Abstract
A 16S rRNA gene approach, including 454 pyrosequencing and quantitative PCR (qPCR), was used to describe the bacterial community in Rhipicephalus turanicus and to evaluate the dynamics of key bacterial tenants of adult ticks during the active questing season. The bacterial community structure of Rh. turanicus was characterized by high dominance of Coxiella and Rickettsia and extremely low taxonomic diversity. Parallel diagnostic PCR further revealed a novel Coxiella species which was present and numerically dominant in all individual ticks tested (n = 187). Coxiella sp. densities were significantly higher in female versus male ticks and were overall stable throughout the questing season. In addition, we revealed the presence of the novel Coxiella sp. in Rh. sanguineus adult ticks, eggs, and hatched larvae, indicating its vertical transmission. The presence of both spotted fever group Rickettsia spp. (SFGR) and non-SFGR was verified in the various individual ticks. The prevalence and density of Rickettsia spp. were very low compared to those of Coxiella sp. Furthermore, Rickettsia sp. densities were similar in males and females and significantly declined toward the end of the questing season. No correlation was found between Coxiella sp. and Rickettsia sp. densities. These results suggest different control mechanisms in the tick over its different bacterial populations and point to an obligatory and facultative association between the two tick species and Coxiella sp. and Rickettsia spp., respectively.
INTRODUCTION
Many pestiferous arthropods carry obligatory and facultative bacterial symbionts, which fundamentally shape their biology and may determine their efficiency as disease vectors and their performance as pests (2, 35). Ticks are considered among the most important vectors of pathogens of medical and veterinary importance (10).
Incidences of arthropod-borne epidemics, such as the Crimean-Congo hemorrhagic fever transmitted by ticks (48), are expected to increase due to global climate changes and other drivers associated with modern life (reviewed in references 9, 21, and 28). Indeed, the prevalence of tick-borne pathogens such as Borrelia burgdorferi and Anaplasma phagocytophilum has been correlated with seasonal climatic changes (14). Therefore, it is expected that other microbial tenants within tick hosts are affected in the same manner.
In studies based on low-throughput techniques such as clone libraries and denaturing gradient gel electrophoresis (DGGE), the bacterial communities of several ixodid tick species, among them Amblyomma americanum, Ixodes scapularis, and Ixodes ricinus, were found to be dominated by a single bacterial genus along with other less dominant but diverse bacterial species, such as Arsenophonus sp., Serratia grimesii, and Klebsiella oxytoca. While each individual tick usually showed relatively few bacteria, a higher diversity of bacterial taxa was found at the tick population level (4, 8, 23, 31, 39, 44). More recent studies based on high-throughput techniques have revealed higher bacterial diversity of individual ticks. However, to the best of our knowledge, a combined quantitative and qualitative assessment of tick bacterial communities has not been performed to date. Such a dual analysis might be important for the evaluation of dynamic interactions within bacterial populations (27, 50). Determining seasonal effects on an entire bacterial community within a tick vector might result in better resolution of host-bacterium interactions and of bacterium-bacterium interactions within the host. This approach could be used to develop future strategies for the prevention and control of tick-borne diseases.
In Israel, the brown dog tick, Rhipicephalus sanguineus (Acari: Ixodidae), is most common and is often found with a second sympatric species, Rhipicephalus turanicus (16, 30). While Rh. sanguineus is the known vector of Rickettsia conorii (9, 22), several other bacterial pathogens, such as Coxiella burnetii, Ehrlichia canis, and Anaplasma platys, have also been found in these two tick species, making them potential vectors (10, 20). In addition, Rh. sanguineus also harbors a Coxiella-like endosymbiont (34).
In this study, we adopted a 16S rRNA gene-based approach to describe the composition and dynamics of the bacterial communities in Rh. turanicus and Rh. sanguineus. PCR-DGGE and 454 pyrosequencing were used to describe the tick bacterial community, and quantitative PCR (qPCR) assays were used to follow the seasonal changes in bacterial populations.
MATERIALS AND METHODS
Tick collection and rearing.
Questing ticks were collected by dragging a 1-m2 white cotton cloth over the vegetation in two consecutive years. In 2009, ticks were collected in March and April from the outskirts of Kibbutz Hulda, Israel (31° 49′ 56.28″ N, 34° 53′ 0.24″ E) and in May from the sand dunes in Caesarea (32° 30′ 0″ N, 34° 54′ 0″ E). In 2010, ticks were collected weekly between March and July from the outskirts of Kibbutz Hulda. Collected ticks were immediately preserved in 100% ethanol and kept at −20°C until further use.
In addition, several fully engorged female ticks were collected in 2010 from dogs located in a dog kennel (Rehovot, Israel [31° 53′ 38.03″ N, 34° 48′ 59.04″ E]). They were kept in a moistened plastic tube at 25°C until eggs were laid. A batch from each tick clutch was then surface sterilized in soapy water and 70% ethanol and preserved in 100% ethanol for DNA extraction. Most of the eggs were incubated until the larvae hatched. The newly hatched larvae were preserved in 100% ethanol for DNA extraction as described further on.
Tick species classification.
Ticks were first classified as Rh. sanguineus or Rh. turanicus according to their morphological characteristics based on the work of Walker et al. (45).
To ascertain the identity of the collected ticks used in this study, a specific gene-based assay was developed. Partial fragments of the 12S mitochondrial rRNA gene were sequenced from 10 randomly selected ticks as previously described (3). Phylogenetic analysis (detailed below) showed that some of the ticks were closely related to Rh. turanicus and others to Rh. sanguineus (see Fig. S1 in the supplemental material). Based on the aligned sequences, a new primer specific to Rh. sanguineus ticks (SNR) (see Table S1) was designed and used together with the T1B forward primer to screen all collected ticks.
Tick surface sterilization and DNA extraction.
For surface sterilization, alcohol-preserved ticks were vortexed for 1 min in 3% sodium hypochlorite followed by 70% ethanol and three washes in sterile 1× Dulbecco's phosphate buffer saline (Lonza Group, Basel, Switzerland). The ticks were then air dried for 10 min before DNA extraction or stored in absolute ethanol at −20°C until further use. Extraction of total DNA from each individual tick was performed with the Qiagen blood and tissue extraction kit (Hilden, Germany) according to the manufacturer's instructions with modifications: ticks were placed in a sterile petri dish containing 20 μl of the supplied ATL buffer (Qiagen, Hilden, Germany) and were cut into four pieces using a sterile scalpel blade. The tick pieces and the fluids were transferred to a sterile 1.5-ml tube containing 160 μl ATL buffer. New sterile petri dishes and scalpel blades were used for each tick. DNA concentration and purity were determined in a NanoDrop ND1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). DNA samples were stored at −20°C until further analysis.
PCR-DGGE of 16S rRNA.
DGGE was performed to compare bacterial community composition among individual ticks. DNA served as the template for PCR amplification with primer pairs 341f (with a GC clamp) and 907r targeting Bacteria, as previously described (32). PCR products were separated on a 6% polyacrylamide gel (1 mm thick) with a top-to-bottom gradient of urea and formamide (20% to 70%). Gels were run for 17 h at 100 V with heating (60°C) in a D-Code system (Bio-Rad, Hemel Hempstead, United Kingdom). Gels were then stained with GelStar nucleic acid gel stain (Biowhittaker Molecular Applications, Rockland, ME) and photographed on a UV transillumination table (302 nm) with a Kodak digital camera. A migration marker was added to each run to enable comparison of bacterial community patterns obtained from different gels. PCR-DGGE community patterns were aligned and analyzed using Fingerprinting II software (Bio-Rad, Hercules, CA).
Cloning and sequencing of excised DGGE bands.
Specific selected bands were excised from the gels with a scalpel blade, placed in a 2-ml sterile tube containing 100 μl Tris-EDTA buffer (pH 8.0), and incubated at 37°C for 2 h to facilitate diffusion of the DNA from the band into the solution; 1 μl of the band extract served as the template for PCR with the 341f–907r primer pair without GC-clamp tail. Positive products were cloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA) and transformed into chemically competent Escherichia coli DH5-α cells. Clones were examined by PCR and agarose gel electrophoresis (1.5%) and by DGGE analysis. Plasmids containing the appropriate inserts (in comparison to the original sample and the excised band) were recovered using the Wizard Plus SV Minipreps DNA purification system (Promega, Madison, WI) and sequenced in an ABI PRISM 3730xl DNA analyzer (Applied Biosystems, Carlsbad, CA) at the Alexander Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Israel.
Mass sequencing of 16S rRNA gene fragments.
To characterize the Rh. turanicus bacterial community, 36 representative samples, 17 male and 19 female ticks collected from Hulda in 2010, were sent to the Research and Testing Laboratory (Lubbock, TX) for mass-sequencing analysis. 16S rRNA bacterial tag-encoded FLX amplicon pyrosequencing was performed using the Roche 454 sequencing system as previously described (12). Amplicons originating from the 530F–1100R (V4–V6) region, numbered in relation to the E. coli 16S rRNA gene, were sequenced. Retrieved sequences were analyzed using MOTHUR (40). Sequences shorter than 200 bp, as well as those of low quality, were omitted. Sequences were then aligned using the Silva-compatible alignment database, and a distance matrix was calculated (36). Sequences were grouped into operational taxonomic units (OTUs) at a 97% sequence similarity threshold (i.e., sequences that differ by ≤3% are clustered in the same OTU). Representatives of each OTU were classified with the MOTHUR classify.seqs module, and affiliation, down to the genus level, was verified by the ARB program package (25), the Ribosomal Database Project (http://rdp.cme.msu.edu/) (46), and NCBI GenBank databases.
Based on the mass-sequencing data classification (genus-based OTU-level similarity), different diversity parameters were calculated. For each sample, the adequacy of the sampling effort was calculated using Good's coverage estimate equation: C = 1 − (n1/N), where n1 is the number of taxa represented by only a single clone sequence, and N is the total number of clone sequences in the specific library examined. The maximum value of C is 1 (18). The Shannon-Weaver index of diversity (H′), which combines species richness and relative abundance of bacterial species within a community, was calculated using the following equation: H′ = −Σpi · ln(pi), where pi is the proportion of the ith clade in the clone library. The more diverse the sample, the higher the H′ value (42). Dominance (D), the estimation of the species relative abundance distribution in a sample, was calculated using the following equation: D = Σ(pi)2 (43). A dominance of 1 represents a community composed with a single population. The Chao1 richness estimate, the number of expected different species in a community, was calculated according to Chao (7).
PCR, primer design, and validation.
PCR was conducted in a T-gradient basic thermocycler (Biometra, Goettingen, Germany) in 0.2-ml polypropylene tubes. Each 50-μl reaction mixture contained 1.5 units of DreamTaq DNA polymerase (Fermentas, St. Leon-Rot, Germany), 1× DreamTaq buffer, 0.2 mM deoxynucleoside triphosphate (dNTP) mix (Larova, Teltow, Germany), 6.25 μg/ml bovine serum albumin (Roche Diagnostics, Mannheim, Germany), and 0.5 μM each primer (see Table S1 in the supplemental material).
New primers used in this study were designed using the ARB probe-design module (25). Specificity of the primers was verified in silico using ARB, RDP, and NCBI databases and checked against fully cloned nontarget 16S rRNA genes, including from Rickettsia and Coxiella, by PCR. PCR products of each newly designed primer pair were cloned, of which 10 randomly selected clones were sequenced as described above. Plasmid sequences were matched against the NCBI database. In all cases, the 10 clones gave the expected target organism result. The different primers used in this study and their respective reaction conditions are listed respectively in Tables S1 and S2 in the supplemental material.
Quantitative PCR.
SYBR green-based qPCR was used for the detection and quantification of Rickettsia spp. and Coxiella sp. in 128 Rh. turanicus DNA samples. In addition, the tick 18S rRNA gene was quantified and used as a reference gene for normalization of the data.
qPCR assays were conducted in polypropylene 96-well plates in a 7300 qPCR system (ABI, Applied Biosystems). Each 25-μl reaction mixture contained 1× Absolute Blue SYBR green ROX mix (Thermo Fisher Scientific, Surrey, United Kingdom), 1 μl of each 10 μM primer (see Table S1 in the supplemental material), 9.5 μl of H2O, and 1 μl of template DNA. The PCR conditions for each assay are listed in Table S2.
Each plate contained triplicate reaction mixtures for each DNA sample (representing an individual tick), the appropriate set of standards, and no-template controls. Melting curves were traced after each assay to confirm that the fluorescence signal had been retrieved from specific PCR products. PCR products were also examined using agarose gel electrophoresis to further confirm the specificity of the amplification. For all target genes, six 10-fold dilutions of the calibration standards were measurable down to 100 copies of the DNA target. The standard curve slopes, correlation coefficients, and amplification efficiencies were calculated using the 7300 system SDS software version 1.4 (ABI) and are listed in Table S3 in the supplemental material.
Phylogenetic analyses.
Coxiella classification was based on the 16S rRNA gene sequence. Nine nearly full-length sequences of the 16S rRNA gene of Coxiella were obtained from five individual ticks. Sequences were aligned with known Coxiella symbionts as well as with Coxiella burnetii. The phylogenetic relationships between the different genotypes of Rickettsia identified in this study were determined based on the sequences of two known genes commonly used for Rickettsia classification: “gene D,” encoding a 17-kDa cell surface antigen, and the ompA gene, encoding outer membrane protein A (15, 41). Molecular classification of ticks was based on mitochondrial 12S rRNA gene sequences (3). Several clones from each gene of interest were sequenced at least twice. Maximum likelihood trees based on Kimura's 2-parameter model were constructed using molecular evolutionary genetics analysis (MEGA) 5.10 software (http://www.megasoftware.net). Sequences were aligned using Muscle algorithm, and bootstrap analyses with 1,000 resamplings were performed to test the robustness of the branching. Similar phylogenic trees were obtained with neighbor-joining trees based on the ClustalW alignment algorithm.
Statistical analyses.
Differences in diversity indices of the communities composition, based on pyrosequencing data, were determined by Mann-Whitney U test. The effects of season and tick sex on bacterial abundance were tested by Kruskal-Wallis test, and the critical range (P < 0.05) was determined. All statistical calculations were performed with STATISTICA (version 7.1) software (StatSoft Inc., Tulsa, OK).
Nucleotide sequence accession numbers.
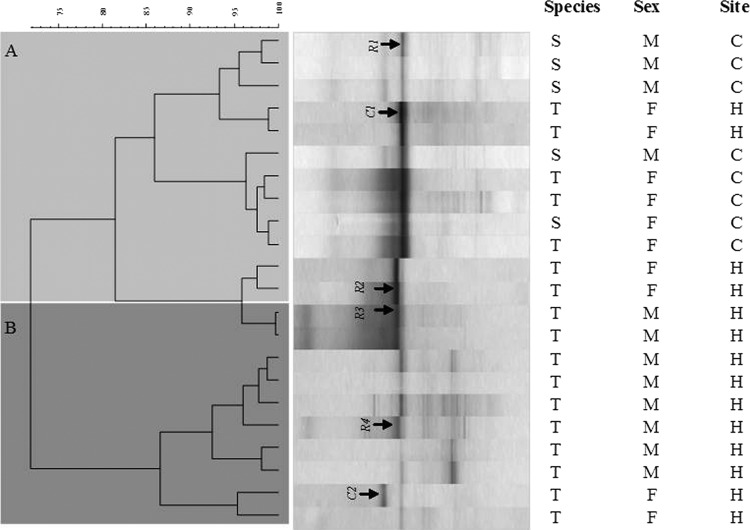
Sequences have been deposited in the NCBI GenBank SRA database under accession no. SRA049091. Accession numbers for bands shown in Fig. 1 are as follows (Coxiella [C] or Rickettsia [R]): R1, JQ480832; C1, JQ480828; R2, JQ480831; R3, JQ480829; R4, JQ480830; and C2, JQ480827.
Fig 1.
Bacterial community composition of 22 ticks based on PCR-DGGE of 16S rRNA gene fragments. Resulting DGGE patterns (each lane represents an individual tick) were aligned based on bands migration distances using Fingerprint II software, and a neighbor-joining tree based on Jaccard distance matrix was constructed. T, Rh. turanicus; S, Rh. sanguineus; F, female; M, male; H, Hulda; C, Caesarea. Bands corresponding to Coxiella (C) or Rickettsia (R) are indicated; see Materials and Methods for their accession numbers. A and B, clusters composed of mostly females and males, respectively.
RESULTS
Tick classification.
DNA extracted from ticks collected in 2009 was used for the PCR-DGGE analysis (Fig. 1). The DNA extracted from ticks collected in 2010 was used for qPCR and for 454-pyrosequencing analyses. A total of 187 ticks were collected and classified based on analysis of the 12S ribosomal rRNA gene (see Fig. S1 in the supplemental material). All ticks collected from Hulda were classified as Rh. turanicus. Ticks collected from Caesarea were classified as Rh. sanguineus or Rh. turanicus. In addition, the five engorged females collected from dogs were classified as Rh. sanguineus.
Bacterial community composition in Rh. turanicus and Rh. sanguineus.
Composition of bacterial communities was determined for 22 individual ticks collected in 2009 using PCR-DGGE of 16S rRNA gene fragments. Cluster analysis comparison of band migration patterning revealed that the bacterial community composition varied among individuals and was not related to site of collection or tick species (Fig. 1). Although sample size is quite small, cluster A was mostly composed of female ticks, and cluster B was mostly composed of males. The main clusters of PCR-DGGE patterns were determined according to the presence or absence of one of three dominant bands. These bands were identified as either Rickettsia spp. or Coxiella sp. Based on their migration distances, at least two different genotypes of Rickettsia and of Coxiella were detected in the different tick individuals. Furthermore, markedly less dominant but diverse populations were also found, showing variations in richness and composition among individual ticks (Fig. 1).
Bacterial diversity in field-collected Rh. turanicus.
The above results suggested that the bacterial community of Rh. turanicus and Rh. sanguineus is dominated by Coxiella sp. and Rickettsia spp., with other rare, albeit diverse bacterial taxa. To further describe the bacterial community of Rh. turanicus, 454 pyrosequencing was performed for ticks collected in 2010. A total of 155,365 high-quality bacterial 16S rRNA gene fragment sequences from 36 individual ticks were obtained and analyzed. These sequences were assigned into 1,466 OTUs at a 97% sequence similarity threshold. According to ARB-SILVA database analysis, most OTUs were classified into two major genera: Coxiella (Gammaproteobacteria) and Rickettsia (Alphaproteobacteria). Further classification based on genus affiliation of the sequences yielded 74 genus-level OTUs, with coverage values greater than 99.9% per individual tick.
Individual ticks were characterized by extremely low bacterial taxonomic richness during the tick collection intervals (Table 1). The Chao1 richness estimates (genus-level OTUs) were similar to the observed values, further supporting low diversity. However, a significantly high taxonomic richness was found for male ticks compared to females (Mann-Whitney U test, P < 0.01). Accordingly, significantly high Shannon H′ and lower dominance (D) were found in males compared to females (Mann-Whitney U test, P < 0.01). Although some variation in diversity parameters was observed among the collection dates, these were not significant between sexes.
Table 1.
Richness and diversity estimates of individual tick bacterial communities based on mass sequencing of 16S rRNA gene fragments
| Sex | Month | Individuals | Reads | Diversitya |
|||
|---|---|---|---|---|---|---|---|
| Genera | Chao1b | Shannon H′c | Dominanced | ||||
| Female | March | 6 | 24,531 | 3.8 | 5.2 | 0.07 | 0.97 |
| April | 4 | 14,084 | 2.8 | 3.1 | 0.15 | 0.92 | |
| May | 4 | 14,498 | 1.7 | 1.7 | 0.06 | 0.97 | |
| June | 5 | 24,362 | 5.0 | 8.0 | 0.13 | 0.93 | |
| Male | March | 6 | 13,717 | 9.0 | 13.0 | 0.43 | 0.79 |
| April | 7 | 35,437 | 6.4 | 8.6 | 0.38 | 0.75 | |
| May | 2 | 12,943 | 4.7 | 4.8 | 0.23 | 0.86 | |
| June | 2 | 15,786 | 9.5 | 10.5 | 0.04 | 0.99 | |
Average values calculated for individual ticks.
Chao1 taxon richness estimate (7), calculated based on genus classification.
Shannon index of diversity H′ = −Σpi · ln(pi); pi is the relative abundance of the ith genus.
Dominance D = Σ(pi)2.
Coxiella comprised almost 90% of the total sequences obtained and its relative abundance showed little seasonal variation in either sex (Table 2). Rickettsia comprised almost 10% of the total sequences obtained. However, in Rickettsia-positive samples, the relative abundance was higher in males than in females. Moreover, seasonal variation in Rickettsia-positive ticks was also more pronounced in males, amounting to almost 50% in April and declining to almost 25% in May.
Table 2.
Relative abundance of Rickettsia and Coxiella based on mass sequencing of 16S rRNA gene fragmenta
| Sample type | Genus | % relative abundance |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Females |
Males |
||||||||
| March | April | May | June | March | April | May | June | ||
| Rickettsia+ | Coxiella | 96.74 | 91.22 | 95.55 | 93.55 | 73.16 | 50.34 | 72.61 | NA |
| Rickettsia | 3.21 | 8.75 | 4.45 | 6.32 | 23.72 | 49.49 | 26.97 | NA | |
| Other | 0.05 | 0.03 | 0 | 0.13 | 3.12 | 0.17 | 0.42 | NA | |
| Rickettsia− | Coxiella | 99.92 | 99.93 | 99.99 | 99.93 | 94.85 | 99.60 | 99.48 | 99.51 |
| Other | 0.08 | 0.07 | 0.01 | 0.07 | 5.15 | 0.40 | 0.52 | 0.49 | |
The average number of sequences obtained divided by the total sequences of the appropriate group are presented for Rickettsia-positive and Rickettsia-negative samples. NA, no ticks collected for that group.
In addition to Coxiella and Rickettsia, diverse, albeit rare (less than 0.01% of all sequences) bacterial taxa were found (Table 3). These bacteria belonged to several phyla, including Actinobacteria, Bacteroidetes, and Firmicutes. Among the rare taxa, the most common were Propionibacter, Serratia, Pseudomonas, and Ralstonia, which appeared in 12, 7, 6, and 6 individual ticks, respectively. Altogether, these rare taxa were represented by 72 genus-level OTUs, explaining 0.45% of the total sequences obtained (700 sequences). These rare taxa appeared mainly in males rather than females (Table 3).
Table 3.
Prevalence of rare bacterial taxa (phyla and next dominant taxa) in female and male Rh. turanicus ticks and the total number of sequences obtained using 454 pyrosequencinga
| Phylum | Taxon | Prevalence (%) |
No. of sequences | |
|---|---|---|---|---|
| Females | Males | |||
| Actinobacteria | 33 | 72 | 95 | |
| Propionibacter | 11 | 56 | 66 | |
| Bacteroidetes | 11 | 44 | 171 | |
| Bacteroides | 0 | 33 | 27 | |
| Parabacteroides | 0 | 6 | 66 | |
| Firmicutes | 17 | 50 | 194 | |
| Uncultured Lachnospiraceae | 0 | 33 | 106 | |
| Alphaproteobacteria | 0 | 22 | 13 | |
| Betaproteobacteria | 28 | 67 | 72 | |
| Ralstonia | 0 | 33 | 21 | |
| Burkholderia | 6 | 11 | 9 | |
| Sutterella | 0 | 6 | 28 | |
| Uncultured Comamonadaceae | 17 | 17 | 9 | |
| Gammaproteobacteria | 33 | 72 | 130 | |
| Serratia | 6 | 33 | 17 | |
| Pseudomonas | 11 | 28 | 34 | |
| Deltaproteobacteria | 6 | 17 | 6 | |
| Other | 16 | 44 | 19 | |
Female Rh. turanicus ticks, n = 19; male Rh. turanicus ticks, n = 17.
A novel Coxiella sp. is found in Rh. turanicus and Rh. sanguineus.
The 16S rRNA gene sequences of the Coxiella sp. found in the selected ticks differed from other published sequences, suggesting the presence of another Coxiella genotype in the tested ticks (Fig. 2).
Fig 2.
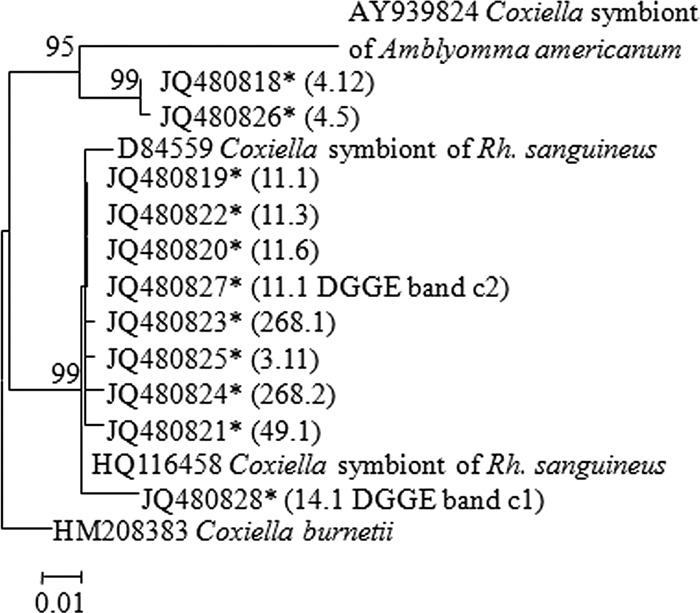
Phylogenetic tree of Coxiella symbionts based on partial 16S rRNA gene sequences. Maximum likelihood trees based on Kimura 2-parameter model were constructed using MEGA software (version 5.10). Bootstrap analyses with 1,000 resamplings were performed to test the robustness of the branching. Bootstrap values higher than 75% are indicated. Sequences obtained in the present work are designated by an asterisk and by tick and clone number in parentheses.
Based on the obtained sequences, species-specific primers were designed to target the specific Coxiella sp. from field-collected ticks. All 187 individual Rh. turanicus and 15 Rh. sanguineus ticks were positive for the Coxiella sp. Eggs and the corresponding larvae of engorged Rh. sanguineus were also positive for the Coxiella sp.
Rickettsia spp. in Rh. turanicus.
Using group-specific primers targeting most known rickettsiae, 26% of the ticks tested (49/187) were found positive. Similar percentages of rickettsial infection were found for both male and female ticks. In addition, sequences obtained from the DGGE analysis suggested that more than one rickettsial genotype is harbored by Rh. turanicus ticks: 36 of the 49 ticks were positive for both “gene D” and ompA, suggesting a spotted fever group (SFG) Rickettsia, and the other 13 were negative for both, suggesting bellii-like Rickettsia. Phylogenetic classification based on fragments of “gene D” (Fig. 3A) showed that the obtained sequences were most closely related to R. massiliae and R. barbariae of the SFG Rickettsia. Phylogenetic classification using ompA gave similar results (Fig. 3B).
Fig 3.
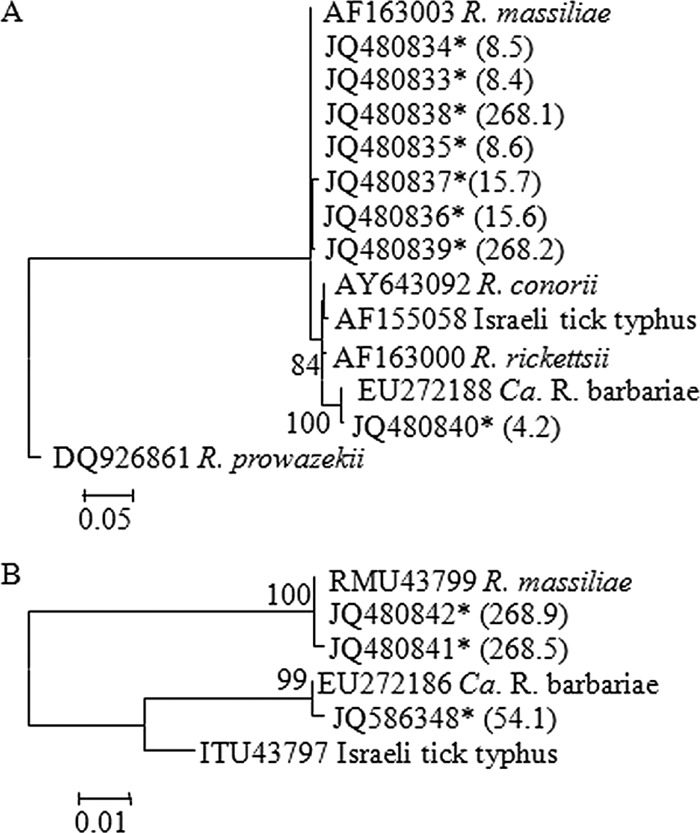
Phylogenetic tree of Rickettsia symbionts based on sequences of “gene D” (A) and ompA (B). Maximum likelihood trees based on Kimura 2-parameter model were constructed using MEGA software (version 5.10). Bootstrap analyses with 1,000 resamplings were performed to test the robustness of the branching. Bootstrap values higher than 75% are indicated. Sequences obtained in the present work are designated by an asterisk and by tick and clone number in parentheses.
Coxiella and Rickettsia dynamics in Rh. turanicus.
The densities of the Coxiella sp. and Rickettsia spp. were determined by qPCR in 60 Rh. turanicus males and 68 females. The density of Coxiella sp. was significantly higher in females than in males in each of the months tested, by an average of 1 order of magnitude (Kruskal-Wallis, P < 0.05) (Fig. 4A). During the seasons, Coxiella sp. densities in females increased slightly from March to April and declined 2-fold from April to June. These fluctuations, however, were not significant. Males showed markedly less seasonal variation in Coxiella sp. densities. In addition, Coxiella densities were similar in Rickettsia-positive and Rickettsia-negative ticks in both sexes.
Fig 4.
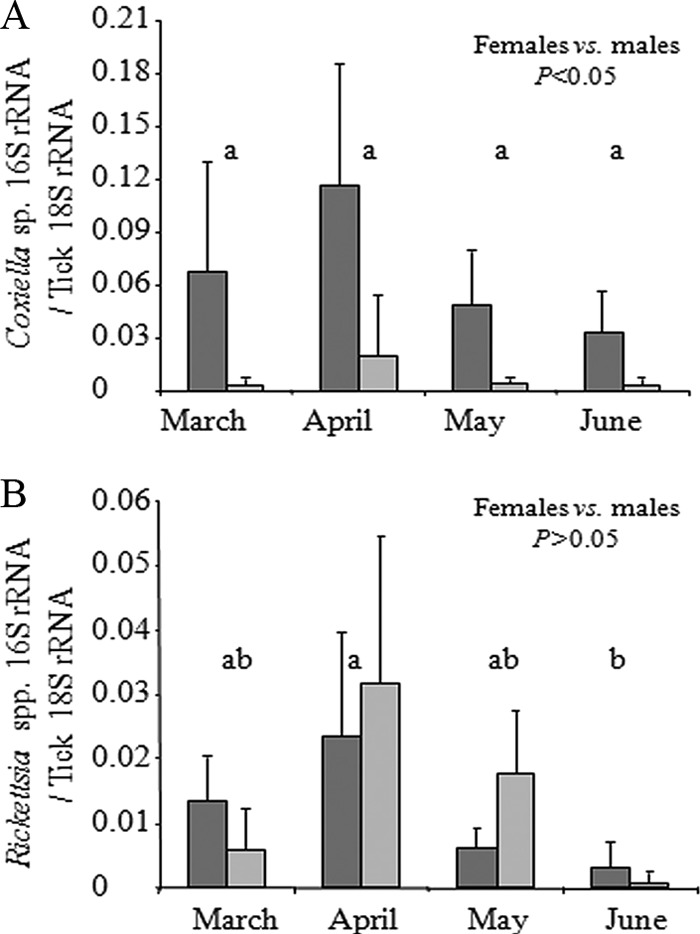
Seasonal change (mean + standard deviation [SD]) of Coxiella sp. densities (A) and Rickettsia sp. densities (B) in individual field-collected ticks as determined by qPCR. Densities were determined as targets of 16S rRNA gene per targets of tick 18S rRNA gene. Dark gray bars, females; light gray bars, males. Letters above columns denote between-month statistical significance at P < 0.05.
The densities of the Rickettsia population were not affected by tick sex (Kruskal-Wallis, P > 0.05) but were affected by the month of collection (Kruskal-Wallis, P < 0.001) (Fig. 4B). Rickettsia densities increased slightly between March and April but were significantly reduced between April and June, by over 7-fold (Kruskal-Wallis followed by multiple comparison, P < 0.001). In addition, during a season, Coxiella sp. densities were 5-fold higher than Rickettsia densities in females but 2-fold lower in males.
DISCUSSION
As obligatory blood feeders, ticks are expected to present a relatively simple and restricted system for studying microbial diversity and interactions. Questing adult ticks may fast for long periods and can thus be considered a closed system in which bacterial interactions can occur between autochthonous populations. The bacterial community of Rh. turanicus, showing average relative abundance values of 89.5% for a single genus (Coxiella) and 9.6% for another single genus (Rickettsia), represents one of the simplest fully described systems involving associations between arthropods and bacteria. This simple bacterial system is expected in specialist arthropods as opposed to generalists (11), although a more diverse bacterial community in Rh. turanicus might be expected due to its interactions with animal hosts while feeding. Our findings in Rh. turanicus are in agreement with previous findings for other tick species. For example, in A. americanum, Coxiella appeared in 89% of the sequences obtained based on clone libraries (8), and in the cattle tick Rhipicephalus (Boophilus) microplus, Coxiella appeared in 98.2% of the sequences obtained from the female ovaries. In the female unfed cattle ticks, however, Coxiella was found in only 2.8% of the sequences (1). Experimental setup, choice of primers, use of different sequence alignment algorithms, and OTU classification methods may result in significant variation in the resulting estimates of diversity parameters (13, 49). Such variations in data acquisition and analysis should be considered when comparing different studies.
The high frequency of the Coxiella sp. in all ticks tested in the current study together with its high dominance in each individual tick may suggest that this Coxiella sp. sustains an obligatory association with its tick hosts. In support of this assumption are the finding of Coxiella sp. in adult laboratory-reared Rh. sanguineus ticks (34) and our identification of Coxiella sp. in eggs and larvae of Rh. sanguineus. The latter suggests vertical transmission of Coxiella in Rh. sanguineus and may explain the high frequency of Coxiella in the tick populations. Although not tested in this study, we hypothesize that the same mechanism holds true for Rh. turanicus. The identification of other Coxiella-like symbionts in Amblyomma cajennense and A. americanum (8, 24, 27) further supports the hypothesis that these bacteria are ubiquitous in various tick species, although they were not found in I. ricinus (6).
The major secondary symbionts identified were of the genus Rickettsia, which are known as secondary symbionts from several systems where they affect their host in various ways (reviewed in reference 19). Here we found Rickettsia in 26% of the ticks tested. Rickettsia comprised less than 10% of the bacterial community in females and up to 50% in males, and its density was significantly lower than that of Coxiella sp. These findings support a facultative association between Rickettsia and the tick. Unlike our findings for Coxiella, the average densities of Rickettsia were similar in males and females and decreased significantly toward the end of the season. Although an opposite trend was found in the pea aphid Acyrthosiphon pisum, where the primary symbiont Buchnera aphidicola is suppressed in the presence of Rickettsia (38), starving ticks may benefit from reduction of facultative tenants to sustain the obligatory ones.
Several factors have been proposed to explain the low frequency or abundance of either primary or secondary symbionts. Among them are competition among symbionts, increased virulence, and bottlenecks experienced by symbionts during vertical transmission (29). Based on the relative abundance of Coxiella sp. and Rickettsia spp., a competitive interaction between the two populations could be suggested in males. However, the quantitative measurements (absolute abundances) suggest that there is no relationship between these populations: Coxiella sp. densities varied between males and females independent of the presence or abundance of Rickettsia spp. and were significantly lower in males than in females. Tick regulation of higher Coxiella densities might explain the reduced densities of Rickettsia. Alternatively, competition among the Rickettsia populations found here (two SFG and one non-SFG Rickettsia populations) might govern their low densities and prevalence. In Dermacentor andersoni, massive occurrence of R. peacockii in the tick ovaries was suggested to prevent transovarial transmission of R. rickettsii (5, 33). This phenomenon was also found in D. variabilis (26). In addition, different bacterial physiological characteristics may govern dynamics and interaction patterns. SFG Rickettsia for example, has the capacity for actin-based motility (17), whereas this ability is not yet known for Coxiella-like symbionts. The lowest values of Rickettsia densities were recorded in ticks that harbored the non-SFG Rickettsia: this might be indirect evidence of competition within the Rickettsia populations. In addition to intrinsic factors, extrinsic factors that relate to climate variation may also affect the composition of arthropod bacterial communities, as shown in the present work.
Along with the dominant Coxiella and Rickettsia, highly diverse but rare bacteria, represented by a few sequences, were also identified. These bacteria included Propionibacter, Serratia, Pseudomonas, and Ralstonia species. Such bacteria are best known as gut inhabitants in other arthropods (11, 47) and have also been detected in other tick species (1, 6). Although numerically rare, the prevalence among individuals was relatively high; for example, 33% in the case of Propionibacter. Thus, the assemblage of rare bacteria might have been underestimated due to PCR bias, especially in a system typified by a high-dominance structure. The contribution of these bacteria to the tick has yet to be elucidated in consideration with the hologenome theory of evolution (37), suggesting that these rare bacteria may be an important source for new genes and may thrive under the right conditions.
In conclusion, the bacterial community of questing Rh. turanicus ticks described in this work was characterized by extremely low bacterial diversity dominated by a novel Coxiella sp. and showed seasonal and sexual variation. Our results support an obligatory relationship between Coxiella and its tick host which is influenced by seasonal changes.
Supplementary Material
ACKNOWLEDGMENTS
We greatly thank Dmitry Apanaskevich for help with the morphological identification of ticks and the Israel Taxonomy Initiative (ITI) for supporting his visit to Israel.
This research was supported by Israel Science Foundation grant 456/10 to Y.G.
Footnotes
Published ahead of print 30 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Andreotti R, et al. 2011. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 11:6 doi:10.1186/1471-2180-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189 [DOI] [PubMed] [Google Scholar]
- 3. Beati L, Keirans JE. 2001. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari : Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J. Parasitol. 87:32–48 [DOI] [PubMed] [Google Scholar]
- 4. Benson MJ, Gawronski JD, Eveleigh DE, Benson DR. 2004. Intracellular symbionts and other bacteria associated with deer ticks (Ixodes scapularis) from Nantucket and Wellfleet, Cape Cod, Massachusetts. Appl. Environ. Microbiol. 70:616–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burgdorfer W, Hayes SF, Mavros AJ. 1981. Nonpathogenic Rickettsiae in Dermacentor andersoni: a limiting factor for the distribution of Rickettsia rickettsii, p 585–594 In Burgdorfer W, Anacker RL. (ed), Rickettsiae and rickettsial diseases. Academic Press, New York, NY [Google Scholar]
- 6. Carpi G, et al. 2011. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6:e25604 doi:10.1371/journal.pone.0025604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265–270 [Google Scholar]
- 8. Clay K, et al. 2008. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol. Ecol. 17:4371–4381 [DOI] [PubMed] [Google Scholar]
- 9. Dantas-Torres F. 2010. Biology and ecology of the brown dog tick, Rhipicephalus sanguineus. Parasite Vector 3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. 2008. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci. 13:6938–6946 [DOI] [PubMed] [Google Scholar]
- 11. Dillon RJ, Dillon VM. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49:71–92 [DOI] [PubMed] [Google Scholar]
- 12. Dowd SE, et al. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125 doi:10.1186/1471-2180-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engelbrektson A, et al. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4:642–647 [DOI] [PubMed] [Google Scholar]
- 14. Estrada-Peña A, et al. 2011. Correlation of Borrelia burgdorferi sensu lato prevalence in questing Ixodes ricinus ticks with specific abiotic traits in the Western Palearctic. Appl. Environ. Microbiol. 77:3838–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fournier PE, Roux V, Raoult D. 1998. Phylogenetic analysis of spotted fever group Rickettsiae by study of the outer surface protein rOmpA. Int. J. Syst. Evol. Microbiol. 48:839–849 [DOI] [PubMed] [Google Scholar]
- 16. Gilot B, Laforge ML, Pichot J, Raoult D. 1990. Relationships between the Rhipicephalus sanguineus complex ecology and Mediterranean spotted fever epidemiology in France. Eur. J. Epidemiol. 6:357–362 [DOI] [PubMed] [Google Scholar]
- 17. Goldberg MB. 2001. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 65:595–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 19. Gottlieb Y, Perlman SJ, Chiel E, Zchori-Fein E. 2011. Rickettsia get around, p 191–207 In Zchori-Fein E, Bourtzis K. (ed), Manipulative tenants—bacteria associated with arthropods. CRC Press, Boca Raton, FL [Google Scholar]
- 20. Harrus S, Baneth G. 2005. Drivers for the emergence and re-emergence of vector-borne protozoal and bacterial diseases. Int. J. Parasitol. 35:1309–1318 [DOI] [PubMed] [Google Scholar]
- 21. Harrus S, Perlman-Avrahami A, Mumcuoglu KY, Morick D, Baneth G. 2011. Molecular detection of Rickettsia massiliae, Rickettsia sibirica mongolitimonae and Rickettsia conorii israelensis in ticks from Israel. Clin. Microbiol. Infect. 17:176–180 [DOI] [PubMed] [Google Scholar]
- 22. Harrus S, et al. 2011. Molecular detection of Ehrlichia canis, Anaplasma bovis, Anaplasma platys, Candidatus Midichloria mitochondrii and Babesia canis vogeli in ticks from Israel. Clin. Microbiol. Infect. 17:459–463 [DOI] [PubMed] [Google Scholar]
- 23. Heise SR, Elshahed MS, Little SE. 2010. Bacterial diversity in Amblyomma americanum (Acari: Ixodidae) with a focus on members of the genus Rickettsia. J. Med. Entomol. 47:258–268 [DOI] [PubMed] [Google Scholar]
- 24. Jasinskas A, Zhong JM, Barbour AG. 2007. Highly prevalent Coxiella sp. bacterium in the tick vector Amblyomma americanum. Appl. Environ. Microbiol. 73:334–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF. 2002. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J. Med. Entomol. 39:809–813 [DOI] [PubMed] [Google Scholar]
- 27. Machado-Ferreira E, et al. 2011. Coxiella symbionts in the Cayenne tick Amblyomma cajennense. Microb. Ecol. 62:134–142 [DOI] [PubMed] [Google Scholar]
- 28. McMichael AJ, et al. 2003. Climate change and human health risks and responses. World Health Organization; http://www.who.int/globalchange/publications/cchhbook/en/ [Google Scholar]
- 29. Mira A, Moran NA. 2002. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb. Ecol. 44:137–143 [DOI] [PubMed] [Google Scholar]
- 30. Moraes-Filho J, Marcili A, Nieri-Bastos FA, Richtzenhain LJ, Labruna MB. 2011. Genetic analysis of ticks belonging to the Rhipicephalus sanguineus group in Latin America. Acta Trop. 117:51–55 [DOI] [PubMed] [Google Scholar]
- 31. Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC. 2006. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ. Microbiol. 8:761–772 [DOI] [PubMed] [Google Scholar]
- 32. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niebylski ML, et al. 1997. Rickettsia peacockii sp. nov, a new species infecting wood ticks, Dermacentor andersoni, in western Montana. Int. J. Syst. Bacteriol. 47:446–452 [DOI] [PubMed] [Google Scholar]
- 34. Noda H, Munderloh UG, Kurtti TJ. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 63:3926–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pais R, Lohs C, Wu YN, Wang JW, Aksoy S. 2008. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74:5965–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosenberg E, Zilber-Rosenberg I. 2011. The hologenome concept, p 323–340 In Rosenberg E, Gophna U. (ed), Beneficial microorganisms in multicellular life forms. Springer, Berlin, Germany [Google Scholar]
- 38. Sakurai M, Koga R, Tsuchida T, Meng XY, Fukatsu T. 2005. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 71:4069–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schabereiter-Gurtner C, Lubitz W, Rolleke S. 2003. Application of broad-range 16S rRNA PCR amplification and DGGE fingerprinting for detection of tick-infecting bacteria. J. Microbiol. Methods 52:251–260 [DOI] [PubMed] [Google Scholar]
- 40. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sekeyova Z, Roux V, Raoult D. 2001. Phylogeny of Rickettsia spp. inferred by comparing sequences of ‘gene D’, which encodes an intracytoplasmic protein. Int. J. Syst. Evol. Microbiol. 51:1353–1360 [DOI] [PubMed] [Google Scholar]
- 42. Shannon C, Weaver W. 1963. The mathematical theory of communication, 5th ed University of Illinois Press, Urbana, IL [Google Scholar]
- 43. Simpson EH. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 44. van Overbeek L, et al. 2008. Diversity of Ixodes ricinus tick-associated bacterial communities from different forests. FEMS Microbiol. Ecol. 66:72–84 [DOI] [PubMed] [Google Scholar]
- 45. Walker JB, Keirans JE, Horak IG. 2000. The genus Rhipicephalus (Acari: Ixodidae): a guide to the brown ticks of the world. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 46. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wong CNA, Ng P, Douglas AE. 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13:1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yilmaz GR, et al. 2009. The epidemiology of Crimean-Congo hemorrhagic fever in Turkey, 2002–2007. Int. J. Infect. Dis. 13:380–386 [DOI] [PubMed] [Google Scholar]
- 49. Youssef N, et al. 2009. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl. Environ. Microbiol. 75:5227–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zanettii AS, Pornwiroon W, Kearney MT, Macaluso KR. 2008. Characterization of rickettsial infection in Amblyomma americanum (Acari: Ixodidae) by quantitative real-time polymerase chain reaction. J. Med. Entomol. 45:267–275 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.