Abstract
Altering gene expression in response to stimuli is a critical mechanism through which organisms execute developmental programs and respond to changes in their environment. Packaging of promoter DNA into chromatin can greatly impact the ability of RNA polymerase II to transcribe a gene. Promoter chromatin environments thus play a central role in establishing transcriptional output appropriate for specific environmental conditions or developmental states. Recent genomic studies have illuminated general principles of chromatin organization and deepened our understanding of how promoter sequence and nucleosome architecture may impact gene expression. Concurrently, pausing of polymerase during early elongation has been recognized as an important event influencing transcription of genes within stimulus-responsive networks. Promoters regulated by pausing are now recognized to possess a distinct chromatin architecture that may facilitate the plasticity of gene expression in response to signaling events. Here we review advances in understanding chromatin and pausing, and explore how coupling Pol II pausing to distinct promoter architectures may help organisms achieve flexible yet precise transcriptional control.
Keywords: chromatin, nucleosome, pausing, transcription
1. Paradigms for Regulation of Inducible Gene Expression by Chromatin
Understanding how gene expression levels are elevated or diminished in response to a signaling event is one of the central challenges of molecular biology. Stimulus-responsive transcription is integral to developmental programs and responses to changing environments. Its dysregulation is linked to a broad range of disease states. Extensive study of gene regulation has revealed that chromatin organization plays a central role in this process. Wrapping of DNA around histone octamers to form the fundamental unit of chromatin, the nucleosome, reduces DNA accessibility to the transcription machinery [1, 2]. Thus, the occupancy and position of nucleosomes with respect to binding sites for transcription activators and general transcription factors can influence these critical DNA-protein interactions and impact both basal gene expression and a gene’s inducibility by specific cues.
1.1 Modulating Gene Expression through Stimulus-induced Chromatin Opening and Pol II Recruitment
An elegant paradigm for chromatin-mediated regulation of inducible transcription has emerged, perhaps best illustrated by the activation pathway of the S. cerevisiae phosphate-repressed PHO5 promoter (reviewed in [3]). Under conditions of plentiful phosphate, positioned nucleosomes limit access to the PHO5 promoter. In response to phosphate starvation, the PHO4 transcription factor binds to activator sequences and nucleosomes are actively removed from the promoter by chromatin remodelers [4-6]. Nucleosome removal allows for recruitment of the transcription machinery and RNA synthesis. Regulation of stimulus-responsive transcription may then be distilled into a series of distinct and sequential events: i) a specific signal triggers a transcription activator to bind its target genes, recruiting chromatin remodelers to increase DNA accessibility, ii) this permits recruitment of the general transcription factors and RNA polymerase II (Pol II) to the promoter, and iii) the polymerase initiates transcription, followed directly by productive RNA synthesis (Figure 1A).
Figure 1. Two Paradigms for Regulation of Inducible Gene Expression.
A) Transcriptional activation at the S. cerevisiae PHO5 promoter proceeds through a series of ordered events: i) a signal (lightning bolt) leads to binding of transcription factor Pho4 to upstream activating sites (UAS) and removal of nucleosomes by chromatin remodelers (REMOD) permitting ii) recruitment of the transcription machinery and transcription initiation, followed directly by iii) productive RNA synthesis. Chromatin opening follows signaling.
B) In contrast, activation of the Drosophila heat shock genes proceeds through a distinct pathway: i) GAGA recruits chromatin remodeler NURF to remove nucleosomes and open promoter chromatin, allowing ii) Pol II initiation and transition to the paused state, whereupon iii) an activating signal (lightning bolt) triggers pause release and productive RNA synthesis. Note that chromatin opening precedes signaling.
1.2 Modulating Gene Expression through Stable Chromatin Opening and Pol II Pausing
Early studies revealed an alternative to this paradigm, where chromatin opening at a stimulus-responsive promoter preceded receipt of an activating signal. Prior to activation, genes encoding the stress-induced Drosophila heat shock proteins (hsps) were found to possess accessible promoter chromatin characterized by DNAseI hypersensitivity [7, 8] and reduced histone occupancy [9]. These accessible hsp promoters were also found to be pre-bound by Pol II and several general transcription factors [10-12]. Prior to heat shock, Pol II transcribes a 20-60 nt RNA before halting in early elongation. Heat shock releases this paused Pol II into the gene and promotes subsequent rounds of transcription, making use of the accessible chromatin structure and pre-assembled transcription apparatus to enable extremely rapid and robust gene activation.
Thus, in contrast to the PHO5 promoter, stimulus-responsive transcription from the paused hsp promoters proceeds through a distinct series of events: i) prior to receipt of a stimulus, nucleosome disassembly and promoter opening occur, followed by ii) recruitment, initiation and pausing of Pol II, subsequent to which iii) receipt of an activating stimulus triggers release of paused Pol II into productive elongation and synthesis of the RNA transcript (Figure 1B).
1.3 Pol II Pausing Is a Prevalent Mechanism Permitting Precise Regulation of Transcription
In the past five years, global surveys of Pol II distribution in numerous metazoan species have revealed that promoter-proximal pausing is not confined to the hsp genes but represents a prevalent mode of transcriptional regulation [13-17]. These studies employed diverse genomic methods for evaluating Pol II occupancy profiles and transcript formation (e.g. Pol II ChIP-chip, ChIP-seq, GRO-seq, short RNA-seq) to reach a common conclusion: gene expression is often limited – not through recruitment of Pol II to a gene promoter - but through the subsequent release of an engaged, paused Pol II downstream into the gene. Importantly, pausing is enriched among genes in stimulus-responsive pathways [13, 14, 18-20]. This has been taken to suggest that pausing’s major function is to facilitate induction of pathway targets like hsp genes. However, it is worth noting that critical components of stimulus-responsive signal transduction cascades, that are not themselves induced in response to stimuli, are also paused [21]. Furthermore, most paused genes are actively transcribed in un-stimulated cells [15, 19]. The de-coupling of promoter chromatin opening and productive transcription (Figure 1B) may therefore also be important for fine-tuning basal gene expression levels [19]. Together, these findings suggest that a substantial portion of transcription is regulated, not through signal-dependent chromatin opening and Pol II recruitment as seen at PHO5, but through pause release.
Understanding metazoan developmental and environmentally-responsive networks will therefore be aided by a detailed knowledge of the mechanisms through which pausing and chromatin cooperate to regulate gene expression. Here we review advances in understanding the relationship between Pol II pausing, promoter chromatin architecture, and precise control of transcription.
2. Pausing and Promoter Chromatin Accessibility
A critical distinction between inducible transcription at paused genes like the hsps and recruitment-regulated genes like PHO5 is the timing of chromatin opening. At PHO5, DNA is made accessible following receipt of a specific signal. At the hsps, DNA is made accessible prior to the receipt of an activating stimulus. Below we examine how promoter DNA attains and remains in an accessible state at paused genes.
2.1 Establishing Accessible Chromatin at Paused Promoters
At the hsp genes in Drosophila, the GAGA factor plays a key role in establishing an accessible promoter environment before a signal activating gene expression arrives. GAGA is a transcription factor that is constitutively bound upstream of the hsp promoters. GAGA recruits the NURF chromatin remodeler, which establishes a nucleosome-deprived, accessible promoter state [22, 23]. GAGA-binding motifs are highly enriched at paused promoters and ChIP-chip in Drosophila S2 cells indicates that GAGA occupies a considerable fraction of these potential binding sites [24]. GAGA-mediated recruitment of chromatin remodelers may therefore represent a general strategy for ‘opening’ the chromatin environment at a subset of paused promoters in the absence of activating signals. Interestingly, NURF initiates chromatin opening but appears not to be required for its maintenance [25], suggesting continuous promoter access might hinge upon other factors.
2.2 Maintaining Accessible Chromatin at Paused Promoters
Paused Pol II itself may play an important role in maintaining the open chromatin environment established by GAGA and NURF. Pausing results in part from the activity of the pause-inducing Negative Elongation Factor (NELF) complex [26]. Reduction of pausing through depletion of NELF in Drosophila cells causes a global decrease in Pol II promoter occupancy [19]. Surprisingly, this decrease in promoter Pol II was not accompanied by an increase in Pol II within gene bodies, or a general increase in RNA synthesis [21]. In contrast, the expression of many genes was down-regulated following NELF-depletion, suggesting that NELF-mediated pausing actually facilitated expression of these genes. The mechanism underlying this phenomenon was simple yet unexpected: the paused Pol II blocked nucleosome assembly and helped maintain an open, accessible promoter chromatin architecture [19, 21]. Loss of pausing allowed nucleosomes to occlude promoter DNA and prevent further recruitment of the transcription machinery.
Together, these findings suggest that after chromatin remodelers make a promoter accessible to the transcription machinery, Pol II pausing enables maintenance of this state. In this view, a promoter can be maintained in a continually accessible, ‘primed’ state as long as polymerase that is released into the gene is rapidly replaced by recruitment and pausing of additional Pol II. Importantly, this suggests a model for gene regulation wherein a diminished recruitment rate or a reduction in pausing could permit nucleosome reassembly and attenuate gene expression.
3. Global Relationships Between Promoter Architecture and Pausing
At the same time that pausing was emerging as a prevalent method of gene regulation, two major breakthroughs revolutionized our understanding of how promoter nucleosome environments are structured. First, rapidly evolving genomic technologies have enabled the creation of high-resolution genome-wide maps of nucleosome location and occupancy [19, 27-30]. These maps have moved us beyond the painstaking dissection of individual promoters, towards the capability to extract global rules governing nucleosome organization. Secondly, great progress has been made in understanding the contributions of DNA sequence to determining in vivo chromatin structure [31-33]. Importantly, these studies have led to the creation of models with significant power to predict the most thermodynamically favored locations for nucleosome binding [31, 34].
When coupled, these two advances permit both the visualization of nucleosome organization across an entire genome, and the ability to predict how specific arrangements of promoter nucleosomes may change in response to stimuli or in different cellular environments. This combination has proven powerful for understanding how Pol II pausing may regulate stimulus-responsive transcription. Below, we first review advances in understanding promoter nucleosome organization, and the contribution of DNA sequence to establishing promoter chromatin environments. Then, we explicitly consider how sequence-specified nucleosome architectures can be linked to Pol II pausing to regulate inducible gene expression.
3.1 Promoter Nucleosome Organization – The Basics
Maps of nucleosome occupancy now exist for many eukaryotic genomes [19, 27-30, 35]. Together they define some basic principles of promoter nucleosome organization, as well as highlighting differences between species. Transcription start sites are often located within or just downstream from so-called nucleosome free regions (NFRs) (Figure 2A). NFRs, which are actually nucleosome-depleted rather than truly nucleosome-free, represent local zones of ‘open’ chromatin that are more permissive for recruitment of transcription factors and the basic transcription machinery. NFRs frequently contain promoter DNA motifs such as the TATA box to facilitate this recruitment. Downstream of NFRs, nucleosomes tend to be relatively closely spaced in ordered arrays. The first nucleosome downstream of the NFR is referred to as the +1 nucleosome, followed by the +2, +3, etc. (Figure 2A). Upstream of the NFR, yeast promoters frequently possess a −1 nucleosome occupying a stereotypical position over the upstream activating sequences [27], while in metazoans, upstream nucleosomes occur in a greater variety of locations.
Figure 2. Influence of Pol II and the Transcription Machinery on Chromatin Architecture.
A) Nucleosome occupancy was summed across 17,116 Drosophila promoters to create a ‘metagene’ profile revealing the stereotypical promoter nucleosome arrangement. The locations of the nucleosome free region (NFR) and +1, +2, and +3 nucleosomes are indicated. Data are from [19].
B) Separating Pol II-bound promoters (purple) from Pol II-unbound promoters (green) reveals that Pol II binding is coincident with lower nucleosome occupancy upstream of the TSS (the NFR), and paradoxically, higher downstream occupancy and more regular nucleosome spacing.
C) Nucleosome occupancy is significantly lower downstream of highly paused promoters (red, quartile 1 of pausing indices, 1,866 promoters) than downstream of the least paused promoters (blue, quartile 4 of pausing indices, 1,867 promoters). Pausing index is calculated as (Pol II occupancy TSS +/−250 bp)/(Pol II occupancy TSS +500 bp to gene end) as in [19].
There are at least two important caveats when considering these basic rules. First, they are derived by averaging together nucleosome architectures at thousands of individual genes. Averaging many genes into a ‘metagene’ profile is a powerful way to detect patterns, but may obscure meaningful differences between individual genes. For example, averaging together nucleosome profiles from genes bound by Pol II with those from genes that lack Pol II binding obscures the major influence of Pol II upon nucleosome occupancy and positioning (Figure 2B, and see [36]). Similarly, averaging together all Pol II bound genes obscures differences in downstream nucleosome occupancy between highly paused genes and genes with little pausing (Figure 2C). Thus, care must be taken to avoid ‘averaging out’ distinct nucleosome organizations that are realized in living organisms [37].
Second, the degree to which the promoter region is actually free of nucleosomes remains unclear, and is highly variable. Whereas some promoters may exist as a region of stable nucleosome exclusion, others are best defined as regions of rapid nucleosome cycling [38]. As such, promoter-associated NFRs may best be thought of as regions of relatively low average nucleosome occupancy. Importantly, the level of nucleosome deprivation is dynamic and can be altered under specific conditions or in response to signaling events. For example, in the absence of Pol II binding, some promoters become stably occupied by nucleosomes [19, 21], and there are clear differences between promoter architecture at a given gene in distinct cell types [39].
3.2 The Contribution of DNA Sequence to Promoter Nucleosome Organization
Great progress has been made towards defining rules that govern whether specific DNA sequences favor or disfavor local nucleosome occupancy [31, 32, 34]. We next consider advances in our understanding of how promoter sequences may have evolved to help establish the specific nucleosome architectures discussed above.
Pioneering studies noted that, while nearly any piece of DNA could be induced to form nucleosomes, different sequences were particularly well or ill-suited to bend regularly around the core histone proteins. Specifically, it was noted that poly-dA:dT tracts were highly rigid and thus ‘nucleosome unfriendly’, generating less stable nucleosomal particles. Poly-dA:dT stretches were found at a number of yeast promoters, where they were shown to intrinsically impede nucleosome formation and favor an ‘open’ chromatin environment [40]. More recently, genome wide comparisons between in vivo nucleosome occupancy and DNA-driven in vitro nucleosome assemblies extended these observations to demonstrate a broad role for poly-dA:dT features in establishing NFRs upstream of yeast promoters [31, 32].
DNA sequence generally favors a different nucleosome organization at metazoan promoters. In stark contrast to the widespread poly-dA:dT-driven nucleosome depletion at yeast promoters, many mammalian promoters have high dC:dG content [33]. dC:dG-rich sequences generally favor nucleosome formation, and the percentage of dC:dG content was found to be a key determinant of sequence-specified nucleosome organization [34]. Accordingly, human promoters intrinsically favor high nucleosome occupancy [33].
Many mammalian promoters are characterized by regions of high CpG dinucleotide occurrence known as CpG islands. Sequence-based models predict particularly high nucleosome occupancy at CpG island-containing promoters [33]. This prediction is controversial, as it has been alternately proposed that the high dC:dG content of CpG islands can actually drive nucleosome depletion [41]. However, although CpG island promoters do generally feature low nucleosome occupancy in vivo, this is coupled with high Pol II occupancy [28, 42].
A paradox has thus emerged. A substantial fraction of mammalian promoters have underlying dC:dG-rich DNA sequences that favor high nucleosome occupancy. In vivo, however, many of these promoters exist in a nucleosome-deprived state. What leads to this striking divergence between the thermodynamically most-favored state (nucleosome occupancy) and the most frequent in vivo state (nucleosome deprivation)? We propose that occupancy of many metazoan promoters by polymerase prevents them from realizing their nucleosome-occupied thermodynamic ground states. Furthermore, we suggest that competition between paused Pol II and nucleosomes for promoter binding is a key event in regulating transcriptional output. Below, we explicitly examine the relationship between Pol II pausing, promoter nucleosome architecture, and promoter DNA sequence.
3.3 Pausing is Coupled to a Distinct Promoter Nucleosome Architecture
Does underlying DNA sequence impact promoter nucleosome architecture at highly paused genes, and at genes that lack pausing? Using the model developed by Kaplan et al., we identified two important features of sequence-specified promoter chromatin that differentiate highly paused and less paused genes.
First, highly paused genes are predicted, on average, to have low nucleosome occupancy in the +1 location and further downstream. In contrast, genes that lack pausing are predicted to have higher downstream occupancy [19]. This finding was surprising, as it was proposed that high +1 nucleosome occupancy physically impedes the progress of the polymerase and may therefore generally ‘cause’ pausing [29]. However, detailed analyses of chromatin architecture at the hsp70 locus indicate that the first downstream nucleosome is ~200 bp away from paused Pol II [43], and in vitro transcription assays demonstrated that pausing occurs in a nucleosome-independent manner [44]. In agreement with this, global analysis of nucleosome occupancy in Drosophila revealed that many highly paused genes resemble the hsp70 promoter in lacking well positioned nucleosomes downstream of the transcription start site [19] and have lower downstream nucleosome levels than genes lacking pausing (Figure 2C). Thus, these findings argue strongly against the nucleosome playing a general role in establishing the paused polymerase. In fact, DNA sequence appears to limit stable nucleosome occupancy downstream of paused promoters, reducing obstacles to productive synthesis upon release of paused Pol II. Interestingly, paused genes have elevated intron content downstream of the TSS [19]. Introns are recognized to have substantially higher dA:dT content and lower nucleosome occupancy than exons [45-48]. Therefore, the relative deficit of nucleosomes downstream of paused promoters may result from the elevated intron content of these regions.
Secondly, paused genes have DNA sequences that favor high nucleosome occupancy over the promoter region relative to genes that lack pausing [19]. In particular, paused promoters have a high intrinsic preference to form nucleosomes just upstream of the transcription start site, with a peak of predicted nucleosome occupancy centered ~35 bp upstream of the TSS . In the absence of Pol II occupancy, DNA sequence preferences may drive these promoters towards the thermodynamically favored, nucleosome-occupied and repressed state. In contrast, less paused promoters, which include a number of housekeeping genes, have DNA sequences that favor constitutive nucleosome depletion, similar to many yeast promoters.
Importantly, in vivo nucleosome architectures closely matched sequence based predictions for both highly paused genes and genes with low levels of pausing, when genes were examined in the absence of Pol II binding [19]. To make this determination, we first identified a group of genes that existed in a highly paused state in Drosophila embryos, but were found in a Pol II-absent state in Drosophila S2 cells. Sequence-based models predict high promoter nucleosome occupancy and low downstream occupancy for this group of genes. When paused (i.e. in embryos), these genes displayed very low promoter nucleosome occupancy. However, in the absence of Pol II binding (i.e. in S2 cells), the same genes displayed high promoter nucleosome occupancy. Thus, in the absence of paused Pol II, these promoters adopted their sequence-predicted nucleosome architectures. Notably, genes with low levels of pausing in embryos adopted a different sequence-specified architecture when Pol II was absent in S2 cells, characterized by low promoter and higher downstream occupancy.
Many paused promoters thus have a distinct sequence-specified nucleosome architecture, characterized by high promoter occupancy and lower nucleosome levels downstream. Subsequent to nucleosome remodeling initiated by factors such as GAGA and NURF, Pol II pausing, in conjunction with other mechanisms, can maintain these promoters in a nucleosome deprived state, thereby facilitating ongoing and future gene expression. In the absence of pausing, DNA sequence can help drive high nucleosome occupancy, functionally limiting gene expression. This competition for promoter binding between paused Pol II and nucleosomes may represent a key regulatory switch used to modulate inducibility of stimulus-responsive transcriptional networks.
3.4 Regulation of Stimulus-responsive Transcription Through Pausing and DNA-specified Nucleosome Organization
We envision that the competition between paused Pol II and nucleosomes creates a switch between high nucleosome occupancy (OFF state) and pausing (PAUSED/ON state). Under conditions where gene activity is desired, or when a promoter must be kept in a stimulus-responsive state, nucleosome remodeling would be initiated, to allow for recruitment of the transcription machinery and establishment of paused Pol II. The extended residence time of paused Pol II at these promoters would effectively ensure that nucleosome reassembly did not occur, maintaining the gene in an active or activate-able state. Importantly, by linking nucleosome deprivation to pausing, rather than gene activity or the presence of a specific transcription factor, even a gene with very low basal expression could be held ‘open’ for an extended period of time.
Under conditions where basal transcription or stimulus-responsiveness was not warranted, the promoter could adopt its thermodynamic ‘default’ architecture, where nucleosomes would occupy the promoter, preventing Pol II recruitment and transcription. Multiple lines of evidence support this model. We find that hormone-responsive promoters in Drosophila can transition between nucleosome occupied OFF states and paused PAUSED/ON states (or vice-versa) within 24 hours of hormone treatment [19]. In addition, during T cell activation, genes repressed over an 18-hour period show increased nucleosome occupancy [28].
It is unknown by what mechanisms promoters might transition between nucleosome-deprived PAUSED/ON states and nucleosome-occupied OFF states. Permitted sufficient time, thermodynamic fluctuations should drive nucleosomes to their most favored positions (e.g. just upstream of the TSS at highly paused genes). However, a promoter could transition from the nucleosome-free to the nucleosome-occupied state much more efficiently when assisted by histone chaperones [49] or ATP-dependent chromatin remodelers.
4. Pausing and Chromatin Landscapes
In addition to modulating DNA accessibility, nucleosomes can regulate transcription by functioning as platforms for covalent histone modifications and binding of transcriptional co-activators. Two recent landmark studies in Drosophila have distilled genome-wide occupancy patterns of numerous histone modifications and chromatin binding proteins into a handful of distinct ‘chromatin landscapes’ [50, 51]. These investigations reveal that highly-regulated genes, such as those controlling developmental programs, are not associated with ‘average’ actively transcribed chromatin. Instead, they are coupled to particular landscapes enriched for specific histone modifications and chromatin binders. Here we briefly review these advances in understanding global chromatin organization and examine the association of pausing with these newly-defined chromatin states. We then explore potential roles for specific chromatin landscapes in determining whether a promoter adopts a POISED/ON state or OFF state.
4.1 Pausing Is Linked to Specific Chromatin Landscapes
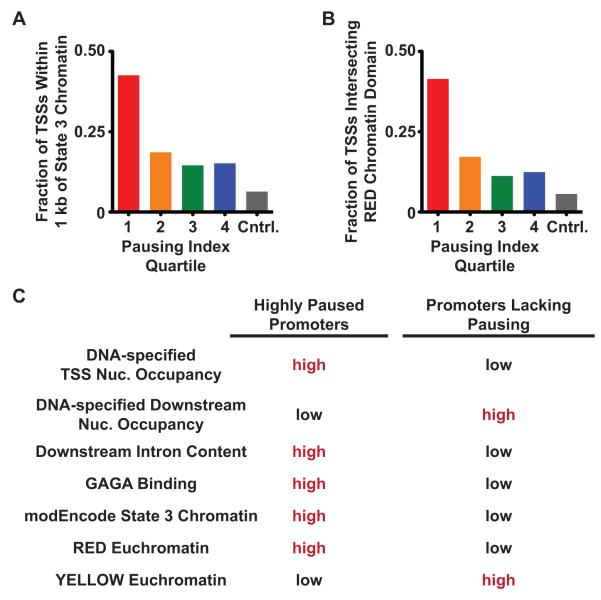
The modENCODE consortium used genomic distributions of many histone modifications and chromatin binding proteins to define 9 distinct chromatin states that occur in Drosophila [51]. Of these, state 3 is similar to chromatin found at mammalian enhancers and is enriched near genes with functions in development and stimulus-responsive transcriptional programs. At many of these genes, the state 3 domain is found within a lengthy 1st intron. Additionally, cell-type specific regions of open chromatin (i.e. those likely associated with stimulus-responsive genes) were also found most frequently in state 3 domains. Highly paused genes are also linked to developmental regulation and display elevated intron content [19]. These shared features suggest that state 3 chromatin and pausing might occur frequently at the same genes. We investigated this possibility and find substantial enrichment of state 3 domains within 0.5 kb of highly paused promoters compared to other Pol II-bound promoters or control genomic regions selected at random (Figure 3A). The cooccurrence of state 3 chromatin and pausing hints at shared regulatory functions, but further studies are necessary to illuminate a functional relationship.
Figure 3. Paused Promoters are Linked to Distinct Chromatin Landscapes.
A) The most highly paused promoters in Drosophila S2 cells (quartile 1 of pausing indices, n=1,866 promoters) are frequently in close proximity to ‘state 3’ chromatin [51]. State 3 is characterized by modifications to histone H3 (H3K4me1, H3K27ac, and H3K18ac) and is associated with developmental regulation. Bars indicate the fraction of TSS regions within 0.5 kb of a region of ‘state 3’ chromatin. Total Pol II ChIP-chip data and pausing index calculations are from [19]. Less paused genes in pausing index quartiles 2-4 are shown for comparison, along with control genomic regions selected at random (Cntrl., n=1867).
B) The most highly paused Drosophila promoters frequently overlap with a region of RED chromatin [50]. RED chromatin is characterized by the presence of GAGA, BRM, and MED31, and is linked to developmentally regulated genes that show tissue-restricted expression patterns. Bars indicate the fraction of TSS regions (TSS +/−250 bp) that intersect with a region of RED chromatin. Total Pol II ChIP-chip data from Kc cells were generated by the modENCODE project [52] and pausing indices were calculated as in [19]. Less paused genes in quartiles 2-4 are shown for comparison, along with random control genomic regions.
C) Highly paused Drosophila promoters and those lacking pausing can be differentiated by the relative frequency of occurrence of specific chromatin features as indicated.
Employing a technically distinct but philosophically similar approach to the modENCODE consortium, Filion et al. employed DAM-ID to determine genomic distributions of a large group of chromatin binding/modifying proteins and key histone modifications [50]. Their efforts resulted in a map of 5 distinct chromatin types (‘chromatin colors’) in Drosophila Kc cells. Notably, they identified two distinct types of transcriptionally active chromatin. RED chromatin is linked to genes that are developmentally regulated and show tissue-restricted gene expression, while YELLOW chromatin typifies genes with broad (less regulated) expression patterns. Interestingly, RED chromatin was distinguished from all other chromatin types by the presence of several proteins including the mediator subunit MED31, the ATP-dependent chromatin remodeler BRM, and GAGA.
The linkage of GAGA to RED chromatin is particularly intriguing. As noted above, GAGA cooperates with NURF to remodel nucleosomes and increase DNA accessibility at a number of paused promoters [22]. GAGA is thus a shared node between pausing and ‘RED chromatin’ regulatory networks. Using Pol II binding data generated by the modENCODE consortium in Kc cells, we determined relative pausing levels for Pol II bound genes (pausing indices, calculated as the ratio of the promoter Pol II signal to the downstream signal, as in [19]) and asked where paused promoters fell in relation to the chromatin colors. This analysis revealed that highly paused genes showed substantially greater overlap with RED chromatin relative to less-paused promoters (Figure 3B). Why might pausing and RED chromatin frequently co-occur? One possibility is that the RED chromatin landscape helps to establish pausing at a specific set of promoters.
4.2 Pausing and Chromatin Landscapes
We propose that RED chromatin may help determine whether a gene is found in either the PAUSED/ON state or the OFF state. GAGA is much more likely to recognize cognate binding sites located within RED chromatin than those found in other chromatin colors [50]. This suggests that as-yet unidentified features of RED chromatin may direct GAGA to bind to a limited subset of all potential sites. In effect, RED chromatin may function as a ‘map’ directing GAGA to appropriate binding sites. This could determine –in a cell-type-specific or condition-specific manner – which promoters are rendered accessible by GAGA and NURF and thus which are able to enter the signal-responsive PAUSED/ON state. Establishing which features of RED chromatin govern GAGA binding may be a key to understanding promoter transitions between the OFF and PAUSED/ON states.
Interestingly, pausing-enriched RED chromatin is distinguished from other chromatin types by the presence of multiple modulators of chromatin structure, including the ATP-dependent nucleosome remodeler BRM, the chromatin assembly factor CAF-1 (a component of the NURF complex), and SU(VAR)2-10 [50]. Understanding how these factors may contribute to the unique architecture of paused promoters is of great interest. Additionally, it will be important to determine if the chromatin color ‘map’ changes during development or in response to stimuli, and in doing so changes the spectrum of genes regulated by pausing. Recently identified patterns of co-occurrence between pausing and specific chromatin features (summarized in Figure 3C) should facilitate the formation and testing of hypotheses regarding their coupled regulation of stimulus-responsive transcription.
5. Conclusions
Both pausing and promoter chromatin are recognized to play important roles in altering gene expression in response to environmental and developmental cues. It has become apparent that, far from simply coexisting at highly regulated promoters, an intricate linkage between pausing and chromatin exists, where perturbation of either impacts the other. Rapid experimental advances have produced many hypotheses regarding the mechanisms through which nucleosome architectures, histone modifications, chromatin remodelers, and paused Pol II may cooperate to control inducible gene expression. Testing these will require a more detailed understanding of both pausing and promoter chromatin environments, and careful consideration of the interplay between these key regulators of inducible transcription.
Highlights.
Pol II pausing and promoter chromatin cooperate to precisely regulate transcription
Pausing is coupled to specialized promoter nucleosome architectures
Distinct chromatin landscapes are enriched in paused promoters
Promoter architecture and chromatin landscapes may enable cell-type specific pausing
Acknowledgements
We thank Adelman lab members and Dr. Paul Wade for critical reading of the manuscript, and David C. Fargo and Adam Burkholder for bioinformatics support. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to K.A. (Z01 ES101987).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- [2].Pina B, Bruggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- [3].Reinke H, Horz W. Anatomy of a hypersensitive site. Biochim Biophys Acta. 2004;1677:24–29. doi: 10.1016/j.bbaexp.2003.09.014. [DOI] [PubMed] [Google Scholar]
- [4].Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Molecular cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- [5].Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Molecular cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- [6].Adkins MW, Williams SK, Linger J, Tyler JK. Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Molecular and cellular biology. 2007;27:6372–6382. doi: 10.1128/MCB.00981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- [8].Keene MA, Corces V, Lowenhaupt K, Elgin SC. DNase I hypersensitive sites in Drosophila chromatin occur at the 5′ ends of regions of transcription. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Karpov VL, Preobrazhenskaya OV, Mirzabekov AD. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5′ region. Cell. 1984;36:423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- [10].Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- [11].Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- [13].Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nechaev S, Fargo DC, Dos Santos G, Liu L, Gao Y, Adelman K. Global Analysis of Short RNAs Reveals Widespread Promoter-Proximal Stalling and Arrest of Pol II in Drosophila. Science. 2009 doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci U S A. 2008;105:7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & development. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- [23].Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- [24].Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Molecular cell. 1997;1:141–150. doi: 10.1016/s1097-2765(00)80015-5. [DOI] [PubMed] [Google Scholar]
- [26].Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- [27].Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- [28].Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tillo D, Hughes TR. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. Embo J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rozenberg JM, Shlyakhtenko A, Glass K, Rishi V, Myakishev MV, FitzGerald PC, Vinson C. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics. 2008;9:67. doi: 10.1186/1471-2164-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Benjamin LR, Gilmour DS. Nucleosomes are not necessary for promoter-proximal pausing in vitro on the Drosophila hsp70 promoter. Nucleic Acids Res. 1998;26:1051–1055. doi: 10.1093/nar/26.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Molecular cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- [47].Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schwartz S, Meshorer E, Ast G. Chromatin organization marks exonintron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- [49].Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr Opin Genet Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- [50].Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, Linder-Basso D, Plachetka A, Shanower G, Tolstorukov MY, Luquette LJ, Xi R, Jung YL, Park RW, Bishop EP, Canfield TK, Sandstrom R, Thurman RE, MacAlpine DM, Stamatoyannopoulos JA, Kellis M, Elgin SC, Kuroda MI, Pirrotta V, Karpen GH, Park PJ. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]