Abstract
Variants associated with meconium ileus in cystic fibrosis (CF) were identified in 3,763 patients by GWAS. Five SNPs at two loci near SLC6A14 (min P=1.28×10−12 at rs3788766), chr Xq23-24 and SLC26A9 (min P=9.88×10−9 at rs4077468), chr 1q32.1 accounted for ~5% of the phenotypic variability, and were replicated in an independent patient collection (n=2,372; P=0.001 and 0.0001 respectively). By incorporating that disease-causing mutations in CFTR alter electrolyte and fluid flux across epithelia into an hypothesis-driven genome-wide analysis (GWAS-HD), we identified the same SLC6A14 and SLC26A9 associated SNPs, while establishing evidence for the involvement of SNPs in a third solute carrier gene, SLC9A3. In addition, GWAS-HD provided evidence of association between meconium ileus and multiple constituents of the apical plasma membrane where CFTR resides (P=0.0002, testing 155 apical genes jointly and replicated, P=0.022). These findings suggest that modulating activities of apical membrane constituents could complement current therapeutic paradigms for cystic fibrosis.
Cystic fibrosis is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene1. CFTR is a chloride channel located on the apical membrane of epithelia, where ion conduction and solute trafficking contribute to the regulation of transepithelial fluid flow. Individuals with the same loss-of-function CFTR mutations have variable disease severity, and differentially affected CF-associated organs including lung, pancreas, liver, intestines, and vas deferens; thus additional features, including other genes (referred to as modifier genes) may affect disease pathophysiology. Approximately 15% of CF patients have severe intestinal obstruction at birth, a complication known as meconium ileus2. Meconium ileus develops in utero, and presents following birth with complete intestinal obstruction that requires either medical or surgical intervention. This neonatal complication is highly indicative of CF, occurs in either sex, displays notable heritability exceeding 88%3, and is likely minimally affected by environmental influences.
The North American CF Gene Modifier Consortium has accumulated 3,763 participants with ‘severe’ (pancreatic exocrine insufficient) CFTR alleles and genome-wide genotype data at 543,927 SNP loci4,5 (Table 1 and Online Methods). The definition of meconium ileus was consistent within the consortium and was recorded following rigorous chart review. A conventional GWAS for meconium ileus used a generalized estimating equations (GEE) model6 to include collected sibling-pairs, and led to five genome-wide significant SNPs (P<5×10−8)7 from two regions that include SLC26A9 on chromosome 1 and SLC6A14 on chromosome X (Fig. 1, Supplementary Fig. 1, Table 2; sex-specific results in Supplementary Table 1). CFTR was not a significant confounder or effect modifier when incorporated in the GWAS (Supplementary Fig. 2 and Supplementary Table 2), indicating SLC6A14 and SLC26A9 are independent contributors to meconium ileus. We then replicated the associations in SLC6A14 (min P=0.001) and SLC26A9 (min P=0.0001) with meconium ileus in an independent combined collection from North America and France (Table 2).
Table 1.
CF consortium participants of the meconium ileus study
| Consortium Center/Site | Sample Sizea | CFTR Genotype | Status of CF Patientsb | ||
|---|---|---|---|---|---|
| n | Phe508del/Phe508del n (%) |
Phe508del/Other or Other/Other n (%) |
MI (M:Fc) n |
Non-MI (M:Fc) n |
|
|
CGS (Canadian Gene Modifier Study Population Samples) |
1661 | 992 (59.7) | 669 (40.3) | 252 (128:124) |
1409 (772:637) |
|
GMS-Lung Study (UNC/CWRU, Extreme Phenotype Study) |
1120 | 1116 (99.6) | 4 (0.4) | 177 (99:78) |
943 (498:445) |
|
GMS-Liver Study (Samples from North America Only) |
80 | 58 (72.5) | 22 (27.5) | 23 (14:9) |
57 (35:22) |
|
TSS (Johns Hopkins University, Family-based Twin and Sibling Study) |
902 | 522 (57.9) | 380 (42.1) | 159 (78:81) |
743 (397:346) |
| Total | 3763 | 2688 (71.4) | 1075 (28.6) | 611 (319:292) |
3152 (1702:1450) |
Size of CF patient sample used for the meconium ileus GWAS and GWAS-HD discovery study is given by consortium site. The TSS site contained sibling pairs. There were 3,199 unrelated individuals among the total of 3,763 studied.
Meconium ileus (MI) status is given separately for each center.
Gender break-down is indicated for each patient category.
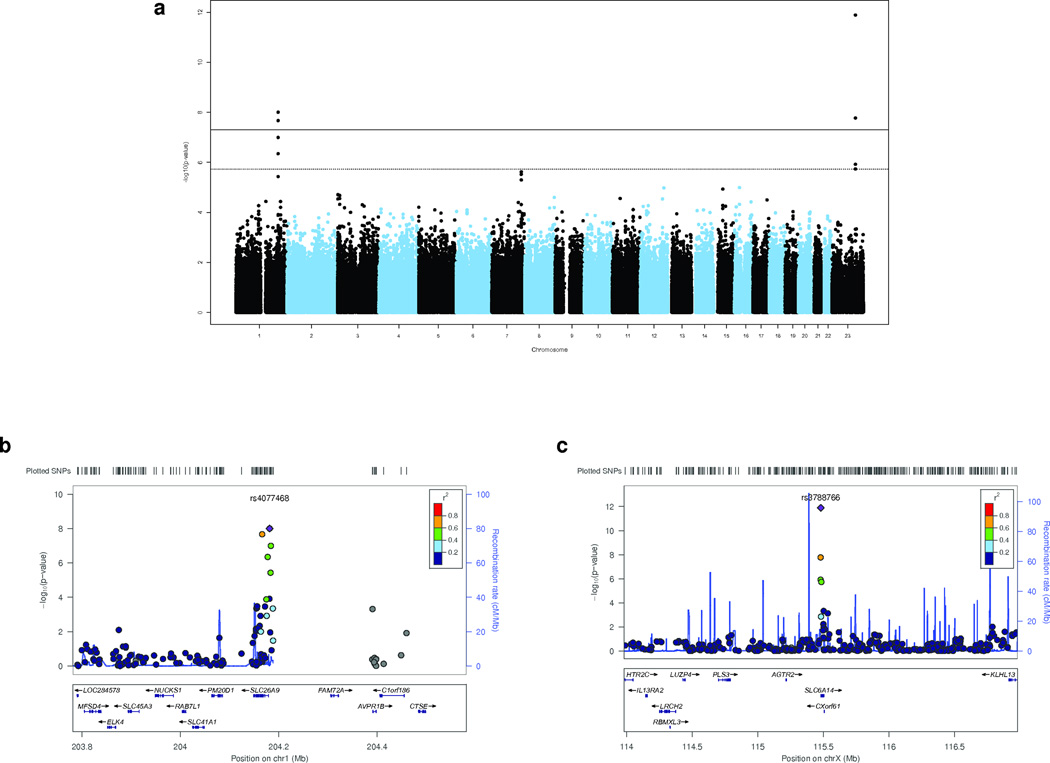
Figure 1.
Meconium Ileus GWAS identifies genome-wide significant SNPs. Association analysis was performed on all SNPs with minor allele frequencies > 2% that passed QC criteria (Online Methods).
(a) Genome-wide Manhattan plot for meconium ileus. The black solid line corresponds to the genome-wide significance threshold7 (P<5×10−8), and the black dashed line to the suggestive association threshold, expected once per genome scan (P <1/543,927=1.84×10−6). A total of five SNPs in two regions (SLC6A14 on chromosome X, and SLC26A9 on chromosome 1) have association evidence exceeding the genome-wide threshold. The SNPs, rs4077468 and rs4077469 are in perfect LD and appear as one SNP as they are separated by only 128bp.
(b) Regional plot for SLC26A9. LocusZoom viewer was used to display the association evidence around SLC26A9 based on NCBI Build 36/hg18. Symbol coloring reflects HapMap CEU LD r2 values with the most significant SNP. The significant SNPs, rs4077468, rs7419153 and rs12047830 (Table 2), occur 2.17 kb, 4.72 kb and 4.12 kb upstream, respectively, of the transcription start site. The significant SNP, rs7512462, occurs in intron 5. A gap occurs in the genomic sequence between the SLC26A9 and FAM72A genes in both NCBI36.3 and GRCh37 primary reference assemblies.
(c) Regional plot for SLC6A14. The association evidence around SLC6A14 was displayed as above. Rs3788766 (Table 2) is positioned 0.95 kb upstream of the transcription start site and is within the binding site of the CEBPB transcription factor as annotated by ENCODE33 (not shown). The mRNA transcript corresponding to CXorf61 is downstream (3’) of SLC6A14.
Table 2.
Discovery and replication of SNPs in SLC26A9 and SLC6A14.
| Discovery Sample (611:3152)d | Replication Samplea | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
N. America (251:889)d |
France (154:1078)d |
Combinedb (405:1967)d |
||||||||||||
| SNP | CHR | POS | GENE | Risk Allele |
Risk Allele Frequency |
OR | P | OR | P | OR | P | OR | P | |
| MI | Non- MI |
|||||||||||||
| rs4077468c | 1 | 204181380 | SLC26A9 | T | 0.66 | 0.57 | 1.45 | 9.88×10−9 | 1.45 | 0.0005 | 1.27 | 0.0575 | 1.37 | 0.0001 |
| rs7512462 | 1 | 204166218 | SLC26A9 | T | 0.66 | 0.57 | 1.45 | 2.14×10−8 | 1.31 | 0.0134 | 1.20 | 0.2120 | 1.27 | 0.0063 |
| rs7419153 | 1 | 204183932 | SLC26A9 | T | 0.44 | 0.36 | 1.42 | 1.01×10−7 | 1.42 | 0.0007 | 1.20 | 0.1290 | 1.33 | 0.0004 |
| rs12047830 | 1 | 204183322 | SLC26A9 | C | 0.56 | 0.49 | 1.34 | 3.72×10−6 | 1.33 | 0.0054 | 1.27 | 0.0510 | 1.31 | 0.0007 |
| rs3788766 | X | 115480867 | SLC6A14 | T | 0.72 | 0.59 | 1.50 | 1.28×10−12 | 1.14 | 0.1328 | 1.47 | 0.0006 | 1.25 | 0.0011 |
| rs5905283 | X | 115479909 | SLC6A14 | C | 0.61 | 0.50 | 1.34 | 1.69×10−8 | 1.04 | 0.6030 | 1.47 | 0.0002 | 1.19 | 0.0074 |
| rs12839137 | X | 115479578 | SLC6A14 | C | 0.82 | 0.75 | 1.39 | 1.20×10−6 | 1.20 | 0.0666 | 1.15 | 0.3275 | 1.18 | 0.0386 |
The North American replication collection consisted of 1,140 individuals that correspond to the continuing enrolment at the North American sites. The French cohort consisted of 1,232 patients collected at 38 French CF centers (Online Methods). The French replication samples were genotyped genome-wide with the Illumina 660W-Quad BeadChip platform while the North American replication samples were genotyped on the six SNPs using Taqman® Assay-on-Demand Assays.
The combined replication P value was calculated using the inverse-variance method, most powerful when different studies have the “same direction” of effect. The corrected type 1 error for the replication study is 0.008, using the conservative Bonferroni correction for the 6 SNPs tested.
There is another significant SNP on chromosome 1 (rs4077469, P=9.88 × 10−9) but not included as it is separated by only 128bp and in perfect LD with rs4077468.
Meconium ileus status (MI:Non-MI) break-down for each site.
The signal intensity plots of the associated SNPs reflected autosomal- and X-associated SNPs at SLC26A9 and SLC6A14, respectively. Imputation analysis using MACH and minimac8,9 identified the same regions of association as the genotyped SNPs (Online Methods, Supplementary Fig. 3). The five associated SNPs in SLC6A14 and two in SLC26A9 (Fig. 1b and 1c) are positioned just upstream of their respective transcription start sites such that binding of activating or repressing transcription factors may be affected as highlighted by ENCODE data10 (data not shown). Neither SLC6A14 nor SLC26A9 coding regions exhibit evidence for CNVs; however, there is a gap in the reference sequence >10 kb upstream of the SLC26A9 locus.
The seven SNPs genotyped (Table 2) in the two genes account for <5% of the meconium ileus variation, estimated by pseudo R-squared11, likely reflecting the common problems in association studies of locus heterogeneity and low power given the available sample size. Whereas conventional GWAS is often considered for complex disease mapping, modifier gene studies could incorporate disease etiology and pathobiology information to increase power and account for heterogeneity. To do so, we proposed application of GWAS with consideration of a hypothesis (and so is hypothesis-driven; GWAS-HD) to systematically prioritize SNPs for genome-wide analysis. The highest priority markers are also evaluated as a set to test the statistical significance of the hypothesis used for prioritization (Supplementary Fig. 4).
The GWAS-HD prioritization in this CF application is based on the knowledge that a major source of CF pathophysiology is impaired fluid and electrolyte flux in epithelia of CF-affected organs. The polarized epithelial layer forms a highly selective barrier between organ and ductal interfaces. Transepithelial ‘function’ is achieved by cell polarization whereby many determinants and regulators of fluid, solute and ion transport reside at the apical membrane alongside CFTR, with contributing features from basolateral surfaces. We have shown in a mouse model that CFTR function in the gastrointestinal epithelium is critical for preventing intestinal obstructions12. Thus, we hypothesized that with loss of CFTR, (genetic) variation in other apical membrane constituents could modify CF phenotypes, such as meconium ileus.
A list of 157 gene products (Fig. 2 and Supplementary Table 3) was annotated as localized to the apical plasma membrane using AmiGO13 with Gene Ontology data14,15. CFTR and many solute transporters were included. However, the brush border membrane protein SLC6A14 was not listed, reflecting the high specialization of its corresponding intestinal cavity and a limitation of the GO annotation that we accepted without additional curation to avoid bias. In total, 3,814 GWAS SNPs were within ±10 kb of the boundaries of 155 genes (NCBI36/hg18); 2 genes were not tagged by any of the GWAS SNPs.
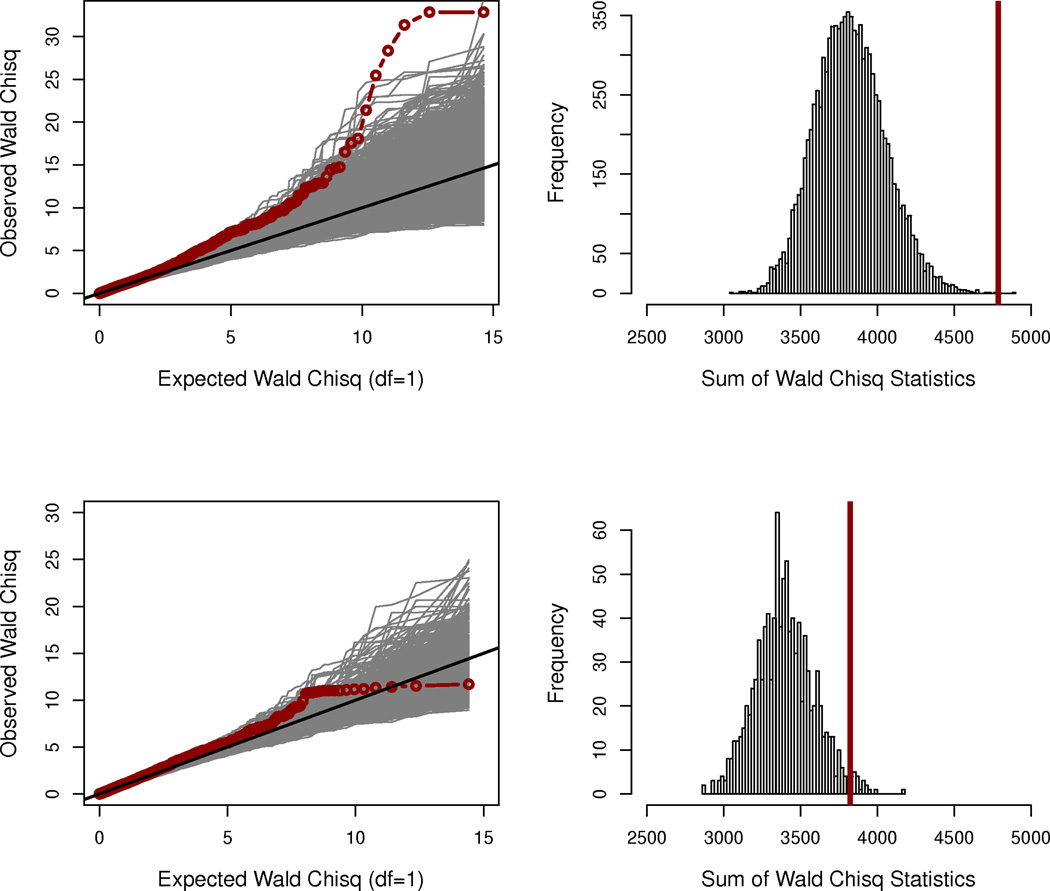
Figure 2.
The apical membrane hypothesis identifies genes associated with meconium ileus. A list of 157 genes was annotated using the AmiGO tool13 (version 1.7; March 28, 2010) based on the Gene Ontology data14 (GO:00163245) using the cell location search phrase “apical plasma membrane” with restriction to Homo sapiens. In total, 3,814 GWAS SNPs are within ±10 kb of the boundaries of 155 genes (NCBI36/hg18). Two genes were not tagged by any of the GWAS SNPs; SLC6A14 was not annotated to the apical plasma membrane.
(a) QQ-plot of the apical SNPs in the discovery sample. The observed association statistics (red), and the statistics calculated from the 10,000 phenotype-permutated replicates are shown (light gray).
(b) Statistical significance of the apical membrane hypothesis in the discovery sample. Statistical significance (permutation P = 0.0002) was established via a sum statistic, summing the association evidence (Wald χ21 statistic) over all the 3,814 SNPs with the observed sum statistic displayed as a vertical line (red), and the 10,000 permutation-based sum statistics displayed as a histogram (light gray).
(c) QQ-plot of the apical SNPs in the replication sample. The SNPs in the French replication cohort were required to have MAF > 6% because of the reduced sample size (1232 × 6% ≈ 3763 × 2%). In total 3,420 GWAS SNPs are within ±10 kb of the boundaries of 154 apical genes.
(d) Statistical significance of the apical membrane hypothesis in the French replication sample (permutation P=22/1,000=0.022).
To implement the GWAS-HD for meconium ileus using the apical hypothesis, we first prioritized the genome-wide markers by assigning the 3,814 SNPs of the apical genes to a high priority group and all remaining genome-wide SNPs to a low priority group. We then performed two statistical analyses (Supplementary Fig. 4 and Online Methods). The first, analogous to a conventional GWAS, was to conduct single-SNP association analysis using all of the 543,927 GWAS SNPs at the genome-wide level, however after up-weighting the 3,814 apical SNPs via the SFDR control.16 The second analysis, focusing only on the 3,814 high priority SNPs using a multi-SNP/gene analysis, tested the prioritization hypothesis itself to determine if multiple proteins present on the apical plasma membrane contribute to meconium ileus susceptibility.
As in the conventional GWAS, SNPs from SLC6A14 showed the strongest evidence for association with meconium ileus in the single-SNP GWAS-HD analysis (Supplementary Fig. 5), despite not being in the high priority group, reflecting the robustness of SFDR17. In addition, SNPs from SLC26A9, and two additional apical genes, ATP2B2 and SLC9A3, showed association evidence with q values <0.05 (Table 3). A gene-based analysis (Online Methods) of ATP2B2 (P=0.0006) and SLC9A3 (P=0.0001) indicated evidence for allelic heterogeneity after comparing results with single-SNP analysis (Table 3). SLC9A3 was replicated in the French cohort (P=0.017) while ATP2B2 was not (P=0.283) (Supplementary Table 3).
Table 3.
Ranked SNPs with q values < 0.05 from GWAS or GWAS-HD
| GWAS | GWAS-HD | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | CH R |
BP | P value | q valuea | rankb | q valuea | rankb | Apicalc | GENE |
| rs3788766 | X | 115480867 | 1.28×10−12 | 0 | 1 | 0 | 1 | 0 | SLC6A14 |
| rs4077468 | 1 | 204181380 | 9.88×10−9 | 0.0018 | 3 | 0 | 2 | 1 | SLC26A9 |
| rs4077469 | 1 | 204181508 | 9.88×10−9 | 0.0018 | 2 | 0 | 3 | 1 | SLC26A9 |
| rs7512462 | 1 | 204166218 | 2.14×10−8 | 0.0023 | 5 | 0 | 4 | 1 | SLC26A9 |
| rs7419153 | 1 | 204183932 | 1.01×10−7 | 0.0091 | 6 | 0.0001 | 5 | 1 | SLC26A9 |
| rs7415921 | 1 | 204177506 | 4.49×10−7 | 0.0346 | 7 | 0.0003 | 6 | 1 | SLC26A9 |
| rs12047830 | 1 | 204183322 | 3.72×10−6 | 0.167 | 12 | 0.0022 | 7 | 1 | SLC26A9 |
| rs5905283 | X | 115479909 | 1.69×10−8 | 0.0023 | 4 | 0.0045 | 8 | 0 | SLC6A14 |
| rs1318819d | 3 | 10413649 | 2.11×10−5 | 0.6309 | 18 | 0.0107 | 9 | 1 | ATP2B2 |
| rs4684689d | 3 | 10423426 | 2.76×10−5 | 0.6828 | 20 | 0.0123 | 10 | 1 | ATP2B2 |
| rs495435d | 3 | 10443723 | 4.80×10−5 | 0.7766 | 30 | 0.0191 | 11 | 1 | ATP2B2 |
| rs12741299 | 1 | 204181139 | 1.23×10−4 | 0.7766 | 81 | 0.0426 | 12 | 1 | SLC26A9 |
| rs1874361 | 1 | 204174809 | 1.31×10−4 | 0.7865 | 90 | 0.0426 | 13 | 1 | SLC26A9 |
| rs17563161d | 5 | 550624 | 1.47×10−4 | 0.796 | 98 | 0.0437 | 14 | 1 | SLC9A3 |
The q value is a genome-wide adjusted P value that controls the false discovery rate.
The rank indicates the genome-wide ordering of the SNPs based on the original association evidence (GWAS) or SFDR q value after incorporating the apical plasma membrane hypothesis (GWAS-HD). The GWAS and GWAS-HD rank results of all 543,927 SNPs are provided in Supplementary Fig. 5.
Apical indicates whether a gene was (= 1) or was not (= 0) on the generated apical membrane constituent list.
The SNPs from ATP2B2 and SLC9A3 identified by GWAS-HD; results in the French replication sample are P=0.1606, 0.1948, 0.0674, 0.0665 for rs1318819, rs4684689, rs495435, and rs17563161 respectively, where rs17563161 was imputed.
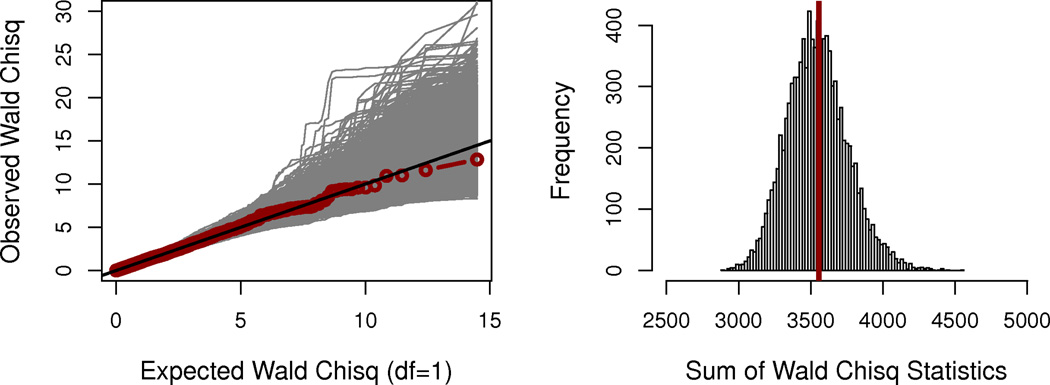
Next, restricting analysis to the 3,814 SNPs annotated to the 155 apical genes (which does not include SLC6A14), we tested the apical prioritization hypothesis as part of the GWAS-HD. Here we observed genome-wide significant evidence for association between meconium ileus and multiple constituents of the apical plasma membrane (permutation P=0.0002, testing all 3,814 SNPs jointly and not subject to multiple hypothesis testing (Fig. 2a and 2b)). Even with the exclusion of SLC26A9 (as well as SLC6A14), the apical hypothesis remained significant (P=0.0058). Thus, GWAS-HD further established the involvement of other genes coding for apical constituents despite insufficient power to detect individual SNPs, even within the context of our prioritized GWAS. For comparison, we also constructed a null hypothesis list of membrane-localized genes. As expected, the 224 GO-annotated nuclear envelope genes tagged by 3,537 GWAS SNPs showed no relationship with meconium ileus (permutation P=0.4639; Fig. 3).
Figure 3.
Assessment of the nuclear envelope null hypothesis. A list of 231 genes was generated from the nuclear envelope as defined by GO annotation (GO:0005635) similarly as for the apical membrane list. In total, 3,537 GWAS SNPs are within ±10 kb of the boundaries of 224 tagged genes (NCBI36/hg18). A priori, the nuclear envelope list should not contain genes associated with meconium ileus (under the null of no association).
(a) QQ-plot of the nuclear envelope gene SNPs in the discovery sample.
(b) Statistical significance of the nuclear envelope hypothesis in the discovery sample. Statistical evaluation indicates that genes listed in the nuclear envelope are, as expected, not significantly associated with meconium ileus (permutation P=4639/10,000=0.4639).
The French cohort with genome-wide data provided independent validation of the apical hypothesis (permutation P=0.022; Fig. 2c and 2d; Online Methods). The statistical significance of this gene set (which excludes SLC6A14) in the French cohort remained after further excluding SLC26A9 (P=0.021) and then both SLC26A9 and SLC9A3 (P=0.023). Although analysis of the apical hypothesis in a larger independent cohort should be considered as part of future efforts, the replication in the smaller French cohort supports a (common) mechanism of contributing genes, even though the responsible gene variants across the two datasets may not be the same.
To determine which apical genes were driving the association, the degree of genetic heterogeneity in meconium ileus, and the common contributors across the French and North American samples, we implemented Lasso18. Using the North American sample, we jointly analyzed all 3,740 SNPs available in the apical genes (which include SLC26A9 and SLC9A3), and SLC6A14 (Online Methods), that were not in perfect linkage disequilibrium. Forty-eight SNPs spanning 36 different genes were retained by Lasso in the multivariate regression model (Supplementary Table 3). These SNPs jointly explained ~17% of the meconium ileus variation in the North American sample. The percentage explained by the same 48 SNPs in the French sample was 8.1% (Online Methods), presumably due to the smaller sample size and genetic heterogeneity. We then tested the significance of a score for each individual in the French cohort constructed from a weighted sum of the number of risk alleles (defined in the North American sample) across the 48 SNPs19. The significant association between meconium ileus and this score (P=0.0137; Online Methods) provided additional complimentary evidence of common contributors between the two cohorts, with SNPs in SLC9A3 and SLC6A14 being two specific examples.
In summary, conventional GWAS identified SNPs in SLC6A14 and SLC26A9 as significantly associated with meconium ileus. GWAS-HD single-SNP analysis identified the same SNPs in SLC6A14 and SLC26A9, as well as SNPs in SLC9A3; and multi-SNP analysis provided evidence that multiple constituents of the apical plasma membrane are collectively associated with meconium ileus, yielding considerable additional information beyond single SNP/gene associations. GWAS-HD can be applied to other Mendelian disorders, or even complex traits provided there is a biologically-based hypothesis and participating relevant genes can be compiled.
Although gene prioritization has been used in other approaches such as pathway or gene enrichment analyses20–22, GWAS-HD involves key differences. First, in contrast to the previous inclusion/exclusion approaches where all genotyped SNPs/genes are not analyzed simultaneously, GWAS-HD performs parallel single-SNP analysis of all GWAS SNPs, and multi-SNP/gene analysis focusing on the set of SNPs/genes of interest. The prioritized single-SNP analysis interrogates all available SNPs via the SFDR control, yet enables increased statistical power for regions favoured a priori. For example, SLC6A14 would be omitted by inclusion/exclusion approaches, yet it remained the highest ranked gene for association with meconium ileus in GWAS-HD. Second, methods such as interactive pathway analysis can be restrictive because contributing genes/proteins must relate to each other via direct or indirect links, which may be disturbed when a cog component (such as CFTR) is dysfunctional in the disease state. While gene products that participate in maturation or delivery of CFTR may be contributory, consideration of local components of processes that may compensate for the ion and fluid flow disturbance in CF is enabled in the apical hypothesis. Third, distinct from an exhaustive search of all plausible interactive pathways, GWAS-HD focuses on a single biological hypothesis and provides statistical significance for all genes involved jointly, alleviating the multiple testing burden.
It should be noted that the specific statistics or models used in our GWAS-HD application such as SFDR, Lasso, and the sum and score statistics may not be the most powerful ones in any specific setting. For example, the adaptive rank truncated product statistic23 could be used to identify the common subset of associated apical genes across two samples; and there are alternative weighting and prioritization approaches24.
The SLC9A3 gene codes for a sodium/hydrogen exchanger that when disrupted has been shown to decrease intestinal obstructions in a CF mouse model25. SLC6A14 codes for a sodium- and chloride-dependant neutral and basic amino acid transporter26,27. SLC26A9 encodes an anion transporter, likely a chloride channel with multiple modes that include chloride/bicarbonate exchanger and sodium-anion cotransporter capabilities28. It has also been reported to physically interact with CFTR29 and be influenced by CFTR activity, at least in lung-related tissues30.
It is notable that SLC9A3 has previously been associated with infections and pulmonary function in CF31. In the consortium discovery sample, rs6864158 (MAF=0.43) in SLC9A3 was associated with both the lung (P=0.0003, analyzed previously5) and meconium ileus (P=0.0001); this provides evidence that some genes may play a role in multiple CF co-morbidities. Both SLC6A14 and SLC26A9 are robustly expressed in human lung epithelia, sweat gland, as well as intestinal epithelia as measured by RT-PCR (not shown). We anticipate that meconium ileus modifier genes may also influence early pathology in other CF-affected organs, and thus could provide important insights into the mechanisms of CF disease severity and co-morbidities.
These findings collectively have important practical implications for CF, where therapeutic strategies should consider pharmacologic modulation of epithelial function, in complementation with paradigms aimed at directly improving function or delivery of the mutated CFTR gene product to the apical membrane32.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the patients and families who participated in this study, the CFF Patient Registry, the University of North Carolina DNA Laboratory, and Genome Quebec and McGill University Innovation Centre; Isidro Wong for technical help with the replication genotyping; the computer specialists and the clinical research assistants, Jean-François Vibert, Malika Mahloul, Delphine Michon, Alexandra Blondel and Pauline Touche involved in the study design and patients’ recruitment of the French sample; Evan Hawbaker and Anthony Dang for data coordination; Drs. Yves Berthiaume, Andrew Sandford and Peter Pare for ascertainment of Canadian phenotype data and DNA; Jennifer Breaton, Mary Christofi, Nicole Anderson, Katherine Keenan, Chelsea Taylor and Julie Avolio for coordinating and verifying the Canadian data collection; Edgar Crowdy for database development and management in Canada; Sarah Norris, Adriane Kohl, Patricia Miller, Leia Charnin, Will Hannah, and Sonya Adams for recruitment, phenotyping and data entry at the University of North Carolina; Whitney Wolf for DNA, and Hemant Kelkar, Thomas Randall and Annie Xu for bioinformatics at the University of North Carolina; Nulang Wang, Kyle Kaniecki, Jenna Bonner and Christopher Watson for technical assistance at Johns Hopkins University.
This work was supported in part by Genome Canada through the Ontario Genomics Institute as per research agreement 2004-OGI-3-05 with the Ontario Research Fund, Research Excellence Program; the University of Toronto McLaughlin Centre; the Ontario Ministry of Research and Innovation Early Researcher (LJS); Cystic Fibrosis Canada (CFC) grants to PD, JZ, and LJS; Natural Sciences and Engineering Research Council (NSERC) to LJS; NSERC (250053-2008) and Canadian Institutes of Health Research (CIHR;MOP 84287) grants to LS; Lloyd Carr-Harris Foundation; the US National Institutes of Health grants: R01HL68927, K23DK083551, R01HL068890, R01DK066368, R01HL095396, P30DK027651, HG-0004314; U.S. Cystic Fibrosis Foundation (CFF) grants: CUTTIN06P0, R025-CR07, KNOWLE00A0, RDP-R026-CR07, DRUMM0A00; Flight Attendant Medical Research Institute grant: FAMRI2006; Lawson Wilkins Pediatric Endocrine Society grant: LWPES Clinical Scholar Award; Institut National de la Santé et de la Recherche Médicale, Assistance Publique-Hôpitaux de Paris, Université Pierre et Marie Curie Paris, Agence Nationale de la Recherche (R09186DS), Direction Générale de la Santé, Association Vaincre La Mucoviscidose, Chancellerie des Universités (Legs Poix), Association Agir Informer Contre la Mucoviscidose, GIS-Institut des Maladies Rares. Funds for genome-wide genotyping of North American participants were generously provided by the U.S. CFF. RD received a Joint Fellowship from the CIHR and Ontario Women's Health Council.
Appendix
Contributing North American CF Centers and Principal Investigators
Aaron,S., Ottawa General Hospital, Ottawa, Canada/Accurso,F., University of Colorado Health Sciences Center, CO/Acton,J., Cincinnati Children's Hospital and Medical Center, OH/Ahrens,R., University of Iowa Hospitals & Clinics, IA/Aljadeff,G., Lutheran General Children's Hospital, IL/Allard,C., Hôpital de Chicoutimi, Chicoutimi, Canada/Amaro,R., University of Texas at Tyler Health Center, TX/Anbar,R., SUNY Upstate Medical University, NY/Anderson,P., University of Arkansas, AR/Atlas,A., Morristown Memorial Hospital, NJ/Bell,S., The Prince Charles Hospital, Australia/Berdella,M., St. Vincents Hospital & Medical Center, NY /Berthiaume,Y., Hôtel-Dieu De Montréal/Biller,J., Children's Hospital of Wisconsin, WI/Bishop,G., Saint John Regional Hospital, Saint John, Canada/Bjornson,C., Alberta Children’s Hospital, Calgary, Canada/Black,H., Asthma & Allergy Specialists, Charlotte, NC/Black,P., Children's Mercy Hospital, MO/Boas,S., Children's Asthma Respiratory&Exercise Specialists, IL/Boland,M., Children’s Hospital of Eastern Ontario, Ottawa, Canada/Borowitz,D., Women & Children's Hospital of Buffalo, NY/Boswell,R., University of Tennessee, TN/Boucher,J., Centre Hospitalier Régional de Rimouski, Rimouski, Canada/Bourke,B., Our Lady’s Children Hospital, Ireland/Bowman,C.M., Medical University of South Carolina, SC/Boyle,M., Johns Hopkins Hospital, MD/Brown,C., California Pacific Medical Center, CA/Brown,D., Pediatric Pulmonary Associates., SC/Brown,N., University of Alberta Hospitals, Edmonton, Canada/Brusky,J., University Hospital, Saskatoon, Canada/Caffey,L.F., University of New Mexico, NM/Cantin,A., Centre hospitalier universitaire de Sherbrooke, Fleurimont, Sherbrooke, Canada/Casciaro,R., Istituto G. Gaslini, Italy/Castellani,C., Azienda Ospedaliera de Verona, Italy/Chatfield,B., University of Utah Health Sciences Center, UT/Chesrown,S., University of Florida, FL/Chilvers,M., B.C. Children’s Hospital, Vancouver, Canada/Chipps,B., Sutter Medical Center, CA/Cipolli,M., Azienda Ospedaliera de Verona, Italy/Clancy,J.P., University of Alabama at Birmingham, AL/Cohen,R., Kaiser Permanente, OR/Colombo,J., University of Nebraska Medical Center, NE/Cronin,J., Women & Children's Hospital of Buffalo, NY/Cruz,M., St. Mary's Medical Center, FL/Cunningham,J., Cook Children's Medical Center, TX/Davidson,G., B.C. Children’s Hospital, Vancouver, Canada/Davies,J, University of New Mexico, NM/Davies,L., University of New Mexico, SOM, NM/Debray,D., Centre Hospitalier Universitaire de Bicêtre, France/DeCelie-Germana,J., Schneider Children's Hospital, NY/Devenny,A., Royal Hospital for Sick Children, Scotland/DiMango,E., Columbia University Medical Center, NY/Doornbos,D., Via-Christi, St. Francis Campus, KS/Dozor,A., New York Medical College-Westchester Medical Center, NY/Dunitz,J., University of Minnesota, MN/Egan,M., Yale University SOM, CT/Eichner,J., Great Falls Clinic, MT/Elliot,G., Virginia Commonwealth University, Virginia/Farrell,J., Janeway Child Health Centre, St. John’s, Canada/Ferkol,T., St. Louis Children's Hospital, MO/Fiel,S., Morristown Memorial Hospital, NJ/Flume,P., Medical University of South Carolina, SC/ Freitag,A.,Chedoke-McMaster Hospital, Hamilton, Canada/ Franco,M., Miami Children's Hospital, FL/Froh,D., University of Virginia Health System, VA/Garey,N., Saint John Regional Hospital, Saint John, Canada/Geller,D., Nemours Children's Clinic Orlando, FL/Gershan,W., Children's Hospital of Wisconsin, WI/Gibson,R., Children’s Hospital & Regional Medical Center, WA/Giusti,R., Long Island College Hospital, NY/Gjevre,J., University Hospital, Saskatoon, Canada/Gondor,M., University of South Florida, FL/Gong,G., Phoenix Children's Hospital, AZ/Goulet,S., Centre Hospitalier Régional de Rimouski, Rimouski, Canada/Guill,M., Medical College of Georgia, GA/Gutierrez,H., University of Alabama at Birmingham, AL/Hadeh,A., Drexel University College of Medicine, PA/Hardy,K., Children's Hospital - Oakland, CA/Henderson,K., Janeway Child Health Centre, St. John’s, Canada/Hiatt,P., Texas Children's Hospital, TX/Hicks,D., Children's Hospital of Orange County, CA/Holmes,B., Regina General Hospital, Regina, Canada/Holsclaw,D., University of Pennsylvania, PA/Holzwarth,P., St. Vincent Hospital - Genetics, WI/Honicky,R., Michigan State University, MI/Howenstine,M., Riley Hospital for Children, IN/Hughes,D., IWK Health Centre, Halifax, Canada/Jackson,M., St. Mary’s Hospital, Kitchener, Canada/James,P., Lutheran Hospital, IN/ Jenneret A., Hôtel Dieu de Montréal, Montréal, Canada/Joseph,P., University of Cincinnati, OH/Kanga,J., University of Kentucky, KY/Katz,M., Baylor College of Medicine, TX/Kent,S., Victoria General Hospital, Victoria, Canada/Kepron,W., Health Sciences Centre, Winnipeg, Canada/Knowles,M., University of North Carolina at Chapel Hill, NC/Konig,P., University of Missouri-Columbia, MO/Konstan,M., Case Western Reserve University, OH/ Kovesi,T., Children’s Hospital of Eastern Ontario, Ottawa, Canada/Kramer,J., Oklahoma Cystic Fibrosis Center, OK/Kraynack,N., Children's Hospital Medical Center of Akron, OH/Kumar,V., Laurentian Hospital, Sudbury, Canada/Lacaille,F., Hôpital Necker-Enfants Malades, France/Lahiri,T., Fletcher Allen Health Care, VT/Landon,C., Pediatric Diagnostic Center, CA/Lands,L., Montréal Children’s Hospital, Montréal, Canada/Lapin,C., Connecticut Children's Medical Center, CT/Larj,M., Wake Forest University Baptist Med. Ctr., NC/Lavoie,A., Hôtel-Dieu De Montréal/Ledbetter,J., TC Thompson Children's Hospital, TN/Lee,R., Naval Medical Center - Portsmouth, VA/Leigh,M., University of North Carolina at Chapel Hill, NC/Lester,L., University of Chicago Children's Hospital, IL/Lever,T., Eastern Maine Medical Center, ME/Levy,H., Children's Hospital Boston, MA/Lieberthal,A., Kaiser Permanente Southern California, CA/Liou,T., University of Utah, UT/Lipton,A., National Naval Medical Center, MD/Lobritto,S., Columbia University Medical Center, New York/Lothian,B., Royal University Hospital, Saskatoon, Canada/Lougheed,D., Hotel Dieu Hospital, Kingston, Canada/Lyttle,B., London Health Sciences Centre, London, Canada/Macek,M., Charles University and University Hospital Motol, Czech Republic/Malhotra,K., Grand River Hospital, Kitchener, Canada/Marcotte,J., Hôpital Sainte-Justine, Montréal, Canada/Matouk,E., Montréal Chest Institute, Montréal, Canada/McCarthy,M., Providence Medical Center, WA/McColley,S., Children's Memorial Hospital & Northwestern University, IL/McCoy,K., Columbus Children's Hospital, OH/McNamara,J., Children's Hospitals and Clinics of Minneapolis, MN/Mencin,A., Columbia University Medical Center, New York/Michael,R., Queen Elizabeth II Health Sciences Centre, Halifax, Canada/Miller,S., University of Mississippi Medical Center, MS/Milot,M., Hôpital de Chicoutimi, Chicoutimi, Canada/Moffett,K., West Virginia University, WV/Montgomery,M., Alberta Children’s Hospital, Calgary, Canada/Moore,P., Vanderbilt University Medical Center, TN/Morgan,W., Tucson Cystic Fibrosis Center, AZ/Morris,R., Janeway Children’s Health & Rehabilitation, St. John’s, Canada/Morse,M., Methodist Children's Hospital, TX/Moskowitz,S., Children’s Hospital & Regional Medical Center, WA/Moss,R., Stanford University Medical Center, CA/Murphy,P., University of Nebraska Medical Center, NE/Nakielna,E., St. Paul’s Hospital, Vancouver, Canada/Nasr,S., University of Michigan Medical Center, MI/Nassri,L., Sparks Regional Medical Center, AR/Naureckas,E., University of Chicago Hospitals, IL/Nielson,D., University of California at San Francisco, CA/Noseworthy,M., Janeway Children’s Health & Rehabilitation, St. John’s, Canada/Noyes,B., St. Louis University, MO/Olivier,K., Wilford Hall USAF Med. Ctr. San Antonio, TX/Olson,E., University of Florida, FL/Omlor,G., Akron Children's Hospital, OH/Orenstein,D., Children's Hospital of Pittsburgh, PA/O'Sullivan,B., University of Massachusetts Memorial Health Care, MA/Parker,H.W., Dartmouth-Hitchcock Medical Center, NH/Passero,M., Brown University Medical School Rhode Island Hospital, RI/Pasterkamp,H., Children’s Hospital of Winnipeg, Winnipeg, Canada/Paterson,N., London Health Sciences Centre, London, Canada/Pedder,L., Chedoke-McMaster Hospital, Hamilton, Canada/Perkett,E., Vanderbilt University Medical Center, TN/Perrault,L., Centre de santé et des services sociaux de Rouyn-Noranda, Quebec, Canada/Perry,G., University of Kansas Medical Center, KS/Petit,N., Center Hospitalier Rouyn-Noranda, Rouyn-Noranda, Canada/Pian,M., University of California San Diego Children's Hospital, CA/Pineau,L., Grand River Hospital, Kitchener,Canada/Platzker,A., Children's Hospital of Los Angeles, CA/Popa,C., Centre de santé et des services sociaux de Rouyn-Noranda, Quebec, Canada/Prestidge,C., Children's Medical Center of Dallas, TX/Price,A., London Health Sciences Centre, London, Canada/Rabin,H., Foothills Medical Centre, Calgary, Canada/Radford,P., Phoenix Children's Hospital, AZ/Ratjen,F., The Hospital for Sick Children, Toronto, Canada/Regelmann,W., University of Minnesota, MN/Ren,C., University of Rochester Medical Center, Strong Memorial Hospital, NY/Repetto,G., Universidad del Desarrollo de Santiago, Chile/Retsch-Bogart,G., University of North Carolina at Chapel Hill, NC/Richards,W., Memphis Lung Physicians, MS/Riva,M., Via-Christi, St. Francis Campus, KS/Rivard,L., Centre hospitalier universitaire de Sherbrooke, Fleurimont, Sherbrook, Canada/Roberts,D., Providence Medical Center, AK/Rock,M., University of Wisconsin Hospital, WI/Rosen,J., Albany Medical College, NY/Rowland,M., Our Lady’s Children’s Hospital, Ireland/Royall,J., Childrens Hospital of Oklahoma, OK/Rubenstein,R., Children's Hospital of Philadelphia, PA/Ruiz,F., University of Mississippi Med. Ctr., MS/Scanlin,T., Children's Hospital of Philadelphia, PA/Schechter,M., Emory University School of Medicine, GA/Schmidt,H.J., Virginia Commonwealth University, VA/Schwartzman,M., Joe DiMaggio Children's Hospital, FL/Scott,P., Georgia Pediatric Pulmonology Assoc., PC, GA/Shay,G., Kaiser Permanente Medical Center, CA/Simon,R., University of Michigan Health System, MI/Smith,P., Long Island College Hospital, NY/Solomon,M., The Hospital for Sick Children, Toronto, Canada/Spencer,T., Children's Hospital of Boston, MA/Stecenko,A., Emory University, GA/Stokes,D., University of Tennessee, TN/Sullivan,B., Marshfield Clinic, WI/Taylor-Cousar,J., University of New Mexico, NM/Taylor,C., University of Sheffield, United Kingdom/Thomas,N., Pennsylvania State University College of Medicine, PA/Thompson,H., St. Luke's CF Clinic, ID/Toder,D., Children's Hospital of Michigan and Harper University Hospital, MI/Tullis,E., St.Michael’s Hospital, Toronto, Canada/Turcios,N., University of Medicine & Dentistry of NJ, NJ/Van Biervliet,S., Ghent University Hospital, Belgium/van Wylick,R., Hotel Dieu Hospital, Kingston, Canada/Varlotta,L., St. Christopher's Hospital for Children, PA/Vauthy,P., Toledo Children's Hospital, OH/Voynow,J., Duke University, NC/Wainwright,C., Royal Children's Hospital, Australia/Walker,P., St. Vincent’s Hospital - Manhattan, NY/Warren,W.S., Hershey Medical Center, PA/Waters,I., Royal Jubilee Hospital, Victoria, Canada/Wilmott,R., St. Louis University, MO/ Wilcox,P., St. Paul’s Hospital, Vancouver, Canada/Wilschanski,M., Hadassah Medical Organization, Israel/Wojtczak,H., Naval Medical Center - San Diego, CA/Yee,W., New England Medical Center, MA/Zacher,C., St. Alexius Heart & Lung CF Clinic, ND/Zanni,R., Monmouth Medical Center, NJ/Zeitlin,P., Johns Hopkins Hospital, MD/Zuberbuhler,P., University of Alberta Hospitals, Edmonton, Canada.
Contributing French CF Centers and Principal Investigators
Abely,M., American Memorial Hospital, Reims/Bassinet,L., Centre Hospitalier Intercommunal de Créteil, Créteil/Belleguic,C., Hôpital Pontchaillou, Rennes/Bellon,G., Hôpital Femme Mère Enfant, Bron/Bessaci,K., American Memorial Hospital, Reims/Bonnel,A.S., Hôpital André Mignot, Le Chesnay/Bremont,F., Hôpital des Enfants de Toulouse, Toulouse/Brouard,J., Centre Hospitalier Universitaire de Caen, Caen/Bui,S., Hôpital Des Enfants Groupe Pellegrin, Bordeaux/Chiron,R., Hôpital Arnaud de Villeneuve, Montpellier/Chumbi-Flores,R., Hôpital de la Tronche, Grenoble/Dalphin,J.C., CNRS-UFC, UMR 6249 Chrono-environnement, Hôpital Jean Minjoz, Besançon/Dalphin,M.L., Centre Hospitalier Universitaire de Besançon, Besançon/David,V., Hôpital Mère-Enfant, Nantes/De Miranda,S., Hôpital Foch, Suresnes/Derelle,J., Hôpital d’Enfants, Vandoeuvre les Nancy/Domblides,P., Hôpital Haut Lévêque, Pessac/Dominique,S., Centre Hospitalier Universitaire Charles Nicolle, Rouen/Dubus,J.C., Hôpital d’Enfants de la Timone, Marseille/Durieu,I., UCBL1,Groupe Hospitalier Lyon Sud - Hospices Civils de Lyon, Pierre Bénite/Dury,S., Hôpital Maison Blanche, Reims/Ellafi,M., Centre Hospitalier Universitaire de Caen, Caen/Epaud,R., Centre Hospitalier Intercommunal de Créteil, Créteil/Fanton,A., Hôpital d’Enfants du Bocage, Dijon/Fayon,M., Hôpital Des Enfants Groupe Pellegrin, Bordeaux/Fleurence,E., Hôpital d’Enfants, Saint-Denis de la Réunion/Foucaud,P., Hôpital André Mignot, Le Chesnay/Ginies,J.L., Centre Hospitalier Universitaire d’Angers, Angers/Godbert,B., Hôpital de Brabois, Vandoeuvre les Nancy/Grenet,D., Hôpital Foch, Suresnes/Guillot,M., Centre Hospitalier Robert Bisson, Lisieux/Heraud,M. C., Centre Hospitalier Estaing, Clermont-Ferrand/Housset,B., Centre Hospitalier Intercommunal de Créteil, Créteil/Hubert,D., Hôpital Cochin, Paris/Huet,F., Hôpital d’Enfants du Bocage, Dijon/Kessler,R., Hôpital Civil, Strasbourg/Labbé,A., Centre Hospitalier Estaing, Clermont-Ferrand/Laurans,M., Centre Hospitalier Universitaire de Caen, Caen/Le Bourgeois,M., Necker Hôpital d’Enfants Malades, Paris/Le Roux,P., Hôpital Jacques Monod, Montivilliers/Llerena,C., Hôpital de la Tronche, Grenoble/Loeuille,G.A., Centre Hospitalier de Dunkerque, Dunkerque/Marguet,C., Centre Hospitalier Universitaire Charles Nicolle, Rouen/Melly,L., Hôpital Renée Sabran, Giens/Moisan-Petit,V., Centre Hospitalier Bretagne Atlantique, Vannes/Munck,A., Hôpital Robert Debré, Paris/Murris-Espin,M., Hôpital Larrey, Toulouse/Nove-Josserand,R., Groupe Hospitalier Lyon Sud - Hospices Civils de Lyon, Pierre Bénite/Pautard,J.C., Hôpital Nord, Amiens/Pin,I., INSERM U823 Université Joseph Fourier,Hôpital de la Tronche, Grenoble/Pramil,S., Centre Hospitalier Universitaire Charles Nicolle, Rouen/Prevota,A., Hôpital Calmette, Lille/Rames,C., Hôpital Nord, Amiens/Rault,G., Centre de Perharidy, Roscoff/Reix,P., Hôpital Femme Mère Enfant, Bron/Remus,N., Centre Hospitalier Intercommunal de Créteil, Créteil/Renouil,M., Groupe Hospitalier Sud Réunion, Saint-Pierre de la Réunion/Reynaud-Gaubert,M., Hôpital Nord, Marseille/Richaud Thiriez,B., Hôpital Jean Minjoz, Besançon/Roussey,M., Université de Rennes 1, Hôpital Sud Annexe Pédiatrique, Rennes/Sermet-Gaudelus,I., Necker Hôpital d’Enfants Malades, Paris/Stremler,N., Hôpital d’Enfants de la Timone, Marseille/Uffredi,M.L., Centre Hospitalier Bretagne Atlantique, Vannes/Urban,T., Centre Hospitalier Universitaire d’Angers, Angers/Vigneron,P., Centre Hospitalier Bretagne Sud, Lorient/Wallaert,B., Hôpital Calmette, Lille/Weiss,L., Hôpital de Hautepierre, Strasbourg.
Footnotes
SOFTWARE AND DATASET URLS
AmiGO, http://amigo.geneontology.org/
CNVs, http://projects.tcag.ca/variation/
ENCODE, http://genome.ucsc.edu/ENCODE/
LocusZoom, http://csg.sph.umich.edu/locuszoom/
-
GWAS-HD Design and ConceptLJS, LS, JMR, PRD
-
GWAS DesignLJS, LS, JMR, PRD, RD, MC, JZ, HC, PYB, AC, MRK, WKO, FAW, GRC, MLD
-
Manuscript PreparationLJS, LS, JMR, XL, WL, PRD, HC, PYB, MRK, FAW, GRC, RD
-
Statistical AnalysisWL, XL, LS, LJS, TAC, PYB, PFB
-
Data CoordinationFAW, LH, SB, VD
-
Recruitment and PhenotypingPRD, JZ, KK, MC, HC, AC, MRK, WKO, RGP, JRS, SDW, LH, SB, KN
-
GenotypingFL, RVP, JMR, DZ, WKO, RGP, JRS, LH, RD
-
BioinformaticsTAC, JMR, RVP, PYB, PFB, RGP, JRS, RD
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Kerem B, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 2.Welsh MJ, Ramsey BW, Accurso FJ, Cutting GR, et al. Cystic fibrosis. In The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill, Inc; 2001. pp. 5121–5188. [Google Scholar]
- 3.Blackman SM, et al. Relative contribution of genetic and nongenetic modifiers to intestinal obstruction in cystic fibrosis. Gastroenterology. 2006;131:1030–1039. doi: 10.1053/j.gastro.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, et al. Understanding the population structure of North American patients with cystic fibrosis. Clin. Genet. 2011;79:136–146. doi: 10.1111/j.1399-0004.2010.01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright FA, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat. Genet. 2011;43:539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang K-Y, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 7.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrika. 1991;78:691–692. [Google Scholar]
- 12.Hodges CA, Grady BR, Mishra K, Cotton CU, Drumm ML. Cystic fibrosis growth retardation is not correlated with loss of Cftr in the intestinal epithelium. Am. J. Physiol. Gastrointest.Liver Physiol. 2011;301:G528–G536. doi: 10.1152/ajpgi.00052.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carbon S, et al. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GO-Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Craiu RV, Paterson AD, Bull SB. Stratified false discovery control for large-scale hypothesis testing with application to genome-wide association studies. Genet.Epidemiol. 2006;30:519–530. doi: 10.1002/gepi.20164. [DOI] [PubMed] [Google Scholar]
- 17.Yoo YJ, Bull SB, Paterson AD, Waggott D, Sun L. Were genome-wide linkage studies a waste of time? Exploiting candidate regions within genome-wide association studies. Genet. Epidemiol. 2010;34:107–118. doi: 10.1002/gepi.20438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B. 1996;58:267–288. [Google Scholar]
- 19.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedroso I, Breen G. Gene set analysis and network analysis for genome-wide association studies' in Genetics of Complex Human Diseases - A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2009. [DOI] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantor R, Lange K, Sinsheimer J. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu K, et al. Pathway analysis by adaptive combination of P-values. Genet. Epidemiol. 2009;33:700–709. doi: 10.1002/gepi.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas DC, et al. Use of pathway information in molecular epidemiology. Hum. Genomics. 2009;4:21–42. doi: 10.1186/1479-7364-4-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford EM, Sartor MA, Gawenis LR, Clarke LL, Shull GE. Reduced NHE3-mediated Na+ absorption increases survival and decreases the incidence of intestinal obstructions in cystic fibrosis mice. Am. J. Physiol. Gastrointest. Liver Physiol.y. 2009;296:G886–G898. doi: 10.1152/ajpgi.90520.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraga S, Serrao MP, Soares-da-Silva P. L-type amino acid transporters in two intestinal epithelial cell lines function as exchangers with neutral amino acids. J. Nutr. 2002;132:733–738. doi: 10.1093/jn/132.4.733. [DOI] [PubMed] [Google Scholar]
- 27.Höglund PJ, Adzic D, Scicluna SJ, Lindblom J, Fredriksson R. The repertoire of solute carriers of family 6: Identification of new human and rodent genes. Biochem. Biophys. Res. Commun. 2005;336:175–189. doi: 10.1016/j.bbrc.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 28.Chang MH, et al. Slc26a9--anion exchanger, channel and Na+ transporter. J. Membr. Biol. 2009;228:125–140. doi: 10.1007/s00232-009-9165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang MH, et al. Slc26a9 is inhibited by the R-region of the cystic fibrosis transmembrane conductance regulator via the STAS domain. J. Biol. Chem. 2009;284:28306–28318. doi: 10.1074/jbc.M109.001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertrand CA, Zhang R, Pilewski JM, Frizzell RA. SLC26A9 is a constitutively active, CFTR-regulated anion conductance in human bronchial epithelia. J. Gen. Physiol. 2009;133:421–438. doi: 10.1085/jgp.200810097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorfman R, et al. Modulatory effect of the SLC9A3 gene on susceptibility to infections and pulmonary function in children with cystic fibrosis. Pediatr. Pulmonol. 2011;46:385–392. doi: 10.1002/ppul.21372. [DOI] [PubMed] [Google Scholar]
- 32.Pedemonte N, et al. Small-molecule correctors of defective ΔF508-CFTR cellular processing identified by high-throughput screening. J. Clin. Invest. 2005;115:2564–2571. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The ENCODE Project Consortium. A User's Guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001046. e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood SD, et al. Inaccuracy of reporting of meconium ileus on case report forms of the gene modifier study. Abstract #200. Pediatr. Pulmonol. Suppl. 2007;42:272. [Google Scholar]
- 35.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dmitromanolakis A, Paterson AD, Sun L. Accurate IBD inference identifies cryptic relatedness in 9 HapMap populations. American Society of Human Genetics - 59th Annual Meeting; 2009. Abstract #1768. [Google Scholar]
- 38.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roeder K, Bacanu S, Wasserman L, Devlin B. Using Linkage Genome Scans to Improve Power of Association in GenomeScans. Am. J. Hum. Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.