Abstract
Enteropathogenic Escherichia coli (EPEC) is a major cause of diarrhea in infants in developing countries. We have identified a functional type II secretion system (T2SS) in EPEC that is homologous to the pathway responsible for the secretion of heat-labile enterotoxin by enterotoxigenic E. coli. The wild-type EPEC T2SS was able to secrete a heat-labile enterotoxin reporter, but an isogenic T2SS mutant could not. We showed that the major substrate of the T2SS in EPEC is SslE, an outer membrane lipoprotein (formerly known as YghJ), and that a functional T2SS is essential for biofilm formation by EPEC. T2SS and SslE mutants were arrested at the microcolony stage of biofilm formation, suggesting that the T2SS is involved in the development of mature biofilms and that SslE is a dominant effector of biofilm development. Moreover, the T2SS was required for virulence, as infection of rabbits with a rabbit-specific EPEC strain carrying a mutation in either the T2SS or SslE resulted in significantly reduced intestinal colonization and milder disease.
INTRODUCTION
The type II secretion system (T2SS) is a multimeric protein translocase used by pathogenic Gram-negative bacteria to secrete virulence determinants. These virulence determinants are commonly toxins, such as cholera toxin of Vibrio cholerae (59) and the heat-labile toxin (LT) of enterotoxigenic Escherichia coli (ETEC) (61). Alternatively, the virulence determinants are degradative enzymes such as elastase and phospholipase C from Pseudomonas aeruginosa (6). Proteins destined to be secreted by the T2SS are first translocated across the cytoplasmic membrane via either the Sec or Tat machinery (52, 66), then folded and assembled in the periplasm, and finally secreted across the outer membrane by the T2SS itself.
The multimeric type II secretion apparatus is composed of at least 12 subunits, all of which are required for activity (34, 58). The complex spans the periplasm and consists of an outer membrane pore (48), a platform of proteins associated with the inner membrane that provide the energy to drive secretion (53), and a set of proteins that assemble into a pilus-like structure (the pseudopilus) in the periplasm (65).
In E. coli, three distinct T2SSs have been identified. A plasmid-encoded T2SSEDL933 in an enterohemorrhagic E. coli (EHEC) strain, EDL933, secretes StcE, a protease that cleaves complement components (39). The T2SSEDL933 also contributes to EHEC adherence and intestinal colonization (31). A second system, T2SSK-12 in E. coli K-12 strain MC4100, is nonfunctional under standard culture conditions but secretes endogenous chitinase when derepressed (24). The third system, T2SSH10407, identified by us in ETEC strain H10407, is responsible for the secretion of LT (61). The T2SSH10407 is highly conserved, as homologs are present in other E. coli pathotypes including EHEC, enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC), and extraintestinal E. coli (ExPEC) (9, 13, 14, 32, 33, 38, 49, 63). Remnants of T2SSH10407 are also present in the E. coli K-12 strains MG1655 and W3110 (54, 61, 63).
To characterize the T2SSH10407, we investigated this system in the prototypical human EPEC strain, E2348/69. EPEC is a leading cause of infantile diarrhea in developing countries (55) but, unlike ETEC, does not appear to secrete an enterotoxin (41, 56). The major virulence determinant of EPEC is an ∼35.6-kb chromosomal pathogenicity island, termed the locus of enterocyte effacement (LEE). The LEE encodes a type III protein secretory system, at least nine type III secreted proteins (e.g., EspB, -F, and -G), and an outer membrane adhesin (intimin) encoded by the eae gene (19, 21, 32), all of which are required to produce distinctive attaching and effacing lesions in the intestinal epithelium that characterize infection with EPEC (25).
Characterization of the T2SSH10407 of E2348/69 showed that this system is functional, that it secretes a lipoprotein, SslE, for secreted and surface-associated lipoprotein from E. coli (formerly YghJ [32, 67]). Both SslE and T2SS mutants were attenuated for biofilm formation. Importantly, T2SSH10407 and SslE mutants of a rabbit-specific EPEC (REPEC) strain, E22, showed a reduced ability to colonize the intestine and to cause disease in a rabbit model of infection. Our results demonstrate that the T2SSH10407 and its substrate SslE are critical virulence determinants of EPEC. As this type II secretion complex and SslE are highly conserved, they may also contribute to the virulence of other pathogenic E. coli strains.
MATERIALS AND METHODS
Bacterial strains, plasmid growth conditions, and primers.
The bacterial strains and plasmids used in this study are listed in Table 1. Strains were propagated in Luria-Bertani (LB) or Casamino Acids-yeast extract-salts (CAYE) medium (23, 57) at 37°C with shaking (225 rpm) unless otherwise noted. The primers used in the construction of plasmids and mutants are listed in Table 2.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E22 | REPEC serotype O103:H2, Rifr | 42 |
| E22ΔgspD | E22 ΔgspD, Rifr | This study |
| E22ΔgspK | E22 gspK::Kanr, Rifr | This study |
| E22ΔsslE | E22 ΔsslE, Rifr | This study |
| E2348/69 | EPEC serotype O127:H6 | 40 |
| E2348/69ΔgspD | E2348/69 ΔgspD | This study |
| E2348/69ΔgspK | E2348/69 gspK::Kanr | This study |
| E2348/69ΔsslE | E2348/69 ΔsslE | This study |
| H10407 | ETEC serotype 078:H11, LT+, ST+ | 22 |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR thiA | 10 |
| MT2 | H10407 gspK::Kanr | 61 |
| Plasmids | ||
| pACBSR | Medium-copy-no. vector, p15A ori, Cmr | 29 |
| pACYC184 | Medium-copy-no. vector, p15A ori, Cmr, Tcr | 12 |
| pBAD24 | Medium-copy-no. vector, ori pMB1, Apr | 26 |
| pBR322 | Medium-copy-no. vector, ori pMB1, Apr, Tcr | 8 |
| pCP20 | Low-copy-no. vector, ori R101, repA101ts, flp, Apr, Cmr | 18 |
| pFT-A | Low-copy-no. vector, ori R101 repA101ts, flp, Apr | 51 |
| pGEM-T Easy | High-copy-no. vector, ori pMB1, Apr | Promega |
| pJP15 | sslEE2348/69 in pSU39, Kanr | This study |
| pJP16 | sslEE22 in pSU39, Kanr | This study |
| pJP33 | hltA promoter and hltB from H10407 in pBR322, Apr | This study |
| pJP49 | gspKE22 in pACYC184, Cmr | This study |
| pJP51 | gspDE22 in pACYC184, Cmr | This study |
| pJP52 | gspDE2348/69 in pACYC184, Cmr | This study |
| pJP115 | micA promoter from E2348/69 in pMU6400, Tpr | This study |
| pJP168 | PBAD promoter and araC replaced by PtetA and tetR gene from Tn10 in pBAD24, Apr | This study |
| pJP169 | sslEE2348/69-tetraCys in pJP168, Apr | This study |
| pJP183 | pBR322 with the promoter and NH terminus of the Tcr gene deleted, Apr | This study |
| pKD4 | FRT-flanked Kanr gene, Kanr, Apr | 18 |
| pKD46 | Low-copy-no. vector, ori R101, repA101ts, PBAD-λ Red, Apr | 18 |
| pMU2386 | Single-copy-no. translational fusion vector, Tpr | 5 |
| pMU6400 | Synthetic RBS with ATG start codon fused in phase with codon 8 of lacZ in pMU2386, Tpr | This study |
| pSU39 | Medium-copy-no. vector, p15A ori, Kanr | 7 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Rifr, rifampin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance.
Table 2.
Primers used in this study
| Primer | Sequence 5′ to 3′ |
|---|---|
| 2386lB4 | GATCCGTCATAGTTGTGTTTCCTGTTCATCTAGTTATCTAGAGCTAGATCTAGGA |
| 2386lT4 | AGCTTCCTAGATCTAGCTCTAGATAACTAGATGAACAGGAAACACAACTATGACG |
| E22sslE.F | ACAGAATTCGCCAGCGTTATCGGCGTTATC |
| E22sslE.R | ACAGGATCCGTGAACGCCTTATCCGGCATAC |
| E2348sslE.F | TCATCTAGAGCCAGCGTTATCGGCATTATCAC |
| E2348sslE.R | ACTAACCTTGAACGCCTTATCCGGCCTA |
| gspD.F | AGAGGATTCGCGCTAACCGCATTTAATCCAG |
| gspD.ISceIF | TAGGGATAACAGGGTAATCACTGATCCACGAGCAATGATTGC |
| gspD.ISceIR | TAGGGATAACAGGGTAATAGACCGCATTCAGCCGCTGTTC |
| gspD.KmF | CTAAGGAGGATATTCATATGGTGACCATTCTGCGTGACGGTATGG |
| gspD.KmR | GAAGCAGCTCCAGCCTACACACCCCTGTGCTTCCAGCAGGTTAAG |
| gspD.R | AGAGTCGACAACTGTAGGGCAGACGCACG |
| gspK.F | AGAGGATCCTTGAACGCATCTGGTTGTTACGC |
| gspK.R | AGAGTCGACGATCAGCGGCGCAAAGC |
| micA.F | GACAAGCTTCGAACACGGTGATTGCGTCG |
| micA.R | GACTCTAGACTCTGAATTCAGGGATGATGATAAC |
| P5 | GCAGCAGGTGACTAACGGC |
| P12 | CAGGGCTTAACCACGGGTC |
| pKD4F | TGTGTAGGCTGGAGCTGCTTC |
| pKD4R | CATATGAATATCCTCCTTAG |
| TetRA.F | GACTAGATCGATCTTGGTTACCGTGAAGTTACCATC |
| TetRA.R | CACGCTAGCATTCATTTCACTTTTCTCTATCACTGATAGG |
| TF | CAGTCAGAGGTTGACATATATAACAG |
| TOXB3′ | GCGGATCCTAGCTTAGTTTTTTCTGTGTG |
| TR | GTCAACCTCTGACTGCATACAAGAAG |
| sslE.ISceIF | TAGGGATAACAGGGTAATCGGATGATCGTTAATATCATCCGGC |
| sslE.ISceIR | TAGGGATAACAGGGTAATAATCCTCCGACGGTTGCCAG |
| sslE.KmF | CTAAGGAGGATATTCATATGGTGGCGGACACGCTGATGCTGTG |
| sslE.KmR | GAAGCAGCTCCAGCCTACACAGCCATCACAACCGGCTAACAGG |
| SslEtct.Fs | TATCCATGGATAAGAAATTTAAATATAAGAAATCGC |
| SslEtct.R | GTGAAGCTTTTAACGGCCGCCCGGTTCCATGCAGCAGCCCGGGCAGCAGTTCAGAAAGCTGCCCGCTTCGGCTGGCATCGAATACTCGG |
| SslE.R1 | TTCGGCTGGCATCGAATACTC |
| SslEtct.Rs | GTGAAGCTTTTAACGGCCGCC |
| YJ001 | TTTGGATCCTAATCAGGTTGCCATGATTC |
Construction of nonpolar gspD, gspK, and sslE mutants.
The deletion mutation in the gene encoding the essential type II pseudopilin, gspK, was constructed in E2348/69 and E22 by allelic exchange with gspK::Kanr, amplified from E. coli strain MT2 using Platinum Taq DNA polymerase (Promega) and primers P5 and P12. These exchanges were facilitated by the λ Red recombinase system carried on plasmid pKD46 (18). Deletion mutations in gspD and sslE of E2348/69 and E22 were constructed using the “gene-gorging” technique (29). Phusion high-fidelity DNA polymerase (New England BioLabs) was used to amplify the FLP recombination target (FRT)-flanked Kanr gene from pKD4 (using primers pKD4F and pKD4R) and ∼0.5 kb of DNA flanking the target genes (using primer pairs gspD.ISceIF/gspD.KmR or sslE.ISceIF/sslE.KmR for the upstream flanks and gspD.ISceIR/gspD.KmF or sslE.ISceIR/sslE.KmF for the downstream flanks) from the genomic DNA. The Kanr fragment was joined to the appropriate upstream and downstream fragments by overlapping extension PCR (11) using Platinum Taq DNA polymerase and primer pairs gspD.ISceIF/gspD.ISceIR or sslE.ISceIF/sslE.ISceIR. The ISceI-flanked PCR products were cloned into pGEM-T Easy to yield the donor plasmids required for gene gorging. The allelic exchange of the wild-type chromosomal gene with the Kanr-disrupted gene was facilitated by the λ Red recombinase system carried on plasmid pACBSR (29). When required, the Kanr gene was excised using the flanking FRT sites and FLP helper plasmids pCP20 (18) and pFT-A (51). All mutations were confirmed by PCR analysis, using primers flanking the targeted regions.
Construction of trans-complementing plasmids.
pJP52 and pJP51, the gspD complementing plasmids, were generated by amplifying 2,151-bp fragments containing gspD from E2348/69 and from E22 genomic DNA, respectively, using primers gspD.F and gspD.R, digesting the DNA with BamHI/SalI, and ligating it to BamHI/SalI-digested pACYC184. pJP49, the gspK-complementing plasmid, was generated by amplifying a 1,057-bp fragment containing gspK from E22 genomic DNA, using primers gspK.F and gspK.R, digesting the DNA with BamHI/SalI, and ligating it to BamHI/SalI-digested pACYC184. pJP15 was generated by amplifying a 5,251-bp fragment containing the sslE gene from E2348/69 genomic DNA using primers E2348sslE.F and E2348sslE.R, digesting the DNA with HindIII/XbaI, and ligating it to HindIII/XbaI-digested pSU39. pJP16 was generated by amplifying a 5,143-bp fragment containing sslE from E22 genomic DNA using primer pair E22sslE.F/E22sslE.R, digesting the DNA with BamHI/EcoRI, and ligating it to BamHI/EcoRI-digested pSU39. Phusion high-fidelity DNA polymerase was used in all these amplification reactions.
Construction of LTB expression vector.
Overlapping PCR was used to generate a 793-bp fragment containing the hltA promoter upstream of the hltB gene. Platinum Taq DNA polymerase and primers YJ001 and TR, which incorporate a BamHI site and an overhang region complementary to htlB, respectively, were used to amplify a 213-bp fragment encompassing the hltA promoter (68) from H10407 genomic DNA. A 595-bp fragment containing hltB was amplified from H10407 genomic DNA using primers TOXB3′ and TF, which incorporate a BamHI site and a 5′ overhang region complementary to the hltA promoter, respectively. The two PCR products were joined using Vent polymerase (New England BioLabs) and primers YJ001 and TOXB3′, and the resultant 793-bp fragment was cloned into the BamHI site of pJP183 to generate plasmid pJP33.
Construction of plasmid expressing SslE-tetraCys.
pJP168, a tetracycline-inducible gene expression vector, was generated by replacing the ClaI-NheI fragment of pBAD24 carrying araC-PBAD with a 769-bp ClaI-NheI fragment containing tetR-PTetA of Tn10, amplified from pFT-A using primers TetRA.F and TetRA.R. Overlapping PCR was used to generate a 4,640-bp fragment containing the coding sequence of sslE-tetraCys flanked by NcoI/HindIII restriction endonuclease sites. This involved joining a 4,574-bp fragment containing the sslE coding sequence, amplified from E2348/69 genomic DNA using primers SslEtct.Fs and SslE.R1, to primer SslEtct.R, using Phusion high-fidelity DNA polymerase and primers SslEtct.Fs and SslEtct.Rs. The sslE-tetraCys fragment was inserted into the NcoI/HindIII site of pJP168, generating plasmid pJP169.
Construction of the lacZ fusion plasmids.
The transcriptional lacZ fusion vector pMU6400 was constructed by ligation of annealed primers 2386lT4/2386lB4 to HindIII/BamHI-digested pMU2386, so that the ATG start codon of the ribosome binding site (RBS) encoded by these oligonucleotides was in phase with the lacZ coding sequence. The micA-lacZ fusion plasmid, pJP115, was constructed by ligating the promoter region of micA, amplified from E2348/69 genomic DNA using primers micA.F and micA.R, to HindIII/XbaI-digested pMU6400.
Assay for LTB.
The GM1 ganglioside enzyme-linked immunosorbent assay (ELISA) used to assay LTB was performed as described previously (61).
Protein identification.
Edman degradation of SslE was performed by the Australian Proteomic Analysis Facility, North Ryde, Australia. Tandem mass spectrometry was performed at the Bio21 Institute, The University of Melbourne, Australia.
Visualization of SslE.
The procedure used to visualize tetraCys-tagged SslE with the biarsenical dye Lumio Green was performed according to the directions of the manufacturer (Invitrogen) with the following changes: Opti-MEM was replaced with phosphate-buffered saline (PBS), and the Lumio labeling reagent contained 5 mM dithiothreitol (DTT). Bacteria were fixed in 1.6% paraformaldehyde prior to labeling and mounted on poly-l-lysine slides using Dako fluorescence mounting medium.
Proteinase K shaving.
Cells from 1-ml aliquots of bacterial culture were harvested by centrifugation at 3,000 × g for 3 min at 4°C. Bacteria were washed with 20 mM Tris-HCl, pH 8, and resuspended in 600 μl of the same solution. Polymyxin B and/or proteinase K were added to final concentrations of 2.0 and 0.02 mg/ml, respectively. Samples containing polymyxin B or proteinase K were incubated on ice for 10 and 30 min, respectively, and 10 μl of treated bacterial cells was processed by SDS-PAGE.
β-Galactosidase assays.
β-Galactosidase activity was determined by the method of Miller as described previously (44, 62).
Biofilm flow cell cultivation.
Biofilms were cultivated at 37°C in three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm (BioCentrum-DTU). A plastic coverslip (ibidi GmbH) was attached to the flow cell with silicon glue and used as substratum for biofilm growth. Bacteria were cultured overnight in LB, diluted 1:100 in 37°C Dulbecco's modified Eagle's medium (DMEM), and inoculated into each channel. After 1 h flow was recommenced and maintained at 451 μl/min for 96 h using a peristaltic pump. At various time points to 96 h, samples were fixed with 4% paraformaldehyde and stained with Syto9 (Invitrogen) to allow visualization.
Microscopy and image analysis of biofilms.
Microscopic image acquisition of biofilms was performed utilizing confocal laser scanning microscopy on a Nikon A1R-A1 inverted microscope. Simulated 3-dimensional (3D) fluorescence blend images of individual panels were generated using the Imaris software package (Bitplane AG). For COMSTAT quantification of biofilm growth (30), each strain was grown in a single channel, and image stacks were acquired from equivalent areas of the flow channel where the flow of medium was calculated to be laminar.
Infection of rabbits.
For in vivo assays of virulence, 4- to 5-week-old New Zealand White rabbits were inoculated with either a wild-type REPEC strain, E22, or an isogenic mutant. Rabbits were weighed daily and examined for clinical signs of illness including weight loss and evidence of diarrhea as described previously (2). Fecal shedding of the infecting strains was determined by culture of rectal swabs on MacConkey agar supplemented with rifampin (50 μg/ml) to distinguish them from the rabbits' microbiota. Animals were euthanized when they lost greater than 15% body weight or demonstrated severe diarrhea. Segments from the distal small intestine were obtained from animals within each experimental group and fixed in 2.5% gluteraldehyde, postfixed in 2.5% osmium tetroxide for 1 h, dehydrated through a graded acetone series, and embedded in Epon-Araldite epoxy resin. Thin sections were cut and stained with 10% uranyl acetate and 2.5% lead citrate before being viewed under a Phillips CM10 electron microscope at 80 kV.
FAS and adherence assays.
Fluorescence actin staining (FAS) was performed as described previously (36) using a 3-h incubation period with the following changes: HEp-2 cells were incubated with 50 μg/ml phalloidin conjugated to tetramethyl rhodamine isocyanate (TRITC; Sigma) for 30 min. To detect adherent bacteria, DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) was applied at a dilution of 1:20,000 in PBS for 5 min. Images were acquired using a Zeiss LSM700 inverted confocal microscope using 405- and 555-nm diode laser lines.
Statistical analysis.
Fisher's exact test was used to compare fecal bacterial shedding and incidence of diarrhea between experimental groups. All other analyses were performed by using Student's t test. A two-tailed P value of <0.05 was taken to indicate statistical significance.
RESULTS
A functional homolog of T2SSH10407 is conserved in EPEC.
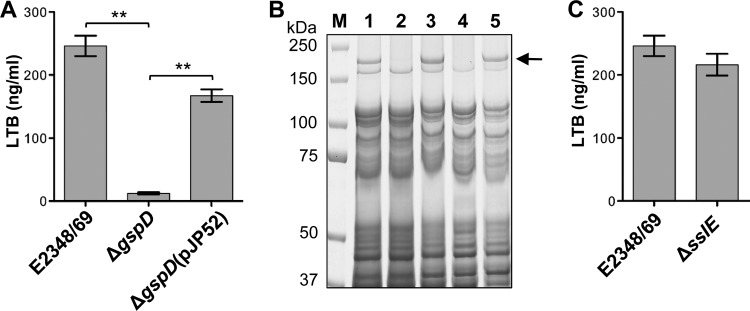
Comparison of the predicted sequences of the 13 components of the T2SS of ETEC strain H10407 and EPEC strain E2348/69 revealed a high degree of sequence conservation, with amino acid identity ranging from 90.7 to 99.3%. To determine if this T2SS is functional in EPEC, we established a cellular system to measure the secretion of LTB, a nontoxic form of LT, which is the substrate of this T2SS in ETEC. LTB, expressed from pJP33, was measured with a GM1 ganglioside enzyme-linked immunosorbent assay (ELISA) (61). The results showed that the wild-type strain, E2348/69, secreted significantly more LTB into the culture medium than an isogenic T2SS mutant, E2348/69ΔgspD (P < 0.005) (Fig. 1A) (gspD encodes the outer membrane secretin that forms the pore of the T2SS). In addition, LTB secretion was significantly restored in a trans-complemented mutant, E2348/69ΔgspD(pJP52) (P < 0.005) (Fig. 1A).
Fig 1.
EPEC strain E2348/69 requires a functional T2SS to secrete LTB and SslE into the culture medium. (A) Concentration of LTB in the 5-hour CAYE culture supernatant of wild-type E2348/69, an isogenic T2SS mutant, E2348/69ΔgspD, and a trans-complemented mutant, E2348/69ΔgspD(pJP52), containing an LTB expression plasmid (pJP33) was measured by using GM1 ELISA. Values are the means (±SEM) of three independent experiments performed in triplicate (**, P < 0.005). (B) SslE is secreted by the T2SS of EPEC strain E2348/69. SslE (arrow) was detected in the supernatant of wild-type strain E2348/69 (lane 1) and trans-complemented mutants, E2348/69ΔgspD(pJP52) (lane 3) and E2348/69ΔgspK(pJP53) (lane 5), but not in the isogenic T2SS mutants, E2348/69ΔgspD (lane 2) and E2348/69ΔgspK (lane 4). Strains were grown in LB for 5 h. Proteins in the supernatants were precipitated with 10% trichloroacetic acid, washed in 25% acetone, separated by SDS-PAGE using 4–12% bis-Tris NuPAGE gels (Invitrogen), and stained with Coomassie brilliant blue R-250. (C) SslE is not required for the secretion of LTB. Concentration of LTB in the 5-hour CAYE supernatant of wild type, E2348/69, and its isogenic ΔsslE mutant containing the pJP33 expression plasmid was measured by using GM1 ELISA. Values represent means (±SEM) of three independent experiments performed in triplicate.
SslE is a substrate of the EPEC T2SS.
To determine the native substrate(s) of the T2SS of EPEC, we compared the secretomes of the wild-type strain and its isogenic gspD and gspK mutants. The gspK gene encodes a subunit of the type II pseudopilus. Analysis of the protein profiles revealed a high-molecular-weight protein of approximately 170 kDa in the secretome of wild-type EPEC that was absent from the secretome of the T2SS mutants (Fig. 1B). This was excised and analyzed by using tandem mass spectrometry, which identified this protein as YghJ, which is henceforth termed SslE. Sequence analysis with Prosite (20) suggested that SslE is a lipoprotein, with a strong prediction of a signal peptide (residues 1 to 23) and Cys24 predicted as being subject to covalent N-palmitoylation and the addition of S-diacylglycerol. Consistent with this, LipoP 1.0 (35) predicted a Signal peptidase II cleavage site (score = 37.4) between residues 23 and 24. When SslE was purified from the extracellular medium and subjected to sequence analysis by Edman degradation, it was blocked, consistent with the acylation of the N-terminal Cys residue.
As SslE is part of the T2SSH10407 operon (67), we investigated if SslE is a functional part of the secretion machinery. However, the amount of LTB in the supernatant of the sslE mutant (E2348/69ΔsslE) was not significantly different (P = 0.3) from that secreted by the wild type (Fig. 1C).
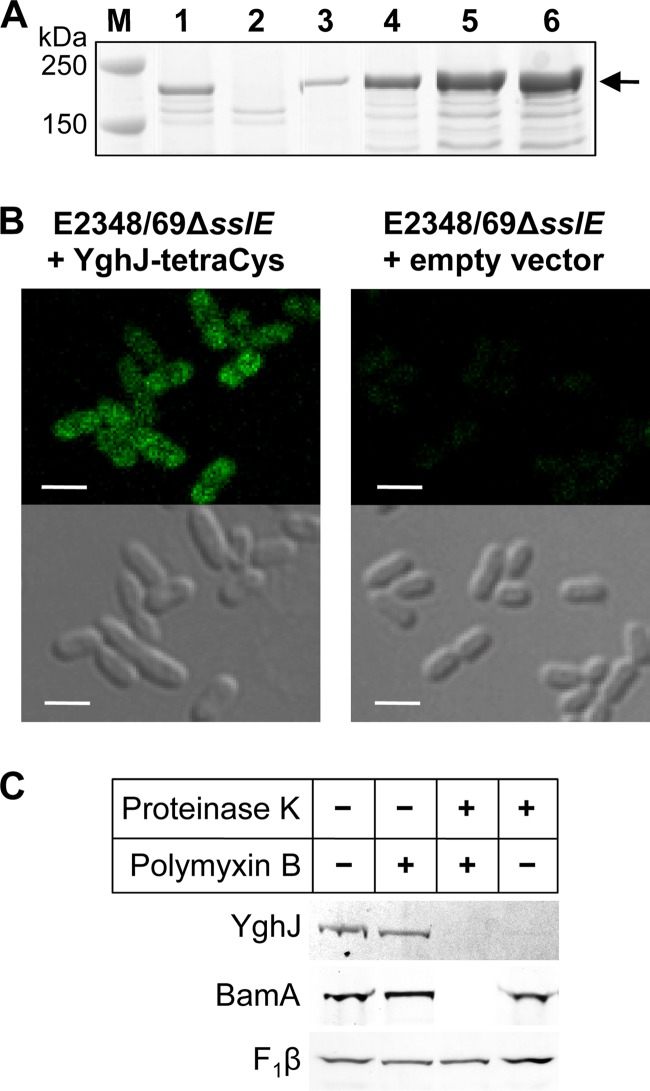
SslE is located on the outer cell surface.
The best-studied T2SS is that of Klebsiella pneumoniae, whose substrate is the lipoprotein pullulanase, which becomes transiently anchored on the cell surface before being released into the medium after the end of exponential growth (37, 43). To determine if SslE behaves in a similar manner, we constructed a plasmid, pJP169, where sslE was tagged at its C terminus with a tetracysteine tag and expressed from an inducible tetA promoter. To establish conditions where the levels of SslE-tetraCys in the cell were similar to those of SslE in the wild-type strain, the expression of SslE-tetraCys was determined in response to various amounts of the inducer, anhydrotetracycline (Fig. 2A). SslE-tetraCys was visualized with Lumio Green by using confocal microscopy, which showed that SslE is localized on the bacterial cell surface (Fig. 2B). Treatment with proteinase K removed the fluorescent signal, confirming that SslE was surface located. In addition, incubation of E2348/69ΔsslE(pJP169) with proteinase K in the presence or absence of polymyxin B (which selectively permeabilizes the outer membrane allowing access of proteinase K to the periplasm [15]) revealed that SslE was cleaved without polymyxin B (Fig. 2C). This result confirmed the outer surface membrane location of SslE.
Fig 2.
SslE-tetraCys is located on the outer surface of the outer membrane of EPEC strain E2348/69. (A) Titration of the inducer, anhydrotetracycline (ATc), to determine the amount required to induce expression of sslE-tetraCys to levels approximate to those of SslE in wild-type E2348/69. E2348/69ΔsslE(pJP169), where sslE-tetraCys was expressed from a tetA promoter, was grown in CAYE to an optical density at 600 nm (OD600) of ∼2 and then induced for 2 h with 0, 30, 35, 40, or 50 ng/ml ATc (lanes 2 to 6, respectively). Proteins were separated by SDS-PAGE using 4–12% bis-Tris NuPAGE gels (Invitrogen) and stained with Coomassie brilliant blue R-250. (B) Fluorescent and phase-contrast images of E2348/69ΔsslE carrying plasmid pJP169 (pJP168::sslE-tetraCys) or the empty vector, pJP168, which was grown in CAYE to an OD600 of ∼2 and then induced for 2 h with 35 ng/ml anhydrotetracycline. Bacteria were visualized by differential interference phase contrast on a Zeiss LSM700 inverted confocal microscope, and Lumio Green was excited using a 488-nm diode laser line. Bar, 2 μm. (C) E2348/69ΔsslE(pJP169) cells were grown as described above and incubated without (−) or with (+) polymyxin B and proteinase K, as indicated. Proteins, separated by SDS-PAGE using 4-12% bis-Tris NuPAGE gels (Invitrogen), were analyzed by Lumio Green detection for SslE and by immunoblotting for the controls, BamA (inner surface, outer membrane protein) and the cytoplasmic membrane protein FoF1 ATP synthase β-subunit (F1β). The latter served as a control for sample loading, as it is protected from proteinase K by the inner membrane.
Cell envelope integrity in sslE and T2SS mutants.
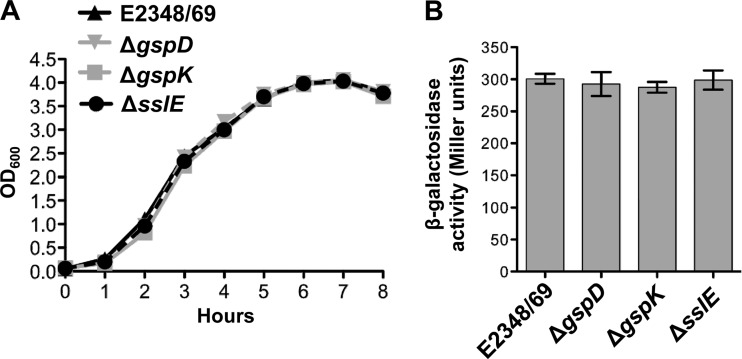
Mutations in the T2SS of V. cholerae result in growth defects and compromised outer membrane integrity, leading to defects in secretion and induction of RpoE activity (RNA polymerase σE) (60). This was not the case in EPEC as comparison of the growth curves of the E2348/69 wild type and the T2SS mutants showed that disruption of gspD or gspK had no effect on growth (see Fig. 4A). Growth of an sslE mutant was also not impaired (Fig. 3A).
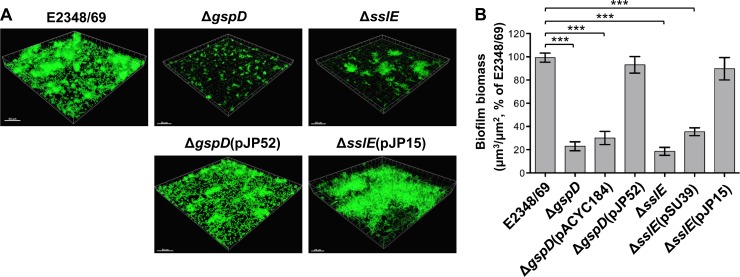
Fig 4.
Analysis of biofilm formation by EPEC E2348/69 and its derivatives. (A) Confocal laser scanning images of wild-type E2348/69 and its isogenic mutants, E2348/69ΔgspD (T2SS mutant) and E2348/69ΔsslE, and trans-complemented mutants, E2348/69ΔgspD(pJP52) and E2348/69ΔsslE(pJP15). Shadow fluorescence projection images are rendered in Imaris. Bar, 50 μm. (B) Quantification of three-dimensional biofilm image stacks of wild-type E2348/69 and its isogenic mutants, E2348/69ΔgspD, and E2348/69ΔsslE, trans-complemented mutants, E2348/69ΔgspD(pJP52) and E2348/69ΔsslE(pJP15), and these mutants complemented with empty vector, E2348/69ΔgspD(pACYC184) and E2348/69ΔsslE(pSU39). T2SS and sslE mutants show significant defects in biofilm formation. Mean biomass was calculated by using COMSTAT analysis (30) of at least eight random image stacks from each strain. Values represent the means (±SEM) of three independent experiments expressed as a percentage of the wild type (***, P < 0.0001).
Fig 3.
Analysis of growth and induction of the global stress response by T2SS and sslE mutants. (A) T2SS and sslE mutants do not display impaired growth. Overnight cultures of wild-type E2348/69 and its isogenic mutants, E2348/69ΔgspD, E2348/69ΔgspK (T2SS mutants), and E2348/69ΔsslE, were grown in LB and diluted 1 in 100 in 10 ml LB in 50-ml Erlenmeyer flasks. (B) T2SS and sslE mutants do not induce RpoE activity. Mid-log-phase cultures (OD600, ∼0.7) of wild-type E2348/69 and its isogenic mutants, E2348/69ΔgspD, E2348/69ΔgspK (T2SS mutants) and E2348/69ΔsslE, containing the micA-lacZ transcriptional fusion plasmid, pJP115, were grown in LB supplemented with trimethoprim and assayed for β-galactosidase activity. Values represent means (±SEM) of three independent experiments performed in quadruplicate.
To assess if inactivation of the T2SS or SslE in E2348/69 led to induction of a σE-mediated stress response, we constructed and used a micA-lacZ transcriptional fusion plasmid (pJP115) as a reporter. In E. coli K-12, micA is transcribed from a single, σE-dependent promoter (64). As the sequence of micA of E2348/69, including its promoter, is identical to that of E. coli K-12, the activity of micA-lacZ provides a good measure of RpoE activity. However, we found no increase in β-galactosidase levels in the E2348/69 T2SS or sslE mutants compared with the wild type, indicating that these mutations did not elicit a global stress response (Fig. 3B).
SslE is required for biofilm formation by EPEC.
Moreira et al. (46) have proposed that the T2SS of EPEC might be involved in the mature stage of biofilm development. After finding that the T2SS played no part in the formation of biofilms in a static assay (data not shown), we used confocal laser scanning microscopy to examine the ability of E2348/69 and an isogenic T2SS mutant, E2348/69ΔgspD, to form biofilms in a continuous-culture flow cell system. No obvious differences between the wild-type and mutant biofilms were noted early during incubation; however, at 96 h clear differences were evident (Fig. 4A), with the T2SS mutant, E2348/69ΔgspD, showing sparse cell attachment to the substratum and the formation of small microcolonies. In comparison, both E2348/69 wild type and a trans-complemented T2SS mutant, E2348/69ΔgspD(pJP52), exhibited mature-stage biofilms with numerous large microcolonies and clearly visible water channels. Quantitative analysis of biofilm biomass indicated a significant reduction in the mean biofilm biomass for the T2SS mutant, E2348/69ΔgspD, compared with the wild type (P < 0.0001) (Fig. 4B). Complementation of the ΔgspD mutant with plasmid pJP52 restored mean biofilm biomass to wild-type levels. These results showed that biofilm formation in the T2SS mutant is attenuated and does not progress past the microcolony stage.
To determine if SslE is required for biofilm formation, we generated a plasmid expressing SslE (pJP15) and tested an E2348/69 isogenic mutant, E2348/69ΔsslE, and a trans-complemented mutant, E2348/69ΔsslE(pJP15), in the continuous-culture flow cell system. These analyses showed that the sslE mutant has defects in biofilm maturation similar to those of the T2SS mutant, E2348/69ΔgspD (Fig. 4A and B).
SslE is a virulence determinant of EPEC.
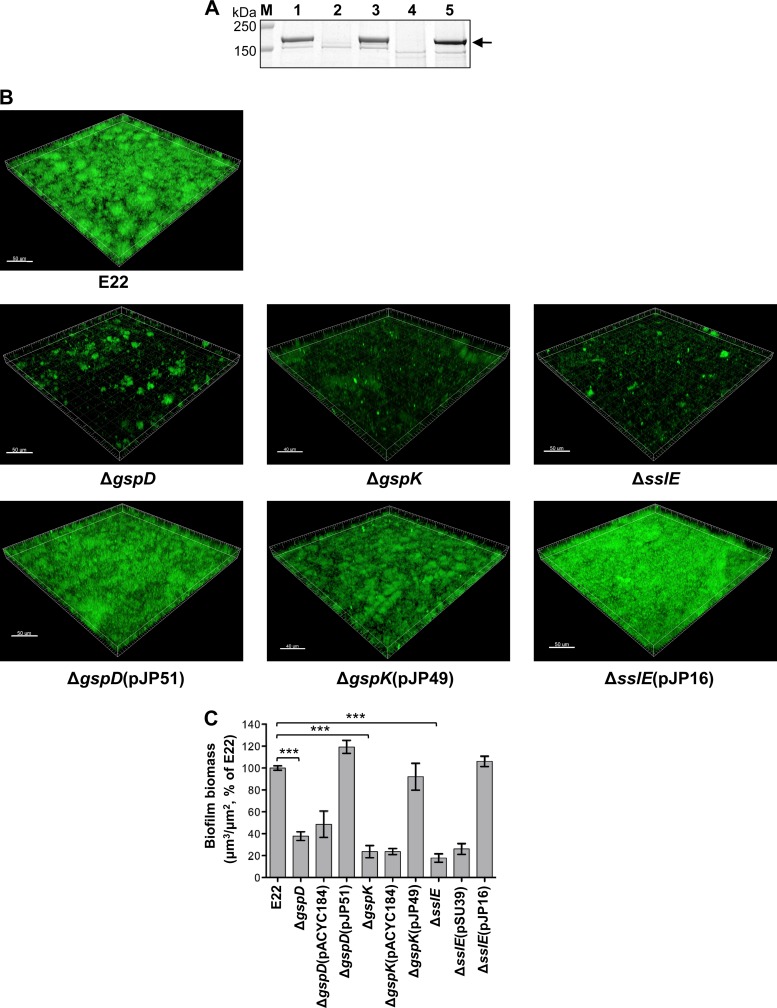
Rabbits infected with REPEC, e.g., strain E22, show the same clinical and pathological features as humans infected with EPEC (1, 42, 69). To ensure that REPEC strain E22 was an appropriate proxy for EPEC strain E2348/69, we constructed T2SS, sslE, and complemented mutant strains of E22 and analyzed their protein secretion profiles and ability to form biofilms. This analysis showed that SslE is a substrate of the T2SS in E22 (Fig. 5A) and that the T2SS and SslE are required for biofilm formation by this strain (Fig. 5B and C), confirming a role for the T2SS and SslE in biofilm formation in REPEC strains, as well as the human-specific strain E2348/69.
Fig 5.
Proteomic and biofilm analysis of REPEC E22 and its derivatives. (A) SslE is secreted by the T2SS of REPEC strain E22. SslE (arrow) was detected in the supernatant of wild-type strain E22 (lane 1) and the trans-complemented mutants, E2348/69ΔgspD(pJP51) (lane 3) and E2348/69ΔgspK(pJP49), but not in the isogenic T2SS mutants, E2348/69ΔgspD (lane 2) and E2348/69ΔgspK (lane 4). Strains were grown in LB for 5 h. Proteins in the supernatants were precipitated in 10% trichloroacetic acid and washed in 25% acetone. Proteins were separated by SDS-PAGE using 4–12% bis-Tris NuPAGE gels (Invitrogen) and stained with Coomassie brilliant blue R-250. (B) Confocal laser scanning images of wild-type E22 and its isogenic mutants, E2348/69ΔgspD, E2348/69ΔgspK (T2SS mutants), and E2348/69ΔsslE, and the trans-complemented mutants, E2348/69ΔgspD(pJP51), E2348/69ΔgspK(pJP49), and E2348/69ΔsslE(pJP16). Shadow fluorescence projection images were rendered in Imaris. Bar, 50 μm. (C) Quantification of three-dimensional biofilm image stacks of wild-type E22 and its isogenic mutants, E2348/69ΔgspD, E2348/69ΔgspK (T2SS mutants), and E2348/69ΔsslE, these mutants complemented with empty vector, E2348/69ΔgspD(pACYC184), E2348/69ΔgspK(pACYC184), and E2348/69ΔsslE(pSU39), and trans-complemented mutants, E2348/69ΔgspD(pJP51), E2348/69ΔgspK(pJP49), and E2348/69ΔsslE(pJP16). T2SS and sslE mutants show significant defects in biofilm formation. Mean biomass was calculated by using COMSTAT analysis of at least eight random image stacks from each strain. Values represent means (±SEM) of three independent experiments expressed as a percentage of the wild type (***, P < 0.0005).
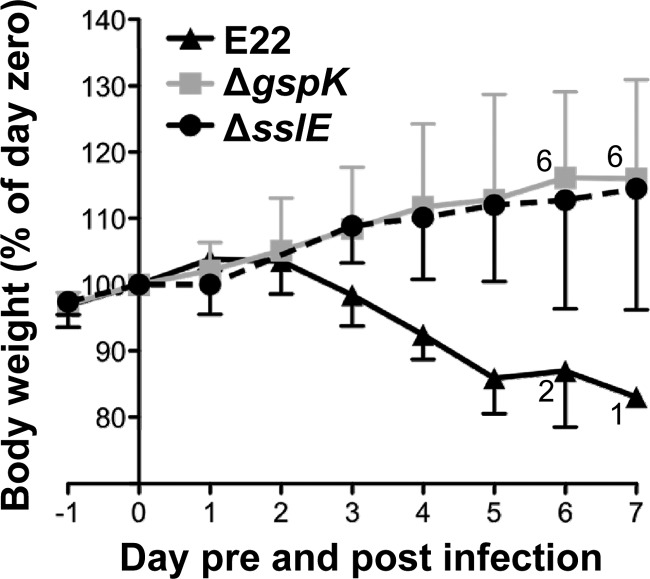
To investigate the effect of the T2SS and SslE on the virulence of EPEC, we infected infant rabbits with wild-type REPEC strain E22 and isogenic T2SS and sslE mutants. Following inoculation of 7 rabbits with 106 CFU of wild-type E22, large numbers of REPEC were shed in stools, such that >105 CFU were recovered from rectal swabs from all rabbits within 3 days of inoculation. Loss of body weight began within 48 h of infection and continued until animals were euthanized (Fig. 6). All rabbits developed clinical illness characterized by diarrhea with weight loss requiring euthanasia. Five rabbits were euthanized on day 5, one on day 6, and one on day 7 (Fig. 6). These results were consistent with previous reports of infection of rabbits with E22 (42).
Fig 6.
Effect of infection with REPEC strain E22 or its isogenic derivatives on weight gain by infant rabbits. Values are the mean (±SD) weight for each group expressed as a percentage of the weight on the day of infection (day zero). Eight rabbits were inoculated with 106 CFU wild-type E22, and seven rabbits were inoculated with 106 CFU of either the T2SS (ΔgspK) or the sslE mutant. When some rabbits in a group were euthanized, due to a body weight loss of greater than 15% or severe diarrhea, the surviving numbers of rabbits are shown adjacent to the data points.
Following inoculation with 106 CFU of isogenic T2SS (ΔgspK) or sslE mutant strains, rabbits shed the challenge strain from the second day after inoculation to 7 days postinoculation but yielded lower bacterial counts in rectal swabs during the entire observation period (31/47 and 39/56 [days with counts of <103 CFU/days of observation] for the ΔgspK and ΔsslE strains, respectively) compared to 6/38 for rabbits infected with the wild-type strain (P < 0.0001; Fisher's exact test). One rabbit inoculated with the ΔgspK mutant began to lose weight on day 2, developed diarrhea on day 3, and was subsequently euthanized on day 5. Nevertheless, rabbits inoculated with either mutant strain experienced significantly fewer days of diarrhea (7/47 and 5/56 for the ΔgspK and ΔsslE mutants, respectively) compared to rabbits infected with the wild-type strain (17/38; P < 0.005).
Body weight is another sensitive indicator of illness in the REPEC/rabbit infection model (1, 69), and during the course of infection, rabbits infected with the mutant strains showed normal weight gain, in contrast to those infected with the wild type (Fig. 6). Taken together, these results indicated that the T2SS and its substrate, SslE, are required for the virulence of REPEC strain E22.
Phenotype of sslE and T2SS mutants is independent of the LEE.
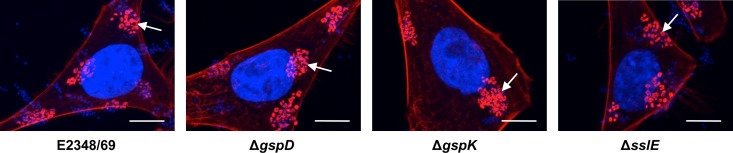
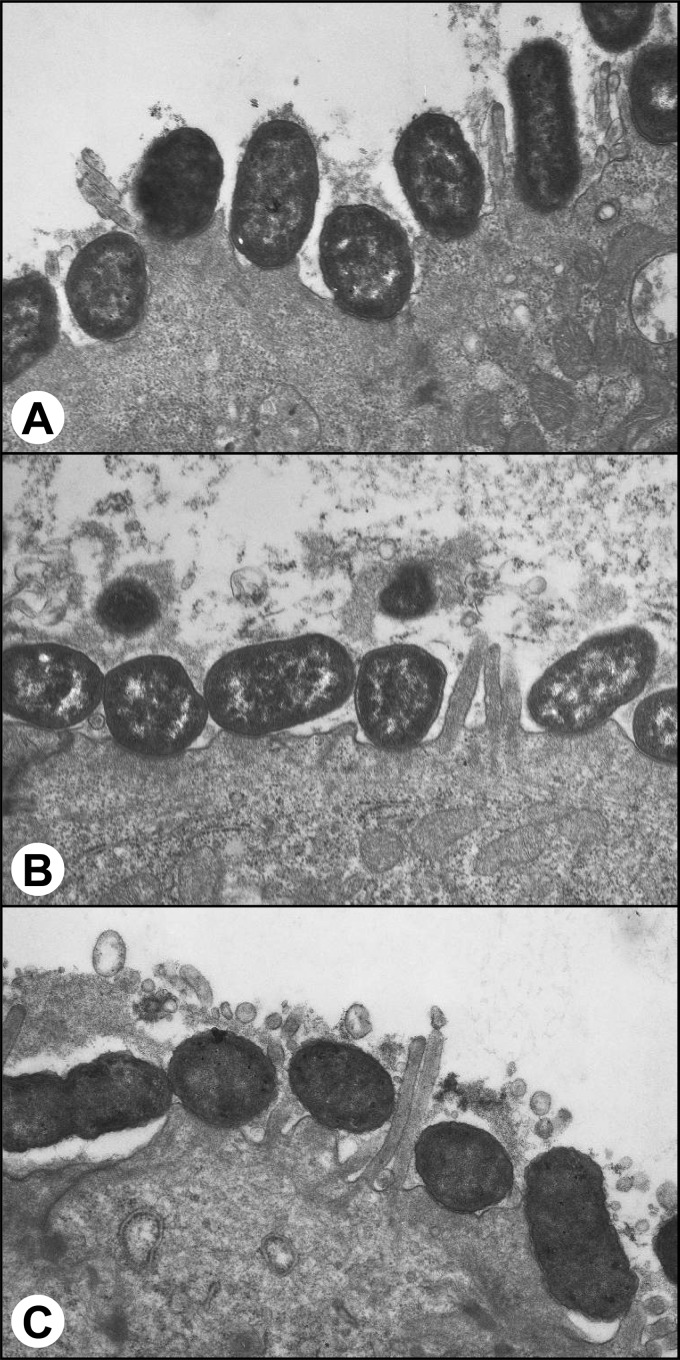
The major virulence factor of EPEC is its ability to produce attaching and effacing lesions in the intestinal epithelium. These are mediated by the products of the genes carried on the LEE. To determine if the phenotypes we observed for the T2SS and sslE mutants of EPEC were directly attributable to the inactivation of these genes and not due to interplay with the LEE or known adherence factors, such as bundle-forming pili, we examined the ability of the T2SS and sslE mutants to elicit fluorescent actin staining (FAS) and to adhere to cultured epithelial cells. FAS is a well-established method to demonstrate the ability of EPEC to evoke attaching and effacing lesions in vitro and is a marker of a functional LEE (36). The results showed that the E2348/69 T2SS and sslE mutants were FAS positive and displayed a localized adherent wild-type phenotype. This indicates that the mutants were not defective in adherence or in expression of the proteins encoded on the LEE (Fig. 7). These data were confirmed by electron microscopic examination of ileal tissue from rabbits which showed that the T2SS and sslE mutants of EPEC strain E22 caused attaching and effacing lesions that were indistinguishable from those elicited by the wild-type strain (Fig. 8).
Fig 7.
Effect of mutations in the T2SS or SslE of E2348/69 on attaching and effacing lesion formation and adherence to HEp-2 cells visualized by using fluorescence actin staining (FAS) and nucleic acid staining, respectively. Representative fluorescent images of HEp-2 cells incubated for 3 h with wild-type E2348/69 or its isogenic mutants, E2348/69ΔgspD, E2348/69ΔgspK (T2SS mutants), and E2348/69ΔsslE, showing the bacteria (blue) adhering to HEp-2 cells (faint red with blue nucleus) in a pattern of localized adherence and FAS-positive lesions (bright red and arrowed) associated with the adherent bacteria. Actin was stained with TRITC-conjugated phalloidin, and bacterial and HEp-2 cell DNA was stained with DAPI. Bar, 10 μm.
Fig 8.
Transmission electron micrographs of sections of rabbit ileum 7 days after inoculation with E. coli E22 (wild type) (A), an isogenic T2SS (ΔgspK) mutant (B), or an isogenic sslE mutant (C). Note the characteristic attaching and effacing lesions caused by all three strains. Original magnification, ×14,000.
DISCUSSION
In this study, we characterized a functional homolog of the T2SSH10407 in EPEC. This T2SS is also present, and highly conserved, in other E. coli pathotypes. Remnants of the T2SSH10407 are also present in E. coli K-12 strains, which possess pppA-gspC and the distal gspL-gspM genes but not the remainder of the operon. Conservation of the entire operon for T2SSH10407 in pathogenic varieties of E. coli suggests that this system performs an essential function: not for planktonic growth, but for pathogenesis. In ETEC, the T2SSH10407 is essential for the secretion of LT, but EPEC and other E. coli pathotypes do not possess LT, suggesting that the T2SSH10407 homologs in these bacteria have alternate substrates.
A proteomics approach revealed that SslE is a major substrate secreted by the T2SS of EPEC strain E2348/69. Use of an LTB reporter, however, demonstrated that this T2SS is capable of secreting other substrates, and we do not rule out that EPEC and other pathotypes of E. coli may secrete other proteins via the T2SS. Nevertheless, several lines of evidence point to SslE as the dominant substrate. First, the sslE gene is located immediately upstream of all sequenced T2SSH10407 operons. We reported previously that the gspCDEFGHIJKLM cluster and three other upstream genes, sslE (yghJ), pppA, and yghG, are cotranscribed and that the promoter of sslE plays a major role in the expression of this 14-gene transcriptional unit (67). Thus, activation of substrate (SslE) expression is coordinated with expression of the entire T2SS. Second, sslE mutants failed to develop as biofilms and showed a reduced ability to colonize the intestine and cause disease in a rabbit model of infection, thus phenocopying type II secretion mutants.
Although the biochemical function of SslE is yet to be determined, this work has established its link to EPEC pathogenesis. Accessory colonization factor D (AcfD), encoded by V. cholerae, is an ortholog of SslE, and we predict that this protein too will be a substrate of the T2SS. Importantly, AcfD also appears to be required for efficient colonization of infant mouse intestine (50).
Unlike V. cholerae where mutations in the T2SS result in membrane perturbation and induction of a σE-mediated stress response (60), our T2SS EPEC mutants showed no signs of growth defects or σE induction, indicating that the attenuated biofilm and virulence phenotypes are not due to diminished bacterial fitness but are caused by the absence of a type II secreted protein(s), specifically SslE.
We also showed that SslE is translocated across the outer membrane via the T2SS and becomes anchored on the cell surface, thus extending the finding of Moriel et al. (47), who used flow cytometry to detect SslE on the surface of an ExPEC strain isolated from a case of neonatal meningitis. The presence of SslE on the cell surface may be important for cell-to-cell or cell-to-surface interactions, which would explain why SslE expression is required for biofilm formation.
Although previously characterized adhesins have been implicated in EPEC biofilms, they contribute to the early stages of biofilm formation. Specifically, type I fimbriae are involved in attachment to the surface of flow cells; also, the LEE-encoded adhesin EspA and the bundle-forming pili of E2348/69 are required for the formation of small microcolonies (46). However, attachment and small microcolony formation are unaffected in T2SS and sslE mutants, demonstrating that the T2SS and SslE are required for biofilm maturation. The involvement of biofilms in EPEC pathogenesis is novel, but not entirely unexpected, as biofilms have been implicated in the pathogenesis of other E. coli pathotypes such as ExPEC (3, 4) and EAEC (45).
The main virulence determinant of EPEC is the LEE; however, inactivation of the T2SS did not affect expression of the LEE proteins. Consequently, the reduced ability of SslE and T2SS mutants of REPEC to cause disease in rabbits may be explained by the inability of these mutants to produce mature biofilms. Biofilm bacteria are resistant to host defenses, as aggregation into biofilms protects them from mechanical removal and phagocytosis and reduces their accessibility to the humoral immune system (16, 17, 27, 28). Consequently, wild-type bacteria may be able to persist due to the formation of mature biofilms, whereas the T2SS and sslE mutants are accessible to immune defenses and are cleared. Interestingly, a recent subtractive reverse vaccinology study selected SslE as one of nine protective antigens in a sepsis mouse challenge model. Active immunization of mice with SslE from ExPEC strain IHE3034 gave 82% protection against systemic infection with the homologous strain, and passive immunization with an SslE antiserum showed almost complete protection against systemic infection, indicating that SslE may be an attractive vaccine target (47).
In conclusion, we have shown that the T2SSH10407 is an important virulence determinant of EPEC. We demonstrated that the T2SS and its substrate, SslE, are essential for the virulence of EPEC for rabbits and that the T2SS and SslE are also required for the formation of mature biofilms, thus suggesting a role for biofilm formation in EPEC pathogenesis. More importantly, as this T2SS and SslE are both highly conserved, they may also play a role in biofilm formation by other E. coli pathotypes, such as ExPEC and EAEC, where biofilm formation is also required for virulence.
ACKNOWLEDGMENTS
This work was supported by Australian National Health and Medical Research Council (NHMRC) project grants (to R.M.R-B., T.L., and M.T.; and to C.B.W., D.L.B., M.T., and L.T.). Both D.L.B. and M.T. were each supported by an NHMRC Peter Doherty Fellowship. C.B.W. was supported by an NHMRC Career Development Award and an NHMRC Senior Research Fellowship, and T.L. is an ARC Federation Fellow.
We thank Sarah Osvath, Danielja Krmek, and Louise Adams for expert technical assistance. Imaging of biofilms was performed at the UTS Microbial Imaging Facility.
Footnotes
Published ahead of print 26 March 2012
REFERENCES
- 1. Abe A, Heczko U, Hegele RG, Brett Finlay B. 1998. Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J. Exp. Med. 188:1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adams LM, Simmons CP, Rezmann L, Strugnell RA, Robins-Browne RM. 1997. Identification and characterization of a K88- and CS31A-like operon of a rabbit enteropathogenic Escherichia coli strain which encodes fimbriae involved in the colonization of rabbit intestine. Infect. Immun. 65:5222–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson GG, Dodson KW, Hooton TM, Hultgren SJ. 2004. Intracellular bacterial communities of uropathogenic Escherichia coli in urinary tract pathogenesis. Trends Microbiol. 12:424–430 [DOI] [PubMed] [Google Scholar]
- 4. Anderson GG, et al. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105–107 [DOI] [PubMed] [Google Scholar]
- 5. Athanasopoulos V, Praszkier J, Pittard AJ. 1995. The replication of an IncL/M plasmid is subject to antisense control. J. Bacteriol. 177:4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ball G, Durand E, Lazdunski A, Filloux A. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475–485 [DOI] [PubMed] [Google Scholar]
- 7. Bartolome B, Jubete Y, Martinez E, de la Cruz F. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75–78 [DOI] [PubMed] [Google Scholar]
- 8. Bolivar F, et al. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113 [PubMed] [Google Scholar]
- 9. Brzuszkiewicz E, et al. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U. S. A. 103:12879–12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541–555 [DOI] [PubMed] [Google Scholar]
- 11. Chalker AF, et al. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chaudhuri RR, et al. 2010. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen SL, et al. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. U. S. A. 103:5977–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clements A, et al. 2009. The reducible complexity of a mitochondrial molecular machine. Proc. Natl. Acad. Sci. U. S. A. 106:15791–15795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 17. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 18. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dean P, Kenny B. 2009. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr. Opin. Microbiol. 12:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Castro E, et al. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34:W362–W365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elliott SJ, et al. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1–4 [DOI] [PubMed] [Google Scholar]
- 22. Evans DG, Evans DJ, Jr, Pierce NF. 1973. Differences in the response of rabbit small intestine to heat-labile and heat-stable enterotoxins of Escherichia coli. Infect. Immun. 7:873–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Evans DJ, Jr, Evans DG, Gorbach SL. 1973. Production of vascular permeability factor by enterotoxigenic Escherichia coli isolated from man. Infect. Immun. 8:725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Francetic O, Belin D, Badaut C, Pugsley AP. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19:6697–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frankel G, et al. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911–921 [DOI] [PubMed] [Google Scholar]
- 26. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108 [DOI] [PubMed] [Google Scholar]
- 28. Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell. Microbiol. 11:1034–1043 [DOI] [PubMed] [Google Scholar]
- 29. Herring CD, Glasner JD, Blattner FR. 2003. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311:153–163 [DOI] [PubMed] [Google Scholar]
- 30. Heydorn A, et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 31. Ho TD, Davis BM, Ritchie JM, Waldor MK. 2008. Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect. Immun. 76:1858–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iguchi A, et al. 2009. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191:347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson TJ, et al. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson TL, Abendroth J, Hol WG, Sandkvist M. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175–186 [DOI] [PubMed] [Google Scholar]
- 35. Juncker AS, et al. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kornacker MG, Boyd A, Pugsley AP, Plastow GS. 1989. Klebsiella pneumoniae strain K21: evidence for the rapid secretion of an unacylated form of pullulanase. Mol. Microbiol. 3:497–503 [DOI] [PubMed] [Google Scholar]
- 38. Kulkarni R, et al. 2009. Roles of putative type II secretion and type IV pilus systems in the virulence of uropathogenic Escherichia coli. PLoS One 4:e4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lathem WW, et al. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45:277–288 [DOI] [PubMed] [Google Scholar]
- 40. Levine MM, et al. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 41. Long-Krug SA, et al. 1984. Does enteropathogenic Escherichia coli produce heat-labile enterotoxin, heat-stable enterotoxins a or b, or cholera toxin A subunits? Infect. Immun. 46:612–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marches O, et al. 2000. Role of Tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 68:2171–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Michaelis S, Chapon C, D'Enfert C, Pugsley AP, Schwartz M. 1985. Characterization and expression of the structural gene for pullulanase, a maltose-inducible secreted protein of Klebsiella pneumoniae. J. Bacteriol. 164:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller JH. 1974. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Mohamed JA, et al. 2007. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J. Clin. Microbiol. 45:121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moreira CG, et al. 2006. Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli. J. Bacteriol. 188:3952–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moriel DG, et al. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nouwen N, et al. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. U. S. A. 96:8173–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ogura Y, et al. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peterson KM, Mekalanos JJ. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56:2822–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Posfai G, Koob MD, Kirkpatrick HA, Blattner FR. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pugsley AP. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Py B, Loiseau L, Barras F. 1999. Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J. Mol. Biol. 289:659–670 [DOI] [PubMed] [Google Scholar]
- 54. Rasko DA, et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Robins-Browne RM. 1987. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev. Infect. Dis. 9:28–53 [DOI] [PubMed] [Google Scholar]
- 56. Robins-Browne RM, Levine MM, Rowe B, Gabriel EM. 1982. Failure to detect conventional enterotoxins in classical enteropathogenic (serotyped) Escherichia coli strains of proven pathogenicity. Infect. Immun. 38:798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 58. Sandkvist M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271–283 [DOI] [PubMed] [Google Scholar]
- 59. Sandkvist M, et al. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sikora AE, Lybarger SR, Sandkvist M. 2007. Compromised outer membrane integrity in Vibrio cholerae type II secretion mutants. J. Bacteriol. 189:8484–8495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:7066–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tauschek M, et al. 2010. Transcriptional analysis of the grlRA virulence operon from Citrobacter rodentium. J. Bacteriol. 192:3722–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Touchon M, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Udekwu KI, Wagner EG. 2007. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 35:1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vignon G, et al. 2003. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J. Bacteriol. 185:3416–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Voulhoux R, et al. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang J, Baldi DL, Tauschek M, Strugnell RA, Robins-Browne RM. 2007. Transcriptional regulation of the yghJ-pppA-yghG-gspCDEFGHIJKLM cluster, encoding the type II secretion pathway in enterotoxigenic Escherichia coli. J. Bacteriol. 189:142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang J, Tauschek M, Strugnell R, Robins-Browne RM. 2005. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology 151:1199–1208 [DOI] [PubMed] [Google Scholar]
- 69. Zhu C, Feng S, Thate TE, Kaper JB, Boedeker EC. 2006. Towards a vaccine for attaching and effacing Escherichia coli: a LEE encoded regulator (ler) mutant of rabbit enteropathogenic Escherichia coli is attenuated, immunogenic, and protects rabbits from lethal challenge with the wild-type virulent strain. Vaccine 24:3845–3855 [DOI] [PubMed] [Google Scholar]