enrichment cultures about 1 month to form the 28 μmol of Fe(II), the amount of Fe(II) that we measured per g dry weight of sediment in the composite sample from the top 10 cm. Even if we assume that the in situ Fe(II) production rates are lower than the ones we calculated for our enrichment cultures, our calculations still show that the microorganisms we enriched have the capability to generate the amount of Fe(II) present in Lake Kasin. Our MPNs showed that Fe(III) reducers were up to an order of magnitude more abundant than anaerobic Fe(II) oxidizers in the sediment of Lake Kasin (Fig. 3). Considering that hardly any NO3−, but more than 1 mM acetate, was detected in the sediment leachate, we conclude that only minor amounts of the Fe(II) that was formed by the Fe(III) reducers in the sediment are readily reoxidized by anaerobic Fe(II) oxidizers in the reduced zones of the sediment. Thus, a great amount of the Fe(II) below the oxygen penetration depth in the sediments of Lake Kasin has been formed by microbial Fe(III) reduction. The fact that we did not detect any FeS mineral phases by Mössbauer spectroscopy does seem to argue for a minor contribution of abiotic Fe(III) reduction by reduced sulfur species.
In summary, we showed that (i) microbial Fe(III) reduction does occur at concentrations of up to 5 M NaCl, extending the natural habitat boundaries of this important microbial process; (ii) the microbial community composition of Lake Kasin sediment has distinct features but overall resembles the community composition of other hypersaline habitats; (iii) the Fe mineral phases found in Lake Kasin are likely the product of microbial activity; (iv) the iron reduction rates quantified for the obtained enrichment cultures explain the Fe(II) concentrations found in the Lake Kasin sediments; and (v) the presence of anaerobic microbial Fe(II) oxidizers and Fe(III) reducers suggests an active microbial Fe cycling in Lake Kasin at NaCl concentrations close to the solubility limit. Further, the occurrence of microbial Fe(III) reduction at 5 M NaCl does have important implications for ancient Earth scenarios and the search for life on other planets, such as Mars, where Fe-rich hypersaline brines have been inferred (48, 90, 91). Comprehensive studies of Fe(III)-reducing and Fe(II)-oxidizing microbial populations in various environments are a prerequisite to systematically unravel the role and intricate interplay of these microbial processes in the biogeochemical cycling of iron.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karsten Kotte for helpful advice during sampling, Sabine Studenroth and Annegret Walz for conducting IC measurements, and Ellen Struve for performing TIC and TOC measurements. We are further indebted to Heinrich Taubald for XRF analysis as well as to Karin Stoegerer for assistance with the molecular work. We thank Kurt Hanselmann for his advice on composing the salt water medium. Emily-Denise Melton and Elisabeth Swanner are acknowledged for critically reading the manuscript.
This study was funded by the DFG research unit 763 “Natural Halogenation Processes in the Environment - Atmosphere and Soil.”
Footnotes
Published ahead of print 13 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Barrero CA, García KE, Morales AL, Kodjikian S, Greneche JM. 2006. New analysis of the Mössbauer spectra of akaganeite. J. Phys. Condens. Matter 49:6827–6840 [Google Scholar]
- 2. Behrens S, et al. 2008. Monitoring abundance and expression of “Dehalococcoides” species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column. Appl. Environ. Microbiol. 74:5695–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boone DR, et al. 1995. Bacillus infernus sp. nov., an Fe(III)-reducing and Mn(IV)-reducing anaerobe from the deep terrestrial subsurface. Int. J. Syst. Bacteriol. 45:441–448 [DOI] [PubMed] [Google Scholar]
- 4. Cameron FJ, Jones MV, Edwards C. 1984. Effects of salinity on bacterial iron oxidation. Curr. Microbiol. 10:353–356 [Google Scholar]
- 5. Cao YP, Green PG, Holden PA. 2008. Microbial community composition and denitrifying enzyme activities in salt marsh sediments. Appl. Environ. Microbiol. 74:7585–7595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caton TM, Caton IR, Witte LR, Schneegurt MA. 2009. Archaeal diversity at the Great Salt Plains of Oklahoma described by cultivation and molecular analyses. Microb. Ecol. 58:519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chambaere DG, Govaert A, De Grave E, Harts G, Robbrecht G. 1979. A Mössbauer effect study of the quadrupole interaction in paramagnetic chlorine and fluorine containing β-FeOOH. J. Phys. Suppl. 3:350–352 [Google Scholar]
- 8. Coby AJ, Shelobolina ES, Zu H, Roden EE, Picardal FW. 2009. Microbially-mediated anaerobic redox cycling of iron and nitrogen in sediments. Geochim. Cosmochim. Acta 73:A232 [Google Scholar]
- 9. Cornell RM, Schwertmann U. 2003. The iron oxides. Wiley-VCH, New York, NY [Google Scholar]
- 10. DasSarma S. 2007. Extreme microbes. Am. Sci. 95:224–231 [Google Scholar]
- 11. De Bruyne K, Camu N, De Vuyst L, Vandamme P. 2009. Lactobacillus fabifermentans sp. nov. and Lactobacillus cacaonum sp. nov., isolated from Ghanaian cocoa fermentations. Int. J. Syst. Evol. Microbiol. 59:7–12 [DOI] [PubMed] [Google Scholar]
- 12. De Koff JP, Anderson MA, Amrhein C. 2008. Geochemistry of iron in the Salton Sea, California. Hydrobiologia 604:111–121 [Google Scholar]
- 13. DeLong EF. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeLong EF, et al. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503 [DOI] [PubMed] [Google Scholar]
- 15. Dong HL, et al. 2006. Microbial diversity in sediments of saline Qinghai Lake, China: linking geochemical controls to microbial ecology. Microb. Ecol. 51:65–82 [DOI] [PubMed] [Google Scholar]
- 16. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed). 2006. The prokaryotes, a handbook of the biology of bacteria, 3rd ed, vol 7 Springer, New York, NY [Google Scholar]
- 17. Emerson D, Floyd MM. 2005. Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Environ. Microbiol. 397:112–123 [DOI] [PubMed] [Google Scholar]
- 18. Feder F, Trolard F, Klingelhöfer G, Bourrié G. 2005. In situ Mössbauer spectroscopy: evidence for green rust (fougerite) in a gleysol and its mineralogical transformations with time and depth. Geochim. Cosmochim. Acta 69:4463–4483 [Google Scholar]
- 19. Genin J-M, et al. 1998. Thermodynamic equilibria in aqueous suspensions of synthetic and natural Fe(II)-Fe(III) green rusts: occurrences of the mineral in hydromorphic soils. Environ. Sci. Technol. 32:1058–1068 [Google Scholar]
- 20. German Institute for Standardization (DIN) (e. V. registered association) (editor) 1997. Legal regulation. DIN ISO 11265: composition of soil and sediments - determination of electrical conductivity. Berlin, Germany [Google Scholar]
- 21. German Institute for Standardization (DIN) (e. V. registered association) (editor) 2005. Legal regulation. DIN ISO 10390: composition of soil and sediments - determination of pH. Berlin, Germany [Google Scholar]
- 22. Reference deleted.
- 23. Haas BJ, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammer UT. 1986. Saline lake ecosystems of the world. Dr W. Junk Publishers, Dordrecht, Germany [Google Scholar]
- 25. Hollister EB, et al. 2010. Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J. 4:829–838 [DOI] [PubMed] [Google Scholar]
- 26. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 27. Ivarson KC, Ross GJ, Mills NM. 1979. The microbiological formation of basic ferric sulfates, II. Crystallization in presence of potassium, ammonium, and sodium salts. J. Soil Sci. Soc. Am. 43:908–912 [Google Scholar]
- 28. Jiang HC. 2007. Geomicrobiological studies of saline lakes on the Tibetan Plateau, NW China: linking geological and microbial processes. Ph.D. thesis Miami University, Oxford, OH [Google Scholar]
- 29. Jiang HC, et al. 2007. Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan plateau. Environ. Microbiol. 9:2603–2621 [DOI] [PubMed] [Google Scholar]
- 30. Jiang HC, et al. 2006. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl. Environ. Microbiol. 72:7430–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanso S, Greene AC, Patel BKC. 2002. Bacillus subterraneus sp. nov., an iron- and manganese-reducing bacterium from a deep subsurface Australian thermal aquifer. Int. J. Syst. Evol. Microbiol. 52:869–874 [DOI] [PubMed] [Google Scholar]
- 32. Kelly DP, Wood AP. 2000. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov. Halothiobacillus gen. nov and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50:511–516 [DOI] [PubMed] [Google Scholar]
- 33. Kirchman DL. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91–100 [DOI] [PubMed] [Google Scholar]
- 34. Klee AJ. 1993. A computer-program for the determination of Most Probable Number and its confidence-limits. J. Microbiol. Methods 18:91–98 [Google Scholar]
- 35. Koizumi Y, Kojima H, Oguri K, Kitazato H, Fukui M. 2004. Vertical and temporal shifts in microbial communities in the water column and sediment of saline meromictic Lake Kaiike (Japan), as determined by a 16S rDNA-based analysis, and related to physicochemical gradients. Environ. Microbiol. 6:622–637 [DOI] [PubMed] [Google Scholar]
- 36. Kulp TR, et al. 2006. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl. Environ. Microbiol. 72:6514–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuno A, et al. 2002. Mossbauer spectroscopic study on vertical distribution of iron species in sediments from Qinghai Lake, China. Hyperfine Interact. 141:321–326 [Google Scholar]
- 38. Lane DJ. 1991. 16S/23S rRNA sequencing. Nucleic acid techniques in bacterial systematics, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics, John Wiley and Sons, New York, NY [Google Scholar]
- 39. Larese-Casanova P, Haderlein SB, Kappler A. 2010. Biomineralization of lepidocrocite and goethite by nitrate-reducing Fe(II)-oxidizing bacteria: effect of pH, bicarbonate, phosphate, and humic acids. Geochim. Cosmochim. Acta 74:3721–3734 [Google Scholar]
- 40. Lazar CS, Parkes RJ, Cragg BA, L'Haridon S, Toffin L. 2011. Methanogenic diversity and activity in hypersaline sediments of the centre of the Napoli mud volcano, Eastern Mediterranean Sea. Environ. Microbiol. 13:2078–2091 [DOI] [PubMed] [Google Scholar]
- 41. Long DT, et al. 1992. Formation of alunite, jarosite and hydrous iron-oxides in a hypersaline system—Lake Tyrrell, Victoria, Australia. Chem. Geol. 96:183–202 [Google Scholar]
- 42. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ma Y, et al. 2004. Bacterial diversity of the Inner Mongolian Baer Soda Lake as revealed by 16S rRNA gene sequence analyses. Extremophiles 8:45–51 [DOI] [PubMed] [Google Scholar]
- 44. Madigan MT, Martinko JM. 2006. Brock Mikrobiologie, vol 11 Pearson, Munich, Germany [Google Scholar]
- 45. Massana R, Murray AE, Preston CM, DeLong EF. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McBeth JM, Little BJ, Ray RI, Farrar KM, Emerson D. 2011. Neutrophilic iron-oxidizing “Zetaproteobacteria” and mild steel corrosion in nearshore marine environments. Appl. Environ. Microbiol. 77:1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mesbah NM, Abou-El-Ela SH, Wiegel J. 2007. Novel and unexpected prokaryotic diversity in water and sediments of the alkaline, hypersaline lakes of the Wadi an Natrun, Egypt. Microb. Ecol. 54:598–617 [DOI] [PubMed] [Google Scholar]
- 48. Möhlmann D, Thomsen K. 2011. Properties of cryobrines on Mars. Icarus 212:123–130 [Google Scholar]
- 49. Murad E, Cashion J. 2004. Mössbauer spectroscopy of environmental materials and their industrial utilization. Kluwer Academic Publishers, Boston, MA [Google Scholar]
- 50. Murad E, Johnston JH. 1987. Iron oxides and oxyhydroxides, p 507–582 In Long G. (ed), Mössbauer spectroscopy applied to inorganic chemistry, vol 2 Plenum Press, New York, NY [Google Scholar]
- 51. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Muyzer G, Teske A, Wirsen CO, Jannasch HW. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel-electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165–172 [DOI] [PubMed] [Google Scholar]
- 53. Reference deleted.
- 54. Newman DK, Poulain AJ. 2009. Rhodobacter capsulatus catalyzes light-dependent Fe(II) oxidation under anaerobic conditions as a potential detoxification mechanism. Appl. Environ. Microbiol. 75:6639–6646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oren A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oren A. 2001. The bioenergetic basis for the decrease of metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466:61–72 [Google Scholar]
- 57. Oren A. 2002. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 28:56–63 [DOI] [PubMed] [Google Scholar]
- 58. Oren A. 2011. Thermodynamic limits to microbial life at high salt concentrations. Environ. Microbiol. 13:1908–1923 [DOI] [PubMed] [Google Scholar]
- 59. Oren A, Gurevich P, Gemmell RT, Teske A. 1995. Halobaculum gomorrense gen. nov., sp. nov., a novel extremely halophilic archaeon from the Dead Sea. Int. J. Syst. Bacteriol. 45:747–754 [DOI] [PubMed] [Google Scholar]
- 60. Pagaling E, et al. 2009. Microbial biogeography of six salt lakes in inner Mongolia, China, and a salt lake in Argentina. Appl. Environ. Microbiol. 75:5750–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Paul S, Bag SK, Das S, Harvill ET, Dutta C. 2008. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 9:R70.1–R70.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Peplies J, Kottmann R, Ludwig W, Glockner FO. 2008. A standard operating procedure for phylogenetic inference (SOPPI) using (rRNA) marker genes. Syst. Appl. Microbiol. 31:251–257 [DOI] [PubMed] [Google Scholar]
- 63. Pollock J, et al. 2007. Alkaline iron(III) reduction by a novel alkaliphilic, halotolerant, Bacillus sp. isolated from salt flat sediments of Soap Lake. Appl. Microbiol. Biotechnol. 77:927–934 [DOI] [PubMed] [Google Scholar]
- 64. Porsch K, Kappler A. 2011. Fe(II) oxidation by molecular O2 during HCl extraction, Environ. Chem. 8:190–197 [Google Scholar]
- 65. Poulton SW, Canfield DE. 2005. Development of a sequential extraction procedure for iron: implications for iron partitioning in continentally derived particulates. Chem. Geol. 214:209–221 [Google Scholar]
- 66. Powers W. 2004. The science of smell part I: odor perception and physiological response. Iowa State University of Science and Technology, Ames, IA [Google Scholar]
- 67. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ramamoorthy S, et al. 2006. Desulfosporosinus lacus sp. nov., a sulfate-reducing bacterium isolated from pristine freshwater lake sediments. Int. J. Syst. Evol. Microbiol. 56:2729–2736 [DOI] [PubMed] [Google Scholar]
- 69. Rees HC, Grant WD, Jones BE, Heaphy S. 2004. Diversity of Kenyan soda lake alkaliphiles assessed by molecular methods. Extremophiles 8:63–71 [DOI] [PubMed] [Google Scholar]
- 70. Refait P, Olowe RDAA, Genin J-MR. 1991. Mössbauer effect study and crystallographic structure of chlorinated green rust compounds. Hyperfine Interact. 69:839–842 [Google Scholar]
- 71. Roden EE, Zachara JM. 1996. Microbial reduction of crystalline iron(III) oxides: influence of oxide surface area and potential for cell growth. Environ. Sci. Technol. 30:1618–1628 [Google Scholar]
- 72. Rothschild LJ, Mancinelli RL. 2001. Life in extreme environments. Nature 409:1092–1101 [DOI] [PubMed] [Google Scholar]
- 73. Rusch B, Genin J-MR, Ruby C, Abdelmoula M, Bonville P. 2008. Mössbauer study of magnetism in FeII-III (oxy-)hydroxycarbonate green rusts: ferrimagnetism of FeII-III hydroxycarbonate. Hyperfine Interact. 187:1093–1098 [Google Scholar]
- 74. Schink B, Benz M. 1999. Microbial metabolism of iron species in freshwater lake sediments, p 228–234 In Schüring J, Schulz HD, Fischer WR, Böttcher J, Duijnsveld WH. (ed), Redox: fundamentals, processes and applications. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 75. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sobolev D, Roden EE. 2002. Evidence for rapid microscale bacterial redox cycling of iron in circumneutral environments. Antonie Van Leeuwenhoek 81:587–597 [DOI] [PubMed] [Google Scholar]
- 77. Sorensen KB, Canfield DE, Teske AP, Oren A. 2005. Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 71:7352–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sorokin DY, et al. 2010. Sulfidogenesis under extremely haloalkaline conditions in soda lakes of Kulunda Steppe (Altai, Russia). FEMS Microbiol. Ecol. 73:278–290 [DOI] [PubMed] [Google Scholar]
- 79. Sorokin DY, Tourova TP, Lysenko AM, Muyzer G. 2006. Diversity of culturable halophilic sulfur-oxidizing bacteria in hypersaline habitats. Microbiology 152:3013–3023 [DOI] [PubMed] [Google Scholar]
- 80. Sorokin DY, et al. 2007. Methylohalomonas lacus gen. nov., sp. nov. and Methylonatrum kenyense gen. nov., sp. nov., methylotrophic gammaproteobacteria from hypersaline lakes. Int. J. Syst. Evol. Microbiol. 57:2762–2769 [DOI] [PubMed] [Google Scholar]
- 81. Stahl DA, Amann RI. 1991. Development and application of nucleic acid probes in bacterial systematics, p 205–248 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England [Google Scholar]
- 82. Stookey LL. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779–7781 [Google Scholar]
- 83. Straub KL, Kappler A, Schink B. 2005. Enrichment and isolation of ferric-iron- and humic-acid-reducing bacteria. Methods Enzymol. 397:58–77 [DOI] [PubMed] [Google Scholar]
- 84. Straub KL, Schonhuber WA, Buchholz-Cleven BEE, Schink B. 2004. Diversity of ferrous iron-oxidizing, nitrate-reducing bacteria and their involvement in oxygen-independent iron cycling. Geomicrobiol. J. 21:371–378 [Google Scholar]
- 85. Stumm W, Morgan JJ. 1996. Aquatic chemistry, 3rd ed John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 86. Swan BK, Ehrhardt CJ, Reifel KM, Moreno LI, Valentine DL. 2010. Archaeal and bacterial communities respond differently to environmental gradients in anoxic sediments of a California hypersaline lake, the Salton Sea. Appl. Environ. Microbiol. 76:757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Switzer Blum J, Bindi AB, Buzzelli J, Stolz JF, Oremland RS. 1998. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19–30 [DOI] [PubMed] [Google Scholar]
- 88. Switzer Blum J, et al. 2009. Ecophysiology of “Halarsenatibacter silvermanii” strain SLAS-1T, gen. nov., sp. nov., a facultative chemoautotrophic arsenate respirer from salt-saturated Searles Lake, California. Appl. Environ. Microbiol. 75:1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Torrento C, Cama J, Urmeneta J, Otero N, Soler A. 2010. Denitrification of groundwater with pyrite and Thiobacillus denitrificans. Chem. Geol. 278:80–91 [Google Scholar]
- 90. Tosca NJ, et al. 2005. Geochemical modeling of evaporation processes on Mars: insight from the sedimentary record at Meridiani Planum. Earth Planet. Sci. Lett. 240:122–148 [Google Scholar]
- 91. Tosca NJ, McLennan SM, Lamb MP, Grotzinger JP. 2011. Physico-chemical properties of concentrated Martian surface waters. J. Geophys. Res. 116:E05004 doi:10.1029/2010JE003700 [Google Scholar]
- 92. Tschesch AP, Pfennig N. 1984. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137:163–167 [Google Scholar]
- 93. Ussher SJ, Achterberg EP, Worsfold PJ. 2004. Marine biogeochemistry of iron. Environ. Chem. 1:67–80 [Google Scholar]
- 94. van Doesburg W, et al. 2005. Reductive dechlorination of beta-hexachlorocyclohexane (beta-HCH) by a Dehalobacter species in coculture with a Sedimentibacter sp. FEMS Microbiol. Ecol. 54:87–95 [DOI] [PubMed] [Google Scholar]
- 95. Waldron PJ, Petsch ST, Martini AM, Nusslein K. 2007. Salinity constraints on subsurface archaeal diversity and methanogenesis in sedimentary rock rich in organic matter. Appl. Environ. Microbiol. 73:4171–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wani AA, et al. 2006. Molecular analyses of microbial diversity associated with the Lonar soda lake in India: an impact crater in a basalt area. Res. Microbiol. 157:928–937 [DOI] [PubMed] [Google Scholar]
- 97. Widdel F. 1980. Anaerober Abbau von Fettsäuren und Benzoesäure durch neu isolierte Arten. Ph.D. thesis Universität Göttingen, Göttingen, Germany [Google Scholar]
- 98. Widdel F, Pfennig N. 1981. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch. Microbiol. 129:395–400 [DOI] [PubMed] [Google Scholar]
- 99. Williams WD. 2002. Environmental threats to salt lakes and the likely status of inland saline ecosystems in 2025. Environ. Conserv. 29:154–167 [Google Scholar]
- 100. Williams WD, Sherwood JE. 1994. Definition and measurement of salinity in salt lakes. Int. J. Salt Lake Res. 3:53–63 [Google Scholar]
- 101. Yoshida N, Ye L, Baba D, Katayama A. 2009. A novel Dehalobacter species is involved in extensive 4,5,6,7-tetrachlorophthalide dechlorination. Appl. Environ. Microbiol. 75:2400–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Youssef NH, Ashlock-Savage KN, Elshahed MS. 2012. Phylogenetic diversities and community structure of members of the extremely halophilic Archaea (order Halobacteriales) in multiple saline sediment habitats. Appl. Environ. Microbiol. 78:1332–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zavarzina DG, et al. 2006. Geoalkalibacter ferrihydriticus gen. nov. sp. the first alkaliphilic representative of the family Geobacteraceae, isolated from a soda lake. Microbiology 75:673–682 [PubMed] [Google Scholar]
- 104. Zhou JZ, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.