Abstract
While studies of alternative pre-mRNA splicing regulation have typically focused on RNA-binding proteins and their target sequences within nascent message, it is becoming increasingly evident that mRNA splicing, RNA polymerase II (pol II) elongation and chromatin structure are intricately intertwined. The majority of introns in higher eukaryotes are excised prior to transcript release in a manner that is dependent on transcription through pol II. As a result of co-transcriptional splicing, variations in pol II elongation influence alternative splicing patterns, wherein a slower elongation rate is associated with increased inclusion of alternative exons within mature mRNA. Physiological barriers to pol II elongation, such as repressive chromatin structure, can thereby similarly impact splicing decisions. Surprisingly, pre-mRNA splicing can reciprocally influence pol II elongation and chromatin structure. Here, we highlight recent advances in co-transcriptional splicing that reveal an extensive network of coupling between splicing, transcription and chromatin remodeling complexes.
Keywords: alternative pre-mRNA splicing, chromatin, transcription elongation, RNA polymerase II
1. Introduction
In 1977, the Sharp and Roberts laboratories independently made the landmark discovery that genes of higher organisms are separated by non-coding sequences [1, 2]. Since their initial description, introns have been the subject of substantial investigation regarding their potential role. As a result, what was once considered “junk DNA” has now been granted the status of an important regulator of eukaryotic gene expression. Amongst other functions, intron-exon architecture serves as a critical platform for transcriptome diversification via alternative pre-mRNA splicing. Indeed, the first examples of alternative splicing emerged only a few years after the identification of introns, highlighting the shear pervasiveness. It is now commonly accepted that alternative splicing of pre-mRNA in higher organisms is the norm, and greater than 90% of human genes engage in alternative splicing [3, 4]. However, coincident with the transcriptome expansion in higher organisms, introns have lengthened and splice site strengths have weakened [5]. Thus, the evolutionary drive for transcript diversification has posed the spliceosome with the increasingly difficult task of identifying short splice site consensus sequences in a sea of non-coding intronic sequence. Alternative pre-mRNA splicing adds an additional layer of complexity in that splice site recognition must be diversified in a context-dependent manner. Consequently, it should not be surprising that pre-mRNA splicing is coordinated at multiple levels. In addition to regulation via RNA-binding protein recognition of cis-elements encoded within pre-mRNA, recent evidence suggests that transcription elongation rate and chromatin structure contribute to splice site recognition. Rather than operating independently, these processes are highly integrated as a result of co-transcriptional pre-mRNA splicing. However, unlike 5’ end capping and 3’ polyadenylation, which exclusively occur co-transcriptionally, recent evidence suggests that splicing can occur co- or post-transcriptionally [6]. As a result of these new findings, the model of pre-mRNA splicing regulation has required some substantial changes. In particular, we can no longer examine mRNA splicing without also examining structural determinants at the corresponding DNA.
2. Spliceosome assembly
The traditional view of alternative pre-mRNA splicing is based on combinatorial binding of trans-acting factors to composite recognition elements encoded within the nascent transcript to enhance or inhibit spliceosome assembly. The spliceosome is a megadalton complex that binds conserved sequences at the 5’ and 3’ ends of introns [7]. The major spliceosome consists of 5 small nuclear ribonucleoproteins (snRNPs), U1, U2, U4, U5 and U6, and as many as 150 associated proteins [8]. Spliceosome assembly involves a series of discrete complexes that catalyze intron removal through two transesterification steps [7, 9]. If the sequence context surrounding the conserved dinucleotide motif at either splice site is suboptimal, alternative splicing of the exon can occur. The net binding of positive acting factors, such as SR proteins, to exonic and intronic splicing enhancers (ESEs and ISEs) and negative acting factors, such as hnRNPs, to exonic and intronic splicing silencers (ESSs and ISSs) can promote or inhibit spliceosome assembly at the weak splice sites, respectively [10].
3. Co-transcriptional splicing
Early models of pre-mRNA splicing envisioned a distinct spliceosome compartment, spatially separated from transcription. However, evidence for direct coupling between transcription and splicing machineries quickly mounted. The first compelling demonstration of co-transcriptional spliceosome assembly extended from electron micrographs of Drosophila melanogaster embryonic transcription units showing intron looping and associated ribonucleoprotein complexes on transcripts tethered to DNA [11, 12], suggestive of mRNA splicing prior to transcript release. Subsequent immunofluorescence of splicing factors showed localization at sites of transcription [13, 14] in intron-containing genes [15] and RNA in situ hybridization with splice junction probes detected spliced mRNAs at their gene loci [16], thus establishing spatial and functional coupling between transcription and splicing. In recent years, advances in chromatin immunoprecipitation (ChIP) assays and quantitative RT-PCR have provided insight into the extent and kinetics of co-transcriptional splicing. Support for co-localization of splicing and transcription machineries extended from ChIP detection of spliced mRNAs associated with chromatin in yeast [17–19] and in mammalian cells at intron-containing genes [20]. Functional coupling was suggested from the observation that mutation of a yeast SR protein homolog, Npl3, reduced occupancy of U1 and U2 at Npl3 target genes [21].
While these studies definitely established pre-mRNA splicing prior to transcript release, the pervasiveness of coupling remained undefined. To quantitatively assess co-transcriptional splicing, analysis of intron removal in c-Src and fibronectin nascent chromatin associated RNA versus free RNA in the nucleoplasm was performed in human cell lines. This analysis revealed that the vast majority of constitutive exons are co-transcriptionally spliced in a general 5’ to 3’ order and that 3’ exons are more likely to be post-transcriptionally processed [22]. Internal introns flanking alternative exons were also removed co-transcriptionally, although to variable extents [22]. These data begin to reveal a kinetic aspect to splicing, which was further supported by the in vitro observation that SR proteins more effectively enhance co-transcriptional splicing than post-transcriptional splicing [23]. Support for kinetic regulation of alternative splicing comes from two recent studies that linked pol II pausing to co-transcriptional splicing in yeast. The Neugebauer laboratory isolated chromatin associated RNA to show that pol II pauses within terminal exons allowing sufficient time for intron excision prior to transcript release [24]. Similarly, the Beggs laboratory utilized a high-resolution splicing reporter system to demonstrate splicing dependent pol II pausing at the 3’ ends of introns coincident with splicing factor recruitment [25]. These studies raise the question whether pol II pausing represents a splicing “checkpoint.” However, a separate genome-wide analysis in yeast indicated that the majority of yeast exons are spliced post-transcriptionally [26]. Despite potential discrepancies in the extent of co-transcriptional splicing, altogether these data suggest that functional coupling of transcription and splicing has evolved to enhance spliceosomal detection of splice sites across large introns.
4. The Molecular Basis of Coupling- pol II CTD
Several lines of evidence suggest that coupling between transcription and splicing is mediated via transcription dependent recruitment of RNA processing factors. Amongst other demonstrations, redistribution of GFP-tagged splicing factors from nuclear speckles to sites of transcription was reduced in the presence of transcriptional inhibitors [13] and splicing factor ChIP indicated transcription-dependent SR protein recruitment to an inducible target RNA [27]. Importantly, with the exception of U1 snRNP [28], spliceosomal proteins are specifically recruited to intron-containing genes [20, 28], bringing the mechanistic basis of transcription dependent spliceosome recruitment into question. Based on the observation that chimeric minigenes of RNA polymerase III promoters fused upstream of pol II dependent genes are deficient in splicing and polyadenylation, pol II itself was implicated as a potential regulator of spliceosome assembly [29]. Indeed, mutational and deletional analysis of the carboxy-terminal domain (CTD) of the RPB1 subunit of pol II revealed multiple defects in mRNA processing [30, 31]. The CTD consists of tandem YSPTSPS repeats, which vary in number from 26 in yeast to 52 in humans [32]. Dynamic phosphorylation of serine residues on CTD heptad repeats is associated with the stages of pol II elongation (detailed in Lewis and colleagues, this issue). Serine 5 phosphorylation peaks near promoters and remains high proximal to the promoter, but declines within a few hundred nucleotides from the transcriptional start site (TSS) [33]. In contrast, serine 2 phosphorylation gradually increases with distance from the promoter [33]. Pol II is also variably phosphorylated on serine 7 [34] at the promoter and into genes bodies [35–37], however the significance to mRNA processing is less well understood. Given the dynamics of CTD phosphorylation as well as the role of CTD in mRNA processing, it was proposed that phosphorylation of CTD functions to physically tether mRNA processing factors to the transcription elongation complex (TEC) in a stage-specific manner [38, 39]. As described below, while substantial evidence for direct recruitment of 5’ and 3’ end processing proteins by CTD has accumulated, the mechanistic basis for CTD in splicing regulation is less clear.
4.1. 5’ Capping
In the first step of transcript synthesis, unphosphorylated pol II is recruited to the TSS via the Mediator coactivator complex [40]. Preinitiation complex (PIC) assembly triggers activation of TFIIH, which catalyzes phosphorylation of serine 5 [33] and serine 7 [35–37] and Mediator complex release [40]. As the nascent transcript reaches 25–40 base pairs in length, through sequential RNA triphosphatase, guanyltransferase and methyltransferase activity, a 7-methyl G5’ppp5’N moiety is added to the 5’ end of the nascent RNA [41, 42]. This modification serves important roles in the export and translation of the mature mRNA, and also helps to protect the nascent RNA from degradation [43]. Capping occurs co-transcriptionally: the enzyme complex directly interacts with serine 5 phosphorylated pol II CTD [33, 44, 45] and appears to be dependent on kinetically linked promoter proximal pausing of pol II [41]. Pausing occurs as a result of cooperative DSIF (for DRB sensitivity inducing factor) and NELF (for negative elongation factor) binding to the TEC [46, 47]. Following productive cap formation, P-TEFb phosphorylates serine 2 of pol II and the C-terminus of DSIF [46, 48]. DSIF phosphorylation results in NELF release, and converts DSIF to a positive acting elongation factor through cooperative function with the Paf complex and Tat-SF1 [48]. DSIF and P-TEFb remain associated with the TEC and productive elongation ensues [49].
4.2. Splicing
While early studies indicating co-association of pol II CTD and splicing proteins suggested a similar direct coupling mechanism [50–52], several mass spectrometry studies failed to identify splicing factors associated with pol II immunoprecipitation (IP) and reciprocal IP of the spliceosome failed to identify pol II [8, 53–56]. Furthermore, fusion of the pol II CTD to T7 or pol III polymerase did not restore splicing [57]. Nevertheless, pol II and, in particular CTD have been clearly implicated in co-transcriptional recruitment of the spliceosome. Deletion of the CTD impaired recruitment of certain splicing factors in immunofluorescence studies [58] and reduced overall splicing efficiency in separate studies involving splicing reporters [31]. In addition, an intact CTD is required for SRp20-mediated alternative splicing [59] and the multi-functional SR-like protein, Npl3, is associated with pol II CTD in yeast [60]. Thus, it would appear that while the interaction may not always be direct, CTD plays a crucial role in recruiting splicing proteins to nascent RNA. In line with indirect recruitment, a recent study utilizing fluorescently tagged SR proteins found that recruitment of SR proteins to the queried reporter was RNA dependent and that RNase treatment abolished interaction between pol II and splicing proteins [27]. It should be noted, however, that recruitment of U1 to mRNA appears to operate via a unique mechanism. U1 consistently co-immunoprecipitates with pol II [23] and is found at intronless [28] and splicing deficient genes [61]. While seemingly conflicting, the sum of these data can be resolved in a model involving CTD-dependent recruitment of a critical splicing initiation factor, possibly U1, thereby setting the stage for subsequent interactions built on the scaffold of the newly synthesized transcript.
4.3. Poly-adenylation
Similar to 5’ capping, 3’ end processing is tightly coupled to pol II transcription [62]. 3’ end processing is generally comprised of endonucleolytic cleavage downstream of the AAUAAA sequence and polyadenylation of the free 3’OH. These activities are achieved through multi-subunit complexes containing cleavage stimulation factor (CstF) and cleavage polyadenylation specificity factor (CPSF), both of which have been demonstrated to directly interact with pol II CTD [31]. Moreover, like other aspects of mRNA synthesis, this process is dynamically regulated through phosphorylation of pol II CTD. During the process of transcript synthesis, there is a gradual increase in serine 2 phosphorylation of pol II CTD culminating in a peak at the 3’ end of the gene [33]. This modification helps to recruit 3’ processing proteins, such as the cleavage factor Pcf11 (a CstF subunit) [63, 64]. In support of a specific role for serine 2 phosphorylation in 3’ end processing, complete disruption through deletion of the serine 2 kinase Ctk1 in yeast did not affect overall transcript elongation. In contrast, polyadenylation was disrupted in the Ctk1 deletion strain due to impaired recruitment of 3’ processing factors to their sites of action [65]. As one final step in co-transcriptional mRNA processing, factors that dismantle the TEC and aid transcript export are recruited through pol II [39]. Altogether, the abundant evidence linking modifications of pol II CTD to mRNA processing events highlight the importance of co-transcriptionality. The pol II CTD can serve as a dynamic platform that ensures the correct processing factors are available at the appropriate stage of mRNA synthesis.
5. Pol II Elongation Through Chromatin
In contrast to the ATP-driven power strokes of DNA helicases, pol II elongation proceeds via Brownian motion, wherein it is able to slide both forward and backward along the DNA template [66]. While, the forward state is preferred due to the stability conferred by hydrolysis of the incoming nucleotide, pol II elongation is inherently subject to frequent pausing [66, 67]. As a further complication, elongation does not occur on a barrier free linear template. Instead, DNA is tightly packaged into nucleosomes, which pose a significant obstacle to pol II elongation in vitro [68–71]. To achieve efficient elongation, nucleosomes are displaced in front of elongating pol II and reformed in its wake in a process that is dependent on elongation rate [72–74]. Nonetheless, transcription through chromatin results in frequent pausing related to backtracking of pol II. Backtracking can cause the 3’OH of the nascent transcript to become misaligned from the pol II active site [75]. Elongation resumes spontaneously at low levels or more efficiently in response to the elongation factor, TFIIS. TFIIS enhances the intrinsic nuclease activity of pol II, which promotes cleavage of the extended transcript and realigns the 3’OH in the active site [76, 77]. Accordingly, TFIIS has been demonstrated to be required for efficient elongation of a chromatinized template [71, 78, 79]. In addition, pol II elongation can be impeded by DNA-binding proteins, or in response to repressive chromatin structure [80, 81].
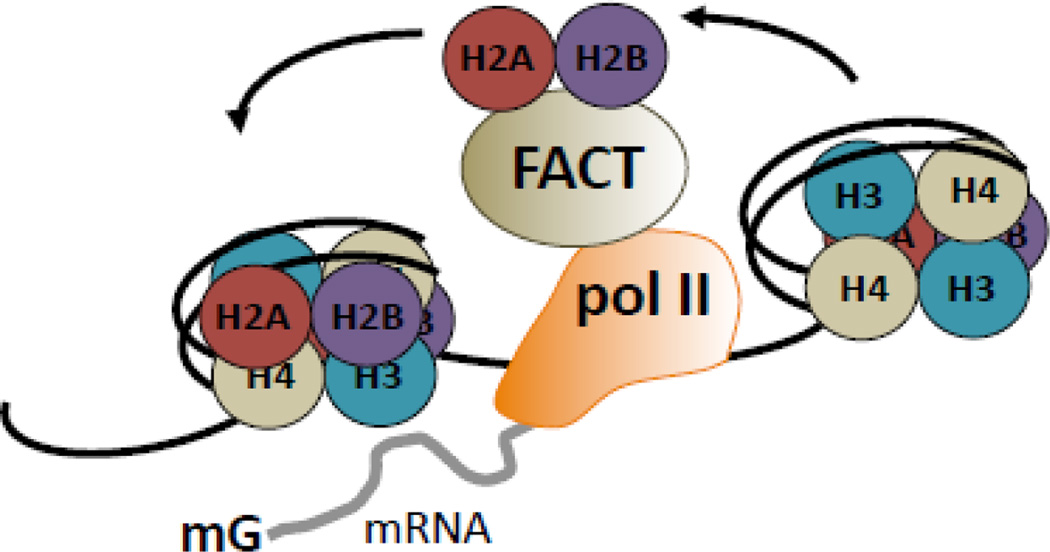
While seemingly inefficient, these barriers to pol II elongation represent important aspects of kinetic regulation of mRNA processing and numerous levels of nucleosome remodeling are in place to ensure chromatin remodeling during elongation [66]. Importantly, recent evidence suggests a high degree of coupling between remodeling machineries and pol II [82]. The ATP-dependent chromatin remodelers SWI-SNF, ISWI, CHD, and INO80/SWR utilize the energy of ATP to remodel chromatin during elongation. For example, the SWI-SNF complex ATPase, RSC, promotes elongation through nucleosomes in vitro [83] and Chd1 has been demonstrated to interact with the elongation factors Paf, DSIF, and FACT [84, 85]. In addition, histone chaperones promote elongation by destabilizing nucleosome structure [66]. Again, this process is mediated by pol II itself. H2A/H2B are located at the exterior of the nucleosome and are rapidly replaced during transcription. H3/H4 are less mobile, and are displaced with slower kinetics [86, 87]. NELF release from the TEC results in H2B monoubiquitination (uH2B) and recruitment of FACT [88, 89]. FACT demonstrates dual-activity wherein it aids both nucleosome destabilization through removal of one H2A/H2B dimer in front of pol II, and reassembly via deposition of histones in the wake of pol II [88, 90] (Figure 1). The H3/H4 chaperones Asf1 and Spt6 demonstrate a similar role with respect to H3 and H4 eviction and reassembly [91, 92]. Importantly, all three factors inhibit spurious transcript production [91–93], suggesting a critical role for co-transcriptional nucleosome reassembly in the ablation of cryptic transcription.
Figure 1. Nucleosomes are co-transcriptionally remodeled.
Nucleosomal arrays pose a barrier to pol II elongation. Phosphorylation of serine 2 of pol II CTD recruits the histone chaperone, FACT. FACT destabilizes nucleosomes in front of pol II through removal of one H2A–H2B dimer. FACT also prevents cryptic transcription by redepositing histones in pol II’s wake, facilitating nucleosome reassembly.
Similar to histone modifications at promoters, intragenic histones can be acetylated, methylated or ubiquitylated with distinct outcomes on gene expression. In contrast to the high levels of acetylation found at promoters, enrichment of histone acetylation in gene coding regions are relatively modest [94]. Nevertheless, histone acetyltransferase (HAT) and histone deacetylase (HDAC) complexes are enriched throughout the coding regions of actively transcribed genes, suggesting significant co-transcriptional histone recycling [66]. This premise is supported by the observation that intragenic HAT and HDAC complexes are distinct from promoter complexes, and show a high degree of coupling to pol II. For example, the Elongator HAT complex stimulates transcription through chromatin and is associated with pol II and with nascent RNA [95–98]. In addition, Gcn5 of the acetylating SAGA (STAGA in humans) co-activator complex interacts with TFIIS and other elongation factors [99, 100] and colocalizes with pol II in transcribed sequences [101]. On the other hand, pol II also recruits HDACs: the Eaf3 subunit of the Rpd3s HDAC complex is assembled at transcribed genes through pol II coupled H3K36 methylation [102–106]. The extensive coupling between HAT and HDAC complexes to pol II suggests an important structural role, wherein HATs are required to promote efficient transcription, and HDACs are required to reestablish repressive chromatin in pol II’s wake. Indeed, deletion of Rpd3s resulted in spurious transcription from cryptic promoters [107], suggesting that rapid reformation of nucleosomes is an integral aspect of regulated gene expression.
While histone methylation-mediated effects on pol II elongation are less well documented, a number of associations to pre-mRNA processing have been described, as detailed in [7.2]. Perhaps this reflects a predominant role for intragenic histone methylation as a scaffold for RNA processing effector proteins (Figure 2). Focusing here on elongation, like acetylation, histone methylation is coupled to pol II. Phosphorylation of pol II CTD on serine 5 results in recruitment of the Set1 histone methylase of the MLL/COMPASS complex, which subsequently directs methylation of H3K4 [108]. Trimethylation of H3K4 peaks at promoters, whereas dimethylation extends into the 5’ region of coding regions and monomethylation persists throughout the gene [109, 110]. H3K36 methylation is considered a hallmark of transcribed DNA, and shows a reverse gradient relative to K4, wherein trimethylation increases towards the 3’ ends of genes [109]. It is thus not surprising that the histone 3 K36 methyltransferase, Set2, binds to pol II phosphorylated on serine 2 [103]. As mentioned above, H3K36 di- and tri-methylation recruits the Rpd3s HDAC complex, and thereby plays a critical role in repressing cryptic transcription [111].
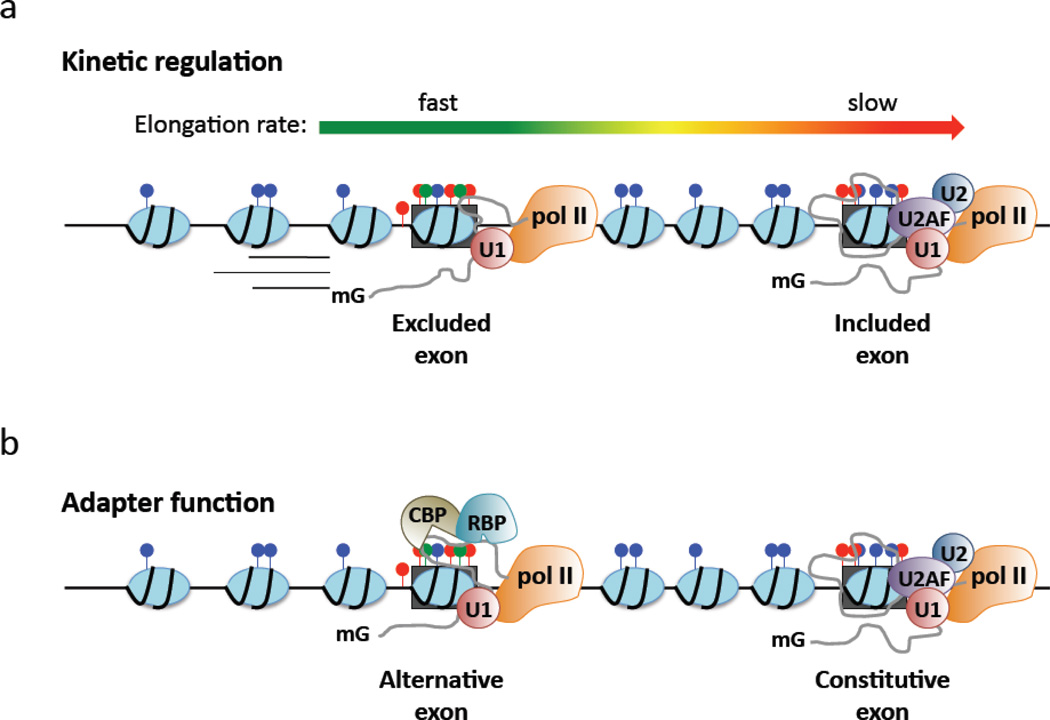
Figure 2. Potential mechanisms for chromatin-associated spliceosome assembly.
Exons are marked by increased nucleosome occupancy, distinct histone modifications and elevated DNA methylation relative to introns. These modifications at the DNA level may influence splice site selection by a) modulating elongation or b) through direct recruitment of auxiliary factors. a) A slow rate of pol II elongation favors spliceosome assembly at weak exons, whereas a rapid rate may not provide a sufficient spatiotemporal window prior to synthesis of competing downstream splice sites. Intragenic chromatin structure may act to locally modulate elongation rate. As shown here, the upstream exon is excluded from mRNA as a result of a locally rapid pol II elongation rate, thereby shifting spliceosome assembly to the downstream exon. b) Chromatin modifications may recruit chromatin binding proteins (CBP) to exonic DNA, which thereby act as adaptor molecules for RNA binding proteins (RBP) that promote or inhibit spliceosome assembly. As shown here, the chromatin context of the upstream exon binds a CBP that recruits an RNA binding splicing repressor, thereby shifting spliceosome assembly to the downstream exon.
Finally, ubiquitylation of histones is also linked to elongation. Elongation is associated with monoubiquitylation of lysine 120 of human H2B (lysine 123 in yeast) (uH2B) [88, 112]. In addition to aiding pol II elongation through mononucleosomes by recruiting FACT, uH2B directs methylation of H3K4 and H3K9 [113, 114] by engaging the relevant methyltransferases Set1/COMPASS and Dot1 [115]. These examples highlight an elegant nexus between elongation and transcription-directed chromatin remodeling, which operates to fine-tune DNA accessibility to pol II against unwanted transcription.
6. Kinetic model of Alternative pre-mRNA splicing
In the previous sections, we detailed multiple barriers to pol II elongation, and described how 5’ and 3’ end processing are related to pausing of pol II. It is thus tempting to speculate that perturbations to elongation rate may similarly influence pre-mRNA splicing. The kinetic model of alternative splicing is dependent on two fundamental bases: 1) the in vivo elongation rate must be variable and 2) modulation of elongation should result in alternative splicing. We will discuss data in support of each of these conditions in turn below.
6.1. In vivo Pol II Elongation Rate
Advances in kinetic analyses have made it possible to assess in vivo elongation rates (detailed in Larson and colleagues, this issue). By using a tandem array reporter system and fluorescence recovery after photo-bleaching to monitor mRNA synthesis in real-time, the pol II elongation rate was calculated to range between 1.9 to 4.3 kb/minute [116, 117]. It was further reported that pol II pauses for an average of 4 minutes during elongation, although it was not possible to determine whether total pausing reflected multiple short pauses or one long pause [116]. In contrast, imagining studies involving single transcription units detected highly variable elongation rates spanning from 0.3–50 kb/minute [118–120]. The wide range of elongation dynamics described in these studies may be a function of the local chromatin environment at the reporter integration site [121], and could parallel rate related variability in co-transcriptional splicing of endogenous genes. Overall, given that genome-wide ChIP sequencing (seq) studies have revealed increased occupancy of pol II at exons relative to introns [122, 123], these kinetic observations fostered an attractive hypothesis involving pol II pausing at exons. Analogous to 5’ or 3’ end processing, this model dictates that pol II pausing at exons provides a sufficient spatiotemporal platform for spliceosome assembly. Conversely, weak exons may lack such pausing signals, reducing the window of opportunity for spliceosome assembly prior to downstream splice site synthesis (Figure 2).
While the kinetic model is very appealing, a recent kinetic study utilizing the reversible inhibitor of new transcription, DRB (5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside) failed to detect exon related pausing. Instead, elongation rate was consistently measured at approximately 3.8 kb/minute, regardless of gene size and exon content [124]. Moreover, splicing was estimated to occur within 5–10 minutes of downstream splice site synthesis irrespective of upstream intron length [124]. In addition, another kinetic study showed that the transcription elongation does not change with splicing of the selected genes [28]. However, as described above, alternative exons show delays and a greater propensity to post-transcriptional splicing relative to constitutive exons [22], suggesting a level of kinetic regulation. Indeed, as detailed below, the strongest endorsement for the kinetic model is yielded from numerous studies linking variable elongation rate to alternative splicing.
6.2. Elongation Kinetics Influence Alternative Splicing
In the past decade, a plethora of studies ranging from focused model genes to genome-wide analyses have provided substantial evidence in support of the kinetic model. Direct proof of an association between elongation and splicing derives from studies utilizing alpha-amanitin resistant RNA pol II bearing a point mutation that lowered elongation rate. Compared to wildtype pol II, the elongation rate mutant caused an increase in the inclusion levels of several tested alternative exons [125]. Likewise, mutation in serine 2 and serine 5 but not in serine 7 of pol II CTD affected transcription elongation and resulted in altered splicing patterns of several genes [126]. Furthermore, artificial introduction of a pol II pausing sequence downstream of an alternative exon correlated with increased inclusion in a tropomyosin minigene [127].
Similarly, promoter structure can influence transcription elongation and reporter systems involving mutant promoters have resulted in alternative splicing. Minigene systems involving promoter swapping altered splicing patterns of weak exons [128] and recruitment of the SF2/ASF SR protein to splicing enhancer sequence was shown to be dependent on the promoter driving transcription [129]. It has further been shown that genes that possess alternative promoters are more likely to yield alternatively spliced transcripts as compared to single promoter genes [130]. While any of the above effects on splicing could theoretically operate independently of elongation, evidence suggests that promoter-mediated effects on splicing are directly due to regulation of elongation rate [131], thereby bolstering the kinetic model.
Perhaps the strongest evidence for the kinetic model derives from studies in which elongation rate was directly modulated through an exogenous stimulus. For example, the topoisomerase I inhibitor, camptothecin stalls elongating pol II, leading to increased spliceosome assembly at FOS mRNA [20]. Conversely, treatment of cells with the histone deacetylase inhibitor trichostatin A, increased elongation and inhibited inclusion of a weak fibronectin EDI exon [132]. Genome-wide analyses provide further support for this association. Camptothecin and DRB treatment yielded increased inclusion of many but not all genes with weak exons [126] and treatment of yeast with the elongation inhibitors mycophenolic acid (MPA) and 6-azauracil increased exon inclusion [133]. Further compelling evidence for kinetic coupling came from a study involving ultraviolet (UV) radiation treatment of cells. UV exposure results in hyperphosphorylation of pol II, leading to reduced pol II elongation and alternative splicing of fibronectin, caspase 9, Bcl-x and additional mRNAs in human cells [134]. This effect was not p53 mediated and was confirmed to be associated with elongation through use of a pol II hyperphosphorylation mutant, which effectively mimicked the UV-mediated effects on reduced elongation and alternative splicing [134]. In sum, these data clearly demonstrate a role for elongation rate in alternative splicing regulation, and support a role for variable elongation as a physiological stimulus for splicing regulation.
7. DNA-mediated alternative pre-mRNA splicing
The above studies linked chromatin-remodeling to elongation and elongation to splicing, raising the question whether chromatin is associated with mRNA splicing. Strong support for this premise arose from genome-wide ChIP-seq studies, which revealed a surprising association between chromatin structure and exon-intron structure. Specifically, exons were found to have elevated nucleosome occupancy, specific histone modifications and increased DNA methylation relative to introns (reviewed in [135]). These associations raised the question whether chromatin modifications aid the spliceosome in the process of exon detection. If this were the case, one could envision that perturbations to chromatin structure could result in alternative splicing of pre-mRNA. The last few years have yielded substantial evidence in support of DNA-mediated regulation of alternative splicing. Examples and their functional relevance are detailed below.
7.1 Nucleosome occupancy
By nature, co-transcriptional splicing suggests that the factors influencing transcription may also affect splicing. One such factor is nucleosome positioning [136]. Two copies of each histone protein, H2A, H2B, H3 and H4, are assembled into an octamer around which 145–147 base pairs (bp) of DNA are wrapped to form a nucleosome core. This highly conserved nucleoprotein complex occurs essentially every 200 40 bp throughout all eukaryotic genomes [137] and, interestingly, the average size of an exon (145bp) [138] approximates the length of nucleosome wrapped DNA.
Recent genomic profiling studies have revealed a non-random distribution of nucleosomes at exons [122, 139–143]. The average size of a human intron is 5.6 kb, and introns range in length from a few base pairs to over 740 kb [144]. Exons that are surrounded by long introns show a higher level of nucleosome occupancy as compared to exons flanked by short introns [143]. As nucleosomes can pose a physical barrier to pol II transcription [145], localization of nucleosomes at exonic sequences may kinetically aid the spliceosome in distinguishing intronic versus exonic sequence. In support of an important role for nucleosome positioning in exon definition, nucleosome occupancy at exons is well-conserved between fruit fly, worms and humans [139–142, 146] and nucleosome occupancy at exons is independent of the expression status of genes [139, 142]. Suggestive evidence for a role for nucleosomes in alternative splicing regulation stems from the observation that exons with weak spice sites show robust nucleosome occupancy, whereas pseudoexons that are not included in mRNAs but are flanked by strong splice sites show nucleosome depletion [139]. In addition, nucleosome positioning within an exon and associated depletion upstream of the acceptor site correlates with exon inclusion, whereas nucleosome occupancy upstream of the acceptor site associated with depletion within the exon correlates with exon exclusion [139]. While these studies highlight a correlation between nucleosome occupancy and exon inclusion, the molecular mechanism underlying this regulation is not clear.
7.2. Exonic Histone Modifications
As briefly described in section [5], characteristic patterns of histone methylation have been associated with active or repressed chromatin. Investigations into these patterns have revealed a great deal of specificity such that mono-, di- or trimethylation of a particular lysine can produce distinct gene expression outcomes. For example, trimethylation of histone 3 K9 and K27 is associated with repressed chromatin whereas monomethylation correlates with active transcription [109]. While studies of the “histone code” were initially limited to transcription initiation, genome-wide ChIP seq data revealed an intriguing potential link to elongation and/or splicing [147, 148]. Like nucleosomes, histone modifications are non-randomly distributed with respect to exon-intron architecture [111, 140, 143]. Genome-wide analysis of 38 histone modifications in human cells demonstrated increased H3K36me3, H3K79me1, H2BK5me1, H3K27me1, H3K27me2, and H3K27me3 over internal exons as compared to the downstream introns. These marks were positively correlated with exon inclusion [140] (Table 1). Reanalysis of human T cell ChIP-seq data [109, 149] further identified enrichment of H3K79me1, H4K20me1, and H2BK5me1 [142], H3K79me1, H2BK5me1, H3K27me1, H3K27me2, H3K27me3 [140], H3K27me2 and H3K4me1 [143] at exons relative to introns (Table 1), which was for the most part independent of transcriptional activity. A few notable exceptions include H3K27me2, which shows lower enrichment over exons in highly transcribed genes, H3K9me3, which is overall depleted at exons [143] and H3K36me3, which is solely detected at transcribed sequences [150]. High-resolution analysis of H3K36me3 had the added effect of revealing a potential link to alternative splicing, wherein reduced levels were detected at alternative exons relative to constitutive exons [111]. However, it should be noted that due to differences in analysis methods, a final verdict has not been issued for most of these associations. For example, chromatin profiling of several model genes of robust alternative splicing indicated equivalent detection of H3K36me3 and H3K79me2 levels in the exclusion versus inclusion state [150]. In addition, normalizing histone modifications to nucleosome levels have yielded conflicting results either confirming exonic enrichment of H3K36me3 [140, 143] or indicating that H3K36me3 mirrors nucleosome occupancy, suggesting a secondary effect to an overall increase in local H3 [139, 142]. Thus, the mechanistic basis for distinct histone modifications at exons remains unclear and will likely be the subject of substantial investigation in the coming years.
Table 1. Exonic histone modifications.
Summary of intragenic histone modifications associated with pre-mRNA splicing genome-wide and in specific model genes.
| Histone modification |
Genome wide association |
Inclusion model | Exclusion model |
|---|---|---|---|
| H3K36me1 | a[182] | ||
| H3K36me3 |
b[140, 142, 182] c [111] |
Inclusion of variant exons of CD44 [172] | Exclusion of exon 18 of NCAM [151]. Exclusion of exon IIIb of FGFR [153]. |
| H3K9me3 | Inclusion of variant exons of CD44[81] | ||
| H3K27me1 |
a[182] b[140] |
||
| H3K27me2 |
b[140] d[143] |
||
| H3K27me3 |
b[140] a[182] |
Exclusion of exon IIIc of FGFR [153] | |
| H3K4me1 | d[143] | Exclusion of exon IIIb of FGFR[153]. | |
| H3K4me3 | Exclusion of exon IIIc of FGFR [153]. | ||
| H3K20me1 | d[142] | ||
| H3K79me1 | b[140] | ||
| H3BK5me1 |
b[140] d[142] |
||
| H3K9ac | Exclusion of exon 18 of NCAM [151]. |
Genome wide association with alternative exon exclusion.
Genome wide association with alternative exon inclusion.
Genome wide reduction at alternative exons as compared to constitutive exons.
Genome wide data increased detection at exons relative to introns.
7.3. Modulation of Chromatin and Alternative Splicing
In contrast to the steady state studies described in [7.2], modulation of histone modifications has uncovered a dramatic role for chromatin in alternative splicing regulation. For example, membrane depolarization of neuronal cells increases H3K36me3 and H3K9 hyperacetylation surrounding exon 18 of neural cell adhesion molecule (NCAM) DNA, and triggers exclusion of the exon from mRNA. Importantly, these changes are restricted to the intragenic DNA and are not found at the promoter. After withdrawal of membrane depolarization, the effects on acetylation and splicing are fully reverted, and inhibition of HDAC function can faithfully recapitulate the physiological stimulus [151, 152]. Furthermore, exons IIIb and IIIc of fibroblast growth receptor 2 (FGFR2) are alternatively spliced in a tissue specific manner. In mesenchymal cells, the alternatively spliced region is enriched in H3K36me3 and H3K4me1, which correlates with exon IIIc inclusion. In contrast, epithelial cells show enrichment of H3K27me3 and H3K4me3, which correlates with exon IIIb inclusion. Strikingly, modulation of H3K36me3 or H3K4me3 levels by up or down regulation of their respective histone methyltransferases results in reciprocal changes in exon IIIb and IIIc inclusion [153]. As further indication of a role for histone modifications in alternative splicing, the proteins that modulate histones have themselves been implicated in splicing regulation. The catalytic subunit of the SWI/SNF chromatin remodeling complex Brm favors inclusion of variant exons in several transcripts [154] and depletion of SWI/SNF subunits in D. melanogaster S2 cells caused changes in the relative abundance of alternative transcripts from a subset of genes [155]. Altogether, these data substantiate a role for histone modifications in splicing regulation.
7.4. Increased DNA methylation at exons
Like histones and nucleosomes, intragenic DNA methylation is enriched at exons relative to introns [156]. This association may partially be attributed to increased nucleosome content at exons, as nucleosomal DNA is methylated at a higher level than flanking regions [122]. In addition, the de novo DNA methyltransferases, DNMT3a and 3b are anchored to a subset of nucleosomes and this interaction is dependent on intact nucleosomes [157].
While DNA methylation at promoters has been convincingly linked to repression of gene expression, the role of gene body methylation is not clear. It has been suggested that DNA methylation induces a dense chromatin structure that inhibits pol II elongation [158], yet separate studies found that intragenic DNA methylation is associated with increased transcription [159]. Thus, intragenic DNA methylation does not appear to have a fixed role with respect to elongation. Instead, methylome analysis in insects revealed a potential role for methylation in pre-mRNA splicing regulation. Analysis of DNA methylation in the honeybee genome found that methylation is almost exclusively restricted to exons, and is altogether absent from intronless, histone-encoding genes. In addition, increased DNA methylation levels at an alternative exon in the GB18602 gene in worker bees relative to the queen bee, was associated with exon skipping [160].
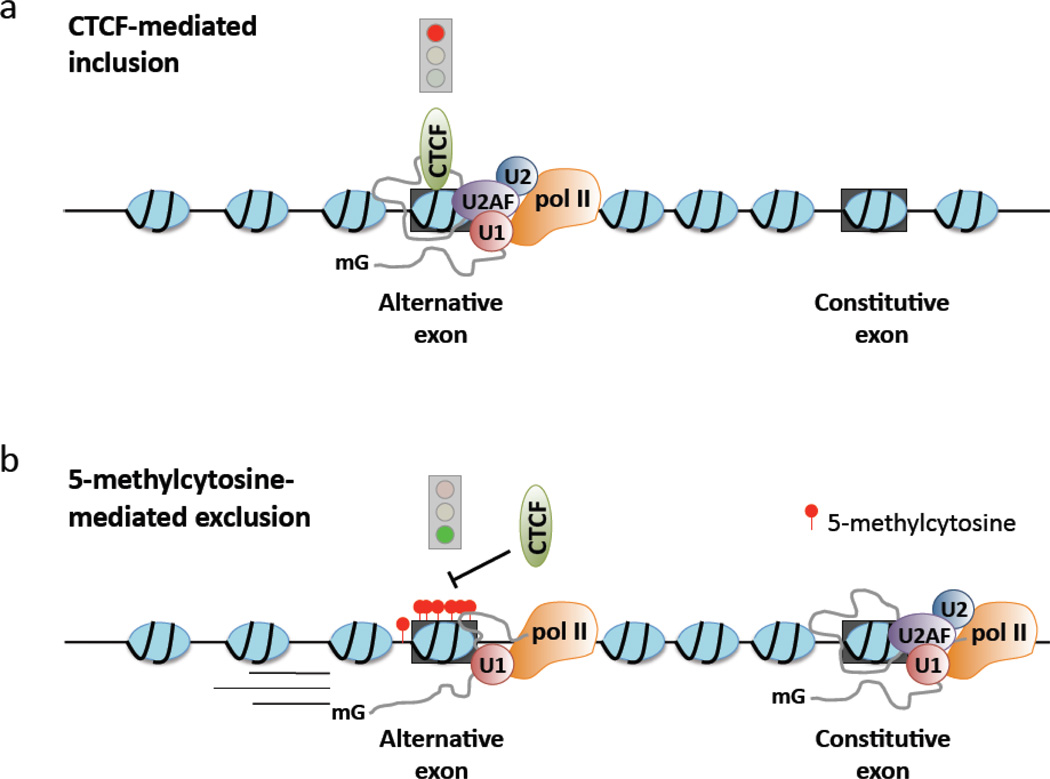
We recently demonstrated a functional association between DNA methylation and alternative splicing through modulation of the zinc-finger DNA-binding protein, CTCF. CTCF binding to exon 5 of the gene encoding CD45 promotes exon inclusion by acting as a barrier to local pol II elongation, and thereby supports the kinetic model of alternative splicing [80]. Genome-wide ChIP-seq and RNA-seq analysis in CTCF depleted cells confirmed that CTCF-mediated pol II pausing globally regulates inclusion of weak upstream exons, but does not influence inclusion of weak downstream exons [80]. Importantly, CTCF binding to DNA is inhibited by methylation of 5-cytosine [161]. Consequently, we found that loss of CTCF binding through methylation of exon 5 DNA resulted in complete loss of exon 5 from CD45 transcripts, thereby establishing the first mechanistic link between DNA methylation and alternative pre-mRNA splicing (Figure 3) [80].
Figure 3. Intragenic CTCF and 5-methylcytosine reciprocally influence exon inclusion in spliced mRNA.
Intragenic assembly of DNA binding proteins can influence co-transcriptional pre-mRNA splicing. a) The zinc-finger DNA binding protein CTCF acts as a direct barrier to pol II elongation, resulting in pol II pausing and spliceosome assembly at weak upstream splice sites. b) DNA methylation inhibits CTCF binding and associated pol II pausing, culminating in reduced spliceosome assembly at weak upstream splice sites and exclusion of exons from spliced mRNA.
Interestingly, genome-wide mapping of CTCF binding sites shows roughly 40–70% conservation between tissues [162], raising the possibility that altered DNA methylation patterns during development may ultimately contribute to tissue-specific alternative splicing patterns. Furthermore, binding of CTCF correlates with increased nucleosome occupancy at adjacent sequences, suggesting that intragenic CTCF may additionally function as an anchor point for nucleosome positioning [163].
7.5. The Adapter Hypothesis
While the above discussion has focused on how determinants at the DNA level can influence splicing through elongation kinetics, DNA binding factors may also affect pre-mRNA processing at the protein network level. DNA-binding proteins may interact with RNA-binding proteins, thereby recruiting them to their site of action. Direct studies in which histone methylation was modulated, as well as indirect studies showing associations between DNA and RNA-binding factors lend support to the “adapter hypothesis” as detailed in the following examples.
The chromatin binding protein, MORF-related gene 15 (MRG15), physically links H3K36me3 and the RNA-binding splicing repressor PTB. MRG15 directly interacts with H3K36me3 and functions to recruit PTB to a subset of PTB-dependent exons. Accordingly, overexpression of MRG15 forced exclusion of PTB-dependent exons, whereas MRG15 depletion led to increased inclusion of PTB-dependent exons [153]. Similarly, chromodomain-helicase-DNA-binding protein 1 (CHD1) recognizes H3K4me3 and facilitates recruitment of splicing factors such as U2 snRNP to active genes [164]. Depletion of either CHD1 and H3K4me3 abrogated the association of U2 snRNP with chromatin and reduced the efficiency of pre-mRNA splicing in vivo [164]. The HAT Gcn5 may also facilitate co-transcriptional recruitment of the U2snRNP to the intron branchpoint via histone acetylation. Deletion of Gcn5 in yeast resulted in the accumulation of unspliced mRNA [165]. In addition, the chromodomain protein, HP1γ associates with H3K9me3 and localizes to DNA corresponding to tandem alternatively spliced exons of the CD44 gene upon activation of protein kinase C (PKC). This increase in HP1γ detection correlates with binding of U2AF65 and PRP8 and is associated with reduced Pol II elongation [81]. In a separate study, HP1γ was also found to interact with the SR protein ASF/SF2 [166], thereby revealing a dual role for HP1γ as both a splicing adaptor and regulator of pol II kinetics.
Similarly, the SWI/SNF protein, Brm, displays a multi-functional role in splicing and elongation. Brm associates with several components of the spliceosome and favors inclusion of alternative exons in E-cadherin, BIM, cyclin D1 and CD44 transcripts. Brm also promotes accumulation of RNA polymerase II (RNAPII) with a modified CTD phosphorylation pattern on regions of the CD44 gene that encode variant exons. Thus, Brm coordinates crosstalk between transcription and RNA processing machinery by decreasing RNAPII elongation rate and facilitating recruitment of the spliceosome at exons with weak splice sites [154].
The numerous studies detailed in this section reveal an emerging theme in mRNA processing. Differential marking of exons through nucleosome occupancy, histone modifications or DNA methylation serve to aid or inhibit spliceosome assembly. This regulation can be at the level of modifying pol II elongation rates and thereby spatiotemporally influencing spliceosome assembly, or at the level of recruiting factors that promote or inhibit co-transcriptional spliceosome recruitment (Figure 2). Importantly, none of the regulatory levels appear to operate independently, and instead display extensive coupling.
8. The spliceosome- more than just splicing?
Several recent papers have raised the intriguing hypothesis that in addition to mediating pre-mRNA splicing, splicing factors may reciprocally affect elongation and chromatin structure, as detailed below.
8.1 Splicing-Promoted Elongation
Unlike the relatively scant evidence for direct interaction between pol II and the spliceosome, numerous associations between splicing proteins and elongation factors have been described (reviewed in [167, 168]). For example, the SR protein SC35 has been shown to have a critical role in pol II elongation. Depletion of SC35 results in increased accumulation of pol II in the gene body of select genes and elongation can be rescued in the presence of recombinant SC35. Impaired elongation in SC35 depleted cells was linked to inefficient recruitment of the elongation factor P-TEFb to the TEC and consequent diminished CTD serine 2 phosphorylation [169]. In addition interaction of the TAT-SF1 elongation factor with U snRNPs was shown to strongly stimulate elongation of an intronless reporter and splicing of intron-containing templates, revealing a dual-function in elongation and splicing regulation [170]. These reports reveal a reciprocal role for the spliceosome in promoting efficient elongation.
8.2 Splicing-Promoted Chromatin Remodeling
The observation that splicing proteins promote elongation of intronless templates, to which they would not normally be targeted, suggests a fundamental secondary activity that is independent of splicing catalysis. Indeed, several recent reports have elucidated a role for splicing proteins in the deposition of histone marks on DNA. For example, two independent studies implicated splicing in H3K36 methylation. Deletion of 3’ splice sites in the upstream introns of an integrated β-globin reporter caused a shift in the relative distribution of H3K36me3 away from the 5’end and toward the 3’ end of the reporter, whereas mutation of the polyadenylation site had no effect on H3K36me3 methylation. In addition, global inhibition of splicing by spliceostatin A resulted in rapid repositioning of H3K36me3 away from the 5’ ends and towards the 3’ ends of genes [171]. Splicing inhibition further impaired recruitment of the H3K36 methyltransferase HYPB/Setd2 and reduced H3K36me3 levels, whereas activation of splicing had the opposite effect. Similarly, intronless genes show lower levels of H3K36me3 as compared to intron-containing genes, irrespective of their expression status [172].
In further support of splicing-mediated chromatin remodeling, the Hu RNA-binding proteins increased histone acetylation at regions corresponding to alternative exons of the NF1 and FAS genes, thereby augmenting the local elongation rate and exclusion of the weak exons from spliced mRNA. The mechanism supporting increased acetylation involved Hu-mediated repression of HDAC2 activity. This suggests that splicing regulators can actively modulate chromatin architecture when co-transcriptionally recruited to their target RNA sequences [173].
8.3. TGS-mediated Chromatin Remodeling
In closing, we will highlight one final association between RNA, chromatin remodeling and pol II elongation. Transcriptional gene silencing (TGS) was initially described as a nuclear mechanism for RNA-mediated gene silencing in which a siRNA or miRNA directed against promoter DNA triggered gene silencing through heterochromatin formation, as characterized by increased H3K9me2/3, H3K27me3, and DNA methylation, as well as decreased histone acetylation [174–179]. In contrast, extension of TGS into intragenic sequences resulted in alternative pre-mRNA splicing: exogenously introduced siRNAs directed against intronic or exonic sequence in the region of the alternative EDI exon of the fibronectin gene stimulated EDI inclusion in mature mRNA. This effect was mediated by Argonaute-1 and was associated with increased H3K9me2, H3K27me3, HP1α and reduced pol II occupancy in the region, thereby implicating the kinetic model of alternative splicing. The effect of siRNA on splicing was abolished by depletion of HP1α and by promoting chromatin relaxation or pol II elongation through drug treatments [180]. Similarly, in C. elegans the Ago-related protein, NRDE-3, in complex with siRNA recruits NRDE-2 to target nascent mRNA, resulting in pol II and H3K9me3 accumulation at the corresponding DNA [181]. While physiologically relevant in vivo examples of TGS-coupled alternative splicing are lacking, these data reveal the depth of the interconnected regulation between DNA and RNA.
9. Conclusions and Perspectives
In this review, we have presented an integrated view of the life cycle of an mRNA while still tethered to DNA. In the process, we have highlighted extensive coupling between RNA-processing, pol II modifying and chromatin remodeling machines. It is increasingly clear that none of these processes operate in isolation, and instead exhibit highly coordinated auto- and cross-regulation. Alternative pre-mRNA splicing is not only influenced by the combined actions of RNA binding proteins, but also by the rate of pol II elongation as well as the nature of the transcribed DNA template. In a somewhat unexpected twist, it is now evident that the spliceosome reciprocally influences elongation kinetics and histone modifications. By extension, it will be interesting to see whether future studies implicate splicing in additional features of transcriptionally active intragenic DNA, such as the deposition of variant histones. Overall, in the coming years, it will be of high interest to develop an integrated model of RNA/DNA-mediated alternative pre-mRNA splicing regulation.
Highlights.
-
➢
The majority of metazoan introns are excised co-transcriptionally.
-
➢
Pre-mRNA processing is coordinated through the carboxy-terminal domain of pol II.
-
➢
Pol II elongation kinetics influence splicing decisions.
-
➢
Exons display distinct chromatin architecture relative to introns.
-
➢
Modulation of chromatin can result in altered pre-mRNA splicing.
Acknowledgements
We apologize to our colleagues whom we could not cite due to space constraints. This research was supported by the Intramural Research Program of the NIH, The National Cancer Institute, The Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berget SM, Moore C, Sharp PA. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 3.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 5.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2011 doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J, Xiong J, Wang D, Fu XD. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 2011;21:336–343. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 9.Rino J, Carmo-Fonseca M. The spliceosome: a self-organized macromolecular machine in the nucleus? Trends Cell Biol. 2009;19:375–384. doi: 10.1016/j.tcb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 11.Beyer AL, Bouton AH, Miller OL., Jr Correlation of hnRNP structure and nascent transcript cleavage. Cell. 1981;26:155–165. doi: 10.1016/0092-8674(81)90299-3. [DOI] [PubMed] [Google Scholar]
- 12.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 13.Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 14.Neugebauer KM, Roth MB. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 17.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5'ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 21.Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 28.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sisodia SS, Sollner-Webb B, Cleveland DW. Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol Cell Biol. 1987;7:3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartolomei MS, Halden NF, Cullen CR, Corden JL. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 32.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 33.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 35.Kim M, Suh H, Cho EJ, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem. 2009;284:26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perales R, Bentley D. "Cotranscriptionality": the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Max T, Sogaard M, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem. 2007;282:14113–14120. doi: 10.1074/jbc.M701345200. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jove R, Sperber DE, Manley JL. Transcription of methylated eukaryotic viral genes in a soluble in vitro system. Nucleic Acids Res. 1984;12:4715–4730. doi: 10.1093/nar/12.11.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat Rev Mol Cell Biol. 2011;12:377–384. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- 44.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 47.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lenasi T, Barboric M. P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 2010;7:145–150. doi: 10.4161/rna.7.2.11057. [DOI] [PubMed] [Google Scholar]
- 50.Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci U S A. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vincent M, Lauriault P, Dubois MF, Lavoie S, Bensaude O, Chabot B. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res. 1996;24:4649–4652. doi: 10.1093/nar/24.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robert F, Blanchette M, Maes O, Chabot B, Coulombe B. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J Biol Chem. 2002;277:9302–9306. doi: 10.1074/jbc.M110516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 56.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Natalizio BJ, Robson-Dixon ND, Garcia-Blanco MA. The Carboxyl-terminal Domain of RNA Polymerase II Is Not Sufficient to Enhance the Efficiency of Pre-mRNA Capping or Splicing in the Context of a Different Polymerase. J Biol Chem. 2009;284:8692–8702. doi: 10.1074/jbc.M806919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 59.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 60.Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spiluttini B, Gu B, Belagal P, Smirnova AS, Nguyen VT, Hebert C, Schmidt U, Bertrand E, Darzacq X, Bensaude O. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J Cell Sci. 2010;123:2085–2093. doi: 10.1242/jcs.061358. [DOI] [PubMed] [Google Scholar]
- 62.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 63.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3' end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 64.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 65.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 66.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 67.Larson MH, Landick R, Block SM. Single-molecule studies of RNA polymerase: one singular sensation, every little step it takes. Mol Cell. 2011;41:249–262. doi: 10.1016/j.molcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 69.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 70.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 72.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat Struct Mol Biol. 2011;18:1394–1399. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell. 2010;38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3'----5' direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 77.Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- 78.Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 79.Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 80.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011 doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 82.Buratowski S, Kim T. The role of cotranscriptional histone methylations. Cold Spring Harb Symp Quant Biol. 2010;75:95–102. doi: 10.1101/sqb.2010.75.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 87.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 89.Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3'-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 91.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 92.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 93.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 95.Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 96.Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 97.Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- 98.Kim JH, Lane WS, Reinberg D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci U S A. 2002;99:1241–1246. doi: 10.1073/pnas.251672198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wery M, Shematorova E, Van Driessche B, Vandenhaute J, Thuriaux P, Van Mullem V. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 2004;23:4232–4242. doi: 10.1038/sj.emboj.7600326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Milgrom E, West RW, Jr, Gao C, Shen WC. TFIID and Spt-Ada-Gcn5-acetyltransferase functions probed by genome-wide synthetic genetic array analysis using a Saccharomyces cerevisiae taf9-ts allele. Genetics. 2005;171:959–973. doi: 10.1534/genetics.105.046557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weake VM, Dyer JO, Seidel C, Box A, Swanson SK, Peak A, Florens L, Washburn MP, Abmayr SM, Workman JL. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev. 2011;25:1499–1509. doi: 10.1101/gad.2046211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 104.Schaft D, Roguev A, Kotovic KM, Shevchenko A, Sarov M, Neugebauer KM, Stewart AF. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 107.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 108.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 109.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 110.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 111.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 114.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 115.Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, Shilatifard A. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maiuri P, Knezevich A, De Marco A, Mazza D, Kula A, McNally JG, Marcello A. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 2011;12:1280–1285. doi: 10.1038/embor.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7:631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- 121.Wang F, Koyama N, Nishida H, Haraguchi T, Reith W, Tsukamoto T. The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol Cell Biol. 2006;26:4028–4040. doi: 10.1128/MCB.02189-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 126.Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci U S A. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- 130.Xin D, Hu L, Kong X. Alternative promoters influence alternative splicing at the genomic level. PLoS One. 2008;3:e2377. doi: 10.1371/journal.pone.0002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kadener S, Fededa JP, Rosbash M, Kornblihtt AR. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc Natl Acad Sci U S A. 2002;99:8185–8190. doi: 10.1073/pnas.122246099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR. Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem. 2002;277:43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- 133.Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, Bird G, Bentley D, Bertrand E, Kornblihtt AR. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 135.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative premRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 137.McGhee JD, Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- 138.Thanaraj TA, Stamm S. Prediction and statistical analysis of alternatively spliced exons. Prog Mol Subcell Biol. 2003;31:1–31. doi: 10.1007/978-3-662-09728-1_1. [DOI] [PubMed] [Google Scholar]
- 139.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 140.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8:3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 142.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 143.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fedorova L, Fedorov A. Puzzles of the Human Genome: Why Do We Need Our Introns? Current Genomics. 2005;6:589–595. [Google Scholar]
- 145.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 146.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 148.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hnilicova J, Hozeifi S, Duskova E, Icha J, Tomankova T, Stanek D. Histone deacetylase activity modulates alternative splicing. PLoS One. 2011;6:e16727. doi: 10.1371/journal.pone.0016727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 155.Tyagi A, Ryme J, Brodin D, Ostlund Farrants AK, Visa N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009;5:e1000470. doi: 10.1371/journal.pgen.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 159.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 162.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O'Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 169.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 171.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, Andrau JC, Ferrier P, Carmo-Fonseca M. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 173.Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci U S A. 2011;108:E627–E635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 175.Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 177.Zhang MX, Zhang C, Shen YH, Wang J, Li XN, Chen L, Zhang Y, Coselli JS, Wang XL. Effect of 27nt small RNA on endothelial nitric-oxide synthase expression. Mol Biol Cell. 2008;19:3997–4005. doi: 10.1091/mbc.E07-11-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang MX, Zhang C, Shen YH, Wang J, Li XN, Zhang Y, Coselli J, Wang XL. Biogenesis of short intronic repeat 27-nucleotide small RNA from endothelial nitric-oxide synthase gene. J Biol Chem. 2008;283:14685–14693. doi: 10.1074/jbc.M801933200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gonzalez S, Pisano DG, Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7:2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- 180.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 181.Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Dhami P, Saffrey P, Bruce AW, Dillon SC, Chiang K, Bonhoure N, Koch CM, Bye J, James K, Foad NS, Ellis P, Watkins NA, Ouwehand WH, Langford C, Andrews RM, Dunham I, Vetrie D. Complex exon-intron marking by histone modifications is not determined solely by nucleosome distribution. PLoS One. 2010;5:e12339. doi: 10.1371/journal.pone.0012339. [DOI] [PMC free article] [PubMed] [Google Scholar]