Abstract
Bacterial gene regulation involves transcription factors (TF) that bind to DNA recognition sequences in operon promoters. These recognition sequences, many of which are palindromic, are known as regulatory elements or transcription factor binding sites (TFBS). Some TFs are global regulators that can modulate the expression of hundreds of genes. In this study we examine global regulator half-sites, where a half-site, which we shall call a binding motif (BM), is one half of a palindromic TFBS. We explore the hypothesis that the number of BMs plays an important role in transcriptional regulation, examining empirical data from transcriptional profiling of the CRP and ArcA regulons. We compare the power of BM counts and of full TFBS characteristics to predict induced transcriptional activity. We find that CRP BM counts have a nonlinear effect on CRP-dependent transcriptional activity and predict this activity better than full TFBS quality or location.
Keywords: transcriptional regulation, transcription factors, binding sites, binding motifs, Escherichia coli, Shewanella oneidensis, CRP, Cyclic-AMP receptor protein, ArcA
Background
The ability of bacteria to adjust gene activity due to variations of environmental stimuli is a critical element of efficient bacterial adaptation. The binding of transcription factors (TFs) to their cognate sites in promoters is the most common cellular mechanism for regulating gene expression in response to stimuli. This regulatory mechanism can induce, using the same TF, a variety of transcriptional activity across the genome. Many global regulators, including CRP (cAMP Receptor Protein), ArcA, and FNR, can simultaneously promote or suppress transcriptional activity on dozens or hundreds of genes. It is believed that different transcriptional activity results from differences in promoter characteristics, such as the location of transcription factor binding sites (TFBSs), their orientation, or their similarity to TFBS consensus sequences.1–3 The molecular mechanisms underlying this quantitative effect are not fully understood. Although a variety of bioinformatic and empirical approaches to TFBS identification and influence, applicable to prokaryotes, have been developed,4–9 models do not yet exist that quantify the level of gene expression due to characteristics of a TFBS in a prokaryotic gene promoter.
In lower eukaryotes, progress in large-scale quantitative modeling of gene expression level based on computationally predicted TFBSs has been made. These models take into consideration not simply the presence or absence of the TFBS in the gene promoter, but also the number of TFBSs. This modeling approach has been applied to establish relationships between mRNA expression levels and TFBS mappings for over a hundred different conditions and different TFs in the yeast Saccharomyces cerevisiae.10–12 The approach is based on Jacob and Monod’s model of transcriptional regulation, which assumes that the log-transformed expression level is the sum of the products of the binding strength of each motif and the activity of its corresponding TF,13 ie, the effect of the TF on gene transcription linearly increases with the number of TFBSs. Recent advances in understanding of gene regulation in eukaryotes confirms the effect of BS copy numbers on the gene transcriptional activity even at locations distant from the gene promoter.14 The effect of the number of BSs on gene expression has been also reported in bacteria. Evidence in support of this effect comes from studies of CRP, a global regulator involved in switching between aerobic and anaerobic metabolism in Escherichia coli (E. coli). CRP can exert a transcriptional effect when binding to DNA at positions that are distant from the RNA polymerase BS.15 Additionally, the binding of two CRP molecules to different BSs in a gene promoter increases the level of gene transcription. Specifically, transcription initiation by CRP at either a class I or a class II promoter can be enhanced by a second CRP molecule bound upstream.16 Thus, an additional BS in a bacterial gene promoter may increase not only the probability of TF binding, but it may also enhance transcription initiation at the promoter.
Modeling the effect of the number of BSs on gene transcription in prokaryotes is complicated by the greater length of bacterial TFBSs relative to eukaryotic TFBSs. Genome mapping of TFBSs in eukaryotes indicates that TFs can bind short stretches of DNA, regulatory motifs, in gene promoters to modulate transcription, 17 and that one promoter may have several BSs for the same TF. An examination of sets of experimentally verified18 and computationally predicted19 TFBSs in yeast suggests an average size of eight for eukaryotic TFBSs. Consensus sequences of bacterial TFBSs are comparatively long. The average binding site in RegulonDB has 17 nucleotides, more than twice as many as the average eukaryotic BS. Many bacterial transcription factors are dimeric proteins, and it is generally believed that their TFBSs must be palindromic or symmetrical. For CRP, the most studied bacterial TF, the TFBS consensus sequence (5′-AAATGTGATCTAGATCACATTT-3′) is palindromic with the consensus half site 5′-A1 A2 A3T4G5T6 G7A8T9C10T11. As a rule, however, position weights of the half sites are not equal, which suggests a dominating transcriptional effect for the half with greater weight and an auxiliary effect for the other half. It is also known that the core three or four bases of the half BS consensus sequence are usually the most conserved and the most important for TF binding. In the case of CRP, for example, the protein makes direct contact only with base pairs G:C5, G:C7, and A:T8 in the core motif T4G5T6G7A8.20,21 The flanking bases are recognized indirectly. Indeed, all experimentally confirmed CRP binding sites in the E. coli genome, listed in either RegulonDB22 or in EcoCyc,23 have mismatches when compared to the consensus sequence. In fact, a perfect match between a TFBS and the consensus sequence may not be biologically useful, since it would result in a very strong affinity. In E. coli, for example, CRP binds very tightly (with long dissociation time) to BSs that closely correspond to the consensus.24 These observations suggest that only one monomer of the CRP dimers may bind to short sequences that match a part of the consensus. Specifically, we suggest that a conserved core motif flanked by weak bases, which represents half of the TFBS and which we will designate a Binding Motif (BM) to distinguish it from the long symmetrical binding site (BS), can exert a biologically relevant effect on gene transcription. We further suggest that the number of BMs in a bacterial promoter may provide a molecular mechanism for quantitative adjustment of the transcriptional effect, and, as in lower eukaryotes, may be used for predictive modeling of transcriptional effects of bacterial regulators.
In this study we examine the relationship between BM counts and transcription level using a variety of experimental data from four previously published microarray experiments designed to identify regulatory networks of two global bacterial regulators, CRP and ArcA, in two organisms, Shewanella oneidenses MR-1 (MR-1) and E. coli. The experiments compared the level of gene expression in the wild type strain and in the regulator negative mutant strain. Both strains were grown in conditions where transcriptional effects of the regulators are crucial for bacterial adaptation, namely, a shift from aerobic to anaerobic respiration and stress imposed by addition of isobutanol.2,25–27 A gene was considered regulated (directly or indirectly) by ArcA or CRP in the studies if its level of expression was significantly different between the wild type strain and the regulator knockout. We propose that the inferred level of change in gene expression is proportional to the transcriptional effect of the studied TF on gene activity. We will refer to these experimentally defined changes in gene expression as Transcription Factor Induced Gene Activity or TF IGA. We use TF IGA as the metric to examine whether the transcriptional activity of a gene correlates with number of short BMs of the TF in the gene promoter and with other known modulators, including the quality of the BS and the BS location relative to transcription start. We find that the number of CRP or ArcA BMs in gene promoters has a statistically significant effect on CRP or ArcA dependent transcriptional activity of genes. This effect of BM counts is nonlinear in the case of CRP and correlates with CRP IGA better than either symmetrical BS quality or BS location. Using step-wise regression, we consider the synergetic effects of CRP, IHF (Integration Host Factor), and ArcA BM counts on CRP IGA. We find a negative effect of ArcA BM counts on CRP IGA, independent of CRP BM counts, and a positive, synergetic effect of IHF BM counts and CRP BM counts. To explain these results, we propose a model that involves control of gene expression through DNA bending by CRP and IHF.
Results
Counts of CRP or ArcA BMs in gene promoters have a nonlinear effect on CRP or ArcA dependent transcriptional activity of genes in MR-1
The effect of the number of TF BMs on TF IGA was evaluated using four different datasets as described in the Methods section. The largest microarray dataset was from a study of a crp− mutant of MR-1 and its wild-type strain, available in the Shewanella Knowledgebase.28 CRP plays a major role in the regulation of anaerobic respiration in MR-1, in addition to its role in catabolic repression and in utilization of carbon sources. It activates hundreds of genes involved in anaerobic metabolism.28,29 In this study of the transition from aerobic growth with lactate to anaerobic growth with lactate and fumarate, CRP IGA was calculated for various time points: 0, 20, 40, 60, 90, 120 min, 4, 8, 12, 24 h, steady-state. This dataset includes 655 genes putatively up-regulated by CRP and 632 genes putatively down-regulated by CRP (Supplementary Table S1). Analysis of these genes affected directly or indirectly by CRP indicated low, but statistically significant correlation (Table 2) between BM counts in the upstream intergenic region of the gene, which for convenience we will call the Gene Promoter, and the CRP IGA. Correlation levels for up-regulated genes were consistent at each time point and across all time spans, including at time zero (R = 0.20, P = 2.55 × 10−8), during the first hour (first four time points) of the experiment (R = 0.21, P = 4.16 × 10−9), and across all time points (R = 0.20, P = 1.73 × 10−8). Correlation levels for down-regulated genes were lower (R = −0.10, P = 1.32 × 10−3 across all time points), but similarly consistent. An analysis that considered the density of BM counts in the gene promoter, that is, the BM counts divided by the promoter length, found no significant correlation (R = 0.06, P = 0.077) with CRP IGA in up-regulated genes.
Table 2.
Statistical characterization of the modulating effects of TF BS or BM counts in the gene promoter on the induced changes in gene expression.
| TF | Org | Technology | Design and conditions | Modulating factor | Reg | Max BS | Num genes | R | P-value |
|---|---|---|---|---|---|---|---|---|---|
| ArcA | MR-1 | Homemade oligonucleotide microarray | arcA+ vs. arcA− anaerobic growth | BM counts in gene promoters |  |
18 | 306 | 0.26 | 4.05 × 10−6 |
| ArcA | MR-1 | “ | “ | “ |  |
18 | 339 | −0.19 | 7.32 × 10−4 |
| ArcA | MR-1 | “ | arcA+ vs. arcA− aerobic growth | “ |  |
18 | 317 | 0.16 | 3.24 × 10−3 |
| ArcA | MR-1 | “ | “ | “ |  |
16 | 335 | −0.16 | 3.22 × 103−3 |
| CRP | MR-1 | Affymetrix microarray | crp+ vs. crp− transition from aerobic growth with lactate to anaerobic growth with fumarate, 11 time points | “ |  |
20 | 754 | 0.20 | 1.73 × 10−8 |
| CRP | MR-1 | “ | “ | “ |  |
10 | 1046 | −0.10 | 1.32 × 10−3 |
| CRP | MR-1 | “ | “ | BM counts in promoters of genes predicted by TractorDB |  |
13 | 70 | 0.35 | 2.85 × 10−5 |
| CRP | MR-1 | “ | “ | “ |  |
9 | 85 | −0.42 | 1.84 × 10−6 |
| CRP | MR-1 | “ | “ | Symmetrical BS counts in promoters of genes predicted by TractorDB |  |
6 | 69 | 0.51 | 4.07 × 10−6 |
| CRP | MR-1 | “ | “ | “ |  |
6 | 85 | −0.46 | 6.9 × 10−6 |
| CRP | E. coli | ROMA | RNA transcripts in wild-type CRP reaction vs. control reaction; the effect of CRP binding to the gene promoter on the gene activity in vitro | BM counts in gene promoters |  |
10 | 167 | 0.60 | 1.27 × 10−3 |
| CRP | E. coli | “ | “ | Quality score of symmetrical BS |  |
122 | 0.16 | 0.37 |
The effect of ArcA BM counts on IGA in MR-1 was examined using data from a comparative study of an MR-1 arcA− mutant and its wild type strain grown aerobically and anaerobically. We compared correlations between BM counts in the promoters and ArcA IGA in aerobic and anaerobic growth conditions. Characteristics of the dataset and correlation coefficients are given in Table 2. Although the analysis produced results similar to those from the CRP study, the relationships inferred were not as significant. As was the case with CRP, the transcriptional activation effect of ArcA was more dependent on BM counts than the transcriptional suppression effect of ArcA.
There may be several reasons for the low correlations between BM counts and IGA inferred from the CRP and ArcA studies in MR-1. The most obvious reason is the complexity of the regulatory network involved in the transcriptional adjustment of the organisms to the conditions studied. The transcriptional effect of global regulators is very often indirect, and the experimental datasets most likely include genes regulated by other transcription factors. These regulators may be activated by CRP and ArcA or they may modulate gene transcription as independent co-regulators. Many CRP-activated promoters in E. coli, for example, are repressed by other transcription factors, like CytR7 or LacI,30 but such co-regulators are not known in MR-1. Low correlation may also result from a secondary role of the regulator under the studied conditions, ie, a different regulator, rather than the one that is knocked out, may play the major role in transcriptional reprogramming of genes under the condition. Finally, other characteristics of the binding sites, such as their quality or position in the promoter, may also be responsible for the low correlation. In our further analysis we have attempted to estimate the effects of these other factors on IGA by selecting MR-1 genes for which there is strong evidence of direct regulation by CRP and by examining additional microarray datasets from studies of CRP and ArcA in E. coli.
Correlation of BM counts with CRP IGA is higher for genes presumed to be directly regulated by CRP
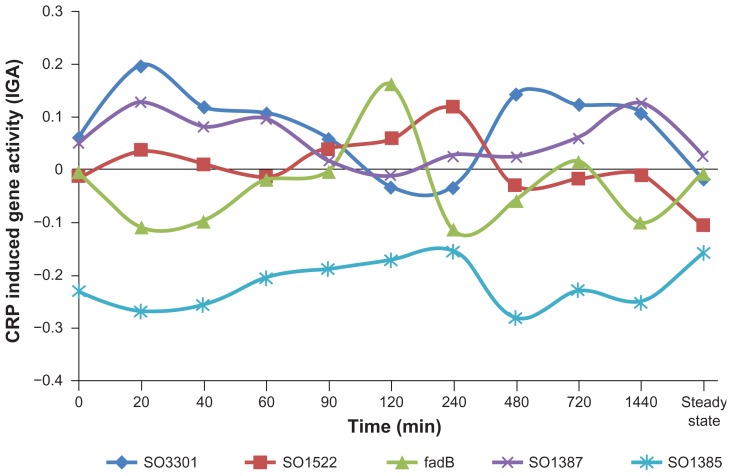
To examine the potential reasons for low correlation in the CRP dataset, we analyzed genes with high BM counts but with low levels of CRP IGA. We observed that although these genes did not have a high level of expression, many showed high variability in the level of transcription across experimental time points, some exhibiting both significant up- and down-regulation (Fig. 1). Such variability in gene expression likely results from co-regulation of the gene by a different regulator and reduces the correlation inferred from the dataset. To limit our analysis to genes that are more likely to be directly regulated by CRP, we selected only those genes predicted by TractorDB31 as up- or down-regulated by CRP in MR-1. TractorDB predictions are made using a comparative genomic approach. A selected MR-1 gene has a known CRP-regulated ortholog in E. coli and a site for which a statistical model of the CRP binding site gives a high score. The orthologous relationship in combination with a predicted BS is assumed to indicate a conserved direct regulatory effect of CRP on gene transcription. This approach results in 121 CRP genes presumed to be down-regulated and 142 CRP genes presumed to be up-regulated. In these sets of genes, we find a higher correlation between BM counts and IGA in both up- and down-regulated genes (Table 2). Among CRP up-regulated genes, the correlation coefficient is 0.35 (P = 2.85 × 10−5), which means that about 13% of the variability in IGA may be attributed to the number of BMs in the gene promoter. For down-regulated genes about 17% (R = −0.42, P = 1.84 × 10−6) of variability in the CRP IGA may be attributed to the number of BMs in the gene promoter. Random selections of the same number of down-regulated genes give an average correlation of −0.09 with a standard deviation of ±0.12. Thus, limiting a dataset to conserved genes that are more likely to be directly regulated by CRP, we find a better correlation between the number of CRP BMs in the gene promoter and the IGA.
Figure 1.
Time dependent CRP-induced gene activity (IGA) of five MR-1 genes with high BM counts and low average CRP IGA.
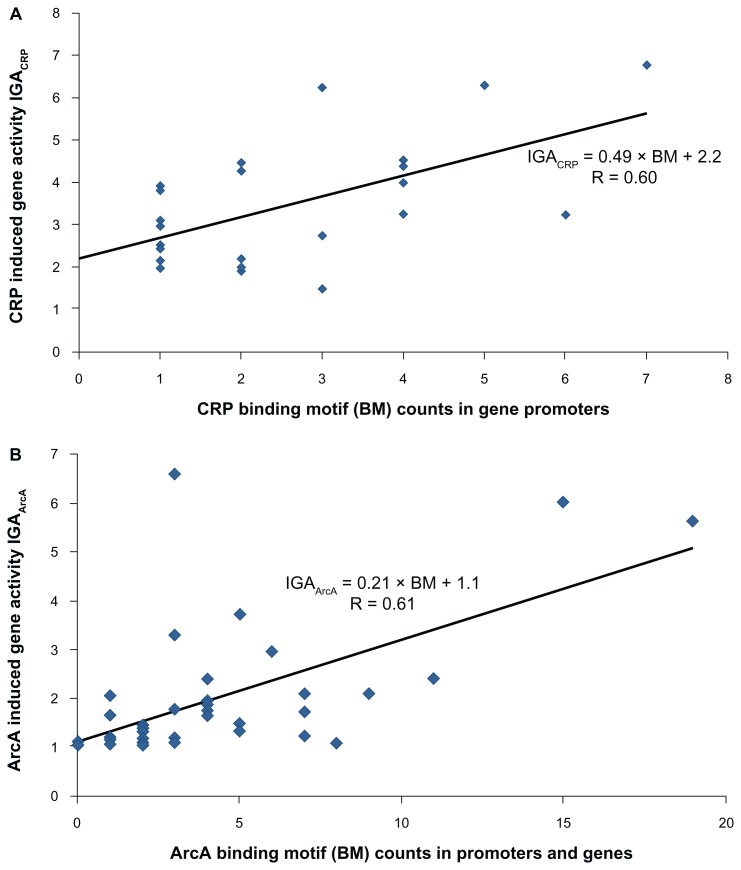
Even better support for a qualitative relationship between CRP BM counts and IGA was found using a study of the regulator in E. coli. In this study, a set of genes regulated by CRP was determined experimentally7 using a microarray-based technique called run-off transcription/microarray analysis (ROMA). This technique was applied in vitro, ie, without interference from other regulators, as contrasted with in vivo studies. The ROMA approach identified 176 operons activated by CRP. As a measure of CRP IGA, we use average ratios of the number of RNA transcripts in the wild type CRP runoff transcription reactions to the control reactions (Supplementary Table S2). The CRP IGA for this set of genes experimentally verified to be regulated by CRP correlates well (R = 0.60, P = 1.27 × 10−3) with the number of CRP BMs in the gene promoters (Fig. 2A and Table 2).
Figure 2.
(A) Effect of total CRP BM counts in a gene promoter on CRP induced gene activity (IGA). (B) Effect of total ArcA BM counts in a gene promoter and gene body on ArcA induced gene activity (IGA).
Correlation of BM counts with ArcA IGA is higher when the activities are measured under conditions regulated primarily by ArcA
The role of ArcA in E. coli suggests that the low correlation between ArcA BM counts and IGA in MR-1 may result from a secondary regulatory role of ArcA. The ArcA protein in E. coli is a typical response regulator that represses aerobic enzymes under anoxic growth conditions.32 ArcA is regulated by an associated sensor kinase, ArcB, which responds to an oxidative state of the cell. Although the ArcA binding motif is highly conserved between E. coli and MR-1, the physiological functions of ArcA in MR-1 are substantially different and not well understood.2 It is likely that ArcA in MR-1 is not a master regulator of the shift from aerobic to anaerobic growth. Thus, changes in IGA in the arcA− MR-1 strain shifting from aerobic to anaerobic growth do not correlate highly with ArcA BM counts because these changes are likely not strongly dependent on ArcA.
To validate the importance of proper experimental conditions for probing ArcA IGA, we analyzed a dataset from a study of the isobutanol response network in E. coli.27 It has been shown that ArcA is a major regulator of this response since isobutanol disrupts the cell membrane, leading to malfunction of the aerobic respiratory chain. This malfunction changes the oxidative state of the cell and necessitates the suppression of aerobic enzymes in the cell, which is regulated by ArcA. The isobutanol response study provided expression ratios (treated/untreated) for ArcA regulon members in the wild type and the arcA− strains. We used these ratios to calculate the ArcA IGA and to compare it with the number of ArcA BMs in the gene promoter and in the body of the gene (Supplementary Table S3). Correlation for this preselected set of up-regulated genes in E. coli is three times greater than the ArcA correlation found in the MR-1 study (Fig. 2B and Table 2). The best correlation (R = 0.60, P = 1.04 × 10−3) was obtained with the total number of ArcA BMs in the gene promoter and in the gene body. This is consistent with the known role of ArcA as a suppressor.32 Reexamining the CRP datasets, we found that the correlation of CRP IGA with BM counts in the gene body was significantly less than with BM counts in the promoter.
Quantity of the BMs in a gene promoter gives a better prediction of the CRP IGA than symmetrical BS quality
In the TractorDB collection, genes that are potentially regulated by CRP have associated symmetrical BSs, in which up to eight mismatches from the consensus CRP binding site are allowed.31 For these predicted symmetrical BSs, we compared the effect of their quality and the effect of BM counts in the promoter on CRP IGA of up- and down-regulated genes in MR-1. BS quality was characterized by a PWM (position weight matrix) model score (Supplementary Table S4), as described in the Methods section. We found that the quality scores of symmetrical BSs were not correlated with transcriptional activity of either up- or downregulated genes. However, in both up- and downregulated genes, CRP IGA correlated significantly with promoter BM counts. BM counts accounted for 13% of the variability (R = 0.36, P = 2.85 × 10−5) in transcriptional activation of up-regulated genes and 17% of the variability (R = −0.42, P = 1.84 × 10−6) in transcriptional suppression of down-regulated genes. Thus, CRP induced changes in transcriptional activity of a gene in MR-1 is more dependent on the number of promoter BM counts than on the quality of the symmetrical BS.
We further examined the effect of BM counts on CRP IGA in E. coli using the E. coli ROMA study (Supplementary Table S5). For E. coli, as for MR-1, BS quality was characterized by a PWM score (see Methods section for details). Both quality of the long symmetrical BS and quantity of the short BMs correlated with CRP IGA (Supplemental Table S5), but correlation was greater with BM counts (R = 0.37, P = 1.08 × 10−8) than with BS quality (R = 0.32, P = 4.56 × 10−7). Differences were more pronounced when only those CPR operons experimentally verified to be activated by CRP were considered. Correlation between BM counts and CRP IGA (R = 0.55, P = 1.02 × 10−3) was significantly greater, but no significant correlation (R = 0.16, P = 0.37) between BS quality and CRP IGA was found. The results of the E. coli in vitro study are consistent with the results from the MR-1 in vivo studies and suggest that BM quantity has a greater modulating effect on CRP IGA than does symmetrical BS quality.
Distribution of BMs in gene promoters and effects of BM locations on CRP IGA in E. coli and MR-1
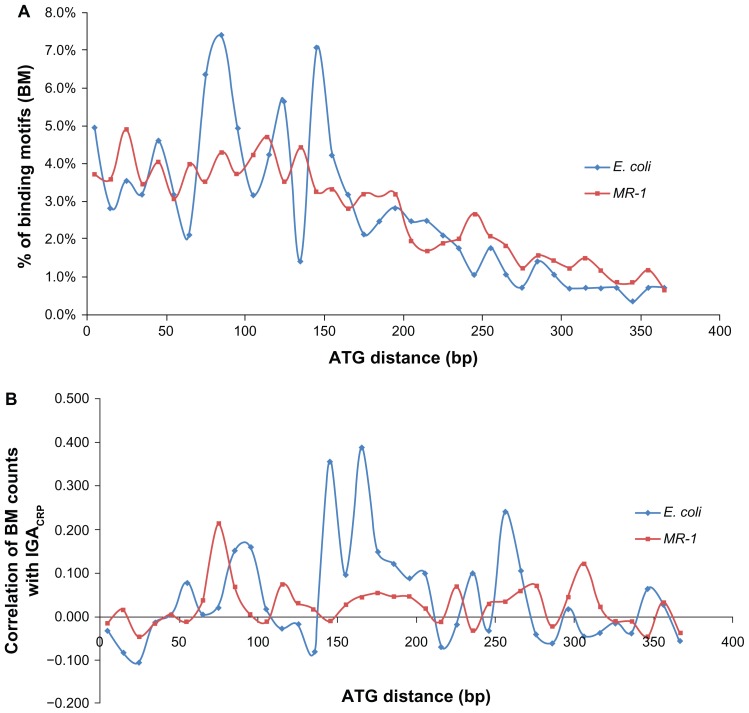
In addition to quality of TF BSs, the location of BSs in gene promoters is another known factor that modulates transcriptional activity of genes. We compared the distribution of BMs in the promoters of E. coli and MR-1 genes that are potentially regulated by CRP. All E. coli genes identified by the ROMA study as members of the CRP regulon and all MR-1 genes identified as activated by CRP in the TractorDB database were examined. For each organism, total promoter BM counts in 10-nucleotide bins were summed across all identified genes (Supplementary Table S6) as described in the Methods section. Figure 3A shows the distribution of BM counts as a function of distance from the transcription start codon (ATG distance) and illustrates that these organisms have a similar distribution of BM counts in promoters of known CRP-regulated genes. There is a significantly high correlation (R = 0.71, P = 8.60 × 10−7) between E. coli and MR-1 promoter BM counts in 10-nucleotide bins. This correlation suggests that E. coli and MR-1 genes regulated by CRP share similarity in both coding regions and promoter structure. It raises the question of whether those bins with high BM counts might contain BMs with a stronger modulating effect on CRP IGA.
Figure 3.
(A) Distribution of binding motifs (BMs) in bins of size 10 (nucleotides) for CRP-activated genes in E. coli and MR-1 as a function of the ATG distance (distance from the transcription start codon). (B) The modulating effect of BM counts in bins of size 10 (nucleotides) on CRP induced gene activity (IGA).
Notes: The modulating effect for each bin is characterized by the correlation between BM counts and CRP IGA, which was calculated using results from the ROMA experiment with E. coli and the microarray study with the MR-1 crp− mutant as described in the Methods and Results sections.
To address this question, we used CRP IGA values from the ROMA experiment in E. coli and from the CRP aerobic to anaerobic transition study in MR-1 to examine the effect of BM location. We characterized the modulating effect of BM counts in each individual 10-nucleotide bin on CRP IGA by the correlation of BM counts to CRP IGA across all selected genes (Supplementary Table S6), as illustrated in Figure 3B. Correlation was significant for three E. coli bins, particularly the bin from −170 to −161 (R = 0.39, P = 4.43 × 10−7). For MR-1 the correlation was relatively low across all bins. Although promoters of CRP-activated genes in MR-1 are enriched with BMs at locations similar to E. coli, the BMs at these locations are not necessary involved in the transcriptional activation under the experimental conditions of the study. In the MR-1 dataset, the correlation of CRP IGA with BM counts in any individual bin was not as strong as the correlation with the total BM counts in the gene promoter. The modulating effect of BM location on CRP IGA was confirmed only in E. coli using data from the ROMA in vitro study.
Synergetic effect of CRP, IHF, and ArcA binding motif counts on CRP IGA
Interaction between different regulators is another known factor modulating gene transcription. Although CRP has a dominant role in catabolite repression in E. coli, several other regulators using different mechanisms are known to be involved at different stages.33 In MR-1, CRP is involved in transcriptional reprogramming of both carbon source utilization and respiration. 29 Sophisticated crosstalk and regulatory coupling exist between transcriptional regulators involved in both processes.34 ArcA is a known regulator of oxygen response and respiration in MR-1,2,35,36 and is, therefore, a plausible co-regulator with CRP in gene transcription. Recent studies also suggest that epigenetic factors involving histone-like proteins, such as FIS, HNS, and IHF, are involved in transcriptional control by CRP.37–39 These observations led us to examine the co-regulatory effects of ArcA and IHF on CPR IGA. For CRP up-regulated genes, we calculated correlations between BM counts for each of the three regulators and CRP IGA (see Methods section for details). The correlation coefficients for ArcA and IHF BM counts are low, R = 0.09 and R = 0.13, respectively, but statistically significant (P = 0.01), suggesting that ArcA and IHF BM counts may have a small positive effect on CPR IGA. These correlation coefficients are about half the size of the correlation coefficient for CRP BM counts with CRP IGA. To evaluate the synergetic effect of CRP, ArcA, and IHF BM counts on CRP IGA we used a step-wise multiple regression analysis that compared linear, non-linear, and interaction effects of the transcription factors on IGA (see Methods section for details). We found that of the nine initial variables representing these effects, only three variables were significant, namely, the positive 2nd degree polynomial effect of CRP BM counts, the positive interaction effect of CRP and IHF BM counts, and the negative linear effect of ArcA BM counts. The analysis produced the following fitting line, with correlation R = 0.25, for CRP IGA:
where IGA is the CRP IGA given as log2 ratio of gene expression in crp+ to crp− strains; BMArcA, BMCRP, and BMIHF are binding motif counts for regulators ArcA, CRP, and IHF, respectively. This regression model confirms a strong non-linear effect of CRP on gene transcription under the conditions of the experimental studies. It also indicates that the histone like protein IHF likely affects CRP-induced transcription of some genes through interaction with CRP, and does not exert an independent transcriptional effect. The transcriptional effect of the global regulator ArcA is likely independent and opposite the effect of CRP for the same genes. The regression model’s correlation coefficient (R = 0.25) indicates that it explains only a portion of CRP IGA. To develop a more robust model, there are other regulatory mechanisms that must be considered.
Explanation of the results in terms of the model of CRP transcriptional regulation
Results of this study indicate that the total number of BMs for various bacterial global regulators, such as CRP and ArcA, are important characteristics of the gene promoter. These numbers predicted levels of CRP IGA better than quality or location of symmetrical BSs, indicating that even BMs that are rather distant from the transcription initiation site may produce a regulatory effect and facilitate gene activity. The study considered two different bacterial organisms, two different experimental technologies, and three different transcription factors. The results in combination with recent reports on biochemical mechanisms of CRP transcriptional activation25 support a regulatory model that involves DNA bending in transcriptional activation. They indicate that specific sets of BMs in the promoter may encode information affecting not only the affinity of CRP binding to DNA, but also the geometry of the CRP/ DNA complex and thus provide additional epigenetic control of gene expression. The importance of distant BMs for transcriptional activity is consistent with observations that flanking sequences can affect the energetics of DNA/CRP complex formation and the geometry of a CRP-induced bend in DNA.40 DNA bending is the most plausible architectural mechanism for bringing distantly located BMs into play.
The computationally predicted synergetic effect of CRP and IHF BMs on CRP IGA in the MR-1 study suggests that IHF may cooperate with CRP in transcriptional regulation. A potential mechanism for this cooperation may be additional bending of the DNA promoter.41 By introducing bending, IHF can affect geometry of the DNA/CRP complex, stabilizing, weakening, or preventing binding of CRP to DNA. In this way IHF may exert an additional level of regulatory control on CRP IGA that is similar to epigenetic control in eukaryotes. Bending of DNA by CRP and IHF may bring distantly bound CRP or other activators into contact with RNA polymerase and thus initiate transcription, or it may change the accessibility of BSs to transcription factors. The synergetic effect of ArcA predicted by the regression model is different from the effect of IHF. ArcA may work independently from CRP under the conditions of the experimental studies, perhaps preventing CRP/DNA complex formation at some promoters. Additional experiments will be necessary to validate the computational predictions.
Conclusions
Transcriptional fine-tuning
Although transcription activation by CRP at the simplest CRP-dependent promoters requires only one DNA binding site and no co-regulators,42 in general gene activation by CRP involves additional molecular mechanisms to fine-tune the expression level of each individual gene for the same concentration of CRP. Our findings indicate that variability in the number of BMs in gene promoters and DNA bending by CRP and IHF may be important genomic mechanisms for this fine-tuning. DNA bending is surprisingly similar to that observed in eukaryotes, which have a complex dynamic chromatin structure. Through remodeling of chromatin structure, eukaryotes change TF accessibility to different BSs and, in this way, achieve variability in the expression of genes responding to the same level of a transcription factor.43 Our results suggest that even though bacteria do not have a sophisticated nucleosome structure, these organisms may utilize bending of DNA by global regulators to change accessibility of BSs and, in this way, adjust the level of gene expression to the environmental stimulus. Another mechanism for fine-tuning the CRP IGA may be binding of cAMP to the TF and the concentration of cAMP in the cell. There are multiple cAMP binding sites in CRP and occupancy of these sites modifies the affinity of CRP for DNA binding sites.44 The more diverse the set of CRP BMs in the gene promoter, the more intricate the control of gene expression implemented by cAMP-related mechanisms.
Why the correlation is low
Results of this study suggest that a set of mechanisms may modulate the effect of a TF on gene expression, and that the influence of each mechanism may vary for different transcription factors, different organisms, and different environmental conditions. Even for the same conditions, we have observed a strong time-dependent effect of CRP at some promoters with high BM counts. Microarray measurements made at different time points, even under similar conditions, may produce opposite results, gene activation or suppression. Such time-dependent patterns of gene expression are not easily quantified and may introduce significant errors in computational predictions. Variation of expression over time may be a reason for the low correlation of BM counts with CRP or ArcA IGA. Time-dependent patterns may also explain contradictory identification of genes regulated by a TF in experimental studies. Different large-scale studies to identify TF BSs often find different sets of genes regulated by the TF. An example is two studies to identify ArcA BSs in E. coli using transcriptional profiling of an ArcA mutant strain.45,46 Although some discrepancies among studies may result from experimental errors, there is a plausible biological explanation of this phenomenon. Some differences may be attributed to different cellular mechanisms of transcription regulation discussed in the previous sections. Inevitable variations in experimental conditions may result in different epigenetic states of bacterial DNA and may, therefore, produce different time-dependent expression patterns for genes regulated by the same level of the TF. In addition to transcriptional regulation by CRP, many genes can be regulated post-transcriptionally. For these genes, their level of expression may be slightly diminished or even unaffected in the crp− mutant strain. Transcription of adenylate cyclase CyaA, for example, which synthesizes cAMP from ATP, is only slightly decreased in crp− mutants, although the reintroduction of CRP increases the transcription four- to five-fold.47 As this example demonstrates, the actual effect of a TF on gene transcription is not accurately characterized by measuring an average ratio of gene expression in wild type versus mutant.
Improved computational modeling
Quantification of gene expression in terms of BM counts may provide a means to improve computational modeling of transcriptional regulatory networks and to reveal principles of transcriptional regulation. Existing computational algorithms combining microarray data for mRNA expression and transcription factor occupancy to identify regulatory networks consider only linear effects of BM counts on gene transcription.10,11 This study demonstrates that for some transcription factors, such as CRP, this effect may be nonlinear. Adding nonlinearity may improve the predictive capability of the computational model. Another potential application of the results may be the development of improved algorithms for locating TF BSs in prokaryotic promoters using TF BMs to supplement direct identification of long BSs.
Methods
Estimation of CRP and ArcA induced gene activity from microarray experiments
Three datasets from microarray experiments were analyzed to evaluate the effects of CRP and ArcA on transcriptional activity in two bacterial species, Shewanella oneidensis MR-1 and E. coli. The number of CRP and ArcA binding sites, their quality, and their locations within gene promoters were considered. In each dataset, the effect of a regulator (CRP or ArcA) on transcriptional activity of a gene was estimated by calculating the log2 ratio of expression levels in the regulator positive (wild type strain) and the regulator negative (mutant strain) for each time point or biological replicate. Overall transcriptional effect for each gene was estimated by averaging the log2 ratios across time points or replicates. This average ratio will be referred to as the CRP/ArcA induced gene activity or IGA. The relationships between IGA and various computationally derived characteristics of gene promoters, including binding site counts, quality, and locations were determined by correlation and regression analysis. For each operon, only the IGA of the first gene was included in the statistical analysis, since unequal levels of expression among genes of the same operon are common, likely because of putative internal promoters or because of alternative regulatory mechanisms controlling gene expression within operons.48 In the following sections we give a brief description of the experimental studies used to characterize CRP/ArcA IGA.
CRP induced gene activity in Shewanella one-idensis MR-1
This is a study of a time-series transition from aerobic growth with lactate to anaerobic growth with fumarate in a crp− mutant strain of S. oneidensis MR-1 and in a wild type strain using an Affymetrix microarray. Growth of the strains was implemented in a bioreactor in modified M1 minimal media in two biological replicates with sampling and transcriptional profiling at various time points: 0, 20, 40, 60, 90, 120 min, 4, 8, 12, 24 h, steady-state.
ArcA induced gene activities in Shewanella one-idensis MR-1
This study of S. oneidensis MR-1 compares the growth of arcA+ and arcA− MR-1 strains in aerobic and anaerobic conditions.2 A homemade microarray with oligonucleotide probes from 99% of all predicted genes in the S. oneidensis genome was used to measure gene expression. Probes were printed in duplicate onto Telechem Superamine slides. Genes that were significantly up- or down-regulated in the ArcA mutant strains were considered putative candidates for activation or suppression by ArcA, respectively. To decrease the rate of false positive candidates for regulation by ArcA, only genes with log2 ratios more than 0.2 or less than −0.2 were included in the analysis.
CRP induced gene activities in E. coli
In this study,7 run-off transcription/microarray analysis (ROMA), was used to identify CRP regulated promoters. This technique found 176 operons activated by CRP in vitro. Using descriptors from the study, 167 genes, each located first in one of the 176 activated operons, were identified. To characterize the effect of CRP binding on transcriptional activity in vitro, average ratios of the number of RNA transcripts in the wild type CRP reaction versus the control reaction were calculated.
Selection of the binding motif consensus for CRP, ArcA, and IHF
Consensus sequences for the binding motifs were selected based on the three most conserved nucleotide bases in the known BS of the TF. To find this core of three base pairs, we used RSAT tools49 to search for occurrences of all possible three nucleotide oligomers in known BSs and in promoters of the corresponding genes listed in RegulonDB.22 Conservation of bases in known long symmetrical BSs and representation of three nucleotide oligomers in BSs and gene promoters were considered in constructing BM consensus sequences (Table 1). Each BM is comprised of a three base pair core and two flanking regions. No substitutions were allowed in the central core sequence and in the weak bases directly adjacent to the core. A maximum of two substitutions were allowed in the remaining bases. Our rule for BM substitution was influenced by recent observations in yeast, namely, that substitutions involving Adenine are unlikely to change expression patterns, while substitutions involving Guanine tend to alter expression patterns.19
Table 1.
Known CRP, ArcA, and IHF binding site consensus sequences and their binding motifs in E. coli derived from analysis of RegulonDB data.
Counting the TF binding motifs
Experimental data from the aforementioned studies on TF IGA were supplemented with counts of the binding motifs (BM counts) in the upstream regions of the genes. FastA nucleotide sequences of MR-1 and E. coli were downloaded from Genbank to use in determining BM counts. Only the first gene of an operon was considered in the analysis. Operons in MR-1 were predicted using the algorithm described by Dam et al.50 RegulonDB22 was used to define operons and TF binding sites for E. coli. For each gene, BM counts across all selected genes were collected in a variety of regions in the promoter (upstream intergenic region of the gene), in the coding sequence, and in different bins relative to the transcription start codon, including 1..30, −370..−1, and in the 37 bins of size 10 from −370..−361 to −10..−1.
Characterization of binding site quality
Position weight matrix (PWM) models were used to characterize the quality of CRP BSs in both MR-1 and E. coli. In MR-1, all full, symmetric CRP BSs from TractorDB were used to construct the PWM model (Supplementary Table S4). For E. coli, CRP BS quality scores reported in the Zheng et al study7 were used. These scores were derived from PWM models described by Tan et al.51 For validation, these PWM models were compared with a PWM model constructed from CRP binding sequences in E. coli experimentally verified by ChIP-chip analysis.52
Statistical analysis
Information on BM counts for each gene in MR-1 and E. coli was supplemented by CRP or ArcA IGA calculated from experimental data. Data for up- and down-regulated genes in each organism and for each TF were analyzed separately. Pairwise Pearson correlations were calculated between the parameters in each dataset to find their relationship. Significance of the correlation was characterized by P-value and by comparison of the calculated coefficient with the coefficient calculated by permutations for the same number of randomly selected genes from the dataset. The R statistical package was used to sample 1000 sets of genes for each dataset in order to calculate the correlation between IGA and BM counts. The distribution of the resulting 1000 correlation coefficients was compared with a normal distribution characterized by the average and the standard deviation of the correlation coefficients. These parameters were then used to calculate a t-statistic to estimate the significance of the correlation in the selected genes.
The synergetic effects of CRP, ArcA, and IHF BM counts (BMCRP, BMArcA, BMIHF) on CRP IGA were determined by stepwise multiple regression (Supplementary Table S7) in which BM counts for these regulators were independent predictor variables and CRP IGA (calculated from the experimental data) was a dependent variable. We compared not only linear effects of the independent variables, but also their interactions and non-linear (2nd degree polynomial) effects on the dependent variable. Specifically, the regression model was
where a0, a1, …, a9 are fitting coefficients. At each step of the analysis, each coefficient of a model term representing a linear, non-linear, or interaction effect was evaluated by its t-statistic value, the ratio of the coefficient to its standard error, and the dependent variable with the minimum input, indicated by the minimum t-statistic, was removed from the set of dependent variables. Multiple regression analysis was repeated until only variables with significant t-statistic values were left in the fitting line.
Supplementary Tables
Supplementary Tables are available from 9357SupplementaryTables.zip
Acknowledgements
The authors would like to thank Tim Gardner and his group for providing unpublished microarray data on crp− mutant strain of S. oneidensis MR-1 for the analysis.
Footnotes
Author Contributions
TVK conceived the study; MRL, TVK, MHS, ASB, and ECB designed the study; ASB contributed experimental data; MRL, TVK, and MHS analyzed the data; MRL, TVK, and ECB wrote the paper. All authors reviewed and approved the final manuscript.
Funding
This research was sponsored by the Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research, as part of the BioEnergy Science Center and the Plant Microbe Interfaces Scientific Focus Area. Oak Ridge National Laboratory is managed by the University of Tennessee-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725. The Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U. S. Department of Energy under contract DE-AC05-76RL01830. This work was supported by Department of Energy.
Disclosures
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Liang W, Silva AJ, Benitez JA. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl Environ Microbiol. 2007;73:7482–7. doi: 10.1128/AEM.01564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao H, Wang X, Yang ZK, Palzkill T, Zhou J. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics. 2008;9:42. doi: 10.1186/1471-2164-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guest JR, Green J, Irvine AS, Spiro S. The FNR modulon and FNR-regulated gene expression. In: Lin ECC, Lynch AS, editors. Regulation of Gene Expression in Escherichia coli. New York: Chapman & Hall; 1996. pp. 317–42. [Google Scholar]
- 4.Narasimhan C, LoCascio P, Uberbacher E. Background rareness-based iterative multiple sequence alignment algorithm for regulatory element detection. Bioinformatics. 2003;19:1952–63. doi: 10.1093/bioinformatics/btg266. [DOI] [PubMed] [Google Scholar]
- 5.Tompa M, Li N, Bailey TL, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23:137–44. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 6.Siggia ED. Computational methods for transcriptional regulation. Curr Opin Genet Dev. 2005;15:214–21. doi: 10.1016/j.gde.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–93. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babu MM, Lang B, Aravind L. Methods to reconstruct and compare transcriptional regulatory networks. Methods Mol Biol. 2009;541:163–80. doi: 10.1007/978-1-59745-243-4_8. [DOI] [PubMed] [Google Scholar]
- 10.Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–71. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 11.Gao F, Foat BC, Bussemaker HJ. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. BMC Bioinformatics. 2004;5:31. doi: 10.1186/1471-2105-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen DH, D’haeseleer P. Deciphering principles of transcription regulation in eukaryotic genomes. Mol Syst Biol. 2006;2:2006.0012. doi: 10.1038/msb4100054. http://www.nature.com/msb/journal/v2/n1/full/msb4100054.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–56. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 14.Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol. 2010;339:250–7. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beatty CM, Browning DF, Busby SJ, Wolfe AJ. Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism. J Bacteriol. 2003;185:5148–57. doi: 10.1128/JB.185.17.5148-5157.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miroslavova NS, Mitchell JE, Tebbutt J, Busby SJ. Recruitment of RNA polymerase to Class II CRP-dependent promoters is improved by a second upstream-bound CRP molecule. Biochem Soc Trans. 2006;34:1075–8. doi: 10.1042/BST0341075. [DOI] [PubMed] [Google Scholar]
- 17.Harbison CT, Gordon DB, Lee TI, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee TI, Rinaldi NJ, Robert F, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 19.Lapidot M, Mizrahi-Man O, Pilpel Y. Functional characterization of variations on regulatory motifs. PLoS Genet. 2008;4:e1000018. doi: 10.1371/journal.pgen.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–7. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Gunasekera A, Zhang X, Kunkel TA, Ebright RH, Berman HM. Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: alteration of DNA binding specificity through alteration of DNA kinking. J Mol Biol. 2001;314:75–82. doi: 10.1006/jmbi.2001.5090. [DOI] [PubMed] [Google Scholar]
- 22.Salgado H, Gama-Castro S, Peralta-Gil M, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–7. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krummenacker M, Paley S, Mueller L, Yan T, Karp PD. Querying and computing with BioCyc databases. Bioinformatics. 2005;21:3454–5. doi: 10.1093/bioinformatics/bti546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaston K, Kolb A, Busby S. Binding of the Escherichia coli cyclic AMP receptor protein to DNA fragments containing consensus nucleotide sequences. Biochem J. 1989;261:649–53. doi: 10.1042/bj2610649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin SH, Lee JC. Determinants of DNA bending in the DNA-cyclic AMP receptor protein complexes in Escherichia coli. Biochemistry. 2003;42:4809–18. doi: 10.1021/bi027259+. [DOI] [PubMed] [Google Scholar]
- 26.Charania MA, Brockman KL, Zhang Y, et al. Involvement of a membrane-bound class III adenylate cyclase in regulation of anaerobic respiration in Shewanella oneidensis MR-1. J Bacteriol. 2009;191:4298–306. doi: 10.1128/JB.01829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brynildsen MP, Liao JC. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol Syst Biol. 2009;5:277. doi: 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karpinets TV, Romine MF, Schmoyer DD, et al. Shewanella knowledgebase: integration of the experimental data and computational predictions suggests a biological role for transcription of intergenic regions. Database (Oxford) 2010:baq012. doi: 10.1093/database/baq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saffarini DA, Schultz R, Beliaev A. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J Bacteriol. 2003;185:3668–71. doi: 10.1128/JB.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, McEwen AE, Crothers DM, Levene SD. Analysis of in-vivo LacR-mediated gene repression based on the mechanics of DNA looping. PLoS One. 2006;1:e136. doi: 10.1371/journal.pone.0000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guía MH, Pérez AG, Angarica VE, Vasconcelos AT, Collado-Vides J. Complementing computationally predicted regulatory sites in Tractor_DB using a pattern matching approach. In Silico Biol. 2005;5:209–19. [PubMed] [Google Scholar]
- 32.Iuchi S, Lin ECC. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci U S A. 1988;85:1888–92. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrenoud A, Sauer U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol. 2005;187:3171–9. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Driscoll ME, Romine MF, Juhn FS, et al. Identification of diverse carbon utilization pathways in Shewanella oneidensis MR-1 via expression profiling. Genome Inform. 2007;18:287–98. [PubMed] [Google Scholar]
- 35.Gralnick JA, Brown CT, Newman DK. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol. 2005;56:1347–57. doi: 10.1111/j.1365-2958.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- 36.Vanrobaeys F, Devreese B, Lecocq E, Rychlewski L, De Smet L, Van Beeumen J. Proteomics of the dissimilatory iron-reducing bacterium Shewanella oneidensis MR-1, using a matrix-assisted laser desorption/ ionization-tandem-time of flight mass spectrometer. Proteomics. 2003;3:2249–57. doi: 10.1002/pmic.200300476. [DOI] [PubMed] [Google Scholar]
- 37.Galán B, Manso I, Kolb A, García JL, Prieto MA. The role of FIS protein in the physiological control of the expression of the Escherichia coli meta-hpa operon. Microbiology. 2008;154:2151–60. doi: 10.1099/mic.0.2007/015578-0. [DOI] [PubMed] [Google Scholar]
- 38.Sclavi B, Beatty CM, Thach DS, Fredericks CE, Buckle M, Wolfe AJ. The multiple roles of CRP at the complex acs promoter depend on activation region 2 and IHF. Mol Microbiol. 2007;65:425–40. doi: 10.1111/j.1365-2958.2007.05797.x. [DOI] [PubMed] [Google Scholar]
- 39.Paul L, Mishra PK, Blumenthal RM, Matthews RG. Integration of regulatory signals through involvement of multiple global regulators: control of the Escherichia coli gltBDF operon by Lrp, IHF, Crp, and ArgR. BMC Microbiol. 2007;7:2. doi: 10.1186/1471-2180-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pyles EA, Lee JC. Escherichia coli cAMP receptor protein-DNA complexes. 2. Structural asymmetry of DNA bending. Biochemistry. 1998;37:5201–10. doi: 10.1021/bi972451a. [DOI] [PubMed] [Google Scholar]
- 41.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 43.Lam FH, Steger DJ, O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–50. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin SH, Lee JC. Communications between the high-affinity cyclic nucleotide binding sites in E. coli cyclic AMP receptor protein: effect of single site mutations. Biochemistry. 2002;41:11857–67. doi: 10.1021/bi026099z. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279:12588–97. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 46.Salmon KA, Hung SP, Steffen NR, et al. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005;280:15084–96. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- 47.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laing E, Mersinias V, Smith CP, Hubbard SJ. Analysis of gene expression in operons of Streptomyces coelicolor. Genome Biol. 2006;7:R46. doi: 10.1186/gb-2006-7-6-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas-Chollier M, Sand O, Turatsinze JV, et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–27. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dam P, Olman V, Harris K, Su Z, Xu Y. Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res. 2007;35:288–98. doi: 10.1093/nar/gkl1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan K, Moreno-Hagelsieb G, Collado-Vides J, Stormo GD. A comparative genomics approach to prediction of new members of regulons. Genome Res. 2001;11:566–84. doi: 10.1101/gr.149301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJW. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. PNAS. 2005;102:17693–8. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables are available from 9357SupplementaryTables.zip