Abstract
6-Nonadecynoic acid (6-NDA), a plant-derived acetylenic acid, exhibits strong inhibitory activity against the human fungal pathogens Candida albicans, Aspergillus fumigatus, and Trichophyton mentagrophytes. In the present study, transcriptional profiling coupled with mutant and biochemical analyses were conducted using the model yeast Saccharomyces cerevisiae to investigate its mechanism of action. 6-NDA elicited a transcriptome response indicative of fatty acid stress, altering the expression of genes that are required for yeast growth in the presence of oleate. Mutants of S. cerevisiae lacking transcription factors that regulate fatty acid β-oxidation showed increased sensitivity to 6-NDA. Fatty acid profile analysis indicated that 6-NDA inhibited the formation of fatty acids longer than 14 carbons in length. In addition, the growth inhibitory effect of 6-NDA was rescued in the presence of exogenously supplied oleate. To investigate the response of a pathogenic fungal species to 6-NDA, transcriptional profiling and biochemical analyses were also conducted in C. albicans. The transcriptional response and fatty acid profile of C. albicans were comparable to those obtained in S. cerevisiae, and the rescue of growth inhibition with exogenous oleate was also observed in C. albicans. In a fluconazole-resistant clinical isolate of C. albicans, a fungicidal effect was produced when fluconazole was combined with 6-NDA. In hyphal growth assays, 6-NDA inhibited the formation of long hyphal filaments in C. albicans. Collectively, our results indicate that the antifungal activity of 6-NDA is mediated by a disruption in fatty acid homeostasis and that 6-NDA has potential utility in the treatment of superficial Candida infections.
INTRODUCTION
Acetylenic acids, a class of fatty acids containing one or more triple bond, are widely distributed in nature and are found in plants, fungi, microbes, and marine organisms (reviewed in reference 3). They have been reported to possess a diversity of pharmacological properties, including antibacterial, antifungal, antiparasitic, and antitumor activities (reviewed in references 6 and 44). We have previously described the antifungal properties of 6-acetylenic acids (triple bond at C-6) that were isolated from the plants Pentagonia gigantifolia and Sommera sabiceoides (21, 22). Among these, 6-nonadecyonic acid (6-NDA), with a chain length of C19 (see the structure shown in Fig. 1A) was the most active, exhibiting strong inhibitory activity against the human fungal pathogens Candida albicans, Aspergillus fumigatus, and Trichophyton mentagrophytes. The MIC values for 6-NDA against these organisms were >100-fold lower than those for undecylenic acid (UDA), a related compound currently used as a topical antifungal agent to treat dermatomycosis and oral thrush (22). 6-NDA also displayed potent activity against patient isolates of C. albicans that are resistant to the widely used antifungal drug fluconazole (21, 22). In addition, 6-NDA demonstrated low in vitro toxicity in mammalian cell lines and also did not produce any obvious toxic effects in mice (22). Thus, 6-NDA is of particular interest as a therapeutic agent, especially as a potential topical antifungal drug candidate for the management of oral candidiasis and dermatomycosis.
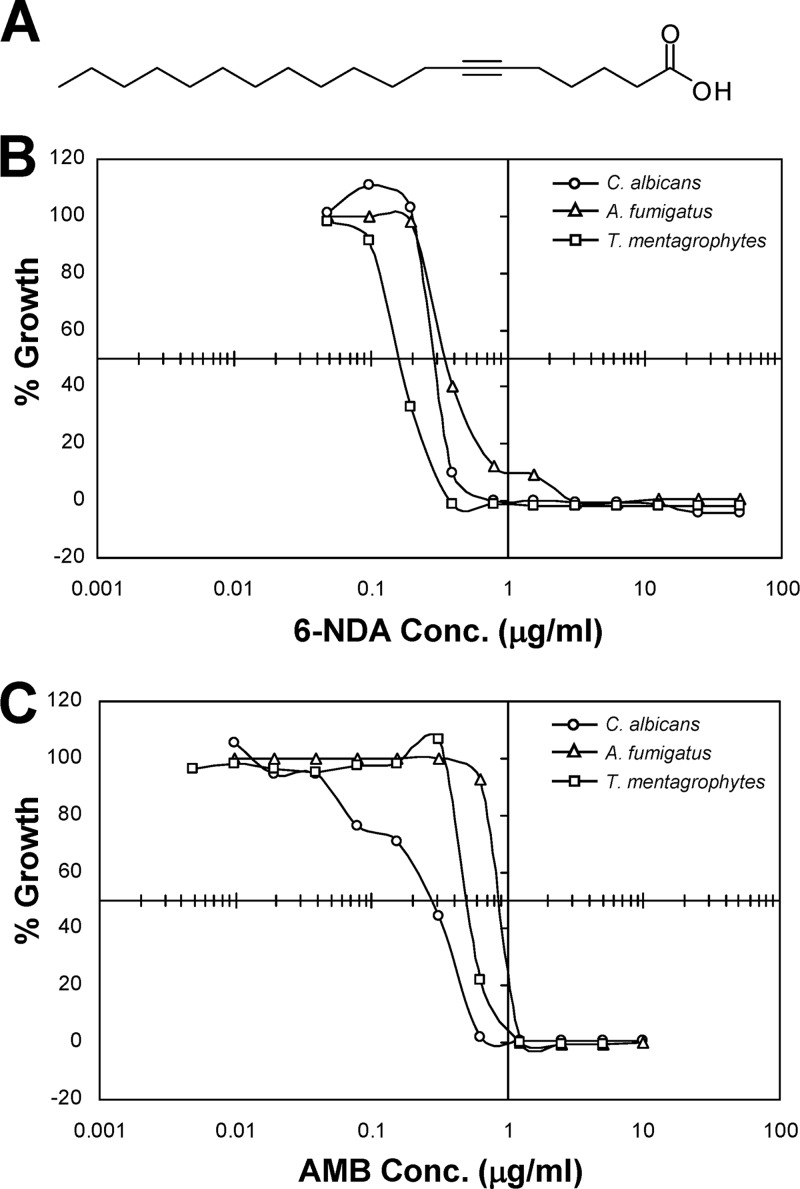
Fig 1.
Structure of 6-NDA and its in vitro antifungal activity compared to amphotericin B. (A) Structure of 6-NDA; (B) dose response curve for 6-NDA; (C) dose response curve for amphotericin B. In panels A and B, broth microdilution assays were performed against the fungal pathogens C. albicans, A. fumigatus, and T. mentagrophytes according to CLSI guidelines (9). The “% Growth” values were calculated from the means from two replicate experiments.
Previous studies have indicated that various mechanisms contribute to the biological activities of acetylenic acids, including the inhibition of (i) fatty acid elongation, (ii) fatty acid synthesis, (iii) fatty acid degradation, (iv) synthesis of eicosanoids (oxygenated derivatives of C20 fatty acids), and (v) DNA synthesis (2, 26, 33, 48, 55). For example, rats fed with 2-hexadecynoic acid (2-HA) overaccumulated palmitate (C16:0) and palmitoleate (C16:1) and showed a corresponding decrease in stearate (C18:0) and oleate (C18:1) (56). Further studies confirmed that 2-HA inhibited the elongation of fatty acids longer than palmitate in rat liver microsomes (55). 2-HA has also been shown to inhibit fatty acid biosynthesis in the malaria parasite Plasmodium falciparum (48). Interestingly, it disrupts not only fatty acid synthesis but also fatty acid degradation in Mycobacterium smegmatis (26). An acetylenic acid with 4 triple bonds, 5,8,11,14-eicosatetraynoic acid, was shown to affect eicosanoid and DNA synthesis in a mammalian cell line (33). Berry et al. (2) have reported that 9-octadecynoic acid binds to DNA and weakly inhibits DNA polymerase activity. Thus, the precise mechanism of action of acetylenic acids is still not clear, and none of the previous work has involved fungal systems. Fungus-specific studies would be particularly useful since fatty acid biosynthesis and degradation in fungi have a few unique features that are different from animal cells; for example, (i) in yeast, two genes instead of one encode the multifunctional type I fatty acid synthase and (ii) fatty acid degradation in yeast cells occurs in the peroxisomes, while the majority of fatty acid breakdown occurs in the mitochondria in mammalian cells (reviewed in references 36 and 49). Thus, further characterization of the mechanism of action of the acetylenic acid compound 6-NDA will not only facilitate its development as an antifungal agent but may also allow the identification of novel fungal target pathways for drug development.
In the present study, we have conducted transcriptional profiling experiments followed by genetic and biochemical analyses to gain insight into the mechanism behind the antifungal activity of 6-NDA. Using Saccharomyces cerevisiae as a model organism, we show that 6-NDA elicits a transcriptome response similar to that obtained when yeast cells are grown in the presence of oleic acid (oleate), causing a significant upregulation of genes involved in peroxisomal functions, especially those required for fatty acid β-oxidation. Mutants of S. cerevisiae lacking transcription factors known to play a role in fatty acid β-oxidation showed increased sensitivity to 6-NDA compared to wild-type cells. Fatty acid profile analysis of S. cerevisiae cells exposed to 6-NDA indicated a significant decrease in the levels of palmitate (C16:0), stearate (C18:0), and oleate (C18:1) and a corresponding increase in laurate (C12:0), myristate (C14:0), and myristoleate (C14:1) levels. In addition, exogenously supplied oleate rescued the inhibitory effect of 6-NDA in S. cerevisiae cells. The human fungal pathogen C. albicans also produced a similar response to 6-NDA, showing a fatty acid profile comparable to that seen in S. cerevisiae and exhibiting reduced sensitivity to 6-NDA upon oleate treatment. In a clinical isolate of C. albicans that is fluconazole resistant, 6-NDA produced a fungicidal effect in combination with fluconazole. Collectively, our results indicate that 6-NDA disrupts fatty acid homeostasis in fungal cells.
MATERIALS AND METHODS
Strains and media.
Synthetic dextrose (SD) medium, containing 2% dextrose and 0.67% yeast nitrogen base without amino acids, was used to grow wild-type S. cerevisiae S288C and C. albicans SC5314 strains. The medium was buffered with 0.165 M 3-(N-morpholino)propanesulfonic acid (MOPS), and the pH was adjusted with NaOH to 7.0 for S. cerevisiae and to 4.5 for C. albicans to maintain yeast morphology. YPD medium (1% yeast extract, 2% peptone, 2% dextrose) at pH 7.0, buffered with MOPS, was used for growing S. cerevisiae deletion mutants obtained from Open Biosystems (Huntsville, AL). The azole-resistant clinical isolate of C. albicans (isolate 17, kindly provided by Theodore C. White) was grown in RPMI 1640 medium, pH 6.0, containing 0.2% dextrose, 0.03% glutamine, and buffered with MOPS. The same medium, adjusted to pH 7.0, was used to grow C. albicans ATCC 90028, A. fumigatus ATCC 9090, and T. mentagrophytes ATCC 9533, with the inclusion of 5% alamarBlue for the latter two strains. Dimethyl sulfoxide (DMSO) and oleate were obtained from Sigma (St. Louis, MO). Fluconazole was obtained from Sequoia Research Products (Pangbourne, United Kingdom). 6-NDA was isolated from the plant S. sabiceoides as described previously (22). All stock solutions were prepared in DMSO.
IC50 determinations for microarray experiments.
S. cerevisiae strain S288C and C. albicans strain SC5314 were used in the microarray experiments, and all procedures, including 50% inhibitory concentration (IC50) determinations, were performed as previously described (1). All experiments were conducted at 30°C for S. cerevisiae and 37°C for C. albicans. For IC50 determinations in small-scale cultures, broth microdilution assays were performed according to the Clinical and Laboratory Standards Institute (CLSI) protocols (9), with the modification that inoculum size was 2 × 106 CFU/ml. This cell density, which is ∼200× greater than the standard protocol, was used in order to mimic the microarray culture conditions.
For determination of IC50s in large-scale cultures, an overnight culture was used to inoculate 50 ml of SD medium to an optical density at 600 nm (OD600) of 0.1 (∼2 × 106 CFU/ml). Multiple cultures were started in order to test 4 to 5 different drug concentrations. The cultures were grown until an OD600 of 0.2 was reached, at which point 6-NDA was added at 2- to 4-fold serial dilutions into each culture. Two rounds of experiments were conducted, with a broad range of 6-NDA concentrations tested in the first round and a narrow range tested in duplicates in the second round. The cultures were grown to late exponential phase (17 h for S. cerevisiae and 9 h for C. albicans), and a final OD600 was measured using an Ultrospec 2000 spectrophotometer (Amersham Biosciences, Piscataway, NJ). The IC50s were determined to be 2.2 μg/ml for S. cerevisiae and 4 μg/ml for C. albicans.
Cell culture and drug exposure for microarray experiments.
Overnight cultures of S. cerevisiae S288C or C. albicans SC5314 were used to inoculate 50 ml of SD medium to an OD600 of 0.1. For S. cerevisiae, four independent 50-ml cultures were grown for each experiment, 2 for treated and 2 for untreated samples; thus, each treatment consisted of 2 biological replicates. For C. albicans, each treatment consisted of 3 biological replicates. The cultures were allowed to recover from stationary phase until an OD600 of 0.2 was reached. 6-NDA was added to each culture at a concentration equivalent to the IC50 (2.2 μg/ml for S. cerevisiae and 4 μg/ml for C. albicans). Control cultures were simultaneously treated with 0.25% DMSO. The cultures were allowed to grow until an OD600 of 0.5 was reached (∼4 h for S. cerevisiae and ∼1.5 h for C. albicans). Cells were harvested by centrifugation, flash frozen in liquid nitrogen, and stored at −80°C.
Microarray hybridization and data analysis.
RNA isolation was performed as described previously (1). For S. cerevisiae, target preparation and hybridization to the Affymetrix GeneChip Yeast Genome 2.0 array were performed according to the Affymetrix GeneChip expression analysis protocol (Affymetrix, Santa Clara, CA) as described previously (1). RNA from each biological replicate sample was hybridized to 2 independent arrays, resulting in 4 hybridizations for each treatment. Image analysis, scaling, and probe set-level data analysis was performed using the default parameters in GeneChip operating software version 1.1 (Affymetrix). For C. albicans, microarrays were constructed from a 70-mer oligonucleotide set (Qiagen, Valencia, CA) which was printed on glass slides (Microarrays Inc., Huntsville, AL). Target preparation and hybridization was performed at the Microarray Core Laboratory of the University of Texas Health Science Center at Houston according to the methods described by Nantel et al. (29). The C. albicans experiment was performed with 3 biological replicate samples and hybridized as dye-swap pairs, resulting in 6 hybridizations for each treatment. Arrays were scanned using the Axon GenePix 4100A scanner (Molecular Devices, Sunnyvale, CA), and image analysis was performed using the Axon GenePix Pro software (Molecular Devices).
Differentially expressed genes were identified using BRB-ArrayTools software developed by Richard Simon and Amy Peng Lam (http://linus.nci.nih.gov/BRB-ArrayTools.html). For S. cerevisiae, data from CHP files generated in GCOS software (background-corrected and normalized signal values after probe set-level data analysis) were used. For C. albicans, data generated in GenePix Pro software (background-corrected signal intensities) were used and normalized using the Lowess algorithm available in BRB-ArrayTools. For both data sets, signal values were log transformed and filtered using BRB-ArrayTools default parameters to remove probe sets or spots with very low signal values, missing signal values, and similar signal values across all the arrays in a given experiment. Differentially expressed genes were identified by performing a two-class comparison between untreated and treated classes using the default statistical parameters available in BRB-ArrayTools, and genes with P values of ≤0.001 were considered to be significant.
Gene annotations were obtained from Saccharomyces Genome Database (SGD) or Candida Genome Database (CGD). Gene Ontology (GO) Term Mapper tool (http://go.princeton.edu/cgi-bin/GOTermMapper) was used to distribute the genes into GO-based biological process categories, and overrepresented GO terms were identified using binomial distribution probability. Hierarchical cluster analysis was performed using Gene Cluster 3.0 (11) using the Pearson correlation similarity metric (uncentered for both genes and arrays) with the average linkage clustering method, and the data were visualized with Java Tree View (40). For comparison, the oleate profile (45) was obtained from the Yeast Functional Genomics Database (YFGdb; http://yfgdb.princeton.edu/).
The transcriptional profiling data described in this article are accessible through accession no. GSE35604 at NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/).
Sensitivity assays.
Sensitivities to 6-NDA of C. albicans ATCC 90028, A. fumigatus ATCC 90906, and T. mentagrophytes ATCC 9533 were assayed as described previously (22). To determine if oleate altered 6-NDA's antifungal activity, broth microdilution assays were performed as follows. An overnight culture started from a single colony of S. cerevisiae S288C or C. albicans SC5314 was diluted in SD medium (identical to the medium used in the microarray experiments) after comparison to the 0.5 McFarland standard to afford a final inoculum of 1 × 104 CFU/ml. After dilution, 180 μl of the inoculum was added to a microplate containing 10 μl of 6-NDA and 10 μl of oleate. The concentration of oleate used was 1.56 μg/ml for S. cerevisiae and 25 μg/ml for C. albicans. At these oleate concentrations, the growth of S. cerevisiae and C. albicans was not inhibited, as determined in pilot checkerboard assays. The microplates were read at 600 nm prior to and after incubation using a BioTek Powerwave XS microplate reader (BioTek Instruments, Winooski, VT). For S. cerevisiae, the temperature of incubation was 30°C, and the time of incubation was 48 h. For C. albicans, the temperature of incubation was 37°C, and the time of incubation was 24 h.
In order to determine if 6-NDA altered the sensitivity of a C. albicans clinical isolate to fluconazole, checkerboard assays were performed according to strict CLSI guidelines (9). Colonies of C. albicans clinical isolate 17 (52), grown on Sabouraud dextrose agar, were resuspended in 0.9% saline and then diluted in RPMI 1640 medium after comparison to the 0.5 McFarland standard to afford a final inoculum of 1.5 × 103 CFU/ml. After dilution, 180 μl of the inoculum was added to a microplate containing 10 μl of 6-NDA and 10 μl of fluconazole at various concentrations. Fluconazole dilutions were placed in consecutive rows of the microplate, and 6-NDA dilutions were placed in consecutive columns. Appropriate controls included solvent control, medium control, a row of only fluconazole dilutions, and a column of only 6-NDA dilutions. The microplate was read at 630 nm prior to and after incubation at 35°C for 48 h using a BioTek Powerwave XS microplate reader. To monitor viability of the cells after drug treatment, the microplate was carefully shaken on a Microplate Genie mixer (Scientific Industries, Bohemia, NY), and a representative aliquot of 2 μl from each well was spotted on a fresh YPD agar plate, which was then incubated at 35°C for 24 h.
To determine the sensitivity of a collection of 166 S. cerevisiae transcription factor deletion mutants against 6-NDA, an agar-based screen was conducted. Single colonies of each mutant were streaked on YPD-7 (MOPS buffered, pH 7.0) agar plates containing either DMSO or 4.5 μg/ml 6-NDA. At this concentration, 6-NDA caused partial growth inhibition of the parental strain BY4742 on YPD-7 agar plates. It should be noted that YPD-7 medium was used for the screen to maintain experimental consistency since all experiments with S. cerevisiae were conducted at pH 7.0. Seven mutants and the parental strain BY4742 were streaked on each screening plate using sterile toothpicks. Plates were incubated at 30°C for 3 days. Strains were identified that showed a marked difference in growth compared with the parental strain on agar plates with 6-NDA but not on plates with DMSO. To confirm the strains identified, agar-based drop-test assays were performed. Overnight cultures at an OD600 of 3.0 were used to prepare 1:5 serial dilutions in YPD-7 medium, which were spotted in 3-μl amounts on YPD-7 agar plates with or without 4.5 μg/ml 6-NDA. Plates were incubated for 2 days at 30°C.
Fatty acid analysis.
Overnight cultures of S. cerevisiae S288C or C. albicans SC5314 were grown in SD broth and used to inoculate fresh medium (10 ml) at an OD600 of 0.1. After one doubling, either 6-NDA (IC50) or DMSO (0.25%) was added to the cultures. The cells were allowed to grow for ∼4.5 doublings (∼15 h for S. cerevisiae and ∼6 h for C. albicans) after treatment (based on DMSO controls, final OD600 of ∼4.0) and harvested by centrifugation. The cells were washed 3 times with sterile distilled water, and cell pellets were flash frozen in liquid nitrogen. The experimental conditions, including media, temperature, aeration, and concentration of 6-NDA, were identical to the conditions used in the microarray experiments. Three independent experiments were performed on independently grown cultures. The cell pellets were processed for gas chromatographic analysis of fatty acid methyl esters (GC-FAME) by Microbial ID, Inc. (Newark, DE) using their established procedures (www.microbialid.com).
Hyphal growth assay.
To examine the effect of 6-NDA on hyphal growth of C. albicans, cells of strain SC5314 were grown overnight in YPD broth, washed with sterile water, and diluted in RPMI 1640 medium (pH 7.0) to afford a final inoculum of 1 × 104 CFU/ml. After dilution, 190 μl of the inoculum was added to a microplate containing 10 μl of 6-NDA at various concentrations (0.195 to 50 μg/ml). After incubation for 6 h at 37°C, cells were visualized by phase-contrast microscopy using an Olympus IX50 inverted microscope (Olympus America Inc., Center Valley, PA) with a 20× objective. Images were obtained with a Nikon DS-Ri1 camera driven by NIS-Elements AR 3.2 software (Nikon Instruments, Inc., Melville, NY).
RESULTS AND DISCUSSION
Transcriptional response to 6-NDA in S. cerevisiae.
In broth microdilution assays, 6-NDA displayed potent antifungal activity, comparable to that of the clinically used drug amphotericin B, against the fungal pathogens C. albicans, A. fumigatus, and T. mentagrophytes (Fig. 1). It showed the strongest activity against T. mentagrophytes, with an IC50 of 0.16 μg/ml, which was approximately 3-fold lower than that obtained with amphotericin B (0.49 μg/ml). While 6-NDA showed similar potency as amphotericin B against C. albicans (IC50 of 0.28 μg/ml for both), its IC50 against A. fumigatus (0.34 μg/ml) was approximately 2.5-fold lower than that of amphotericin B (0.86 μg/ml). To determine the cellular effects of 6-NDA in fungal cells, we made use of S. cerevisiae, a well-established model organism that has been successfully utilized in identifying the molecular pathways targeted by antifungal and therapeutic compounds (34, 47). A transcriptional profiling study was conducted in S. cerevisiae cells (strain S288C) that were exposed for a period of approximately one doubling time (∼4 h) to 2.2 μg/ml of 6-NDA, a concentration resulting in 50% growth inhibition. Genes exhibiting significant differential expression between 6-NDA-treated and solvent-treated cells (P value of ≤0.001, fold change of ≥2) were identified as described in Materials and Methods. A total of 815 6-NDA-responding genes were identified, with 538 showing increased expression and 277 showing decreased expression (see Table S1 in the supplemental material).
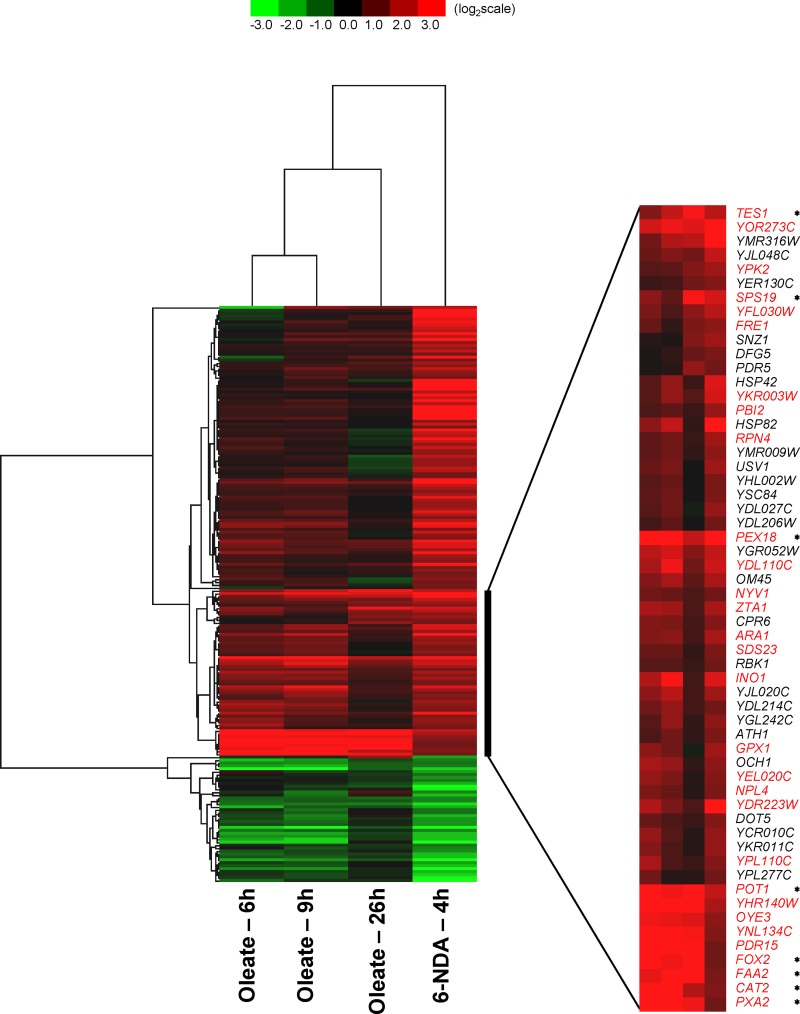
Hierarchical cluster analysis (Fig. 2) revealed significant parallels between the global transcriptional response to 6-NDA and the previously described transcriptional response shown by S. cerevisiae cells grown in the presence of oleate as a sole carbon source (45). Among the 538 genes induced by 6-NDA, 153 of these were also induced by 0.12% oleate (in at least two of the three oleate exposures shown in Fig. 2). Similarly, of the 277 genes repressed by 6-NDA, 44 were also repressed by 0.12% oleate. Of particular significance is a set of 30 genes present within a cluster of 57 genes strongly induced by oleate and 6-NDA (shown in red text in Fig. 2) that are involved in peroxisome function and biogenesis (45). Within this set of 30 genes are present 8 genes (marked by asterisks in Fig. 2) that are known to be regulated by the transcription factors Oaf1 and Pip2 (based on the YEASTRACT database) (50) and that activate the expression of genes required for fatty acid β-oxidation and peroxisomal function (16, 39). A similar induction in peroxisomal genes was observed by Koerkamp et al. (17) when yeast cells were grown on oleate for shorter periods of time within a 90-min time course. Thus, exposure of yeast cells to 6-NDA causes an induction in the expression of peroxisomal genes, many of which are required for fatty acid β-oxidation, as is seen when yeast cells are grown in the presence of oleate.
Fig 2.
Gene expression response to 6-NDA. Data are shown for a subset of 6-NDA-responding genes that are also responsive to oleate treatment (in at least two of the three oleate treatments shown). The oleate profile (described in reference 45) was obtained from the Yeast Functional Genomics Database. Hierarchical cluster analysis was performed using Gene Cluster 3.0, and the data were visualized with Java Tree View. A cluster of 57 6-NDA-induced genes that are also induced by oleate treatment is shown in expanded form to indicate their identities. Genes known to be involved in peroxisome function and biogenesis (described in reference 45) are highlighted in red text. Genes known to be regulated by the fatty acid β-oxidation pathway-activating transcription factors Oaf1 and Pip2 (based on the YEASTRACT database) are marked with asterisks.
The transcriptional responses to 6-NDA were further analyzed by organization into Gene Ontology (GO)-based functional categories using the GO Term Mapper tool (http://go.princeton.edu/cgi-bin/GOTermMapper), as shown in Table 1. Several overrepresented functional categories (P ≤ 0.05) for both 6-NDA-upregulated and -downregulated genes are of particular interest with respect to how yeast cells respond to fatty acid-mediated stress. For example, the enrichment of 6-NDA-responding genes within the functional categories “transport” and “lipid metabolism” is suggestive of membrane dysfunction. Within the lipid metabolism category are present the previously mentioned fatty acid β-oxidation genes (Fig. 2) and also genes involved in phospholipid (INO1, INO2, OPI10, SCS3) and sphingolipid metabolism (IPT1, SUR1, YSR3), indicative of damage to endomembranes such as the endoplasmic reticulum (ER) and Golgi, where these lipids are known to be synthesized. Given that membrane disruption can influence the cell wall, this would explain the induction of cell wall organization genes and the downregulation of conjugation genes in response to 6-NDA (Table 1). As expected, “response to stress” represented an additional major functional category for genes induced in response to 6-NDA toxicity (Table 1). In particular, several genes involved in oxidative stress response were induced by 6-NDA (CTT1, GAD1, GPX1, GRX1, PRX1, SRX1, TRX3, TSA2), a result that is in agreement with the induction of oxidative stress by oleate in yeast cells (17). This induction has been attributed to a redox imbalance in the cells due to a transient uncoupling of the respiratory chain by oleate (17). Additionally, the induction of several autophagy-related genes by 6-NDA (ATG1, ATG5, ATG8, ATG20, ATG29) in the “response to stress” category is significant, given the role of autophagy in the repair of damaged cell membranes (18). Taken together, our results show that there are significant similarities between 6-NDA and oleate transcriptional profiles and that the majority of the transcriptional responses to 6-NDA are due to fatty acid-mediated stress.
Table 1.
Functional distribution of 6-NDA-responding genes in S. cerevisiae
| GO terma | 6-NDA data set frequency (%) | Genome frequency (%) | P value | Genes annotated to the GO termb |
|---|---|---|---|---|
| Upregulated genes | ||||
| Response to stress | 17.29 | 11.04 | 4.53E−06 | ATG1, ATG20, ATG29, ATG5, ATG8, CTT1, DAK1, DDR2, DDR48, GAD1, GPX1, GRE1, GRE3, GRX1, HIM1, HOR2, HSP104, HSP12, HSP26, HSP42, HSP78, HSP82, IMP2', MAG1, ORM2, POL4, PRX1, PTP2, RAD28, REV3, RPH1, RPN4, RTA1, SRX1, SSA1, SSA4, SSE2, STE11, TDP1, TPS1, TPS2, TRX3, TSA2, UBC5, UBI4, XBP1, ZTA1 |
| Carbohydrate metabolism | 7.71 | 5.40 | 5.52E−03 | AMS1, ARA1, ATH1, CAX4, DAK1, ERR3, GCY1, GDE1, GND2, GRE3, GSC2, HOR2, KTR2, MNT4, NTH1, OCH1, PCK1, PFK26, PYK2, RBK1, RMD5, SGA1, SOR2, TKL2, TPS1, TPS2, UBC8, VID28, VID30, YPI1 |
| Cell wall organization | 7.33 | 4.01 | 1.38E−04 | ADY3, CDA1, CHS1, CRH1, CRR1, CWP1, DFG5, DIT1, DIT2, ECM27, ECM4, ECM8, FLC1, FLC2, FMP45, GAS4, GIP1, GSC2, HPF1, HRD1, MPC54, OSW2, PIR3, PST1, RCR1, SED1, SPR28, SPR3, SPS100, SPS22, SVS1, YPK2, YPS3 |
| Lipid metabolism | 5.45 | 3.93 | 1.76E−02 | ATF1, CAT2, FAA2, FOX2, GDE1, INO1, INO2, IPT1, ISC1, IZH2, IZH4, MGA2, OPI10, PLB1, PLB3, POT1, ROG1, SCS3, SPO1, SPS19, SUR1, TES1, TGL2, YDC1, YSR3 |
| Downregulated genes | ||||
| Transport | 26.74 | 20.97 | 4.12E−03 | AGP1, AQR1, ATO3, ATR1, AUS1, CAN1, DAL5, DIP5, DUR3, FCY2, FET3, FTR1, FUI1, GAP1, GNP1, HXT4, LYP1, MCH4, MEP1, MEP2, MEP3, MMP1, MUP1, MUP3, OPT1, OPT2, PDR12, QDR2, SAM3, SEO1, SSU1, SUL2, VHT1, ZRC1, ZRT1, ZRT2 |
| Translation | 19.78 | 16.39 | 2.04E−02 | AGP1, ARG3, CPA2, HOM3, MAK16, MET13, MET14, MET2, MET3, MET32, MUP1, RLI1, RPL18B, RPL22B, RPM2, SAM3, SER33 |
| Amino acid metabolism | 15.75 | 4.99 | 2.3E−11 | AAT1, ACO1, ACO2, ARG3, ARG5,6, ARG8, ASN1, CPA2, DUR1,2, ECM17, GAP1, GCV2, GDH1, GLT1, HIS6, HOM3, LEU9, MET1, MET10, MET13, MET14, MET16, MET2, MET22, MET28, MET3, MET32, MET8, MHT1, SER33, SSU1 |
| Cell cycle | 12.82 | 10.40 | 3.22E−02 | AMN1, BUD9, CDC47, CLB1, CLB6, CSM2, DAD3, DSE1, DSE3, EGT2, FAR1, FKH1, HOF1, KAR4, KAR9, KCC4, MCM3, MCM6, NIS1, NNF1, PAP2, PDS1, RMD6, SDA1, SGO1, SPO16, SPS4, SUN4, TRF5, UME6 |
| Conjugation | 5.86 | 1.93 | 7.2E−05 | AGA1, AGA2, DSE1, FAR1, FIG1, FIG2, FUS1, FUS3, KAR4, MF(ALPHA)2, MFA1, PRM1, PRM2, SAG1, SST2, STE2 |
Data (538 upregulated genes and 277 downregulated genes) were organized into GO-based biological process categories using the GO Term Mapper tool (http://go.princeton.edu/cgi-bin/GOTermMapper). Significantly overrepresented categories (P ≤ 0.05) with a 6-NDA data set frequency of ≥5% are listed.
Within each GO category, a subset of genes are shown that are relevant to the overall biological response to 6-NDA.
Effect of 6-NDA on S. cerevisiae mutants.
To further validate the transcriptional profiling data and to identify key molecular pathways that are required for 6-NDA sensitivity, we analyzed 166 yeast mutants carrying deletions in selected transcription factors (TFs) for their susceptibility to 6-NDA. We selected 135 genes encoding TFs from a list of 203 DNA-binding transcriptional regulators described by Harbison et al. (14) in a study that mapped the DNA sequences bound by these regulators. An additional 31 genes annotated as TFs in the SGD database were also selected due to their role in regulating some of the significant cellular pathways that responded to 6-NDA in our transcriptional profiling study. A complete list of the 166 selected genes is shown in Table S2 in the supplemental material. Since the 166 mutants were generated in the BY4742 parent strain, we conducted a transcriptional profiling study to determine whether this strain responds to 6-NDA in a manner similar to that of the S288C strain. We did indeed find a great deal of similarity between the 6-NDA profiles of the two strains. Of the 460 genes upregulated in the BY4742 strain, 316 were also upregulated in the S288C strain, and of the 128 genes downregulated in the BY4742 strain, 64 were also downregulated in the S288C strain (see Table S3 in the supplemental material). In particular, genes involved in fatty acid β-oxidation were induced by 6-NDA in both strains (see Fig. S1 in the supplemental material). Several additional genes were similarly up- or downregulated in the two strains in various functional categories, including response to stress, carbohydrate metabolism, cell wall organization, lipid metabolism, and cell cycle (see Fig. S1).
Of the 166 mutants analyzed, 11 mutants showed a strong increase in 6-NDA sensitivity and 3 mutants showed a significant decrease in 6-NDA sensitivity compared to the BY4742 parent strain (Fig. 3). All 14 mutants were analyzed by PCR and confirmed to contain deletions in the appropriate genes (see Fig. S2 in the supplemental material). The strong hypersensitivity of the oaf1Δ and pip2Δ strains to 6-NDA (Fig. 3) is consistent with our transcriptional profiling data where we observed an induction of several Oaf1- and Pip2-regulated peroxisomal genes in response to 6-NDA treatment (see Fig. 2). This result is in agreement with a previous study (23), which showed that growth of the oaf1Δ mutant as well as several peroxisomal mutants was strongly inhibited on medium that contained 0.1% oleate as the carbon source compared to growth on medium containing 3% glycerol as the carbon source.
Fig 3.
Effect of 6-NDA on S. cerevisiae transcription factor mutants. The parent strain (BY4742) and mutant strains harboring deletions in 14 different transcription factors genes were grown as described in Materials and Methods. Dilutions (5-fold) were prepared from each culture, inoculated onto YPD agar (MOPS buffered, pH 7.0), and incubated for 2 days at 30°C. Mutants known to have altered sensitivity to oleate (described in reference 23) are marked with asterisks. −6-NDA, medium containing solvent (DMSO); +6-NDA, medium containing 6-NDA at 4.5 μg/ml.
Three additional mutants that showed increased sensitivity to oleate in the Lockshon et al. (23) study also showed increased sensitivity to 6-NDA (Fig. 3). First, hypersensitivity to 6-NDA and oleate (23) was observed in the mutant lacking Rpn4 (Fig. 3), a TF that interacts with Snf1, an AMP-activated protein kinase that regulates glucose-repressed genes as well as genes required for various cellular processes, including peroxisome biogenesis. We also observed 6-NDA hypersensitivity in the mutant lacking Sip3 (Fig. 3), another Snf1-interacting protein. Second, the mutant deficient in Skn7, which plays a role in oxidative stress response and osmoregulation, also showed hypersensitivity to 6-NDA (Fig. 3) and oleate (23). Similarly, increased 6-NDA sensitivity was seen in the mutant strain with a deletion in Yap3 (Fig. 3), another TF involved in osmoregulation. Third, increased sensitivity to 6-NDA and oleate (23) was seen in the mutant deficient in Ppr1 (Fig. 3), a TF involved in the regulation of pyrimidine metabolism. In contrast, the mutant lacking Hir2, a TF that regulates histone gene transcription, showed decreased sensitivity to both 6-NDA (Fig. 3) and oleate (23). Our analysis also showed that deletions in two additional TFs, Hir1 and Hir3, involved in histone gene transcription resulted in decreased sensitivity to 6-NDA (Fig. 3). In addition to the above-mentioned mutants, we also detected increased 6-NDA sensitivity in mutants lacking Ndt80 and Pog1 (Fig. 3), TFs involved in regulating cell cycle-related genes, many of which showed altered expression in the presence of 6-NDA (see Table 1). Finally, we observed strong hypersensitivity to 6-NDA in mutants with deletions in Gcr2 and Sok2 (Fig. 3), TFs that regulate glycolytic genes and cAMP-protein kinase A signal transduction, respectively. Given that glycolytic genes are downregulated in response to oleate (17) and that the cAMP protein kinase A pathway is required for nutrient sensing in yeast (58), it is possible that Gcr2 and Sok2 play important regulatory functions in fatty acid utilization. Taken together, these results further confirm that the molecular pathways involved in mediating sensitivity to 6-NDA are also required for oleate sensitivity and that the majority of these pathways are required for tolerating the consequences of fatty acid-imposed cellular stress.
Effect of 6-NDA on the fatty acid profile of S. cerevisiae.
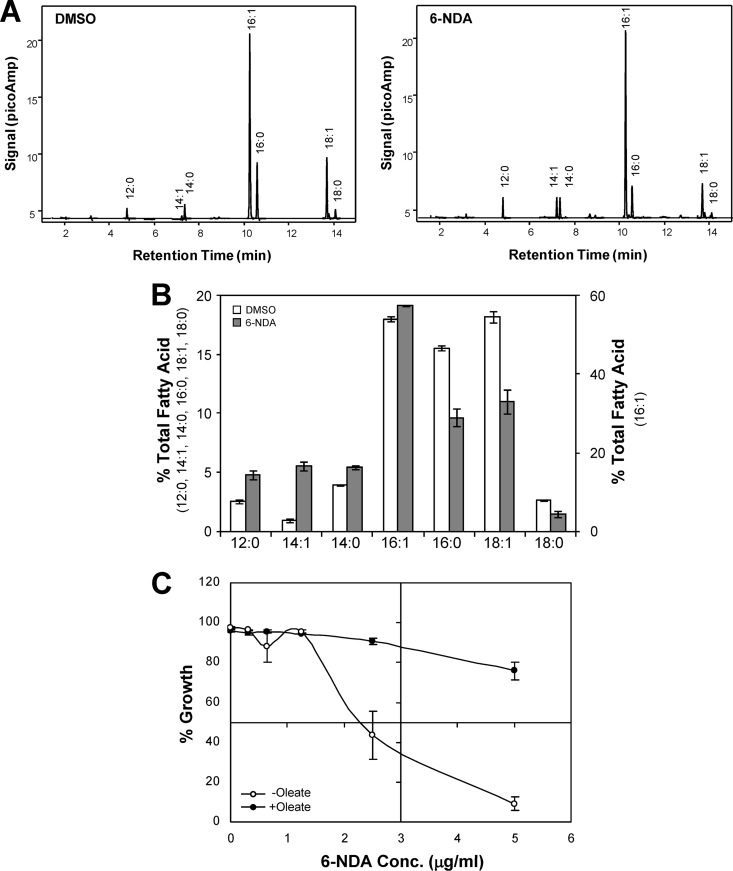
Previous studies have shown that acetylenic acids such as 2-HA inhibit fatty acid synthesis or elongation in prokaryotic as well as eukaryotic cells (26, 48, 55). To determine if 6-NDA similarly affects fatty acid metabolism in yeast cells, we investigated the effect of 6-NDA on the fatty acid profile of S. cerevisiae. Treatment with 6-NDA resulted in a significant decrease in the levels of palmitate (C16:0), stearate (C18:0), and oleate (C18:1) and a corresponding increase in the levels of laurate (C12:0), myristate (C14:0), and myristoleate (C14:1) in wild-type S. cerevisiae cells (Fig. 4A and B). The amounts of palmitate, stearate, and oleate decreased by 38%, 44%, and 39%, respectively, and the amount of laurate, myristate, and myristoleate increased by 90%, 36%, and 482%, respectively, in 6-NDA-treated cells relative to DMSO-treated cells. These results are comparable to a previous study in which rats fed with 2-HA overaccumulated palmitate and palmitoleate and showed a corresponding decrease in the levels of stearate and oleate (56). Follow-up biochemical studies demonstrated that 2-HA inhibited the elongation of fatty acids longer than palmitate in rat liver microsomes (55). Collectively, the results shown in Fig. 4A and B suggest that 6-NDA could similarly inhibit the elongation of fatty acids longer than myristate in yeast cells, although the slight increase in palmitoleate (C16:1) levels observed would appear to be somewhat counter to this notion. Palmitoleate, however, comprises the majority of unsaturated fatty acids found within cellular membranes of several yeast strains (including S288C) and likely plays a critical role in adaptation to stress (10, 13, 24). Therefore, the possibility cannot be discounted that regulatory pathways exist for the maintenance of adequate palmitoleate levels during stress exposure, thus compensating for the inhibitory effects of 6-NDA.
Fig 4.
Effect of 6-NDA on fatty acid homeostasis in S. cerevisiae. (A) Cells of S. cerevisiae strain S288C were grown under experimental conditions identical to those used for microarray analysis. Cells were treated with either DMSO (0.25%) or 6-NDA (2.2 μg/ml) for ∼15 h, and their fatty acid profile was determined by GC-FAME analysis. Three independent experiments were performed on independently grown cultures. Gas chromatograms from one representative experiment are shown. (B) Percent total fatty acid values shown are the means from the three experiments ± standard deviations. The chain length and level of desaturation of the fatty acids detected are shown on the x axis. The differences in fatty acid levels between DMSO-treated and 6-NDA-treated samples were statistically significant (P value of <0.05) based on an independent two-tailed t test, with equal variance. (C) Broth microdilution assays were performed in triplicate using S. cerevisiae strain S288C grown in the presence of various concentrations of 6-NDA, with 1.56 μg/ml oleate (+oleate) or without oleate (−oleate). Percent growth is shown as the mean ± standard error of the mean.
It is worth noting that unlike mammalian cells which synthesize palmitate in the cytoplasm during de novo fatty acid synthesis and further elongate palmitate to stearate using elongases in the endoplasmic reticulum (ER), yeast cells can synthesize both palmitate and stearate in the cytoplasm via fatty acid synthase, which has the ability to elongate C14 and C16 fatty acids (reviewed in reference 49). Thus, it is possible that 6-NDA could target the elongation function of fatty acid synthase. Interestingly, S. cerevisiae can also elongate medium-chain fatty acids using the ER-localized enzyme Elo1, which is responsible for the specific elongation of myristate to palmitate (51). Therefore, another possibility is that 6-NDA might inhibit the function of the Elo1 enzyme. This potential inhibitory effect on two different enzymes is reminiscent of the mechanism of action of the fatty acid synthase inhibitor cerulenin and its analogs. While the primary target of cerulenin is fatty acid synthase, analogs of cerulenin that contain 16 or 18 carbon atoms are also capable of inhibiting ER-localized plant elongases (41). The activity of the analogs is dependent upon their differential partitioning between the cytoplasm and internal membranes, and a similar partitioning of 6-NDA in S. cerevisiae cells could allow it to inhibit fatty acid elongation via the cytoplasmic fatty acid synthase and/or an ER-localized elongase.
Given that 6-NDA caused a significant increase in the levels of laurate, myristate, and myristoleate (90%, 36%, and 482% increase, respectively), it is possible that this overaccumulation of C12 and C14 fatty acids could also play a potential role in the induction of fatty acid β-oxidation genes in our transcriptional profiling analysis (see Table 1 and Fig. 2). Similarly, this excessive buildup of C12 and C14 fatty acids could also contribute, in part, to the 6-NDA hypersensitivity of mutants lacking TFs that regulate fatty acid β-oxidation (see Fig. 3). Interestingly, short- and medium-chain fatty acids that are prematurely released from fatty acid synthase are directed toward the peroxisomal β-oxidation pathway in S. cerevisiae (25).
Since 6-NDA treatment resulted in a 39% decrease in the levels of oleate, one of the most abundant fatty acids in yeast cells, we were also interested in determining whether exogenously supplied oleate would restore growth in 6-NDA-inhibited cells. To address this question, checkerboard assays were first performed using cells exposed to a range of 6-NDA (0.078 to 5 μg/ml) and oleate (0.039 to 100 μg/ml) concentrations to determine whether conditions could be identified for the rescue of yeast cells. Significant restoration of growth by oleate was indicated in these preliminary tests, with maximal rescue effects occurring at 1.56 μg/ml oleate, and oleate was not found to be inhibitory at concentrations of ≤3.12 μg/ml. We therefore performed broth microdilution assays using oleate at a concentration of 1.56 μg/ml to analyze the effect of oleate on 6-NDA activity in more detail. As shown in Fig. 4C, growth inhibition by 6-NDA was significantly reversed in the presence of oleate at this concentration. For example, yeast cells treated with 5 μg/ml of 6-NDA exhibited 9% growth compared to DMSO-treated cells; however, the presence of oleate restored their growth to 76% (Fig. 4C). It is worth noting that the growth effects against S. cerevisiae of the fatty acid synthase inhibitor cerulenin can be overcome by the addition of fatty acids such as oleate to the medium (reviewed in reference 49). Similarly, yeast mutants lacking either of the two subunits of fatty acid synthase, Fas1 or Fas2, are inviable on standard media but can be rescued by supplementation with long-chain fatty acids such as oleate (reviewed in reference 49). Taken together, the results shown in Fig. 4 indicate that 6-NDA prevents the formation of long-chain fatty acids (>14 carbons) and disrupts fatty acid homeostasis in yeast cells.
Effect of 6-NDA on the fatty acid profile of C. albicans.
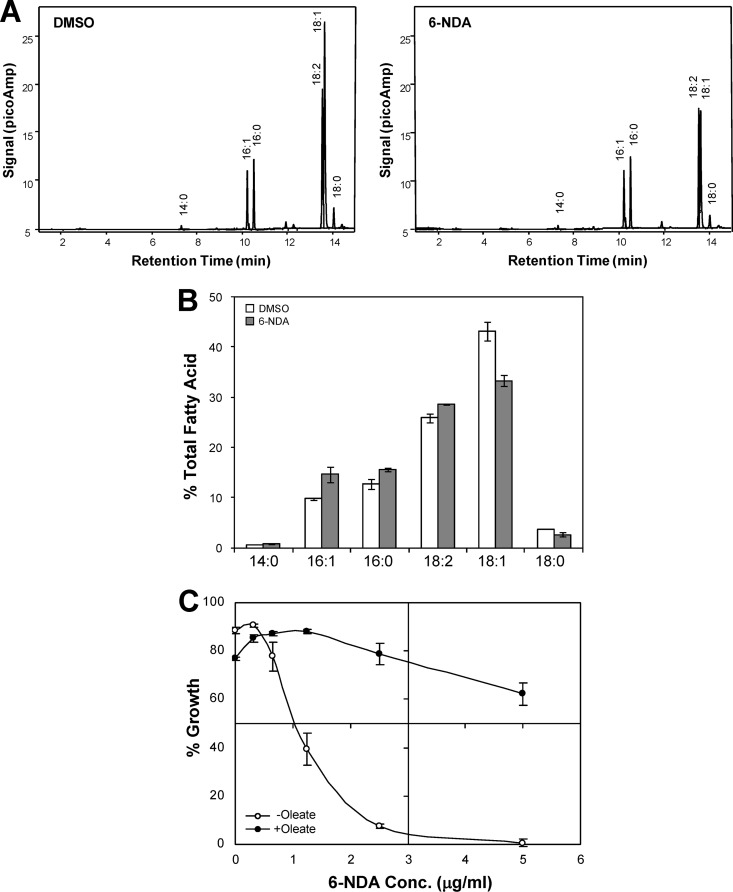
To extend our work from a model organism such as S. cerevisiae to a fungal pathogen, we also conducted biochemical and genomic studies in C. albicans, which is the most frequently encountered Candida species in opportunistic infections in immunocompromised hosts as well as in hospital-acquired bloodstream infections (20, 27, 54). First, to determine how 6-NDA affects fatty acid metabolism in C. albicans, we examined the effect of 6-NDA on its fatty acid profile. Treatment of C. albicans cells with 6-NDA resulted in a significant decrease in the levels of stearate (C18:0) and oleate (C18:1) and a corresponding increase in the levels of palmitate (C16:0) and palmitoleate (C16:1) (Fig. 5A and B). The amounts of stearate and oleate decreased by 26% and 23%, respectively, and the amounts of palmitate and palmitoleate increased by 22% and 49%, respectively, in 6-NDA-treated cells relative to those of DMSO-treated cells. This result is in agreement with previous studies in which 2-HA caused a decrease in stearate and oleate levels in rats and also led to a simultaneous overaccumulation of palmitate and palmitoleate (55, 56). Thus, similar to the effects of 2-HA in mammalian cells, 6-NDA may inhibit elongation of fatty acids longer than palmitate in C. albicans. However, as was also the case in S. cerevisiae with palmitoleate (C16:1) increasing slightly following 6-NDA exposure (Fig. 4B), a small increase in the level of linoleate (C18:2) was actually observed (Fig. 5B), counter to what would be predicted if 6-NDA were to inhibit the formation of fatty acids longer than palmitate in C. albicans. As discussed above, this increase in linoleate could be attributed to compensatory mechanisms counteracting the effects of 6-NDA, which could be associated with, for example, the maintenance of optimal membrane fluidity under unfavorable environmental conditions. Prior studies with C. albicans have shown that cells grown at 25°C (a condition that decreases membrane fluidity) contain a larger proportion of linoleate in their total lipids than cells grown at 37°C (4), thus linoleate may play a particularly critical role in adaptive responses in C. albicans.
Fig 5.
Effect of 6-NDA on fatty acid homeostasis in C. albicans. (A) Cells of C. albicans strain SC5314 were grown under experimental conditions identical to those used for microarray analysis. Cells were treated with either DMSO (0.25%) or 6-NDA (4 μg/ml) for ∼6 h, and their fatty acid profile was determined by GC-FAME analysis. Three independent experiments were performed on independently grown cultures. Gas chromatograms from one representative experiment are shown. (B) Percent total fatty acid values shown are means of the three experiments ± standard deviations. The chain length and level of desaturation of the fatty acids detected are shown on the x axis. The differences in fatty acid levels between DMSO-treated and 6-NDA-treated samples were statistically significant (P value of <0.05) based on an independent two-tailed t test, with equal variance. (C) Broth microdilution assays were performed in triplicate using C. albicans strain SC5314 grown in the presence of various concentrations of 6-NDA, with 25 μg/ml oleate (+oleate) or without oleate (−oleate). Percent growth is shown as the mean ± standard error of the mean.
The results shown in Fig. 5B further suggest that unlike S. cerevisiae, fatty acid synthesis in C. albicans is more similar to mammalian systems, with palmitate being the end product of de novo fatty acid synthesis and further elongation taking place in the ER. Interestingly, conditional shutoff strains of C. albicans for the genes encoding fatty acid synthase subunits Fas1 and Fas2 are inviable under standard growth conditions (57). The growth of these strains is restored only by palmitate and not by stearate, suggesting that the C. albicans fatty acid synthase is responsible for the synthesis of palmitate, and further conversion of palmitate to stearate is catalyzed by other enzymes (57).
Since 6-NDA exposure resulted in a 23% decrease in the levels of oleate in C. albicans cells, we also examined if exogenous oleate would rescue the growth inhibitory effects of 6-NDA. As described above for S. cerevisiae (Fig. 4C), checkerboard assays were first performed to determine appropriate oleate concentrations that could produce a rescue effect in C. albicans. Significant restoration of growth by oleate was also indicated for C. albicans, with maximal rescue occurring at 25 μg/ml oleate. Furthermore, exogenous oleate was not found to be inhibitory to the growth of C. albicans at any of the concentrations tested (up to 100 μg/ml). Broth microdilution assays were therefore performed using oleate at a concentration of 25 μg/ml to analyze the effect of oleate on 6-NDA activity in C. albicans. Similar to our observations in S. cerevisiae, growth inhibition by 6-NDA was significantly reversed in the presence of oleate in C. albicans cells (Fig. 5C). For example, C. albicans cells treated with 5 μg/ml of 6-NDA exhibited 0.6% growth compared to DMSO-treated cells; however, the presence of oleate restored their growth to 62% (Fig. 5C). Taken together, the results shown in Fig. 5 indicate that 6-NDA prevents the formation of long-chain fatty acids (>16 carbons) and disrupts fatty acid homeostasis in C. albicans cells. It is important to note that the observed rescue effect by exogenously provided oleate in C. albicans does imply that 6-NDA may not be suitable for treating systemic infections involving this pathogen, given that the lipid-rich environment in the host may suppress 6-NDA's antifungal activity in vivo. 6-NDA may, however, have considerable efficacy for the treatment of superficial Candida infections, such as oral thrush and vaginitis, and its potential clinical applications along these lines merit further investigation.
Interestingly, the optimal concentration of oleate required for rescuing 6-NDA activity in C. albicans was substantially higher than that required in S. cerevisiae (Fig. 4C and 5C), suggesting that S. cerevisiae is more sensitive to external oleate than C. albicans. It is possible that specific aspects of fatty acid synthesis and utilization in the two organisms could contribute to this effect. For example, in S. cerevisiae, the addition of exogenous fatty acids such as oleate reduces the enzyme activity of acetyl coenzyme A (acetyl-CoA) carboxylase, which catalyzes the first step in fatty acid biosynthesis (15). Oleate also represses the transcription of OLE1, which codes for Δ-9 desaturase, the enzyme responsible for the synthesis of monounsaturated fatty acids in S. cerevisiae (8). Thus, these effects of oleate on fatty acid homeostasis may explain why S. cerevisiae cells cannot tolerate high levels of external oleate. The reduced oleate sensitivity of C. albicans could be due to the fact that this organism can utilize diverse lipids as carbon sources, including saturated and unsaturated fatty acids, complex lipids such as olive oil, and polyethoxylene sorbate (Tween) compounds (38). Given that C. albicans frequently encounters carbon-poor conditions in the host, it has developed strategies to efficiently utilize alternative carbon sources for its survival, which could explain, at least in part, its relative insensitivity to exogenous oleate (37).
Transcriptional response to 6-NDA in C. albicans.
To further explore the cellular effects of 6-NDA in C. albicans, we performed a transcriptional profiling study in this organism. As described above for the S. cerevisiae study, C. albicans cells were treated with a concentration of 6-NDA that resulted in 50% growth inhibition (4 μg/ml) for a period of approximately one doubling time (∼1.5 h). Genes that exhibited significant differential expression between cells treated with 6-NDA and those treated with solvent (P value of ≤0.001, fold change of ≥2) were identified as described in Materials and Methods. A total of 410 genes showed expression changes in response to 6-NDA, with 139 showing increased expression and 271 showing decreased expression (see Table S4 in the supplemental material). The transcriptional changes to 6-NDA were analyzed by organizing the responding genes into GO-based functional categories using the GO Term Mapper tool (http://go.princeton.edu/cgi-bin/GOTermMapper), as shown in Table 2.
Table 2.
Functional distribution of 6-NDA-responding genes in C. albicans
| GO terma | 6-NDA data set frequency (%) | Genome frequency (%) | P value | Genes annotated to the GO termb |
|---|---|---|---|---|
| Upregulated genes | ||||
| Response to stress | 10.48 | 8.59 | 4.57E−02 | CAT1, CDR1, DDR48, GAL10, HAC1, NCE103, RPN4, RTA3, SOD2, SOD4, SNQ2, SVF1, orf19.2175, orf19.2458, orf19.251 |
| Carbohydrate metabolism | 8.87 | 4.58 | 1.57E−02 | ADH2, AMS1, CWH8, GAL10, GCA1, INO1, MNN1, PCK1, UTR2, orf19.3994, orf19.753 |
| Lipid metabolism | 6.45 | 4.21 | 2.11E−02 | AUR1, CWH8, GIT1, INO1, LAG1, MIT1, PLB1, SEC14, orf19.6007, orf19.7441 |
| Downregulated genes | ||||
| Amino acid metabolism | 22.44 | 3.77 | 1.19E−27 | AAT1, ARG4, ARG8, ARO1, ARO3, ARO4, ARO8, ASN1, GCV2, GCV3, HIS1, HIS4, HIS5, HIS7, HOM2, HOM3, HOM6, ILV1, ILV2, ILV3, ILV6, LEU4, LEU42, LYS1, LYS2, LYS4, MET13, MET2, MET6, MIS11, PRO1, PRO2, SAM4, SER1, SER33, STR2, THR1, TRP2, TRP3, TRP4, TRP5, URA2, VAS1 |
| Translation | 20.08 | 6.71 | 1.43E−12 | RPL10A, RPL11, RPL13, RPL14, RPL15A, RPL16A, RPL18, RPL19A, RPL2, RPL20B, RPL21A, RPL24A, RPL25, RPL27A, RPL28, RPL3, RPL32, RPL37B, RPL43A, RPL4B, RPL5, RPL9B, RPP1A, RPP2A, RPP2B, RPS13, RPS14B, RPS15, RPS17B, RPS19A, RPS21, RPS22A, RPS23A, RPS24, RPS27, RPS28B, RPS3, RPS7A |
| Chromosome organization | 6.69 | 4.93 | 4.65E−02 | HHF1, HHF22, HHO1, HHT2, HHT21, HTA1, HTA2, HTB1, NAT4, orf19.1052 |
| Lipid metabolism | 4.72 | 4.21 | 4.71E−02 | ACS1, ATF1, ERG1, ERG3, ERG5, ERG6, ERG9, ERG11, ERG13, ERG251, FET3, HMO1, YMC1, orf19.85, orf19.4581 |
Data (139 upregulated genes and 271 downregulated genes) were organized into GO-based biological process categories using the GO Term Mapper tool (http://go.princeton.edu/cgi-bin/GOTermMapper). Significantly overrepresented categories (P ≤ 0.05) with a 6-NDA data set frequency of ≥4.5% are listed.
Within each GO category, a subset of genes are shown that are relevant to the overall biological response to 6-NDA.
As was observed with S. cerevisiae, several genes in the lipid metabolism functional category were induced by 6-NDA in C. albicans, most likely due to 6-NDA's disruptive effect on fatty acid homeostasis (Table 2). Within the lipid metabolism category, genes involved in phospholipid and sphingolipid biosynthesis were induced, indicating damage to endomembranes such as ER and Golgi. However, unlike the response observed in S. cerevisiae, genes involved in fatty acid β-oxidation were not induced in response to 6-NDA in C. albicans cells. This raises the question of whether this induction may require a longer exposure to fatty acid stress in C. albicans since cells were exposed to 6-NDA for only one doubling time for the transcriptional profiling experiments. To examine this possibility, we treated C. albicans cells with 6-NDA at the IC50 for one doubling time (OD600 of 0.4), two doubling times (OD600 of 0.8), and four doubling times (OD600 of 3.2), and RNA isolated from these samples was subjected to quantitative real-time reverse transcription (RT)-PCR (see Methods in the supplemental material for further details). The genes analyzed consisted of five genes involved in fatty acid β-oxidation (FAA2, POX1, FOX2, POT1, and TES1) and one gene involved in sphingolipid biosynthesis (LAG1), included as a positive control since it was 5.3-fold induced by 6-NDA in the one doubling microarray experiment (see Table 2 and Table S4 in the supplemental material). The primer sequences of the genes analyzed are shown in Table S5 in the supplemental material. While the LAG1 gene did show significant induction at the first two time points, none of the β-oxidation genes tested were significantly induced at any of the time points tested (see Fig. S3 in the supplemental material). It is possible that differences in the regulatory networks required for fatty acid metabolism between C. albicans and S. cerevisiae may account for the discrepancy in the β-oxidation gene response to 6-NDA in the two organisms. Transcription factors required for regulating fatty acid metabolism in S. cerevisiae (Oaf1, Pip2, Adr1, and Cat8) are either absent or have divergent functions in C. albicans. For example, Oaf1 and Pip2 are missing, and Adr1 and Cat8 do not affect the expression of key β-oxidation genes in C. albicans (38). Another possibility is that the large overaccumulation of C12 and C14 fatty acids (see Fig. 4B) likely makes a significant contribution to the induction of β-oxidation genes in 6-NDA-exposed S. cerevisiae cells. In contrast, 6-NDA treatment in C. albicans cells caused a small increase in the levels of C16 fatty acids (see Fig. 5B), which is perhaps not sufficient for induction of the β-oxidation pathway in this organism.
Genes involved in amino acid metabolism and translation were downregulated in response to 6-NDA in C. albicans (Table 2). Several genes in these two functional categories were also downregulated by 6-NDA in S. cerevisiae (Table 1). Interestingly, genes in these functional categories are simultaneously downregulated in C. albicans cells in response to ER stress mediated by tunicamycin and dithiothreitol (53). Thus, this response is consistent with the inhibitory effect of 6-NDA on fatty acid homeostasis and consequently on endomembranes such as the ER. Finally, 6-NDA treatment resulted in the downregulation of 8 genes involved in ergosterol biosynthesis in C. albicans (Table 2, lipid metabolism category). Three ergosterol biosynthetic genes (ERG3, ERG4, and ERG5) were also downregulated in response to 6-NDA in S. cerevisiae (see Table S1 in the supplemental material). Given that ergosterol biosynthesis occurs in the ER, it is possible that this pathway is affected due to 6-NDA-mediated ER damage. It is also possible that the disruption in fatty acid homeostasis caused by 6-NDA could have an effect on sterol homeostasis given that acetyl-CoA is used as a common precursor in both pathways.
Effect of 6-NDA on fluconazole activity.
Since 6-NDA affected the expression of ergosterol biosynthetic genes in C. albicans, we were also interested in determining if 6-NDA could improve the activity of the fungistatic drug fluconazole, which inhibits ergosterol biosynthesis. A broth microdilution checkerboard assay was performed according to CLSI guidelines (9) using a fluconazole-resistant clinical isolate (isolate 17) of C. albicans (52). The presence of 6-NDA improved fluconazole activity in cells of this clinical isolate; for example, the cells exhibited >80% growth in the presence of 200 μg/ml fluconazole, and the addition of a subinhibitory dose of 6-NDA (1.48 μg/ml) resulted in no visible growth (Fig. 6A). More importantly, when aliquots of these cells were spotted on a fresh YPD agar plate to assess whether the cells were viable, cells with no visible growth at high fluconazole concentrations (200 μg/ml and 66.66 μg/ml) in the presence of 6-NDA exhibited no growth after 24 h of incubation on the solid medium (Fig. 6B). Thus, the combination of 6-NDA and fluconazole exhibited fungicidal activity in this clinical isolate of C. albicans.
Fig 6.
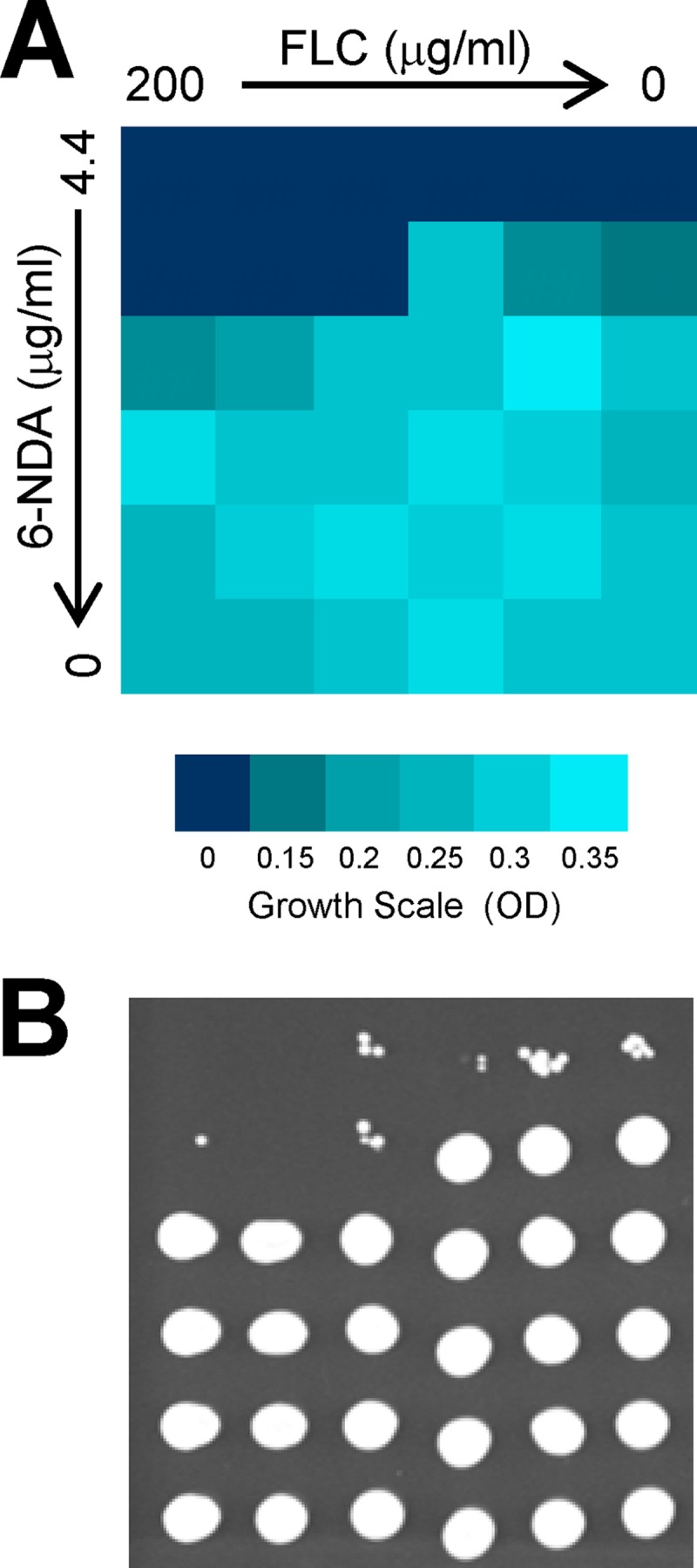
Effect of 6-NDA on fluconazole activity in a clinical isolate of C. albicans. (A) A checkerboard assay was performed to evaluate the combined effects of 6-NDA and fluconazole in a fluconazole-resistant clinical isolate (isolate 17) of C. albicans (described in reference 52). Cells were grown in the presence of 3-fold serially diluted concentrations of 6-NDA and fluconazole for 48 h at 35°C, and OD readings were measured on a microplate reader. (B) To assess viability, 2-μl aliquots from each well were spotted on a fresh YPD agar plate, which was incubated for 24 h at 35°C.
It is worth noting that the 6-NDA-mediated improvement in fluconazole activity was observed only in the fluconazole-resistant clinical isolate of C. albicans and was not observed in the matched fluconazole-susceptible clinical isolate (isolate 1), nor was it observed in the wild-type C. albicans strain SC5314 (data not shown). Thus, the mechanism behind this improvement in fluconazole activity by 6-NDA is probably not related to a general membrane stress that would be exerted due to a disruption in fatty acid homeostasis. This effect is also most likely not due to an alteration in ergosterol levels because of the downregulation of ergosterol biosynthetic genes by 6-NDA. Instead, it could be related to the fact that one of the key molecular factors responsible for fluconazole resistance in clinical isolate 17 is the overexpression of Cdr1, a member of the ABC transporter family of efflux pumps that is involved in the efflux of azole drugs from C. albicans cells (52). Interestingly, Cdr1 is sensitive to changes in the membrane environment (35, 46). For example, Cdr1 function was inhibited when it was expressed in yeast cells with defects in ergosterol biosynthesis (46). In addition, Cdr1 localization to the plasma membrane was disrupted when it was expressed in yeast cells defective either in the ergosterol or in the sphingolipid biosynthesis pathway (35). A similar disruption in Cdr1 function or localization may occur in the presence of 6-NDA causing the resistant clinical isolate to become susceptible to fluconazole.
Effect of 6-NDA on C. albicans hyphal growth.
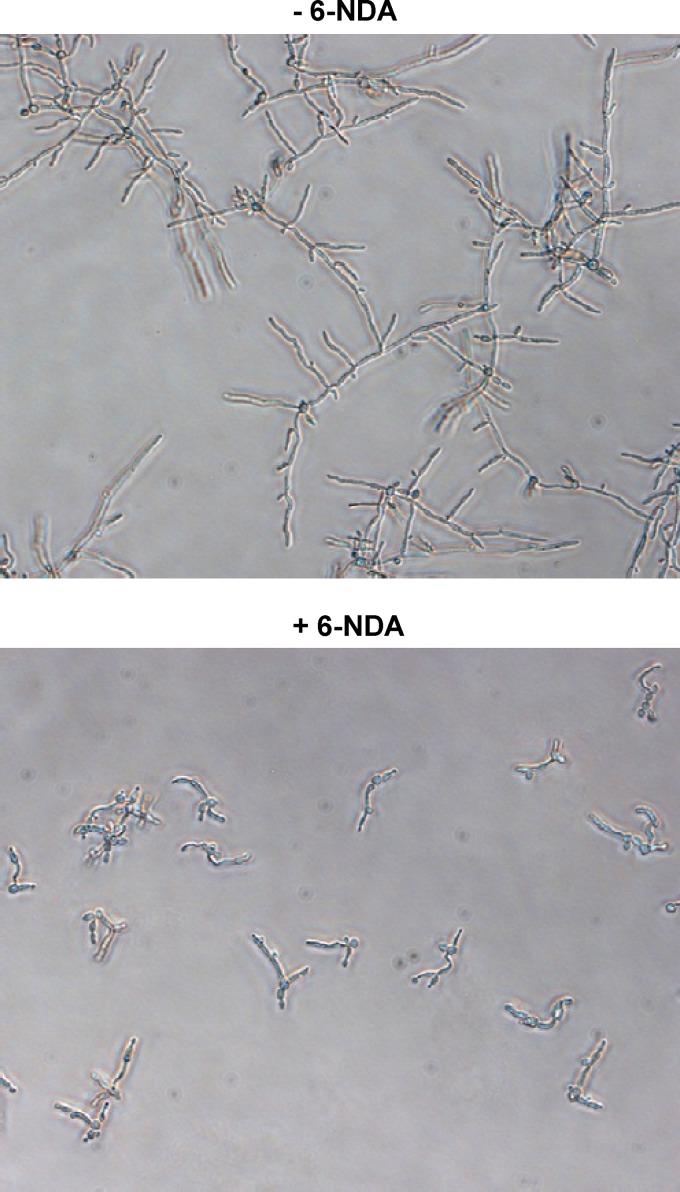
C. albicans has the capacity to exist in both yeast and filamentous forms, and the morphogenetic switch from yeast to hyphae is a major virulence factor, important for both tissue adhesion and invasion (reviewed in reference 5). Since fatty acids have been previously shown to inhibit hyphal formation in C. albicans (reviewed in reference 42), we conducted experiments to determine whether 6-NDA has any effect on C. albicans hyphal growth. C. albicans cells were grown in RPMI 1640 medium (pH 7.0) at 37°C for 6 h in the presence of various concentrations of 6-NDA (0.195 to 50 μg/ml). At all concentrations tested, 6-NDA treatment resulted in the formation of short hyphal filaments, whereas long filaments were seen in solvent control samples. Even at the lowest test concentration of 0.195 μg/ml, a strong inhibition of long hyphae formation was observed (Fig. 7). Unlike other fatty acids such as butyric, capric, lauric, palmitoleic, oleic, linoleic, and undecylenic acids (reviewed in reference 42), 6-NDA did not appear to inhibit yeast-to-hypha transition under the experimental conditions used in our study, since short hyphal filaments were visible in the majority of cells in each of the 6-NDA-treated samples. Thus, it is possible that 6-NDA inhibits hyphal elongation as has been observed for conjugated linoleic acid and caprylic acid (28, 43). Since inhibitors of hyphal growth such as farnesol and nisin Z have been shown to give a protective effect in mucosal candidiasis (reviewed in reference 42), the hyphal inhibitory activity of 6-NDA would be useful in clearing superficial Candida infections. In addition, given the importance of hyphal development in C. albicans biofilm formation for the colonization of medical devices (32), and given that Candida biofilms are highly resistant to most antifungal agents (reviewed in reference 12), 6-NDA may also have potential utility in the treatment of device-related Candida infections.
Fig 7.
Effect of 6-NDA on C. albicans hyphal growth. Cells of C. albicans strain SC5314 were grown in microplates in RPMI 1640 medium (pH 7.0) for 6 h at 37°C in the absence or presence of various concentrations of 6-NDA (0.195 to 50 μg/ml). While long hyphal filaments were visible in the DMSO-treated samples, only short filaments were visible in all of the 6-NDA-treated samples. Digital images were captured with a 20× objective. −6-NDA, medium containing solvent (DMSO); +6-NDA, medium containing 6-NDA at 0.195 μg/ml.
Conclusions.
Using a combination of genomic, genetic, and biochemical approaches, we have shown that 6-NDA, a plant-derived acetylenic acid, mediates its antifungal activity by interfering with fatty acid homeostasis, a molecular pathway distinct from that targeted by antifungal drugs currently in clinical use. Given the requirement for the accurate regulation of intracellular fatty acid levels for fungal cell viability and pathogenesis (7, 19, 30, 31, 57, 59), fatty acid homeostasis represents a promising target pathway for antifungal drug development. The acetylenic acid 6-NDA could therefore serve as a valuable tool for the further characterization of this cellular process as an antifungal target, as well as a novel pharmacological probe for exploring fatty acid homeostasis in fungal cells and gaining insight into its regulation. Our work also shows that 6-NDA abolishes azole resistance in a clinical isolate of the leading human fungal pathogen, C. albicans, causing a fungicidal effect in combination with fluconazole in this isolate. In addition, 6-NDA inhibited C. albicans hyphal growth, thus there is potential utility for this compound in the treatment of mucosal as well as device-related Candida infections.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants from the Public Health Service, National Institute of Allergy and Infectious Diseases, grant no. R01 AI27094 and R21 AI67873, and the USDA-ARS specific cooperative agreement no. 58-6408-2-0009.
Footnotes
Published ahead of print 19 March 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Agarwal AK, et al. 2008. Role of heme in the antifungal activity of the azaoxoaporphine alkaloid sampangine. Eukaryot. Cell 7:387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berry DE, Chan JA, MacKenzie L, Hecht SM. 1991. 9-Octadecynoic acid: a novel DNA binding agent. Chem. Res. Toxicol. 4:195–198 [DOI] [PubMed] [Google Scholar]
- 3. Bohlmann F. 1988. Naturally occurring acetylenes. Bioactive Mol. 7:1–19 [Google Scholar]
- 4. Brondz I, Olsen I. 1990. Fatty acid contents of the yeast and mycelial phase of Candida albicans. J. Chromatogr. 533:152–158 [DOI] [PubMed] [Google Scholar]
- 5. Calderone RA, Fonzi WA. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327–335 [DOI] [PubMed] [Google Scholar]
- 6. Carballeira NM. 2008. New advances in fatty acids as antimalarial, antimycobacterial and antifungal agents. Prog. Lipid Res. 47:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chayakulkeeree M, Rude TH, Toffaletti DL, Perfect JR. 2007. Fatty acid synthesis is essential for survival of Cryptococcus neoformans and a potential fungicidal target. Antimicrob. Agents Chemother. 51:3537–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi JY, Stukey J, Hwang SY, Martin CE. 1996. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem. 271:3581–3589 [DOI] [PubMed] [Google Scholar]
- 9. CLSI 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A3, 3rd ed Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Daum G, et al. 1999. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast 15:601–614 [DOI] [PubMed] [Google Scholar]
- 11. de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454 [DOI] [PubMed] [Google Scholar]
- 12. d'Enfert C. 2006. Biofilms and their role in the resistance of pathogenic Candida to antifungal agents. Curr. Drug Targets 7:465–470 [DOI] [PubMed] [Google Scholar]
- 13. Ding MZ, Tian HC, Cheng JS, Yuan YJ. 2009. Inoculum size-dependent interactive regulation of metabolism and stress response of Saccharomyces cerevisiae revealed by comparative metabolomics. J. Biotechnol. 144:279–286 [DOI] [PubMed] [Google Scholar]
- 14. Harbison CT, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamiryo T, Parthasarathy S, Numa S. 1976. Evidence that acyl coenzyme A synthetase activity is required for repression of yeast acetyl coenzyme A carboxylase by exogenous fatty acids. Proc. Natl. Acad. Sci. U. S. A. 73:386–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karpichev IV, Small GM. 1998. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol. Cell Biol. 18:6560–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koerkamp MG, et al. 2002. Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays. Mol. Biol. Cell 13:2783–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kourtis N, Tavernarakis N. 2009. Autophagy and cell death in model organisms. Cell Death Differ. 16:21–30 [DOI] [PubMed] [Google Scholar]
- 19. Krishnamurthy S, et al. 2004. Dosage-dependent functions of fatty acid desaturase Ole1p in growth and morphogenesis of Candida albicans. Microbiology 150:1991–2003 [DOI] [PubMed] [Google Scholar]
- 20. Lewis RE. 2009. Overview of the changing epidemiology of candidemia. Curr. Med. Res. Opin. 25:1732–1740 [DOI] [PubMed] [Google Scholar]
- 21. Li XC, et al. 2003. Acetylenic acids inhibiting azole-resistant Candida albicans from Pentagonia gigantifolia. J. Nat. Prod. 66:1132–1135 [DOI] [PubMed] [Google Scholar]
- 22. Li XC, et al. 2008. Potent in vitro antifungal activities of naturally occurring acetylenic acids. Antimicrob. Agents Chemother. 52:2442–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lockshon D, Surface LE, Kerr EO, Kaeberlein M, Kennedy BK. 2007. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics 175:77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mannazzu I, et al. 2008. Behaviour of Saccharomyces cerevisiae wine strains during adaptation to unfavourable conditions of fermentation on synthetic medium: cell lipid composition, membrane integrity, viability and fermentative activity. Int. J. Food Microbiol. 121:84–91 [DOI] [PubMed] [Google Scholar]
- 25. Marchesini S, Poirier Y. 2003. Futile cycling of intermediates of fatty acid biosynthesis toward peroxisomal beta-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 278:32596–32601 [DOI] [PubMed] [Google Scholar]
- 26. Morbidoni HR, et al. 2006. Dual inhibition of mycobacterial fatty acid biosynthesis and degradation by 2-alkynoic acids. Chem. Biol. 13:297–307 [DOI] [PubMed] [Google Scholar]
- 27. Morgan J. 2005. Global trends in candidemia: review of reports from 1995–2005. Curr. Infect. Dis. Rep. 7:429–439 [DOI] [PubMed] [Google Scholar]
- 28. Murzyn A, Krasowska A, Stefanowicz P, Dziadkowiec D, Lukaszewicz M. 2010. Capric acid secreted by S. boulardii inhibits C. albicans filamentous growth, adhesion and biofilm formation. PLoS One 5:e12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nantel A, Rigby T, Hogues H, Whiteway M. 2006. Microarrays for studying pathogenicity in Candida albicans. Wiley Press, Hoboken, NJ [Google Scholar]
- 30. Nguyen LN, Gacser A, Nosanchuk JD. 2011. The stearoyl-coenzyme A desaturase 1 is essential for virulence and membrane stress in Candida parapsilosis through unsaturated fatty acid production. Infect. Immun. 79:136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen LN, Trofa D, Nosanchuk JD. 2009. Fatty acid synthase impacts the pathobiology of Candida parapsilosis in vitro and during mammalian infection. PLoS One 4:e8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nobile CJ, Nett JE, Andes DR, Mitchell AP. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ondrey F, Harris JE, Anderson KM. 1989. Inhibition of U937 eicosanoid and DNA synthesis by 5,8,11,14-eicosatetraynoic acid, an inhibitor of arachidonic acid metabolism and its partial reversal by leukotriene C4. Cancer Res. 49:1138–1142 [PubMed] [Google Scholar]
- 34. Parsons AB, Geyer R, Hughes TR, Boone C. 2003. Yeast genomics and proteomics in drug discovery and target validation. Prog. Cell Cycle Res. 5:159–166 [PubMed] [Google Scholar]
- 35. Pasrija R, Panwar SL, Prasad R. 2008. Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob. Agents Chemother. 52:694–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK. 2006. Peroxisomal beta-oxidation—a metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763:1413–1426 [DOI] [PubMed] [Google Scholar]
- 37. Ramírez MA, Lorenz MC. 2007. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot. Cell 6:280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramírez MA, Lorenz MC. 2009. The transcription factor homolog CTF1 regulates β-oxidation in Candida albicans. Eukaryot. Cell 8:1604–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rottensteiner H, Kal AJ, Hamilton B, Ruis H, Tabak HF. 1997. A heterodimer of the Zn2Cys6 transcription factors Pip2p and Oaf1p controls induction of genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Eur. J. Biochem. 247:776–783 [DOI] [PubMed] [Google Scholar]
- 40. Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248 [DOI] [PubMed] [Google Scholar]
- 41. Schneider F, Cassagne C. 1995. Specific inhibition of plant fatty acid elongation by a long-chain cerulenin analogue. Eur. J. Biochem. 228:704–709 [DOI] [PubMed] [Google Scholar]
- 42. Shareck J, Belhumeur P. 2011. Modulation of morphogenesis in Candida albicans by various small molecules. Eukaryot. Cell 10:1004–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shareck J, Nantel A, Belhumeur P. 2011. Conjugated linoleic acid inhibits hyphal growth in Candida albicans by modulating Ras1p cellular levels and downregulating TEC1 expression. Eukaryot. Cell 10:565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siddiq A, Dembitsky V. 2008. Acetylenic anticancer agents. Anticancer Agents Med. Chem. 8:132–170 [DOI] [PubMed] [Google Scholar]
- 45. Smith JJ, et al. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smriti Krishnamurthy SS, Prasad R. 1999. Membrane fluidity affects functions of Cdr1p, a multidrug ABC transporter of Candida albicans. FEMS Microbiol. Lett. 173:475–481 [DOI] [PubMed] [Google Scholar]
- 47. Sturgeon CM, Kemmer D, Anderson HJ, Roberge M. 2006. Yeast as a tool to uncover the cellular targets of drugs. Biotechnol. J. 1:289–298 [DOI] [PubMed] [Google Scholar]
- 48. Tasdemir D, et al. 2010. 2-Hexadecynoic acid inhibits plasmodial FAS-II enzymes and arrests erythrocytic and liver stage Plasmodium infections. Bioorg. Med. Chem. 18:7475–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tehlivets O, Scheuringer K, Kohlwein SD. 2007. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta 1771:255–270 [DOI] [PubMed] [Google Scholar]
- 50. Teixeira MC, et al. 2006. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 34:D446–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Toke DA, Martin CE. 1996. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J. Biol. Chem. 271:18413–18422 [DOI] [PubMed] [Google Scholar]
- 52. White TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wimalasena TT, et al. 2008. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet. Biol. 45:1235–1247 [DOI] [PubMed] [Google Scholar]
- 54. Wisplinghoff H, et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 55. Wood R, Lee T. 1981. Metabolism of 2-hexadecynoate and inhibition of fatty acid elongation. J. Biol. Chem. 256:12379–12386 [PubMed] [Google Scholar]
- 56. Wood R, Lee T, Gershon H. 1980. Effect of methyl 2-hexadecynoate on hepatic fatty acid metabolism. Lipids 15:141–150 [DOI] [PubMed] [Google Scholar]
- 57. Xu D, et al. 2009. Chemical genetic profiling and characterization of small-molecule compounds that affect the biosynthesis of unsaturated fatty acids in Candida albicans. J. Biol. Chem. 284:19754–19764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zaman S, Lippman SI, Zhao X, Broach JR. 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42:27–81 [DOI] [PubMed] [Google Scholar]
- 59. Zhao XJ, McElhaney-Feser GE, Sheridan MJ, Broedel SE, Jr, Cihlar RL. 1997. Avirulence of Candida albicans FAS2 mutants in a mouse model of systemic candidiasis. Infect. Immun. 65:829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.